Diverse land plant species possess similar proteins that function in transcriptional regulation of aluminum tolerance.
Abstract
Aluminum (Al) and proton (H+) tolerances are essential traits for plants to adapt to acid soil environments. In Arabidopsis (Arabidopsis thaliana), these tolerances are mediated by a zinc-finger transcription factor, SENSITIVE TO PROTON RHIZOTOXICITY1 (AtSTOP1), which regulates the transcription of multiple genes critical for tolerance to both stressors. Here, the functions of orthologous proteins (STOP1-like proteins) in other plant species were characterized by reverse genetics analyses and in planta complementation assays. RNA interference of a gene for NtSTOP1 repressed Al and H+ tolerances of tobacco (Nicotiana tabacum) roots. Tobacco roots released citrate in response to Al, concomitant with the up-regulated transcription of an ortholog of an Al tolerance gene encoding a citrate-transporting multidrug and toxic compound extrusion protein. The RNA interference repression of NtSTOP1 blocked this process and also repressed the transcription of another orthologous gene for Al tolerance, ALUMINUM SENSITIVE3, which encodes a prokaryote-type transporter. These results demonstrated that NtSTOP1 regulates Al tolerance in tobacco through the transcriptional regulation of these genes. The in planta complementation assays revealed that other plant species, including woody plants, a legume, and a moss (Physcomitrella patens), possess functional STOP1-like proteins that can activate several H+ and Al-tolerance genes in Arabidopsis. Knocking out the gene encoding the STOP1-like protein decreased the Al tolerance of P. patens. Together, our results strongly suggest that transcriptional regulation by STOP1-like proteins is evolutionarily conserved among land plants and that it confers the ability to survive in acid soils through the transcriptional regulation of Al- and H+-tolerance genes.
Worldwide, more than 30% of land is affected by acid soils (pH < 5.5). The area of acid soils is approximately 4 billion ha, accounting for approximately one-half of the world’s potential arable lands (Macdonald and Martin, 1988; Uexküll and Mutert, 1995). The growth of crop plants in acid soil is severely suppressed because of acid soil syndrome, which includes rhizotoxicities of solubilized ions such as aluminum (Al) and deficiencies of nutrients such as inorganic phosphorus (Kochian et al., 2004). Because root growth inhibition is the main symptom of acid soil syndrome, many crops become susceptible to drought stress in acid soil regions, including those in African countries (Eswaran et al., 1997). Thus, it is believed that resistance to rhizotoxicities in acid soil is one of the most important traits to protect food production around the world against drought stress.
In acid soils, Al3+ is the main ion responsible for rhizotoxicity, but proton (H+) is also harmful to sensitive plant species (Kinraide, 1998). Although research on crop plants has been limited, the growth of H+-sensitive genotypes of Arabidopsis (Arabidopsis thaliana) accessions was impaired when they were grown in acid soil (Ikka et al., 2007). Al3+ and H+ also interact with each other as a result of their chemical and biological properties. Low pH (i.e. high H+ concentration and toxicity) increases the solubility of Al and the proportion of the most toxic Al ion species, Al3+, in soil solutions, while it decreases the negativity of the plasma membrane surface, decreasing its ability to attract Al3+ (Kinraide, 1998, 2003). H+ rhizotoxicity disturbs ion homeostasis and destabilizes the cell wall, causing severe damage to the growing root tips (Koyama et al., 1995, 2001). This might be alleviated by Al, because Al can solidify pectin in the cell wall (Blamey et al., 1993), although plant species must have functional Al-tolerance mechanisms to employ this strategy. In this situation, plants need to develop a series of strategies to tolerate Al and H+ rhizotoxicities to survive in acid soil environments.
In Arabidopsis, resistance mechanisms to acid soil conditions are transcriptionally regulated by the zinc-finger transcription factor STOP1 (for sensitive to proton rhizotoxicity1). The stop1 mutant was first selected as a mutant showing hypersensitivity to H+ rhizotoxicity, but it also showed hypersensitivity to Al3+ rhizotoxicity (Iuchi et al., 2007). The dual hypersensitivities could be explained by the loss of the ability to induce the expression of a series of genes related to Al3+ and H+ tolerance (Sawaki et al., 2009). The STOP1 gene regulates the Al-inducible expression of ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (AtALMT1; Hoekenga et al., 2006) and ALUMINUM SENSITIVE3 (ALS3; Larsen et al., 2005), which encodes a prokaryote-type ATP-binding cassette transporter with a possible role in UDP-Glc transport. The Arabidopsis stop1 mutant also showed repressed transcription of genes with roles in ion homeostasis, nitrogen and carbohydrate metabolism, and those encoding proteins that stabilize the cell wall, all of which contribute to H+ tolerance (Sawaki et al., 2009). As a result, the stop1 mutant showed hypersensitivity in acid soil (Sawaki et al., 2009). This indicated that additional research on the STOP1 system would provide further details about the complex system of Al and H+ tolerances in plants.
Homologs of the STOP1 gene in Arabidopsis (hereafter AtSTOP1) exist in a wide range of plant species (Iuchi et al., 2008). In rice (Oryza sativa), a functional homolog of AtSTOP1 was identified from studies on an Al-sensitive mutant (Huang et al., 2009; Yamaji et al., 2009). The mutant carried a dysfunctional ART1 transcription factor with the conserved zinc-finger domains of STOP1. ART1 regulates the expression of the putative ALS3 homolog in rice, SENSITIVE TO ALUMINUM RHIZOTOXICITY2 (STAR2; Huang et al., 2009). In addition, transcriptome analysis of near-isogenic wheat (Triticum aestivum) lines with contrasting Al tolerances suggested that the AtSTOP1 homolog is involved in determining variations of Al tolerance (Houde and Diallo, 2008), although its exact function remains unknown. AtSTOP1 regulates the expression of a type of citrate-transporting transporter MATE (for multidrug and toxic compound extrusion), which makes a minor contribution to Al tolerance in Arabidopsis (i.e. knocked down transgenic lines showed small but significant increases in Al sensitivity; Liu et al., 2009). However, MATE-dependent citrate excretion is an important Al-tolerance mechanism in some plant species (Magalhaes et al., 2007). Together, all of these findings suggest that STOP1 transcription factors are ubiquitous in a wide range of plants and regulate Al-tolerance mechanisms, even though the Al-tolerance mechanisms may differ among plant species.
A reverse genetics approach is one way to analyze the function of STOP1 homologs from different plant species. In addition, in planta complementation assays using the Atstop1 mutant can be used to evaluate the transcription factor activities of the homologs in a plant system. In this study, we performed RNA interference (RNAi) suppression of the STOP1-like protein in tobacco (Nicotiana tabacum) and analyzed the phenotypes of complemented transgenic Atstop1 carrying homologous genes isolated from various plant species, including dicots, woody plants, and a bryophyte. These analyses strongly suggested ubiquitous and important roles of STOP1 in acid-soil resistance in a wide range of land plant species.
RESULTS
Isolation of a Gene Encoding a STOP1-Like Protein from Tobacco
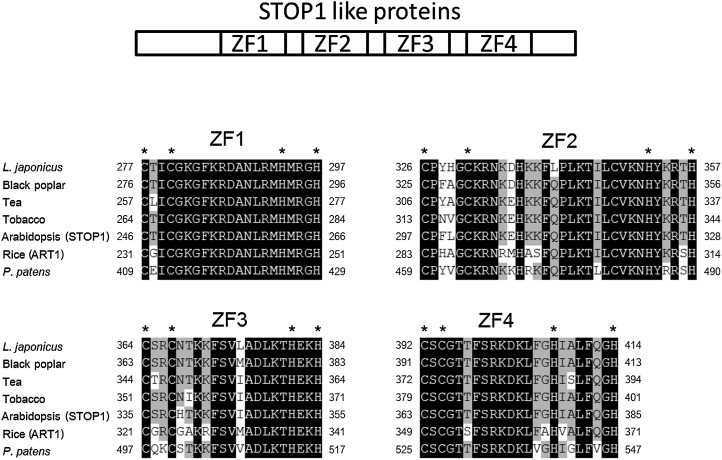
A complementary DNA (cDNA) encoding an ortholog of AtSTOP1 (i.e. a gene encoding a STOP1-like protein) was isolated from tobacco by degenerate PCR followed by RACE-PCR. The degenerate primers were designed from sequences of putative STOP1-like proteins from various plants available in the GenBank database. We conducted sequencing analysis using approximately 30 independent partial cDNA clones (all sharing the same sequence). Based on the results of that analysis, we isolated a unique cDNA encoding a protein with 506 amino acids and containing four Cys-2-His-2 zinc-finger domains that were homologous (Fig. 1) to those of AtSTOP1 and other STOP1-like proteins (i.e. ART1 and other putative STOP1-like proteins that were used for in planta complementation assays in our study; for details, see Fig. 7 below). The tobacco ortholog, hereafter referred to as NtSTOP1, was characterized by producing an RNAi-suppressed line.
Figure 1.
Deduced amino acid sequences of zinc-finger domains of the tobacco NtSTOP1 protein. Zinc-finger domains (predicted in Arabidopsis by Englbrecht et al. [2004]) were compared among AtSTOP1 (Arabidopsis), ART1 (rice STOP1 ortholog), PgSTOP1 (black poplar), LjSTOP1 (L. japonicus), CsSTOP1 (tea), and PpSTOP1 (P. patens). Strictly conserved amino acids are highlighted in black, and residues belonging to conserved amino acid groups are highlighted in gray. Asterisks indicate Cys and His of C2H2 motifs.
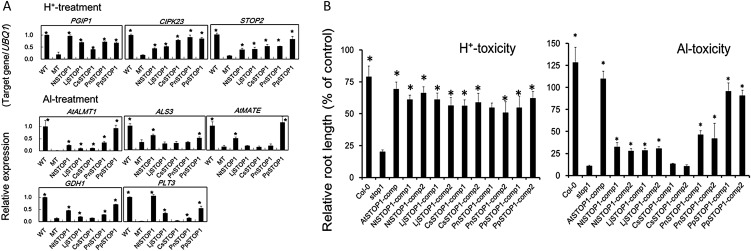
Figure 7.
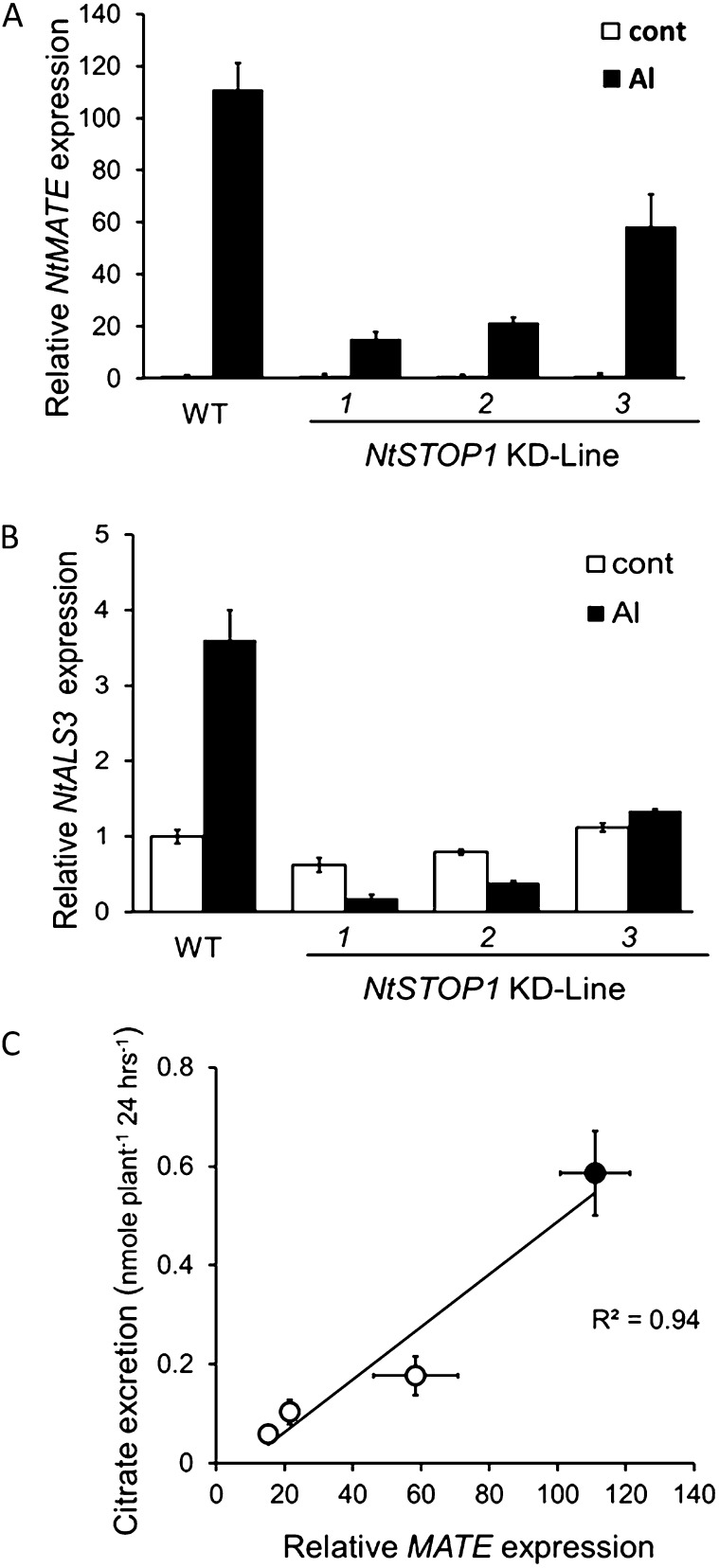
Recovery of gene transcription and H+- and Al-sensitive phenotypes of Atstop1 after complementation with homologous genes encoding STOP1-like proteins. A, Recovery of the transcription of suppressed genes in the Atstop1 mutant (Sawaki et al., 2009) was analyzed in complemented lines after exposure to H+-toxic (pH 4.5 for 24 h) and Al-toxic (10 μm Al, pH 5.0 for 24 h) conditions. Transcript levels of PGIP1, CIPK23, STOP2, AtALMT1, ALS3, AtMATE, GDH1, and PLT3 were quantified by real-time PCR. Mean values ± se (n = 3) are shown. Asterisks indicate significant difference between the complemented lines and Atstop1 (MT; Student’s t test, P < 0.05). WT, Wild type. B, Growth of complemented lines and the wild type (Columbia [Col-0]) in Al-toxic (pH 5.0, 4 μm) and H+-toxic (pH 4.7) conditions. The relative root length (percentage of that in the wild type) of complemented lines is shown. Values are means ± se (n = 3). Complemented lines carried AtSTOP1 or an ortholog of AtSTOP1 (from tobacco [NtSTOP1], black poplar [PnSTOP1], tea [CsSTOP1], L. japonicus [LjSTOP1], or P. patens [PpSTOP1]).
Growth Response of RNAi Transgenic Tobacco Suppressing NtSTOP1
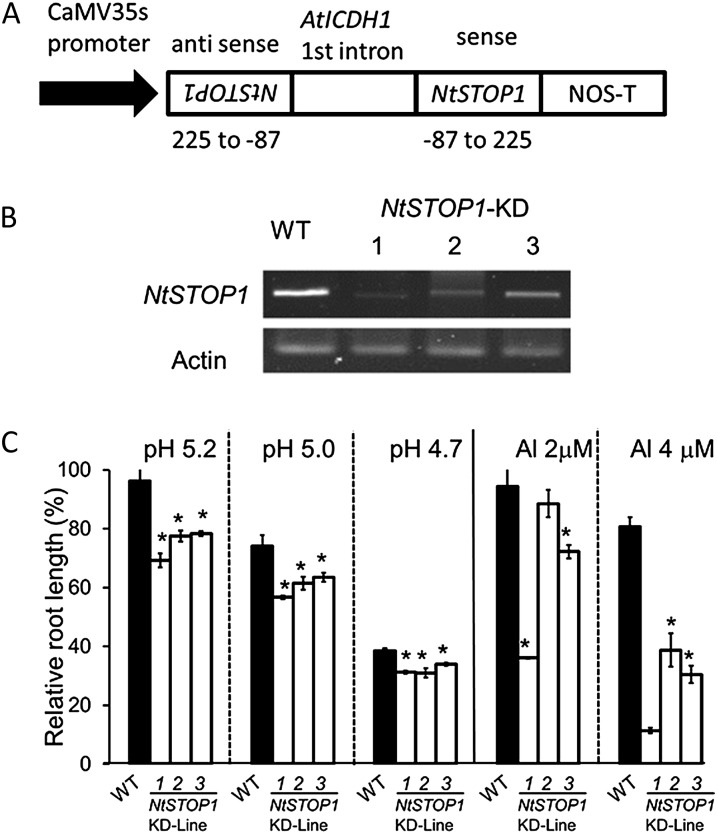
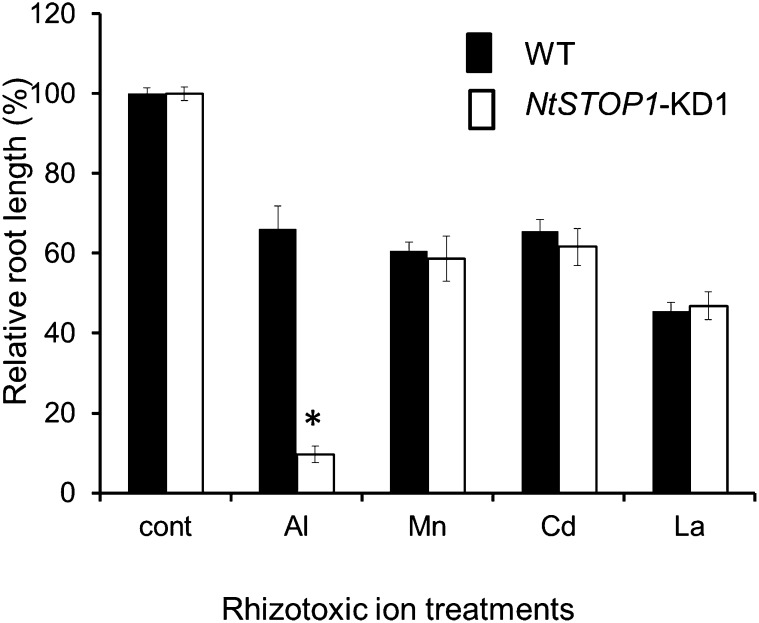
We generated transgenic tobacco plants suppressing NtSTOP1 by Agrobacterium tumefaciens-mediated transformation using a vector carrying an RNAi that targeted sequences of NtSTOP1 (Fig. 2A). These transgenic plants showed repressed expression of NtSTOP1 (Fig. 2B), and plants in the T3 generation were sensitive to Al and H+ rhizotoxicities (Fig. 2C). We conducted root growth assays for three independent RNAi lines (Knock Down1 [KD1], KD2, and KD3) in low-pH and -Al3+ conditions and compared them with the wild type. All KD lines showed significantly inhibited root growth, compared with the wild type, at pH 5.2, 5.0, and 4.7, when relative values (percentage of root growth at pH 5.5) were compared (Fig. 2C). At pH 5.5, there were no significant differences in root growth among the various lines, indicating that repression of NtSTOP1 enhanced H+ sensitivity. Al tolerance was evaluated at pH 5.0 by comparing relative values for root growth (percentage of root growth in medium without Al at pH 5.0; Supplemental Fig. S1). Al sensitivity was significantly enhanced in KD1 and KD3 in the presence of 2 μm Al and in all KD lines in the presence of 4 μm Al (Fig. 2C). The line showing the greatest repression of NtSTOP1 (KD1) was the most sensitive to Al3+ rhizotoxicity (Fig. 2C); however, KD1 did not show increased sensitivity to other rhizotoxic ions (Fig. 3). This suggested that NtSTOP1 regulates both H+ and Al tolerance in tobacco, but it does not regulate tolerance to other stressors.
Figure 2.
Al and H+ tolerance of transgenic tobacco plants with NtSTOP1 suppressed by RNAi inhibition. A, Transgenic tobacco was obtained by A. tumefaciens-mediated transformation using a vector carrying an NtSTOP1 RNAi construct. A partial cDNA of NtSTOP1 sandwiched the first intron of NADP-dependent isocitrate dehydrogenase of Arabidopsis, and the gene construct was driven by the cauliflower mosaic virus 35S promoter (CaMV35s). B, Suppression of NtSTOP1 was analyzed in T3 seed progeny of three independent transgenic lines (NtSTOP1-KD1, NtSTOP1-KD2, and NtSTOP1-KD3) by RT-PCR using ACTIN as the internal control. C, Seedlings of T3 seed progeny of NtSTOP1-KD and the wild type (WT) were grown hydroponically for 1 week in Al-toxic solutions (pH 5.0, 2 and 4 μm) and different levels of H+ toxicity (pH 5.2, 5.0, and 4.7). Means and sd of relative values (percentage of the control; pH 5.0, 0 Al for Al, pH 5.5 for pH; absolute root lengths are shown in Supplemental Fig. S2) from five seedlings are shown.
Figure 3.
The tolerance of NtSTOP1-KD1 to rhizotoxic metals was compared with that of the wild type (WT). Means and sd of relative root length (+toxicants/control) are shown (n = 5). Tolerance to toxic metals was tested by adding the following compounds to control (pH 5.0, no toxicant) solution: 4 μm CdCl2, 0.4 μm LaCl3, 200 μm MnSO4, or 4.0 μm AlCl3. The asterisk indicates a significant difference as compared with the wild type (Student’s t test, P < 0.05).
Repression of Al-Activated Citrate Excretion and Al-Tolerance Genes in Tobacco KD1
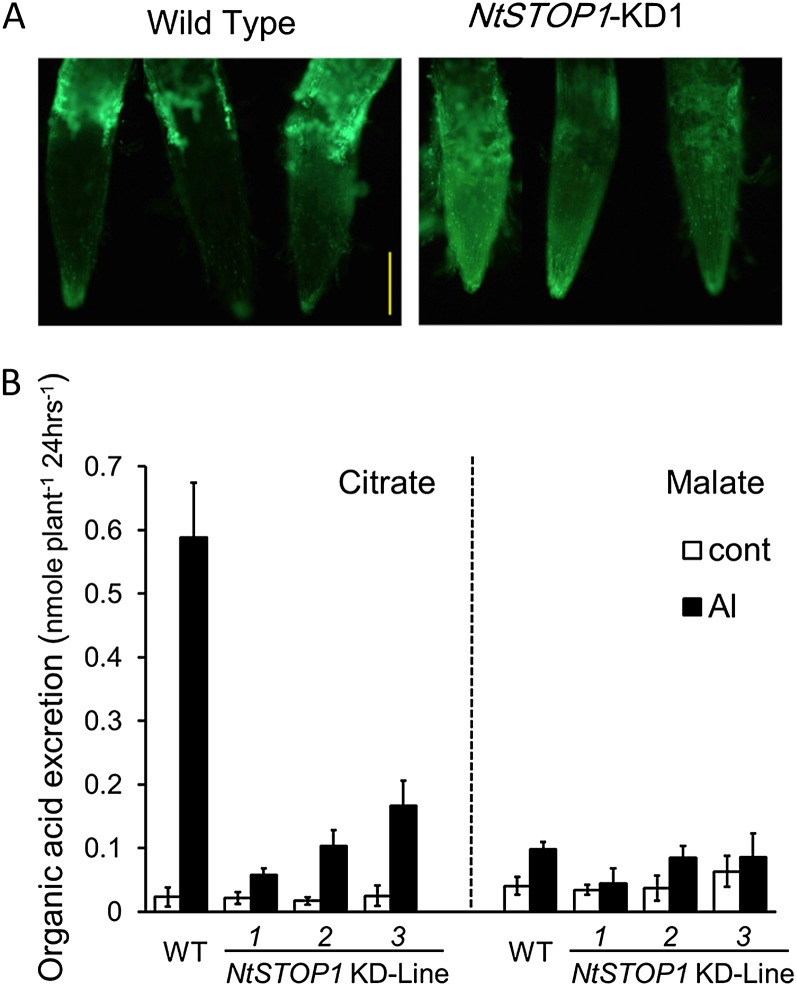
After incubation in Al-rhizotoxic conditions, the root tips of KD1 showed brighter fluorescence than those of the wild type when stained with morin (an Al-detecting fluorescent probe; control plants grown in Al-free medium showed no staining; Fig. 4A). This result showed that KD1 accumulated higher levels of Al than did the wild type, suggesting that RNAi inhibition of NtSTOP1 decreased the ability of the root tips to exclude Al. We also analyzed the concentrations of the major organic acids (malate, citrate, and oxalate) in Al-toxic culture solution (24-h incubation), because excretion of these organic acids is one of the most common mechanisms of Al exclusion from the root tip. In the wild type, the organic acid with the highest concentration was citrate, while oxalate was not detected. Citrate showed an Al-inducible excretion pattern (Fig. 4B). All KD lines showed suppressed Al-inducible citrate excretion (Fig. 4B). These results suggested that the repression of Al-induced citrate excretion is one mechanism underlying the enhanced Al sensitivity in KD lines, accompanied by increased accumulation of Al in the root tip (Fig. 4A).
Figure 4.
Al accumulation in root tips and organic acid excretion in response to Al. A, Root tips of NtSTOP1-KD1 and the wild type were stained with morin after incubation in Al-containing solution (pH 5.0, 20 μm) for 24 h. Fluorescence images of root tips are shown. Bar = 50 μm. B, Citrate (left) and malate (right) excretion from roots of the wild type (WT) and NtSTOP1-KD lines in control (pH 5.0, no Al) and Al-containing (pH 5.0, 20 μm) solutions for 24 h. White bars, Mean values ± sd in control solution; black bars, mean values ± sd in Al solutions (n = 3).
Repressed Transcription of Al-Responsive MATE and ALS3
Previous studies identified that the citrate-transporting MATE protein regulates Al-inducible citrate excretion in various plant species (Magalhaes et al., 2007). Studies on mutants of AtSTOP1 and rice ART1 identified that both mutants showed repressed transcription of orthologous genes (ALS3 in Arabidopsis, STAR2 in rice) that play roles in excluding Al from the root tip. It is possible that repression of these genes could enhance the Al susceptibility of KD lines. To test this possibility, we isolated orthologous genes from tobacco (i.e. orthologous genes encoding citrate-transporting MATE and ALS3-like proteins) and analyzed their transcript levels.
We used degenerate PCR followed by RACE-PCR to isolate genes homologous to citrate-transporting MATE (hereafter NtMATE) and ALS3/STAR2 (hereafter NtALS3). They were homologous to previously isolated genes (see phylogenetic trees in Supplemental Figs. S2 and S3). These two genes were Al inducible, but their transcription was repressed in the KD lines (Fig. 5, A and B). The Al-induced transcript level of NtMATE was much lower in KD1 than in other KD lines, and the transcript level was correlated with citrate excretion and Al tolerance levels (Fig. 5C). In addition, NtALS3 transcript levels were repressed in Al-treated KD lines (ranging from 0.13- to 0.35-fold that in the wild type). These findings suggested that NtSTOP1 regulates the expression of a set of genes that could explain the enhanced Al susceptibility of KD lines.
Figure 5.
Transcript levels of genes orthologous to those associated with Al tolerance in Arabidopsis in NtSTOP1-KD lines. A and B, Seedlings were incubated in Al-toxic (pH 5.0, 20 μm Al) or control (pH 5.0, no Al) solution for 24 h, and root samples were used for transcript analyses. Transcript levels of citrate-transporting MATE, NtMATE (homologous to AtMATE; A) and NtALS3 (homologous to AtALS3; B), were compared between NtSTOP1-KD lines and the wild type (WT). Transcript levels were relatively quantified by real-time PCR using ACTIN as the internal control, and then fold change (relative to the wild-type control) values were calculated. Means and sd of three replications are shown. C, Relationship between NtMATE expression (identical to those in B) and citrate release (identical to Fig. 4B) in different genotypes (white circles, KD; black circle, the wild type). Means ± sd of three replications are shown.
In Planta Complementation Assays with Genes Encoding STOP1-Like Proteins from Various Plant Species
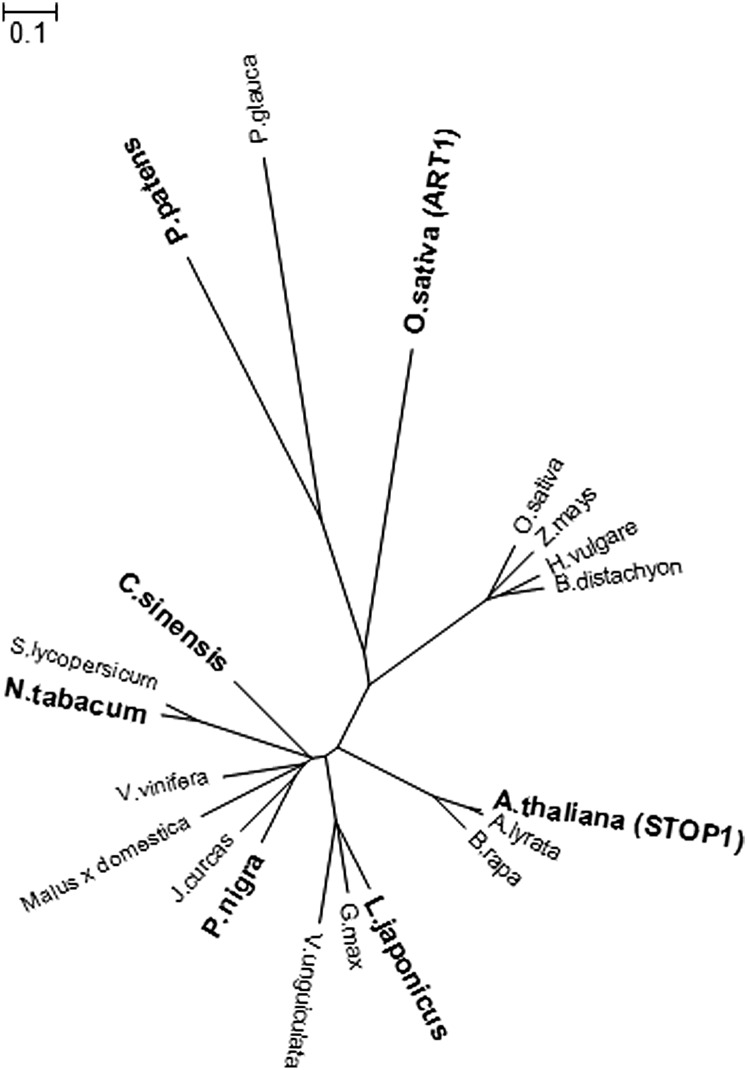
Database searches revealed putative STOP1 genes (i.e. genes encoding STOP1-like proteins) in other plant species (Fig. 6), although the transcription factor functions have been analyzed only for AtSTOP1 and ART1. To analyze the transcription factor activity of other putative STOP1-like proteins (shown in boldface in Fig. 6), we performed in planta complementation assays using the Atstop1 mutant as the complementation host. A homologous gene from tea (Camellia sinensis; CsSTOP1) was newly isolated by degenerate PCR, while a cDNA was isolated by reverse transcription (RT)-PCR from the model legume Lotus japonicus (LjSTOP1) based on the genomic DNA sequence. Two other homologous genes (PnSTOP1 from black poplar [Populus nigra] and PpSTOP1 from the moss Physcomitrella patens) were obtained from the full-length cDNA collection of the RIKEN BioResource Center. These cDNAs and NtSTOP1 were used for the complementation assays.
Figure 6.
Phylogram of STOP1-like proteins in various plant species. The distance indicator shows the relatedness of proteins. Boldface font indicates that the function of the protein has been supported by experiments using mutants (AtSTOP1 [GenBank accession no. NM_103160] and ART1 [NM_001072803]), RNAi suppression (NtSTOP1 [AB811781]), gene KO (PpSTOP1 [AB811778]), and/or in planta complementation assays (CsSTOP1 [AB811780], PnSTOP1 [AB811779], and LjSTOP1 [AB811782]). Putative orthologs are shown in regular font: B. rapa (AC232513.1), A. lyrata (XM_002891054.1), soybean (Glycine max [XM_003556158]), grape (Vitis vinifera [XM_002270160]), white spruce (BT117929), Vigna unguiculata (TC13125), tomato (AK320912), Jatropha curcas (Jcr4S27000.20), rice (NM_001051470), apple (Malus × domestica; HM122494), Brachypodium distachyon (XM_003564671), and barley (Hordeum vulgare [AK252406]).
The relatedness among the orthologous proteins (AtSTOP1, ART1, and other putative STOP1 proteins from various plant species) is shown in Figure 6. AtSTOP1 was grouped with putative STOP1 proteins from Arabidopsis lyrata and Brassica rapa, while LjSTOP1, NtSTOP1, CsSTOP1, and PnSTOP1 were grouped with those from closely related plant species. For example, LjSTOP1 was grouped with putative STOP1 proteins from other legumes; NtSTOP1 was grouped with an ortholog from tomato (Solanum lycopersicum); tea and black poplar were grouped with putative STOP1 proteins from other broadleaf trees. Rice ART1 and PpSTOP1 (moss) were grouped with a putative STOP1 ortholog from white spruce (Picea glauca; a coniferous tree), while the other rice homologous proteins were grouped with putative orthologs from monocots. This analysis suggested that genes for STOP1/ART1-like proteins are conserved among a wide range of plant species and that rice has multiple copies.
Each isolated cDNA was introduced into the Atstop1 mutant, and then transcript levels of several genes that are known to be repressed in the Atstop1 mutant were analyzed (Sawaki et al., 2009; Fig. 7A). All of the putative orthologs introduced into Atstop1 activated the transcription of several genes, suggesting that each of these STOP1-like proteins functioned as a transcription factor. However, there were differences among the orthologous genes in terms of which genes they activated. For example, under low-pH treatment, the transcript levels of POLYGALACTURONASE-INHIBITING PROTEIN1 (PGIP1), CBL-INTERACTING PROTEIN KINASE23 (CIPK23), and AtSTOP2 (a unique homolog of AtSTOP1) were generally recovered by all of the orthologous genes. For all of these genes, their transcript levels were recovered to at least 0.5 compared with their respective transcript levels in the wild type. In contrast, in the Al treatment, the homologous genes showed different abilities to recover the transcript levels of genes. AtALMT1 transcription was activated by many of the homologous genes, but only PpSTOP1 recovered the transcription of AtALMT1 to a level comparable to that in the wild type. The ability to recover the expression of ALS3 and AtMATE also varied among the orthologs (Fig. 6A). These results revealed that all of the orthologous genes have transcription factor activity in planta, but their encoded proteins did not have the full function of AtSTOP1 to activate all of the regulated genes in Arabidopsis. All these orthologous genes conferred H+ tolerance, as determined by root growth assays (Fig. 6B). In contrast, the recovery of Al3+ tolerance varied among the complemented lines (Fig. 6B) because of the lower ability of most of the transformed genes to activate the transcription of Al tolerance genes.
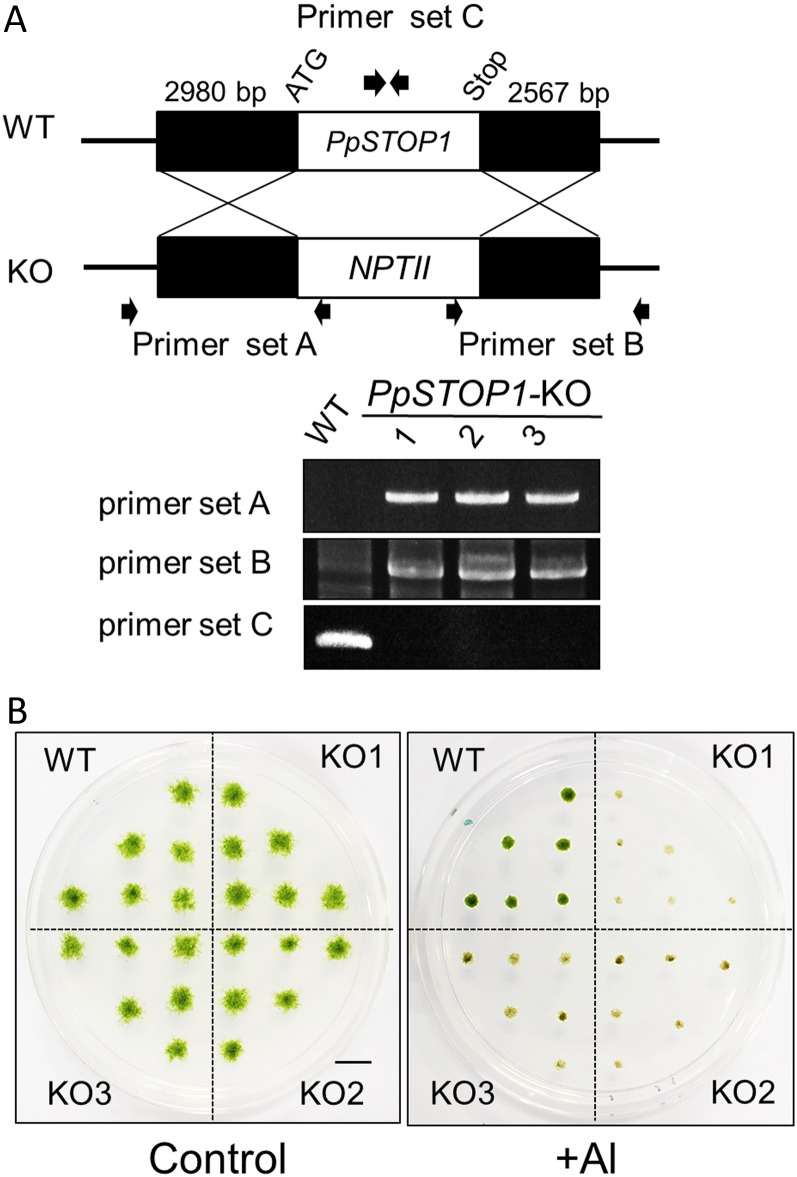
Suppression of Al Tolerance in a Moss by Gene Knockout
To evaluate the function of PpSTOP1 in moss, PpSTOP1 was knocked out by homologous recombination. In the recombinant lines, the PpSTOP1 genomic region was replaced by a kanamycin resistance gene (neomycin phosphotransferase II; Fig. 8A). The growth of knockout (KO) lines was comparable to that of the wild type on control plates (without Al) but was severely suppressed on Al-containing medium (Fig. 8B). Growth inhibition was associated with the reduction of chlorophyll (green color), which is one of the most sensitive indicators of cellular damage in this plant. These results showed that the STOP1 ortholog functioned in regulating Al tolerance in this host. The H+ sensitivity was unclear in the KO lines evaluated under these experimental conditions. Wild-type and KO lines grew comparably on control medium (pH 4.2).
Figure 8.
Al tolerance of a KO line of P. patens (PpSTOP1). A, Genomic PCR of the wild type (WT) and PpSTOP1-KO lines was conducted using primer pairs to amplify target sites (i.e. PpSTOP1 gene, NPTII gene). B, The wild-type and KO lines (KO1–KO3) were grown for 2 weeks on Al-toxic medium containing 400 μm Al at pH 4.2. Bar = 1 cm.
DISCUSSION
Acid soils are associated with both Al3+ and H+ rhizotoxicities, which cause severe damage to many crops. Plant species have developed various strategies to tolerate acidity, including internal tolerance and Al-exclusion mechanisms (Kochian et al., 2004). Recent studies identified that these tolerance mechanisms are regulated by the transcription factors AtSTOP1 (in Arabidopsis; Sawaki et al., 2009) and ART1 (in rice; Yamaji et al., 2009), which have homologous Cis-2-His-2 zinc-finger domains. In this study, we identified functional orthologous proteins (STOP1-like proteins) in other plant species. We conducted in planta complementation of the Atstop1 mutant of Arabidopsis using each of the genes encoding STOP1 proteins. The complemented lines showed recovered transcription of various genes (Fig. 7A) that are suppressed in the mutant due to the dysfunctional AtSTOP1 gene (Sawaki et al., 2009). Repression of the STOP1 ortholog in tobacco enhanced its sensitivity to H+ and Al3+ toxicities (Fig. 2C). RNAi inhibition of NtSTOP1 resulted in repressed transcription of a set of putative orthologs of genes reported to play roles in Al tolerance in other plant species. We evaluated the transcript levels of a gene encoding the citrate-transporting MATE protein (NtMATE), which is homologous to other previously identified citrate-transporting MATEs (Supplemental Fig. S2). The transcript level of NtMATE was correlated with the capacity to excrete citrate, which determined the Al sensitivity of NtSTOP1 RNAi lines (Fig. 5C). This strongly suggested that Al-inducible citrate excretion, which is regulated by NtMATE and NtSTOP1, is an Al-tolerance mechanism of tobacco. This is consistent with the previous finding that Al-tolerant cultured tobacco cells showed enhanced citrate excretion and Al tolerance (Ojima et al., 1989).
Al tolerance was repressed in Arabidopsis Atstop1 (Iuchi et al., 2007) and rice art1 (Yamaji et al., 2009) mutants, both of which carried dysfunctional STOP1-ART1-type transcription factors. However, these two plant species have different Al-tolerance mechanisms. Almost all of the Al-tolerance mechanisms of rice can be categorized as internal Al tolerance (Kochian et al., 2004), whereas Arabidopsis uses a typical Al-excretion mechanism, mainly Al-activated malate excretion through the AtALMT1 malate transporter (Hoekenga et al., 2006). In this study, we identified that a different type of Al tolerance (i.e. MATE-dependent citrate excretion, another Al-exclusion mechanism) is also regulated by the same transcription factor family in different plant species, such as tobacco. This finding suggests that the RNAi approach could be used to find genes that play critical roles in Al-tolerance mechanisms in different plant species. For example, an in planta complementation assay indicated that tea contains a functional CsSTOP1, while physiological studies indicated that it has Al-tolerance mechanisms different from those in other species. Tea plants and cultured cells released oxalic acid in response to Al (Morita et al., 2008, 2011). The release of oxalic acid is one of the major Al-tolerance mechanisms previously identified in other plants, including buckwheat (Fagopyrum esculentum; Zheng et al., 1998). A similar approach in tea would be useful to identify the oxalate excretion system at the molecular level. Although we have not yet characterized the Al-tolerance mechanisms of P. patens, disruption of the homologous gene PpSTOP1 enhanced its Al susceptibility (Fig. 7). Further research on P. patens will uncover as yet unknown mechanisms of Al tolerance in basal land plants.
In other studies, the rice art1 mutant showed enhanced Al sensitivity but no significant changes in H+ sensitivity (Yamaji et al., 2009), whereas the Arabidopsis Atstop1 mutant showed enhanced H+ sensitivity (Iuchi et al., 2007). In this study, we found similar conflicting results in two different plant species using reverse genetics approaches. Tobacco showed a small but significant increase in H+ sensitivity after suppression of NtSTOP1 (Fig. 2C), while the P. patens KO line (PpSTOP1-KO) did not show enhanced H+ sensitivity, at least on a gelled medium at pH 4.2 (Fig. 8). These results suggest that the contribution of STOP1 orthologs to H+ resistance varies among different plant species. A previous study revealed that multiple genes for H+ resistance are regulated by AtSTOP1 in Arabidopsis. However, other genes (i.e. those not strictly regulated by AtSTOP1) and proteins associated with H+ tolerance have also been reported. For example, ROF2 encoding a peptidyl-prolyl cis-trans-isomerase (Bissoli et al., 2012) and RAPID-ALKALIZATION FACTOR proteins (Srivastava et al., 2009) were identified to regulate cellular pH homeostasis. Such systems and other unknown mechanisms of H+ tolerance that are not regulated by STOP1-like proteins may operate in some plant species such as rice and P. patens.
In planta complementation assays showed that all of the tested STOP1-like proteins were able to activate the transcription of several genes in Arabidopsis. This finding can be explained by the highly conserved zinc-finger domains that activate the transcription of genes with the same target cis-motif in their promoter regions. The sequence of the cis-motif would have been conserved among plant species during evolution. However, the differences in activation of AtALMT1 expression among the different complemented lines (Fig. 7A) suggested that the protein structure of AtSTOP1, which is critical for activating the expression of AtALMT1, may not be completely conserved in some of the STOP1-like proteins. In fact, the zinc-finger domain was highly conserved among the STOP1-like proteins, but both the N and C termini showed much lower homologies (Supplemental Fig. S4). Protein phosphorylation/dephosphorylation processes are involved in AtSTOP1-dependent AtALMT1 expression (Kobayashi et al., 2007), but it remains unclear whether AtSTOP1 is directly phosphorylated or whether it interacts with other proteins via protein phosphorylation. Such processes would be affected by differences in protein sequence and/or structure, possibly in the N or C terminus, among the various homologous proteins from different plant species. The recovery of the transcription of other Al-tolerance genes (ALS3 and AtMATE) also varied among the complemented lines (Fig. 5A). As a result, the Al-tolerant phenotype was not conferred by many of the orthologous proteins. On the other hand, the H+-sensitive phenotype was conferred by almost all of the tested orthologous proteins. This finding suggested that the transcriptional activation of Al-tolerance genes is more sensitive to the protein structure of STOP1 than is the transcriptional activation of H+-tolerance genes. To explore this topic, further research is required to examine the structural and functional differences among STOP1-like proteins and the conservation of cis-elements in the promoters of target genes.
Database searches of GenBank (www.ncbi.nlm.nih.gov/genbank) and individual plant databases (e.g. PHYSCObase for P. patens [http://moss.nibb.ac.jp/]; miyakogusa.jp for L. japonicus [http://www.kazusa.or.jp/lotus/]; for black poplar, see Nanjo et al. [2004]) identified that most of the dicots and P. patens contain two or three copies of genes encoding putative STOP1-like proteins. In Arabidopsis, AtSTOP2 is a unique homolog with homologous zinc-finger domains (Englbrecht et al., 2004) that is expressed at much lower levels than AtSTOP1 (at least 10 times less; Y. Kobayashi and H. Koyama, unpublished data) and whose expression is regulated by AtSTOP1 (Fig. 6A). Suppression of Al tolerance by knocking down orthologs suggested that AtSTOP1 orthologous proteins, but not AtSTOP2 orthologs, may play a critical role in Al tolerance in a wide range of plant species. The searches identified that monocots, especially rice, may have a greater number of homologous genes, while the function of each homolog has not yet been analyzed. The increased copy number of STOP1-like proteins in rice may explain its higher Al resistance compared with that of other crops. On the other hand, all tested plants carrying a dysfunctional STOP1-like protein (Arabidopsis and rice; Sawaki et al., 2009; Yamaji et al., 2009) or those with highly reduced STOP1 levels as a result of RNAi (tobacco, our ongoing research) showed repressed expression of ALS3 orthologs. The origin of this protein in plants remains unclear, but it has the structural characteristics of a prokaryotic transporter (Larsen et al., 2005). In cancer cells in animals, many proteins of prokaryotic origin play critical roles in adaptation to cellular acidification (hypoxia; Semenza, 2001; Fernandes et al., 2012). Further research on the STOP1 regulation system may reveal how plants acquired and evolved H+- and Al-tolerance systems that include STOP1-like and ALS3-like proteins.
MATERIALS AND METHODS
Plant Materials
Tobacco (Nicotiana tabacum ‘Xanthi’) was used as the host for Agrobacterium tumefaciens-mediated transformation for RNAi suppression of the AtSTOP1 ortholog. The suppressed lines were designated as NtSTOP1-KD. Arabidopsis (Arabidopsis thaliana) wild-type Columbia (Col-0) and the stop1 mutant were identical to those used in a previous study (Iuchi et al., 2007). The stop1 mutant was used as the host for in planta complementation assays with STOP1 orthologs. Physcomitrella patens patens (Gransden) was the wild-type strain for the gene KO experiment.
Isolation of cDNAs Encoding STOP1 Orthologs, NtMATE, and NtALS3
Full-length cDNAs of AtSTOP1 orthologs were obtained for a model tree (black poplar [Populus nigra]; PnSTOP1) and the model moss P. patens (PpSTOP1) from the plant full-length cDNA library collection of the RIKEN BioResource Center (clone nos. pdp41649 for PnSTOP1 and pds27803 for PpSTOP1, developed by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science, and Technology, Japan). Putative orthologs of AtSTOP1 in tobacco (cv Xanthi), designated as NtSTOP1, and tea (Camellia sinensis ‘Yabukita’), designated as CsSTOP1, and homologs of the Al-inducible citrate-transporting MATE (NtMATE) and ALS3 (NtALS3) in tobacco were isolated by degenerate PCR (for primers, see Supplemental Table S1) followed by 3′ and 5′ RACE procedures as described previously (Kihara et al., 2003). Total RNA isolated from root samples (see below) was used as the template for RT-PCR and degenerate PCR. A cDNA of the STOP1 homolog from the model legume Lotus japonicus (LjSTOP1) was obtained by RT-PCR based on sequence information from the L. japonicus database (http://www.legumebase.brc.miyazaki-u.ac.jp/lotus/top/top.jsp). All newly isolated cDNAs were deposited into GenBank under the following accession numbers: PnSTOP1 (AB811778), PpSTOP1 (AB811779), NtSTOP1 (AB811781), CsSTOP1 (AB811780), LjSTOP1 (AB11782), NtMATE (AB811784), and NtALS3 (AB811783).
STOP1-Suppressing Tobacco Transgenic Lines
The NtSTOP1-RNAi vector for A. tumefaciens-mediated transformation was prepared by replacing the GUS gene of the binary vector pBI121 (Toyobo) with the NtSTOP1-RNAi sequence. The NtSTOP1-RNAi sequence consisted of the partial cDNA of NtSTOP1 (312 bp; −87 to +225, where 0 is first ATG) joined to the 3′ and 5′ ends of the first intron of the Arabidopsis isocitrate dehydrogenase (At1g65930) sequence in the sense (3′ end) and antisense (5′ end) orientations. This intron is efficiently spliced in transformants and can produce double-stranded RNA to suppress the target gene (Nakagawa et al., 2007). The vector was introduced into A. tumefaciens LBA4404 and then into tobacco by leaf disc transformation (Horsch et al., 1985). Regenerated plants showing kanamycin resistance were grown in soil in a growth room (20°C ± 3°C, 12 h of illumination per day, 200 μmol m−2 s−1) until seeding. T2 and T3 generations were obtained by the single-seed descent method. T3 seeds were used for organic acid excretion assays, growth assays, and transcript analyses. Integration of T-DNA was confirmed by genomic PCR as described previously (Kihara et al., 2006).
In Planta Complementation Assays of STOP1 Orthologs
All putative STOP1 orthologs (genes encoding STOP1-like proteins) were connected to the AtSTOP1 promoter (−2,848 from the first ATG) and downstream region (+626 from the stop codon) by overlap extension PCR (Horton et al., 1989). All PCRs were carried out using Prime STAR MAX (Takara), a very-high-fidelity Taq polymerase. The STOP1 ortholog constructs were introduced into the T-DNA of the pBE2113 binary vector and transformed into A. tumefaciens strain GV3101 (a supervirulent strain). Arabidopsis transformation was carried out by the floral dip method (Clough and Bent, 1998), and T2 progeny were used for assays.
Hydroponic Culture
Arabidopsis and tobacco seedlings were grown hydroponically according to methods for evaluating the Al3+ and H+ tolerance of Arabidopsis seedlings (Kobayashi et al., 2007). Briefly, 10 seeds of each line were placed on a plastic mesh supported by a polystyrene float on culture solution. The basal solution lacked inorganic phosphorus and contained 2% (w/v) MGRL nutrients and 200 μm CaCl2 as described by Kobayashi et al. (2007). The H+- and Al3+-toxic solutions were prepared by adjusting the pH with HCl and adding AlCl3 to obtain concentrations known to be toxic to Arabidopsis (pH 4.7 for H+ and Al3+ toxicity, 2 μm at pH 5.0) and tobacco (pH 5.2, 5.0, and 4.7 for H+ and Al3+ toxicity, 2 and 4 μm at pH 5.0). Culture solution at pH 5.0 without Al served as the control to evaluate Al3+ toxicity, while solutions at pH 5.0 (Arabidopsis) and pH 5.5 (tobacco) were used as the controls to evaluate H+ toxicity. All solutions were renewed every 2 d, and seedlings were grown for 7 d at 25°C with 12 h of illumination per day at 37 μE m−2 s−1). Root length was measured using a video microscope, and relative root length (%; stressed/control) values were calculated.
Morin Staining
Staining with the Al-specific fluorescent probe morin (Sigma) was performed following the method described by Tice et al. (1992). Briefly, roots of tobacco plants hydroponically grown in control solution for 7 d were incubated in a solution containing Al (20 μm, pH 5.0) and 2% (w/v) MGRL nutrients for 24 h. The roots were stained with 100 μm morin for 15 min, rinsed with distilled water, and then observed with a fluorescence microscope (AXIO Imager System; Carl Zeiss).
Collection of Root Exudates, and Quantification of Citrate and Malate
Tobacco seedlings were grown for 2 weeks in 150 mL of 1/50 MGRL nutrient (CaCl2 increased to 200 μm) solution containing 1% Suc under aseptic conditions at pH 5.5 in plastic pots. Each plant was incubated for 24 h in 2 mL of 1/50 MGRL solution containing 1% Suc on a six-well plate in the presence or absence of Al (20 μm; pH 5.0). Citrate and malate were quantified by the enzyme reaction (citrate lyase [EC 4.1.3.8] for citrate, malate dehydrogenase [EC 1.1.1.82] for malate)-coupled NADH/NAD+ cycling method developed by Hampp et al. (1984) with minor modifications as described by Kihara et al. (2003). Oxalate in the medium was quantified calorimetrically using the Enzytec kit for oxalate (R-Biopharm) according to the manufacturer’s protocol.
RNA Extraction, RT, and Real-Time RT-PCR
Total RNA was extracted from Arabidopsis and other plant species according to the methods described previously (Suzuki et al., 2001, 2004). For transcript analyses in tobacco and Arabidopsis, total RNA was isolated from the roots of plants subjected to Al-toxic (pH 5.0, 10 μm Arabidopsis, 20 μm tobacco) or control (pH 5.0, no Al) conditions for 24 h. Seedlings that were grown in preculture solution (pH 5.6, no Al) for 10 d (Arabidopsis) or 14 d (tobacco) were used for these analyses. The preculture solution contained 2% (w/v) MGRL nutrients and 200 μm CaCl2, and the same solution without inorganic phosphorus was used to incubate roots for Al treatments and the control. Total RNA was reverse transcribed to cDNA using ReverTra Ace (Toyobo) with oligo(dT) primers. Real-time RT-PCR for quantitative analysis was carried out using Power SYBR Green PCR master mix and an ABI PRISM 7000 instrument (Applied Biosystems) according to the manufacturer’s protocol for the standard curve method. For all experiments, a diluted cDNA series was used to obtain standard curves of target genes and the internal control. The internal controls were the UBQ1 gene (ubiquitin extension protein [At3g52590] for Arabidopsis) and the ACTIN gene (for tobacco). Primer information is shown in Supplemental Table S1. The transcript levels of PGIP1 (At5g06860), CIPK23 (At1g30270), STOP2 (At5g22890), AtALMT1 (At1g08430), ALS3 (At2g37330), AtMATE (At1g51340), GLUTAMATE DEHYDROGENEASE1 (GDH1; At5g18170), and PROBABLE POLYOL TRANSPORTER3 (PLT3; At2g18480) were quantified in Arabidopsis. The transcript levels of NtMATE (GenBank accession no. AB811784) and NtALS3 (GenBank accession no. AB811783) were quantified in tobacco.
DNA Sequencing, Sequence Analysis, and Database Analysis
DNA sequencing analysis was performed with the ABI PRISM 3130xl DNA Sequencer and the ABI BigDye Terminator System (version 3.1) according to the manufacturer’s recommended protocols. Orthologous genes were identified by searches of GenBank (www.ncbi.nlm.nih.gov/genbank/), Miyakogusa.jp (http://www.kazusa.or.jp/lotus/), and National Institute of Basic Biology PHYSCObase (http://moss.nibb.ac.jp/). Sequence analysis, amino acid alignment, and phylogenetic tree analysis (neighbor-joining method and construction of a nonrooted phylogram) were carried out using GENETYX software version 11.01 (Genetyx). The domains described by Englbrecht et al. (2004) were used to identify the zinc-finger domains of AtSTOP1 and other STOP1-like proteins.
KO of PpSTOP1 and Growth Test of P. patens
PpSTOP1 was disrupted using the homologous recombination method as described by Schaefer and Zrÿd (1997). The targeting sequence was synthesized by overlap extension PCR by sandwiching the neomycin phosphotransferase II gene cassette with flanking genomic DNA of PpSTOP1. PpSTOP1 disruptants were obtained as described previously (Komatsu et al., 2009) and confirmed by genomic PCR (for primers, see Supplemental Table S1). Disruptants were grown on full-strength MGRL medium lacking Pi (pH 4.2) containing 0.7% (w/v) agar and 3% (w/v) Suc in the presence or absence of 400 μm AlCl3. Plants were maintained at 25°C under continuous light (50–80 μE m−2 s−1).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers. AB811778 to AB811785.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root growth of tobacco seedlings in hydroponic culture at pH 5.5 and 5.0.
Supplemental Figure S2. Phylogram of citrate-transporting MATE proteins in various plants.
Supplemental Figure S3. Phylogram of ALS3-like proteins in various plants.
Supplemental Figure S4. Deduced amino acid alignment of STOP1-like proteins.
Supplemental Table S1. Sequence information for PCR primers used for degenerate PCR, RT-PCR, and genomic PCR.
Acknowledgments
We thank Atsuko Iuchi, Fumie Mori, and Setsuko Kawamura (RIKEN BioResource Center) for technical support. We thank the RIKEN BioResource Center and the Arabidopsis Biological Resource Center for providing Arabidopsis seeds.
Glossary
- Al
aluminum
- H+
proton
- RNAi
RNA interference
- cDNA
complementary DNA
- RT
reverse transcription
- KO
knockout
References
- Bissoli G, Niñoles R, Fresquet S, Palombieri S, Bueso E, Rubio L, García-Sánchez MJ, Fernández JA, Mulet JM, Serrano R. (2012) Peptidyl-prolyl cis-trans isomerase ROF2 modulates intracellular pH homeostasis in Arabidopsis. Plant J 70: 704–716 [DOI] [PubMed] [Google Scholar]
- Blamey FP, Asher C, Kerven G, Edwards D. (1993) Factors affecting aluminium sorption by calcium pectate. Plant Soil 149: 87–94 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Schoof H, Böhm S. (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran H, Almaraz R, van den Berg E, Reich P. (1997) An assessment of the soil resources of Africa in relation to productivity. Geoderma 77: 1–18 [Google Scholar]
- Fernandes J, Guedes PG, Lage CLS, Rodrigues JCF, Lage CdA. (2012) Tumor malignancy is engaged to prokaryotic homolog toolbox. Med Hypotheses 78: 435–441 [DOI] [PubMed] [Google Scholar]
- Hampp R, Goller M, Füllgraf H. (1984) Determination of compartmented metabolite pools by a combination of rapid fractionation of oat mesophyll protoplasts and enzymic cycling. Plant Physiol 75: 1017–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, Cançado GM, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, et al. (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103: 9738–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R, Fry J, Hoffmann N, Eichholtz D. (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Houde M, Diallo AO. (2008) Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genomics 9: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF. (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikka T, Kobayashi Y, Iuchi S, Sakurai N, Shibata D, Kobayashi M, Koyama H. (2007) Natural variation of Arabidopsis thaliana reveals that aluminum resistance and proton resistance are controlled by different genetic factors. Theor Appl Genet 115: 709–719 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi Y, Koyama H, Kobayashi M. (2008) STOP1, a Cys2/His2 type zinc-finger protein, plays critical role in acid soil tolerance in Arabidopsis. Plant Signal Behav 3: 128–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M. (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104: 9900–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara T, Wada T, Suzuki Y, Hara T, Koyama H. (2003) Alteration of citrate metabolism in cluster roots of white lupin. Plant Cell Physiol 44: 901–908 [DOI] [PubMed] [Google Scholar]
- Kihara T, Zhao CR, Kobayashi Y, Takita E, Kawazu T, Koyama H. (2006) Simple identification of transgenic Arabidopsis plants carrying a single copy of the integrated gene. Biosci Biotechnol Biochem 70: 1780–1783 [DOI] [PubMed] [Google Scholar]
- Kinraide TB. (1998) Three mechanisms for the calcium alleviation of mineral toxicities. Plant Physiol 118: 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinraide TB. (2003) Toxicity factors in acidic forest soils: attempts to evaluate separately the toxic effects of excessive Al3+ and H+ and insufficient Ca2+ and Mg2+ upon root elongation. Eur J Soil Sci 54: 323–333 [Google Scholar]
- Kobayashi Y, Hoekenga OA, Itoh H, Nakashima M, Saito S, Shaff JE, Maron LG, Piñeros MA, Kochian LV, Koyama H. (2007) Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol 145: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Pineros MA. (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, Sakata Y. (2009) Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol 70: 327–340 [DOI] [PubMed] [Google Scholar]
- Koyama H, Toda T, Hara T. (2001) Brief exposure to low-pH stress causes irreversible damage to the growing root in Arabidopsis thaliana: pectin-Ca interaction may play an important role in proton rhizotoxicity. J Exp Bot 52: 361–368 [PubMed] [Google Scholar]
- Koyama H, Toda T, Yotoka S, Dawair Z, Hara T. (1995) Effects of aluminum and pH on root growth and cell viability in Arabidopsis thaliana strain Landsberg in hydroponic culture. Plant Cell Physiol 36: 201–205 [Google Scholar]
- Larsen PB, Geisler MJ, Jones CA, Williams KM, Cancel JD. (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41: 353–363 [DOI] [PubMed] [Google Scholar]
- Liu J, Magalhaes JV, Shaff J, Kochian LV. (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 57: 389–399 [DOI] [PubMed] [Google Scholar]
- Macdonald TL, Martin RB. (1988) Aluminum ion in biological systems. Trends Biochem Sci 13: 15–19 [DOI] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT, Lana UG, Alves VM, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39: 1156–1161 [DOI] [PubMed] [Google Scholar]
- Morita A, Yanagisawa O, Maeda S, Takatsu S, Ikka T. (2011) Tea plant (Camellia sinensis L.) roots secrete oxalic acid and caffeine into medium containing aluminum. Soil Sci Plant Nutr 57: 796–802 [Google Scholar]
- Morita A, Yanagisawa O, Takatsu S, Maeda S, Hiradate S. (2008) Mechanism for the detoxification of aluminum in roots of tea plant (Camellia sinensis (L.) Kuntze). Phytochemistry 69: 147–153 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nanjo T, Futamura N, Nishiguchi M, Igasaki T, Shinozaki K, Shinohara K. (2004) Characterization of full-length enriched expressed sequence tags of stress-treated poplar leaves. Plant Cell Physiol 45: 1738–1748 [DOI] [PubMed] [Google Scholar]
- Ojima K, Koyama H, Suzuki R, Yamaya T. (1989) Characterization of two tobacco cell lines selected to grow in the presence of either ionic Al or insoluble Al-phosphate. Soil Sci Plant Nutr 35: 545–551 [Google Scholar]
- Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, et al. (2009) STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol 150: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd JP. (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Semenza GL. (2001) HIF-1, O2, and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107: 1–3 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koizumi N, Sano H. (2001) Screening of cadmium-responsive genes in Arabidopsis thaliana. Plant Cell Environ 24: 1177–1188 [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H. (2004) RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37: 542–544 [DOI] [PubMed] [Google Scholar]
- Tice KR, Parker DR, DeMason DA. (1992) Operationally defined apoplastic and symplastic aluminum fractions in root tips of aluminum-intoxicated wheat. Plant Physiol 100: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uexküll H, Mutert E (1995) Global extent, development and economic impact of acid soils. Plant Soil 171: 1–15 [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SJ, Ma JF, Matsumoto H (1998) High aluminum resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol 117: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]