A new class of lipid droplet-associated proteins in nonseed tissues is identified by integrated omics approaches.
Abstract
Lipid droplets in plants (also known as oil bodies, lipid bodies, or oleosomes) are well characterized in seeds, and oleosins, the major proteins associated with their surface, were shown to be important for stabilizing lipid droplets during seed desiccation and rehydration. However, lipid droplets occur in essentially all plant cell types, many of which may not require oleosin-mediated stabilization. The proteins associated with the surface of nonseed lipid droplets, which are likely to influence the formation, stability, and turnover of this compartment, remain to be elucidated. Here, we have combined lipidomic, proteomic, and transcriptomic studies of avocado (Persea americana) mesocarp to identify two new lipid droplet-associated proteins, which we named LDAP1 and LDAP2. These proteins are highly similar to each other and also to the small rubber particle proteins that accumulate in rubber-producing plants. An Arabidopsis (Arabidopsis thaliana) homolog to LDAP1 and LDAP2, At3g05500, was localized to the surface of lipid droplets after transient expression in tobacco (Nicotiana tabacum) cells that were induced to accumulate triacylglycerols. We propose that small rubber particle protein-like proteins are involved in the general process of binding and perhaps the stabilization of lipid-rich particles in the cytosol of plant cells and that the avocado and Arabidopsis protein members reveal a new aspect of the cellular machinery that is involved in the packaging of triacylglycerols in plant tissues.
Lipid droplets are subcellular organelles found in essentially all eukaryotic organisms (Chapman et al., 2012; Murphy, 2012). Their role in sequestering neutral lipids within the cytosol of cells has led to the concept that lipid droplet compartments serve as a stable depot for the temporary and efficient storage of high-energy carbon reserves. However, in the last decade, it has become clear that while there are specialized lipid-storing tissues in many multicellular organisms, including the adipose tissues of mammals and seed tissues of higher plants, essentially all cell types have the capacity to synthesize and store triacylglycerols (TAGs) in lipid droplets, even if they are dynamic and short lived (Chapman and Ohlrogge, 2012; Murphy, 2012; Walther and Farese, 2012). In addition to an increasing appreciation for the prevalence of lipid droplets throughout biological systems, a broader range of functions associated with this hydrophobic compartment, beyond energy and carbon storage, has been reported, including lipid signaling (Bozza et al., 2011; van der Schoot et al., 2011; Chapman et al., 2012; Zechner et al., 2012), trafficking of intracellular components (Goodman, 2008; Murphy et al., 2009), and host-pathogen interactions (Saka and Valdivia, 2012).
To understand the diverse functions of lipid droplets, an array of proteins have been identified that are associated with lipid droplets from different cell types and organisms (Cermelli et al., 2006; Hodges and Wu, 2010; Yang et al., 2012; Jolivet et al., 2013). In higher plants, oleosins, caleosins, steroleosins, and other seed-specific lipid droplet proteins have been widely recognized for their role in lipid compartmentalization in oilseeds and some floral tissues, such as anther and pollen (Huang, 1992; Tzen and Huang, 1992; Frandsen et al., 2001; Lin et al., 2005; Huang et al., 2013). While an important function of oleosins in lipid droplets of developing seeds is to stabilize the droplets and prevent fusion during seed desiccation and rehydration (Leprince et al., 1998; Siloto et al., 2006; Schmidt and Herman, 2008; Shimada et al., 2008), there remains a general lack of information about other lipid droplet proteins in plants. For example, despite the widespread occurrence of cytosolic lipid droplets in various plant organs, the proteins associated with lipid droplets from vegetative tissues remain largely unexplored (Chapman and Ohlrogge, 2012; Murphy, 2012). We hypothesize that lipid droplets in nonseed tissues represent a compartment with protein compositions and functions different from the oleosin-coated lipid droplets found in seed tissues. Indeed, oleosin isoforms are highly expressed in seed tissues during lipid accumulation and in some floral tissues (anther and pollen) but are generally absent from most vegetative and fruit tissues of plants, including the oil-rich mesocarp of oil palm (Elaeis guineensis), olive (Olea europaea), and other fruits (Murphy, 2012; Umate, 2012; Huang et al., 2013).
To better understand the subcellular proteins and mechanisms potentially involved in lipid droplet formation, stability, and/or turnover in nonseed plant tissues, we examined the oil-rich mesocarp of avocado (Persea americana ‘Haas’). Lipid-rich avocado fruit tissues have long been used as a model system for biochemical studies of lipid synthesis (Harwood and Stumpf, 1972). Furthermore, anatomical descriptions of the fruit tissues have emphasized the prevalence of numerous lipid droplets in parenchyma cells and specialized idioblast cells throughout the mesocarp tissue of the fruit (Cummings and Schroeder, 1942; Platt-Aloia and Thomson, 1981). Here, we used a combination of lipidomic, proteomic, and transcriptomic approaches to identify a new class of lipid droplet-associated proteins (LDAPs) in plants. Visualization of TAGs in tissue prints of avocado fruit by mass spectrometry imaging was used to select mesocarp regions for lipid droplet analysis and for comparisons of the lipid composition of isolated lipid droplets with that of whole fruit. Multidimensional protein identification technology (MudPIT; Delahunty and Yates, 2007) was used to determine the protein composition of isolated avocado lipid droplets, and the in silico translated transcriptome of avocado mesocarp provided a peptide database for protein identification. Transcriptional profiling of developing avocado mesocarp also revealed that the mRNAs for several of the most abundant proteins were highest during fruit maturation and lipid accumulation. Two of these proteins, which we term LDAP1 and LDAP2, exhibited homology to each other and to the small rubber particle proteins (SRPPs; Oh et al., 1999) of rubber-accumulating plants. The Arabidopsis (Arabidopsis thaliana) homolog (At3g05500) of these avocado LDAP proteins, when transiently expressed in tobacco (Nicotiana tabacum) suspension-cultured cells, was found to associate specifically with the lipid droplet surface. Collectively, using this multiomics approach, we identified a novel class of lipid droplet proteins in nonseed plant tissues, an important step in further characterization of the subcellular machinery involved in general lipid droplet ontogeny and stability in plants.
RESULTS
Imaging Lipids in Avocado Mesocarp
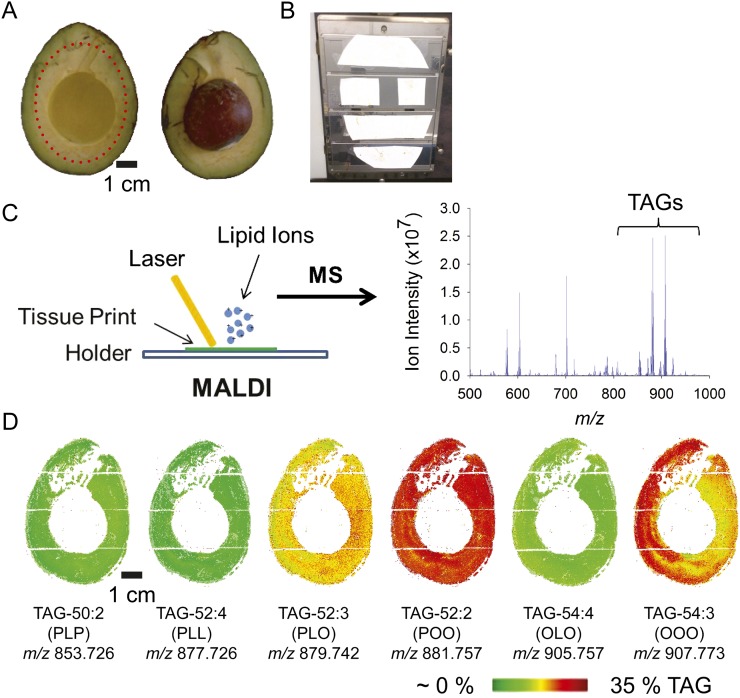
To characterize the lipid content and composition of avocado mesocarp tissues, the distribution of TAG molecular species in avocado tissue slices was visualized using a novel, matrix-assisted laser desorption/ionization (MALDI)-mass spectrometry imaging (MSI) approach (Horn et al., 2012; Fig. 1; Supplemental Table S1). Lipids were visualized by imprinting a slice of mature avocado fruit onto a nitrocellulose membrane, which was subsequently coated with ionization matrix. Step-wise rastering of the laser allowed reconstruction of a high-resolution, two-dimensional map of mesocarp lipids. The mesocarp is especially rich in monounsaturated fatty acids (mostly oleic acid, 18:1), and the molecular species of TAGs with two oleic acids and one palmitic acid, TAG-52:2 at mass-to-charge ratio (m/z) 881.757, or with oleic acid at each position, TAG 54:3 at m/z 907.773, were most prevalent in fruit tissue and were distributed throughout the mesocarp (Fig. 1D). TAG species were distributed relatively uniformly throughout the fruit tissue, except for the species with oleic acid at each position, TAG-54:3, which was relatively more abundant in certain regions of the mesocarp. Because this TAG-54:3 appeared to be the only molecular species with a marked heterogeneous distribution pattern, this heterogeneity is likely a real phenomenon, and although the physiological relevance is unclear at this point, it suggests that lipid droplets isolated from different parts of the fruit may have a slightly different TAG composition, at least for TAG-54:3. For biochemical isolation of lipid droplets (described further below), we excised the central region of the mesocarp, avoiding the chloroplast-containing tissues at the mesocarp periphery. The mass spectrometry maps of individual TAG species (Fig. 1) indicated that this region should indeed provide an abundant and representative source of lipid droplets for protein and lipid compositional analysis.
Figure 1.
In situ lipidomics by MALDI-MSI of avocado fruit reveals the distribution of TAG molecular species throughout the mesocarp tissue of mature fruit. A and B, Avocado was obtained from a local market, and the mesocarp was sliced in half (A) and pressed/printed onto nitrocellulose membrane (B). The tissue print was coated with 2,5-dihydroxybenzoic acid matrix and subjected to MALDI-MSI. C, The laser rastered over the specimen in a 300-μm step size to generate a mass spectrum at each location. D, Images of TAG molecular ions were reconstructed such that the tissue distribution and abundance of each molecular species could be visualized. The heat map for each of the six major TAG species in the avocado mesocarp is shown. The scale for relative TAG abundance is shown at the bottom (based on mol % of total TAG). Each TAG molecular species is denoted below the respective image according to the total acyl carbons and numbers of double bonds (e.g. TAG 54:3 is a TAG molecule with three 18-carbon acyl groups, each with one double bond). L, Linoleic acid; O, oleic acid; P, palmitic acid. The bottom number is the m/z of the parent ion of the K+ adduct (for m/z analysis, see Supplemental Table S1).
Characterization of Isolated Avocado Lipid Droplets
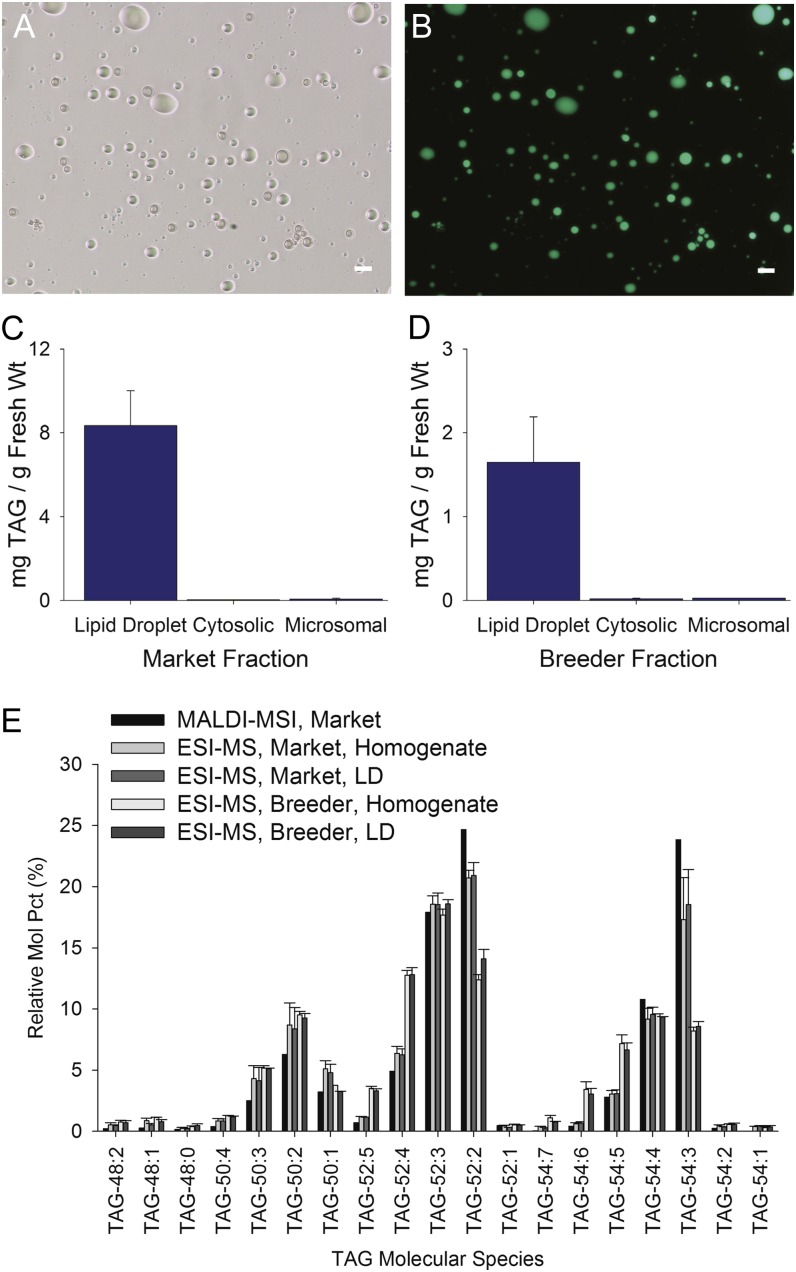
Lipid droplets are relatively simple to isolate from mixtures of most other subcellular components because they are the least dense of all the subcellular organelles, and unlike any other subcellular component, they float in aqueous solutions (Murphy, 2012). Lipid droplets were isolated from avocado mesocarp and enriched by flotation centrifugation. Then they were examined for relative purity by differential interference contrast and fluorescence microscopy (Fig. 2; BODIPY 493/503 stains neutral lipids specifically [Listenberger et al., 2007]). By comparing differential interference contrast and BODIPY 493/503 fluorescence, we established that particles in the isolated lipid droplet fractions were BODIPY positive and generally free from obvious visible contaminants (Fig. 2, A and B). Of course, purity is relative, and it was clear from the sensitive proteomics analysis (described below) that other proteins or subcellular materials likely adhered to or were trapped within this fraction as well. Nonetheless, quantitative analysis of the TAG present in various biochemical fractions confirmed that essentially all of the fractionatedTAGs were associated with the lipid droplet fractions and not with the cytosolic or microsomal fractions (Fig. 2, C and D). The distribution of TAG content in the various fractions isolated was similar irrespective of the developmental stage of the fruit tissue used; preharvest fruits were obtained directly from trees (Fig. 2D, breeder), and postharvest fruits were purchased from a local market (Fig. 2C, market). Both sources of avocados were used for most experiments, with the exception of transcriptomics analysis.
Figure 2.
Isolation of lipid droplets from avocado mesocarp. A and B, Lipid droplets were isolated by differential centrifugation and flotation through Suc, then the “fat-pad” fraction was stained with the neutral lipid-specific fluorescent dye BODIPY 493/503 and imaged using differential interference contrast (A) or fluorescence (B) microscopy. Bars = 25 µm. C and D, The TAG content of each fraction was also quantified by mass spectrometry, confirming the enrichment of TAGs in isolated lipid droplets but not in other subcellular fractions. Similar results were obtained for lipid droplets isolated from mature fruit obtained from either a local market (C) or from the breeder, whereby the fruit was harvested directly from the tree at the orchard (D). E, Comparison of the molecular species profiles (in mol %) of TAGs in isolated lipid droplets (LD) from avocado mesocarp (store-bought versus breeder-harvested fruit) or in tissue prints of store-bought whole fruits analyzed by MADLI-MSI (Fig. 1). The lipid molecular species are indicated by total number of acyl carbons and numbers of double bonds, as is Figure 1. ESI-MS, Electrospray ionization-mass spectrometry.
Lipid droplets have a simple structural organization; typically, they contain a TAG core covered by a monolayer of phospholipids and associated proteins (Huang, 1992; Chapman et al., 2012; Murphy, 2012). We analyzed the TAGs of mesocarp lipid droplets isolated from preharvest and postharvest fruits by direct infusion electrospray ionization-mass spectrometry (Fig. 2E). The TAG composition of the postharvest tissue slices, determined by MALDI-MSI, closely matched the compositions quantified by electrospray ionization-mass spectrometry in total lipid extracts of the tissue homogenates and isolated lipid droplets (Fig. 2E). Differences between TAG composition of lipid droplets isolated from preharvest and postharvest samples likely reflect variations in age, growth conditions, maturity, and/or postharvest treatments (Fig. 2E). Generally, the TAGs in the homogenates and lipid droplet fractions from the preharvest fruits were lower in TAG species with oleic acid and higher in species with linoleic acid, the two fatty acids that change markedly during fruit development and ripening (Eaks, 1990).
Proteomics of Lipid Droplets and Peptide Identification from Mesocarp Transcriptome
The protein composition of lipid droplets isolated from preharvest and postharvest avocado fruits was determined using MudPIT analysis. Specifically, individual peptide sequences were identified by comparing the obtained masses of the peptides with a reference polypeptide database derived from Illumina- and 454-based sequencing of complementary DNA (cDNA) from developing mesocarp of avocado. Annotated lists of matched proteins were compiled and rank ordered by relative abundance (based on spectral counts) within each subcellular fraction. Percentage coverage, representing the portion of an encoded polypeptide sequence matched by independent peptide sequences, was calculated for all proteins. The Arabidopsis protein with the highest sequence similarity to each avocado protein was determined using BLASTX.
Comparison of proteins identified in lipid droplet fractions isolated from either postharvest or direct, tree-harvested avocado fruits revealed a common core list of 70 proteins (Supplemental File S1). Further comparison of these 70 proteins with proteins identified in corresponding microsomal or cytosolic fractions revealed proteins that were specifically enriched in isolated fractions (Supplemental File S1). Proteins were considered “enriched” if they were present in more than 3-fold abundance (spectral counts) in a given subcellular fraction in comparison with other subcellular fractions. Based on these criteria, 17 proteins were enriched in lipid droplet fractions relative to microsomal or cytosolic fractions. A table summarizing the results for these 17 proteins from one representative analysis is provided in Supplemental Table S2. LDAP1 and LDAP2 were the first and fourth most abundant proteins, respectively, in lipid droplets based on spectral counts.
LDAP1 and LDAP2
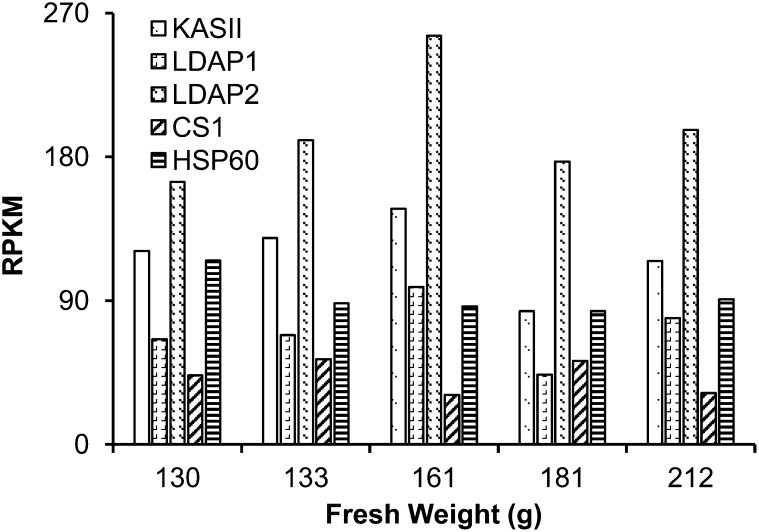
Transcript expression profiles were determined for several proteins identified in the avocado lipid droplet fractions (Fig. 3). Based on RNA-Seq data from five stages of developing mesocarp (130–212 g fruit weight), transcript levels for LDAP1 and LDAP2 were highest during the mid stages of fruit development (approximately 160 g fresh weight) when oil accumulation was greatest (Eaks, 1990). For comparison, the gene expression profiles and transcript levels for the two LDAPs were similar to those of ketoacyl-acyl carrier protein synthase II (Fig. 3), a plastidial enzyme involved in fatty acid biosynthesis (Carlsson et al., 2002). One protein recovered in lipid droplet fractions, annotated as oxidoreductase, also was expressed at high levels in mesocarp at mid stages of development (data not shown), but the closest Arabidopsis homolog, At4g13010, was shown previously to be associated with inner chloroplast envelopes (Miras et al., 2002), so this protein was not investigated further as a lipid droplet protein. Other proteins were enriched in avocado lipid droplet fractions (Supplemental Table S2; e.g. HEAT SHOCK PROTEIN60 and ATCYS1), but their transcript levels did not appear to change much with fruit development (Fig. 3), suggesting that these proteins likely were trapped or adhered to lipid droplets during isolation and were unlikely to be bona fide LDAPs.
Figure 3.
Transcript profile for the four most abundant proteins in lipid droplets was determined by the transcriptome analysis of developing mesocarp of avocado. Expression for genes encoding LDAP1 and LDAP2 peaked at the mid stage of mesocarp development and was similar to that of KETOACYL-ACYL CARRIER PROTEIN SYNTHASE II (KASII), a plastidial fatty acid synthesis gene. Transcripts for CYSTEINE SYNTHASE C1 (CS1) and HEAT SHOCK PROTEIN60 (HSP60) remained about 2-fold lower than that of KASII. RPKM, Reads per kilobase per million mapped reads.
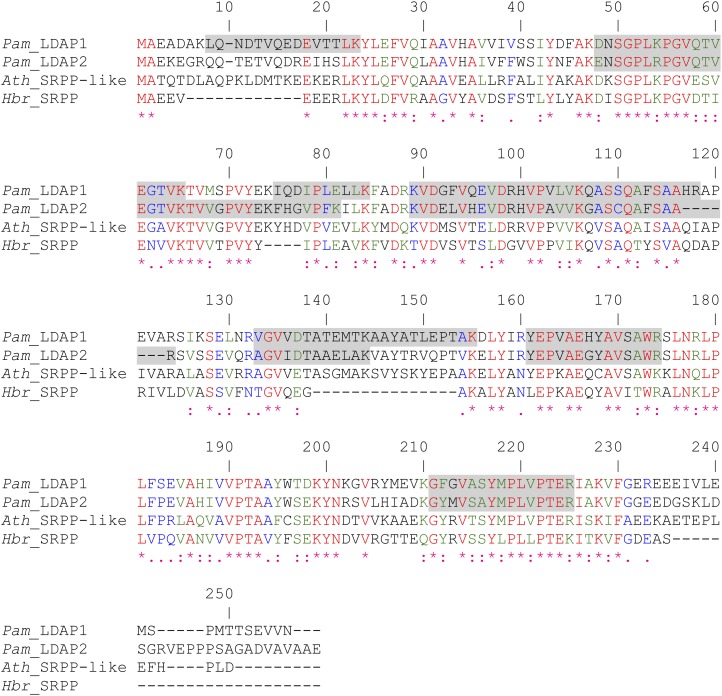
The most intriguing proteins enriched in the lipid droplet fractions of avocado were LDAP1 and LDAP2 (first and fourth in the list, respectively; Supplemental Table S2). These two proteins share 62% identity (86% similarity) at the deduced amino acid level and show extensive regions of similarity and identity with the SRRP of the rubber tree (Hevea brasiliensis; Oh et al., 1999) and the SRPP-like protein At3g05500 of unknown function from Arabidopsis (Fig. 4). Given the level of polypeptide sequence identity and the general similarity of rubber particles and lipid droplets in the storage of hydrophobic compounds in plant cells, we hypothesized that avocado LDAP1 and LDAP2 and Arabidopsis At3g05500 might similarly associate with and stabilize TAG-containing lipid droplets analogous to how SRPPs bind to the surface and stabilize polyisoprenoid-containing rubber particles (Collins-Silva et al., 2012; Hillebrand et al., 2012).
Figure 4.
Polypeptide sequence alignment of avocado LDAP1 and LDAP2 (Pam_LDAP1 and Pam_LDAP2) with the closest homolog from Arabidopsis (Ath_SRPP-like; At3g05500) and the rubber tree SRPP (Hbr_SRPP). Sequences were aligned using the ClustalW algorithm. Peptide regions of the two avocado proteins identified by MudPIT are highlighted in gray. Percentage coverage is 50% for LDAP1 and 41% for LDAP2.
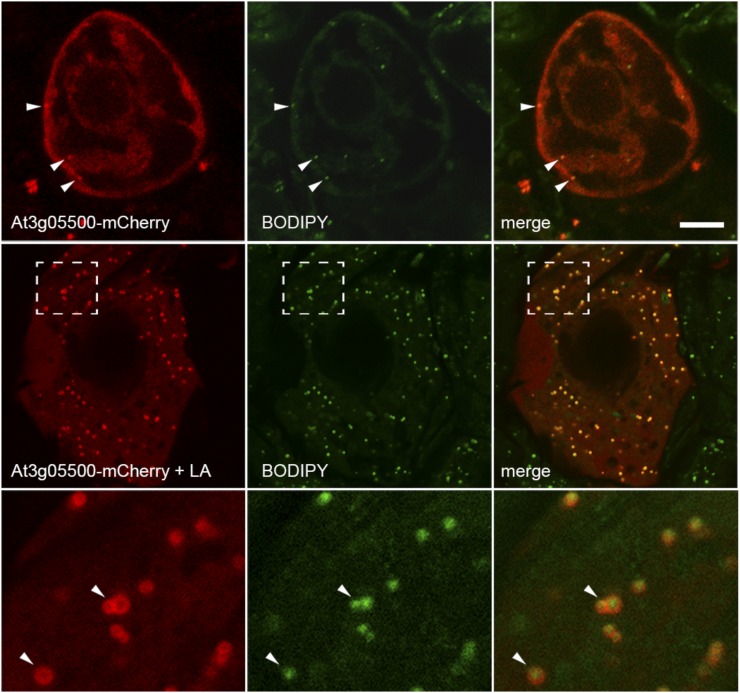
To determine whether the Arabidopsis At3g05500 protein could associate with lipid droplets in plant cells, the protein was tagged at its C terminus with the mCherry fluorescent reporter protein and expressed transiently in tobacco suspension-cultured cells, which are a well-characterized model plant cell system for studying protein localization (Brandizzi et al., 2003; Miao and Jiang, 2007). Notably, these cells typically do not contain high numbers of lipid droplets, but lipid droplet formation can be induced by the addition of free fatty acids to the growth medium (Fig. 5). Expression of the At3g05500-mCherry fusion protein in Bright Yellow-2 (BY-2) cells cultivated in normal growth medium lacking exogenous fatty acids resulted in a predominantly cytosolic localization, with only a few BODIPY-stained lipid droplets present in these cells (Fig. 5, top row). By contrast, incubation of BY-2 cells with linoleic acid after transient transformation resulted in a significant increase in lipid droplet abundance, with the majority of At3g05500-mCherry localized to these BODIPY-stained organelles (Fig. 5, middle row). Upon closer inspection, the fluorescence attributable to the fusion protein was often torus shaped and enclosed the punctate fluorescent signal attributable to the BODIPY-stained lipid droplets, suggesting that the fusion protein was localized to the surface of the lipid droplets (Fig. 5, bottom row). These data confirmed that the SRPP-like protein of Arabidopsis, At3g05500, can indeed localize specifically to lipid droplets in nonseed plant cell types.
Figure 5.
The SRPP-like protein from Arabidopsis (At3g05500) localizes to lipid droplets in tobacco BY-2 cells. The top row shows the predominantly cytosolic localization of transiently expressed At3g05500-mCherry in BY-2 cells that were stained with BODIPY 493/503. The second row shows the localization of the same fusion protein in BY-2 cells that were incubated with linoleic acid (LA) after transient transformation, which induces a dramatic increase in the number and size of lipid droplets in these cells and which reveals extensive colocalization of the fusion protein with lipid droplets. The region of the cell denoted by the hatched boxes is shown in the bottom row at higher magnification, which reveals that the fusion protein exhibits a torus-shaped fluorescence pattern that encloses the BODIPY 493/503-stained lipid droplets. Bar = 10 μm.
DISCUSSION
To date, proteomes of lipid droplets from plant sources (e.g. Brassica spp., Arabidopsis, sesame [Sesamum indicum], and Jatropha spp.) have focused almost entirely on seed-derived organelles and have been dominated by oleosins, caleosins, steroleosins, and only a few other proteins (Jolivet et al., 2004, 2009; Lin et al., 2005; Katavic et al., 2006; Popluechai et al., 2011). Here, we took an alternative approach to identify additional LDAPs by analyzing the proteome of lipid droplets from nonseed tissues. We asked whether there are specialized proteins associated with nonseed lipid droplets or, alternatively, whether the proteins identified would overlap with those already identified in seed lipid droplets. This approach is less likely to be overwhelmed by the well-known, hyperabundant proteins known to associate with lipid droplets in seeds (e.g. oleosins) and provides scope for the identification of additional proteins. These additional proteins may represent a more universal set of proteins for lipid compartmentalization in plant cells and offer new avenues for probing the regulation of TAG accumulation in vegetative tissues of plants. Avocado fruit tissue was selected as a source for lipid droplets for this study for several reasons: (1) avocado mesocarp is readily available and well known to be a rich source of lipid droplets (Cummings and Schroeder, 1942; Platt-Aloia and Thomson, 1981); (2) oleosins generally are not expressed outside of seed tissues (Chapman and Ohlrogge, 2012; Murphy, 2012; Huang et al., 2013) and thus were not expected to be a major protein in avocado mesocarp; and (3) deep sequencing analysis of the avocado mesocarp transcriptome provided a reference database for the identification of proteins by peptide mass fingerprinting.
Although lipid droplets are easily fractionated from tissues by flotation centrifugation, they do have the potential to carry a variety of adventitious subcellular components with them, such as hydrophobic proteins, membrane fragments, and other lipid-soluble compounds that perhaps were not associated with this compartment in situ. Obviously, there were a number of proteins identified in isolated lipid droplet fractions that are most likely not associated with lipid droplets in planta. Vigorous purification techniques such as high-salt washes may be able to partially remove such “contaminants” but also may remove proteins intrinsic to lipid droplets. In this study, our interest focused on discovering new LDAPs. By avoiding proteins that were prevalent in other subcellular fractions or that were not highly expressed in fruit tissues, our approach may have resulted in the exclusion of some important proteins from consideration (or may have included proteins not actually found in lipid droplets). Nevertheless, we were successful in identifying a new class of LDAPs not previously known to bind TAG-containing lipid droplets in plants. Specifically, we identified two homologs of SRPP-like proteins in avocado (i.e. LDAP1 and LDAP2) that were tightly associated with TAG-containing lipid droplets (Fig. 2; Supplemental Table S2) and whose gene expression patterns correlated with TAG accumulation during fruit development (Fig. 3). Similarly, in oil palm mesocarp, which stores up to 90% oil by dry weight, transcript levels for an ortholog of LDAP1 and LDAP2 also were associated with the timing of oil accumulation during fruit development and were substantially higher in oil palm than in fruit tissue from date palm (Phoenix dactylifera), which stores very little oil (Bourgis et al., 2011). Furthermore, we demonstrated that an Arabidopsis protein (At3g05500) with closest amino acid sequence similarity to the avocado LDAP1 and LDAP2 proteins (Fig. 4) could also target to lipid droplets in nonseed plant cells (Fig. 5).
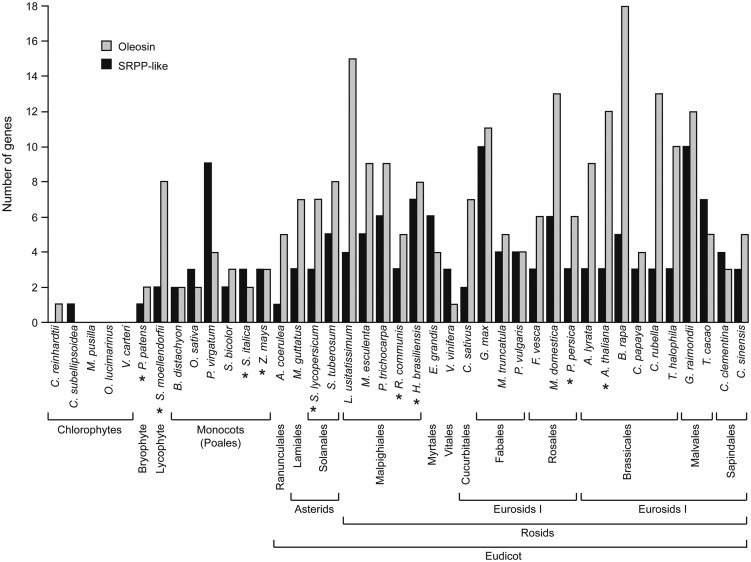
While publicly available gene expression databases indicate that At3g05500 is expressed in all Arabidopsis tissues, expression levels for this gene are approximately 8- to 10-fold higher in tissues with abundant lipid, including developing seeds, stamens, and pollen (Schmid et al., 2005), consistent with a broad-based role for At3g05500 in lipid droplet ontogeny, structure, or stability. While the three major oleosin isoforms in Arabidopsis (i.e. OLE1, OLE2, and OLE3) are expressed at levels more than 100-fold higher in developing seeds than in other tissues (Schmid et al., 2005), the broader gene expression pattern of At3g05500 suggests a more general function for At3g05500 in lipid droplet biogenesis rather than the major role proposed for oleosins in providing stability during seed desiccation/rehydration (Leprince et al., 1998; Siloto et al., 2006; Shimada et al., 2008). It is also notable that, like oleosins, the SRPP-like proteins appear to be plant specific, with the earliest SRPP-like genes present in algae and with rapid expansion of the gene families during radiation of the angiosperms (Fig. 6; Huang et al., 2013). No SRPP-like homologs were detected in other distantly related organisms, such as yeast (Saccharomyces cerevisiae), Drosophila melanogaster, mouse, or human.
Figure 6.
Comparison of gene copy number for oleosin and SRPP-like proteins in distantly related plants and algae. Oleosin and SRPP-like proteins of organisms whose genomes had been sequenced were identified using the BLASTP algorithm available at www.phytozome.net, using Arabidopsis SRPP-like protein (At3g05500) or OLEOSIN1 (At4g25140) as protein “queries.” Organisms with SRPP-like sequences that were also included in the phylogenetic analysis presented in Figure 7 are marked with asterisks.
The SRPP protein family is named for the association of these proteins with rubber particles in laticifer cells of plants that synthesize rubber, such as rubber tree, guayule (Parthenium argentatum), and Russian dandelion (Taraxacum brevicorniculatum; Collins-Silva et al., 2012; Hillebrand et al., 2012). The SRPPs share homology with the rubber elongation factor and rubber allergen protein, and there is evidence that SRPP isoforms may play a role in rubber synthesis (Collins-Silva et al., 2012). For instance, RNA interference suppression of SRPPs in Russian dandelion caused a significant reduction in overall rubber production and rubber synthase activity in comparison with wild-type plants (Hillebrand et al., 2012). Furthermore, rubber particles isolated from the RNA interference dandelion plants appeared to be less stable than those from wild-type plants, with a heterogeneity in size suggestive of rubber particle fusion (Hillebrand et al., 2012). It is unclear at present how the SRPP and rubber elongation factor proteins assemble on the rubber particle surface and influence the biosynthesis of polyisoprenoids from isopentyl pyrophosphate, but they apparently play an important role in stabilizing the surface of the rubber particle and help promote the synthesis of the hydrophobic components stored within.
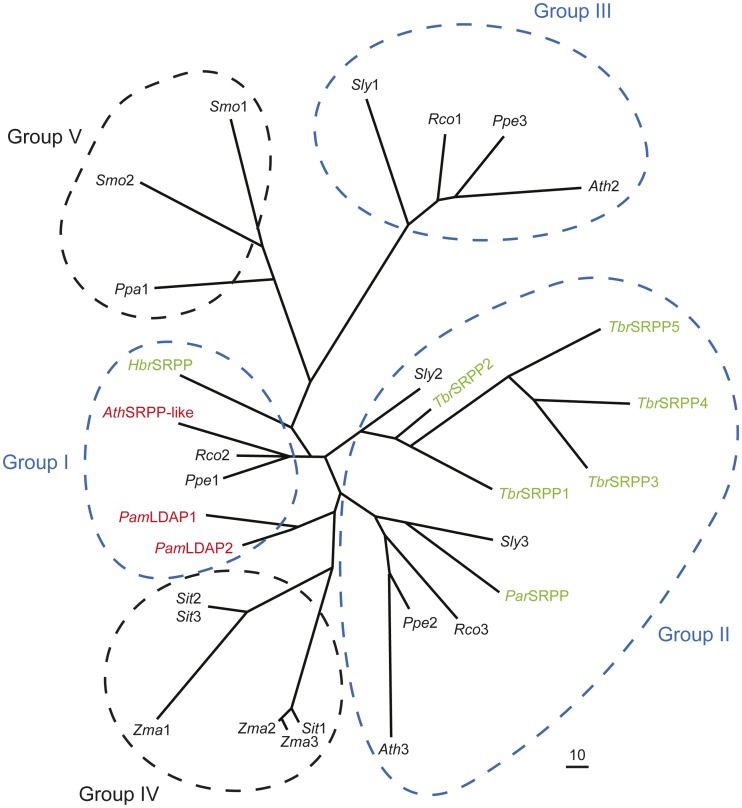
The SRPP proteins, however, may not always be essential for rubber production in plants and may have other functions in addition to stabilizing rubber particles. For instance, rubber particles isolated from Ficus carica and Ficus benghalensis did not contain SRPP proteins that cross reacted with rubber tree SRPP antisera, despite their ability to produce significant amounts of rubber (Singh et al., 2003). Russian dandelion, on the other hand, contains five SRPP-like genes, but only three of the encoded proteins have been detected in high amounts in purified rubber particles, and one of the genes shows a distinct organ and temporal gene expression pattern that is inconsistent with a role in rubber synthesis (Schmidt et al., 2010). A recent draft genome of the rubber tree (Rahman et al., 2013) indicates that there are seven SRPP genes in this genome (Fig. 6). It is possible, therefore, that the broader family of SRPP-like proteins that includes the SRPPs in rubber-producing plants, rubber elongation factor proteins, avocado LDAP1 and LDAP2, and the Arabidopsis SRPP-like protein (At3g05500) plays a more general role in the stability or formation of hydrophobic lipid particles in plant cells. It is also of interest to point out the similarity in the structure of TAG-containing lipid droplets and rubber particles (Schmidt et al., 2010), since both have a monolayer of phospholipid surrounding a hydrophobic core of reduced carbon. As such, the more widespread role of SRPP-like proteins might be to modulate the compartmentalization of TAGs in cytosolic lipid droplets, which essentially all cells must do, while those species of plants that synthesize rubber might have evolved a specialized use of SRPP for this purpose. Evidence in support of this premise comes from a phylogenetic analysis of the SRPP-like proteins from distantly related rubber-producing and non-rubber-producing plants (Fig. 7), which shows that the SRPPs from rubber-producing plants do not form a separate clade but rather are interspersed with other SRPP-like proteins from non-rubber-producing plants, such as Arabidopsis and avocado. Notably, most of the eudicots examined contained three SRPP-like sequences (Figs. 6 and 7), and these sequences were generally grouped into one of three different clades (Fig. 7, groups I, II, and III), suggesting early diversification and perhaps functional specialization. The monocot and bryophyte/lycophyte sequences, on the other hand, formed distinct clades that were separate from these groups (Fig. 7, groups IV and V, respectively).
Figure 7.
Phylogenetic analysis of SRRP-like proteins from rubber-producing and non-rubber-producing plants. Each protein is labeled with the respective genus and species, and bona fide SRPP proteins from rubber-producing plants are highlighted in green, while the SRPP-like proteins shown to associate with lipid droplets in this study (i.e. avocado [Pam] LDAP1 and LDAP2 and Arabidopsis [Ath] SRPP-like At3g05500) are highlighted in red. Other SRPP-like proteins from non-rubber-accumulating plants are labeled numerically and shown in black. Note that most of the eudicots examined (e.g. tomato, R. communis, P. persica, and Arabidopsis) contained three SRPP-like proteins that were found in three distinct clades (groups I, II, and III; circled in blue), while the sequences of the monocots (S. italica and maize) and bryophyte/lycophyte (P. patens and S. moellendorffii) formed other distinct clades (groups IV and V, respectively). The accession numbers of all sequences are provided in “Materials and Methods.” BioEdit version 7.1.3.0 (Hall, 1999) was used for sequence alignment and reconstruction. The phlyogram was generated using the program TreeView (version 1.6.6). [See online article for color version of this figure.]
Future experiments should focus on elucidating the functional role(s) of SRPP proteins in relation to lipid droplet formation/stability. Since the SRPP-like proteins appear to be mostly hydrophilic in nature, it will also be important to determine how these proteins associate with the lipid droplet surface, although recent studies of SRPP from Hevea spp. have yielded some important insight into this process (Berthelot et al., 2012). Furthermore, it will be important to identify proteins, if any, that interact with the SRPPs, as this may provide an avenue into a better understanding of the process of lipid droplet biogenesis in plant cells. By extension, this information might offer clues about the general cellular processes required for efficient packaging of reduced carbon in lipid droplets, a process that will be increasingly important for many ongoing efforts to accumulate large quantities of energy-dense lipids in vegetative tissues of plants.
MATERIALS AND METHODS
MALDI-MSI
MALDI-MSI of avocado (Persea americana) tissue prints was performed on a MALDI LTQ Orbitrap-XL (Thermo Fisher Scientific) system in a manner similar to that described for analysis of cotton (Gossypium hirsutum) seed tissue sections (Horn et al., 2012) with some modifications. Briefly, avocado purchased from a local market was sliced in half and pressed for 2 min against a 0.45-μm pore size, Protran nitrocellulose membrane to generate a tissue print. The tissue print was cut into five pieces, and each piece was adhered to a stainless steel slide with double-sided tape. The ionization matrix 2,5-dihydroxybenzoic acid (99%; Acros Organics) was sprayed onto tissue prints at 20 mg mL−1 in 70% methanol using a SunChrom SunCollect MALDI spotter (Verhaert et al., 2010). Raw data were acquired using Thermo Xcalibur (version 2.1) in positive ionization mode with automatic gain control on, laser energy of 14 µJ, and two successive scan events (m/z 400–1,200 at 60,000 resolution and m/z 50–2,000 at 30,000 resolution). Images of individual molecular species were generated using the Metabolite Imager application as described (Horn et al., 2012), except with constant ppm tolerance (±10 ppm) for selected peaks identified as TAG molecular species based on direct-infusion, shotgun lipidomics of total lipid extracts (see below).
Lipid Droplet Isolation
Lipid droplets were isolated from mesocarp tissues of avocado (cv Haas) purchased from a local market or harvested from the orchards at the University of California, Riverside (a kind gift of Dr. Mary Lu Arpaia). Lipid droplets were isolated from tissue homogenates by flotation centrifugation similar to Chapman and Trelease (1991), with some modifications. Excised mesocarp tissue (approximately 7.5 g) was finely chopped with a single-edge razor blade on ice in a 1:2 (w/v) homogenization solution of 600 mm Suc, 1 mm EDTA, and 100 mm KCl in 100 mm potassium phosphate buffer (pH 7.2). Homogenates were filtered through four layers of Miracloth and centrifuged in a Sorvall HB-6 swinging-bucket rotor at 13,000g for 30 min at 4°C. The top 5 mL of the samples was removed using a Pasteur pipette and stainless steel spatula and used to further purify lipid droplets. The remaining supernatant was used as a source for cytosolic and microsomal fractions. Lipid droplets were purified by two additional flotations through 15 mL of the homogenization solution containing 400 mm Suc. Cytosol and microsomes were fractionated by ultracentrifugation at 150,000g (Beckman TI-75 rotor at 4°C in a Sorvall Discovery 90 ultracentrifuge). Proteins in isolated fractions were precipitated in cold acetone (−20°C) overnight (at least 4:1, v/v). Parallel fractionation experiments were carried out for lipidomics analysis with total lipids extracted from each respective fraction (Horn et al., 2011).
Lipid Analysis
Total lipid extracts were spiked with the internal standard tripentadecanoin (Tri15:0-TAG; Nu-Chek Prep) for quantification. Aliquots of each extract were diluted into an infusion solution of 1:1 (v/v) chloroform:methanol plus 500 mm ammonium acetate. Samples were infused at flow rates of 7.5 µL min−1 into an electrospray ionization source and analyzed on a Thermo TSQuantum triple quadrupole mass spectrometer. Typical instrument conditions for full-scan mode included a 4-kV spray voltage, with three microscans per acquired scan, sheath gas of 25 arbitrary units, capillary temperature of 270°F, and capillary offset of 35 V. Acyl chains within TAGs were determined using a series of neutral loss scans for abundant fatty acids.
MudPIT Proteomics
Acetone-precipitated proteins were dissolved in 2× SDS-PAGE sample buffer and heated at 70°C for 15 min to resolubilize proteins before loading on freshly prepared SDS-polyacrylamide vertical slab gels (12% resolving gel). Protein samples were electrophoresed just past the stacking/resolving gel interface. Gels were stained with Coomassie Brilliant Blue R-250 to visualize proteins. Protein bands were excised from the separating gel and stored in 10% acetic acid until proteomics analysis at the Michigan State University proteomics core facility.
Proteins were trypsin digested in gel (Shevchenko et al., 1996), and peptides were purified by solid-phase extraction (Waters nanoAcquity ultra-performance liquid chromatography system and Waters Symmetry C18 peptide trap; 5 μm, 180 μm × 20 mm). Eluted peptides were sprayed directly into a Thermo Fisher Scientific LTQ-FT Ultra MS device, and survey scans were taken in the Fourier transform mode (25,000 resolution determined at m/z 400) with the top 10 ions in each survey scan subjected to collision-induced dissociation. Tandem mass spectra were analyzed by BioWorks Browser version 3.3.1 (Thermo Fisher Scientific) and compared with the avocado clc4 protein sequence database using Mascot version 2.3. Scaffold version 3.3.3 software was used to analyze and validate protein identifications with the ProteinProphet (Nesvizhskii et al., 2003) algorithm.
Avocado Deep Sequencing and Informatics
Avocado fruits (cv Hass) were harvested monthly, from October 2009 to February 2010, from a tree (44-15-11 Hass Scion on D7 Clonal rootstock) located at the University of California South Coast Research and Extension Center in Irvine. Fruits were shipped overnight at 4°C to Michigan State University. Fruits were weighed, and mesocarp was isolated and flash frozen in liquid N2 and stored at −80°C until used.
Total RNA was extracted from 3 g of mesocarp tissue that had been ground finely in liquid N2 and incubated for 10 min in 30 mL of TRIzol reagent (Life Technologies) and for an additional 5 min with 6 mL of CHCl3. After centrifugation at 12,000g for 15 min at 4°C, the aqueous phase was transferred to a fresh tube and incubated overnight with one-third volume of 8 m LiCl. Samples were then centrifuged at 12,000g for 30 min at 4°C, the pellet was resuspended in 1,000 μL buffer of the RNEasy kit, and RNA was eluted following the manufacturer’s protocol (Qiagen).
Transcriptome data for developing mesocarp was generated using 454 and Illumina sequencing techniques. For 454 sequencing, mRNA was isolated from total RNA using Sera-Mag Oligo(dT) Magnetic Beads (Thermo Fisher Scientific), and cDNA libraries were created using the Roche cDNA Rapid Library Prep Kit (Roche Diagnostics). Sequences were obtained on the Roche 454 GS FLX sequencer using the Titanium chemistry (Roche Diagnostics). For Illumina sequencing, libraries were created using an Illumina prerelease protocol for directional mRNA-seq library preparation (version 1.0). A single-read, 75-cycle run was then performed on the Illumina GAIIx sequencer, following the manufacturer’s protocols. De novo assembly of reads obtained from 454 and Illumina sequencing was accomplished with Trinity RNA-Seq software version 2. Contigs obtained were identified by BLASTX annotation against The Arabidopsis Information Resource 10 database. Finally, the coding sequences for proteins of interest were verified against genome sequencing data provided by the avocado genome sequencing group at the Laboratorio de Servicios Genómicos of Langebio in Cinvestav, Mexico.
Lipid Droplet Targeting of At3g05500
The plasmid pRTL2/At3g05500-mCherry, encoding the full-length open reading frame of At3g05500 fused to the N terminus of the monomeric cherry (Prunus avium) autofluorescent protein mCherry (Shaner et al., 2004), was constructed in the following manner. First, gene-specific forward and reverse primers with NheI overhangs (Fp, 5′-CCGGCCGCTAGCATGGCTACTCAAACGGATC-3′; Rp, 5′-CCGGCCGCTAGCATCAAGTGGATGGAACTCC-3′) were used to amplify (via PCR) the At3g05500 open reading frame from a cDNA library obtained from isolated Arabidopsis (Arabidopsis thaliana) suspension-cultured cell mRNA. The resulting PCR products were digested with NheI and ligated into NheI-digested pRTL2/mCherry, a plant expression vector containing the 35S cauliflower mosaic virus promoter, followed by a multiple cloning site and the mCherry open reading frame (Gidda et al., 2011).
Tobacco (Nicotiana tabacum ‘Bright Yellow-2’) suspension cell cultures were maintained and prepared for bombardment with pRTL2/At3g05500-mCherry plasmid DNA as described previously (Lingard et al., 2008), with the exception that cells were resuspended in BY-2 growth medium rather than transformation buffer prior to bombardment. Induction of lipid droplets in BY-2 cells was carried out as follows: approximately 30 min after bombardment, cells were transferred to a culture flask containing BY-2 growth medium, the cells were then maintained at regular growth conditions for 1 h, and thereafter, linoleic acid-albumin conjugate (Sigma) was added to the cell suspension at a final concentration of 150 µm. Approximately 4 h later, the cells were fixed in 4% (w/v) formaldehyde and stained with BODIPY 493/503 (Molecular Probes) at a final concentration of 0.1 µg mL−1.
Confocal laser-scanning microscopy images of BY-2 cells were acquired using a Leica DM RBE microscope with a Leica 63× Plan Apochromat oil-immersion objective, a Leica TCS SP2 scanning head, and the Leica TCS NT software package (Leica). Fluorophore emissions were collected sequentially in double-labeling experiments; single-labeling experiments showed no detectable crossover at the settings used for data collection. Confocal images were acquired as a z-series of representative cells, and single optical sections were saved as 512- × 512-pixel digital images. All fluorescence images of cells shown in individual figures are representative of more than 50 independent (transient) transformations from at least two independent transformation experiments. Figure compositions were generated using Adobe Photoshop CS and Illustrator CS2 (Adobe Systems).
Accession numbers for avocado LDAP1 and LDAP2 (Fig. 4) transcripts are KF031141 and KF031142, and their nucleotide sequences will be available upon publication of this paper. Other accession numbers are as follows: for Arabidopsis (AthSRPP-like, At3g05500; Ath2, At1g67360; Ath3, At2g47780), for rubber tree (HbrSRPP, AJ223389), for Parthenium argentatum (ParSRPP, AAQ11374), for Physcomitrella patens (Ppa1, EDQ55949), for Prunus persica (Ppe1, EMJ06970; Ppe2, EMJ02455; Ppe3, EMJ25061), for Ricinus communis (Rco1, XP_002514917; Rco2, XP_002512427; Rco3, XP_002531884), for Selaginella moellendorffii (Smo1, XP_002969776), for Setaria italica (Sit1, UniProt K3Z8Z8; Sit2, UniProt K3ZVR0; Sit3, UniProt K3ZW99), for tomato (Solanum lycopersicum; Sly1, XP_004239210; Sly2, XP_004230235; Sly3, XP_004247432), and for maize (Zea mays; Zma1, DAA41722; Zma2, AFW82611; Zma3, AFW82612). The full-length sequences of SRRP proteins from Russian dandelion were derived from Schmidt et al. (2010) and included TbrSRPP1 (DR401025), TbrSRPP2 (DR401208), TbrSRPP3 (DR400748), TbrSRPP4 (DR403071), and TbrSRPP5 (DR401554).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Summary information for avocado triacylglycerol molecular species.
Supplemental Table S2. Proteins enriched in isolated avocado lipid droplets.
Supplemental File S1. Proteins identified in avocado subcellular fractions.
Acknowledgments
We thank Curtis Wilkerson and Doug Whitten of the Michigan State University Proteomics core for MudPIT analysis and peptide identification; Keithanne Mockaitis (Indiana University) for assembly of 454 and Illumina contigs; Nick Thrower (Michigan State University) for read mapping and bioinformatic analysis of Avocado RNASeq data; Mary Lu Arpaia (University of California at Riverside) for providing avocados and advice regarding fruit development during these studies; Drs. Luis Herrera-Estrella and Enrique Ibarra Laclette (Laboratorio de Servicios Genómicos' of Langebio, Cinvestav, Mexico) for sharing avocado genomics information to confirm the coding sequences for LDAP1 and 2. We thank Vladimir Shulaev (University of North Texas) and Kerstin Strupat (Thermo-Fisher Scientific) for instrument access and technical support, respectively, for MS imaging experiments; Drs. Jennifer Saito and Alam Maqsudul (University of Hawaii) for access to the Hevea genome browser; and Dr. Reynald Tremblay (University of Guelph) for his assistance with the phylogentic analysis of SRPPs. Dr. Charlene Case (University of North Texas) assisted with assembly of the manuscript.
Glossary
- MudPIT
multidimensional protein identification technology
- SRPP
small rubber particle protein
- TAG
triacylglycerol
- MALDI
matrix-assisted laser desorption/ionization
- MSI
mass spectrometry imaging
- m/z
mass-to-charge ratio
- cDNA
complementary DNA
- BY-2
Bright Yellow-2
References
- Berthelot K, Lecomte S, Estevez Y, Coulary-Salin B, Bentaleb A, Cullin C, Deffieux A, Peruch F. (2012) Rubber elongation factor (REF), a major allergen component in Hevea brasiliensis latex has amyloid properties. PLoS ONE 7: e48065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue G-F, Drira N, Ohlrogge JB, Arondel V. (2011) Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci USA 108: 12527–12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza PT, Bakker-Abreu I, Navarro-Xavier RA, Bandeira-Melo C. (2011) Lipid body function in eicosanoid synthesis: an update. Prostaglandins Leukot Essent Fatty Acids 85: 205–213 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Irons S, Kearns A, Hawes C. (2003) BY-2 cells: culture and transformation for live cell imaging. Curr Protoc Cell Biol Chapter 1: Unit 1.7. [DOI] [PubMed] [Google Scholar]
- Carlsson AS, LaBrie ST, Kinney AJ, von Wettstein-Knowles P, Browse J. (2002) A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J 29: 761–770 [DOI] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. (2006) The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol 16: 1783–1795 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT. (2012) Biogenesis and functions of lipid droplets in plants. Thematic review series. Lipid droplet synthesis and metabolism: from yeast to man. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB. (2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Trelease RN. (1991) Acquisition of membrane lipids by differentiating glyoxysomes: role of lipid bodies. J Cell Biol 115: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Silva J, Nural AT, Skaggs A, Scott D, Hathwaik U, Woolsey R, Schegg K, McMahan C, Whalen M, Cornish K, et al (2012) Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, result in qualitative and quantitative changes in rubber metabolism. Phytochemistry 79: 46–56 [DOI] [PubMed] [Google Scholar]
- Cummings K, Schroeder CA. (1942) Anatomy of the avocado fruit. California Avocado Society Yearbook 27: 56–64 [Google Scholar]
- Delahunty CM, Yates JR., III (2007) MudPIT: multidimensional protein identification technology. Biotechniques 43: 563–, 565, 567. [PubMed] [Google Scholar]
- Eaks IL. (1990) Change in the fatty acid composition of avocado fruit during ontogeny, cold storage and ripening. Acta Hortic 269: 141–152 [Google Scholar]
- Frandsen GI, Mundy J, Tzen JT. (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112: 301–307 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Falcone M, Kim PK, Rothstein SJ, Andrews DW, Dyer JM, Mullen RT. (2011) Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12: 452–472 [DOI] [PubMed] [Google Scholar]
- Goodman JM. (2008) The gregarious lipid droplet. J Biol Chem 283: 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Harwood JL, Stumpf PK. (1972) Fat metabolism in higher plants. LI. Palmitic and stearic synthesis by an avocado supernatant system. Arch Biochem Biophys 148: 282–290 [DOI] [PubMed] [Google Scholar]
- Hillebrand A, Post JJ, Wurbs D, Wahler D, Lenders M, Krzyzanek V, Prüfer D, Gronover CS. (2012) Down-regulation of small rubber particle protein expression affects integrity of rubber particles and rubber content in Taraxacum brevicorniculatum. PLoS ONE 7: e41874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges BDM, Wu CC. (2010) Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res 51: 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Korte AR, Neogi PB, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee YJ, Chapman KD. (2012) Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell 24: 622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Ledbetter NR, James CN, Hoffman WD, Case CR, Verbeck GF, Chapman KD. (2011) Visualization of lipid droplet composition by direct organelle mass spectrometry. J Biol Chem 286: 3298–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AHC. (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 43: 177–200 [Google Scholar]
- Huang NL, Huang MD, Chen TL, Huang AH. (2013) Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol 161: 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet P, Acevedo F, Boulard C, d’Andréa S, Faure J-D, Kohli A, Nesi N, Valot B, Chardot T. (2013) Crop seed oil bodies: from challenges in protein identification to an emerging picture of the oil body proteome. Proteomics (in press) 10.1002/pmic.201200431 [DOI] [PubMed] [Google Scholar]
- Jolivet P, Boulard C, Bellamy A, Larré C, Barre M, Rogniaux H, d’Andréa S, Chardot T, Nesi N. (2009) Protein composition of oil bodies from mature Brassica napus seeds. Proteomics 9: 3268–3284 [DOI] [PubMed] [Google Scholar]
- Jolivet P, Roux E, D’Andrea S, Davanture M, Negroni L, Zivy M, Chardot T. (2004) Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 42: 501–509 [DOI] [PubMed] [Google Scholar]
- Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ. (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- Leprince O, van Aelst AC, Pritchard HW, Murphy DJ. (1998) Oleosins prevent oil-body coalescence during seed imbibition as suggested by a low-temperature scanning electron microscope study of desiccation-tolerant and -sensitive oilseeds. Planta 204: 109–119 [Google Scholar]
- Lin LJ, Liao PC, Yang HH, Tzen JT. (2005) Determination and analyses of the N-termini of oil-body proteins, steroleosin, caleosin and oleosin. Plant Physiol Biochem 43: 770–776 [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. (2008) Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA. (2007) Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res 48: 2751–2761 [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang L. (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Miras S, Salvi D, Ferro M, Grunwald D, Garin J, Joyard J, Rolland N. (2002) Non-canonical transit peptide for import into the chloroplast. J Biol Chem 277: 47770–47778 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. (2009) Lipid droplet-organelle interactions: sharing the fats. Biochim Biophys Acta 1791: 441–447 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Oh SK, Kang H, Shin DH, Yang J, Chow K-S, Yeang HY, Wagner B, Breiteneder H, Han K-H. (1999) Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem 274: 17132–17138 [DOI] [PubMed] [Google Scholar]
- Platt-Aloia KA, Thomson WW. (1981) Ultrastructure of the mesocarp of mature avocado fruit and changes associated with ripening. Ann Bot (Lond) 48: 451–466 [Google Scholar]
- Popluechai S, Froissard M, Jolivet P, Breviario D, Gatehouse AM, O’Donnell AG, Chardot T, Kohli A. (2011) Jatropha curcas oil body proteome and oleosins: L-form JcOle3 as a potential phylogenetic marker. Plant Physiol Biochem 49: 352–356 [DOI] [PubMed] [Google Scholar]
- Rahman AY, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, Hou S, Ong SY, Ng FL, Lee LS, et al. (2013) Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics 14: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka HA, Valdivia R. (2012) Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol 28: 411–437 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM. (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Schmidt T, Lenders M, Hillebrand A, van Deenen N, Munt O, Reichelt R, Eisenreich W, Fischer R, Prüfer D, Gronover CS. (2010) Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochem 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Wi SG, Chung GC, Kim YS, Kang H. (2003) The micromorphology and protein characterization of rubber particles in Ficus carica, Ficus benghalensis and Hevea brasiliensis. J Exp Bot 54: 985–992 [DOI] [PubMed] [Google Scholar]
- Tzen JT, Huang AH. (1992) Surface structure and properties of plant seed oil bodies. J Cell Biol 117: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umate P. (2012) Comparative genomics of the lipid-body-membrane proteins oleosin, caleosin and steroleosin in magnoliophyte, lycophyte and bryophyte. Genomics Proteomics Bioinformatics 10: 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schoot C, Paul LK, Paul SB, Rinne PL. (2011) Plant lipid bodies and cell-cell signaling: a new role for an old organelle? Plant Signal Behav 6: 1732–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaert PEM, Pinkse MH, Strupat K, Conaway MP (2010) Imaging of similar mass neuropeptides in neuronal tissue by enhanced resolution MALDI MS with an ion trap-Orbitrap hybrid instrument. In SS Rubakhin, JV Sweedler, eds, Mass Spectrometry Imaging, Vol 656. Humana Press, Totawa, NJ, pp 433–449 [DOI] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr (2012) Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81: 687–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, et al (2012) The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res 53: 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. (2012) Fat signals: lipases and lipolysis in lipid metabolism and signaling. Cell Metab 15: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]