Disrupting fumarylacetoacetate hydrolase leads to cell death in Arabidopsis, indicating that the Tyr degradation pathway is essential for plant survival under short-day conditions.
Abstract
Fumarylacetoacetate hydrolase (FAH) hydrolyzes fumarylacetoacetate to fumarate and acetoacetate, the final step in the tyrosine (Tyr) degradation pathway that is essential to animals. Deficiency of FAH in animals results in an inborn lethal disorder. However, the role for the Tyr degradation pathway in plants remains to be elucidated. In this study, we isolated an Arabidopsis (Arabidopsis thaliana) short-day sensitive cell death1 (sscd1) mutant that displays a spontaneous cell death phenotype under short-day conditions. The SSCD1 gene was cloned via a map-based cloning approach and found to encode an Arabidopsis putative FAH. The spontaneous cell death phenotype of the sscd1 mutant was completely eliminated by further knockout of the gene encoding the putative homogentisate dioxygenase, which catalyzes homogentisate into maleylacetoacetate (the antepenultimate step) in the Tyr degradation pathway. Furthermore, treatment of Arabidopsis wild-type seedlings with succinylacetone, an abnormal metabolite caused by loss of FAH in the Tyr degradation pathway, mimicked the sscd1 cell death phenotype. These results demonstrate that disruption of FAH leads to cell death in Arabidopsis and suggest that the Tyr degradation pathway is essential for plant survival under short-day conditions.
Programmed cell death (PCD) has been defined as a sequence of genetically regulated events that lead to the elimination of specific cells, tissues, or whole organs (Lockshin and Zakeri, 2004). In plants, PCD is essential for developmental processes and defense responses (Dangl et al., 1996; Greenberg, 1996; Durrant et al., 2007). One well-characterized example of plant PCD is the hypersensitive response occurring during incompatible plant-pathogen interactions (Lam, 2004), which results in cell death to form visible lesions at the site of infection by an avirulent pathogen and consequently limits the pathogen spread (Morel and Dangl, 1997).
To date, a large number of mutants that display spontaneous cell death lesions have been identified in barley (Hordeum vulgare), maize (Zea mays), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana; Marchetti et al., 1983; Wolter et al., 1993; Dietrich et al., 1994; Gray et al., 1997). Because lesions form in the absence of pathogen infection, these mutants have been collectively termed as lesion-mimic mutants. Many genes with regulatory roles in PCD and defense responses, including LESION SIMULATING DISEASE1, ACCELERATED CELL DEATH11, and VASCULAR ASSOCIATED DEATH1, have been cloned and characterized (Dietrich et al., 1997; Brodersen et al., 2002; Lorrain et al., 2004).
The appearance of spontaneous cell death lesions in some lesion-mimic mutants is dependent on photoperiod. For example, the Arabidopsis mutant lesion simulating disease1 and myoinositol-1-phosphate synthase1 show lesions under long days (LD; Dietrich et al., 1994; Meng et al., 2009), whereas the lesion simulating disease2, lesion initiation1, enhancing RPW8-mediated HR-like cell death1, and lag one homolog1 display lesions under short days (SD; Dietrich et al., 1994; Ishikawa et al., 2003; Wang et al., 2008; Ternes et al., 2011).
Blockage of some metabolic pathways in plants may cause cell death and result in lesion formation. For example, the lesion-mimic phenotypes in the Arabidopsis mutants lesion initiation2 and accelerated cell death2 and the maize mutant lesion mimic22 result from an impairment of porphyrin metabolism (Hu et al., 1998; Ishikawa et al., 2001; Mach et al., 2001). Deficiency in fatty acid, sphingolipid, and myoinositol metabolism also causes cell death in Arabidopsis (Mou et al., 2000; Liang et al., 2003; Wang et al., 2008; Meng et al., 2009; Donahue et al., 2010; Berkey et al., 2012).
Tyr degradation is an essential five-step pathway in animals (Lindblad et al., 1977). First, Tyr aminotransferase catalyzes the conversion of Tyr into 4-hydroxyphenylpyruvate, which is further transformed into homogentisate by 4-hydroxyphenylpyruvate dioxygenase. Through the sequential action of homogentisate dioxygenase (HGO), maleylacetoacetate isomerase (MAAI), and fumarylacetoacetate hydrolase (FAH), homogentisate is catalyzed to generate fumarate and acetoacetate (Lindblad et al., 1977). Blockage of this pathway in animals results in metabolic disorder diseases (Lindblad et al., 1977; Ruppert et al., 1992; Grompe et al., 1993). For example, human FAH deficiency causes hereditary tyrosinemia type I (HT1), an inborn lethal disease (St-Louis and Tanguay, 1997). Although the homologous genes putatively encoding these enzymes exist in plants (Dixon et al., 2000; Lopukhina et al., 2001; Dixon and Edwards, 2006), it is unclear whether this pathway is essential for plant growth and development.
In this study, we report the isolation and characterization of a recessive short-day sensitive cell death1 (sscd1) mutant in Arabidopsis. Map-based cloning of the corresponding gene revealed that SSCD1 encodes the Arabidopsis putative FAH. Further knockout of the gene encoding the Arabidopsis putative HGO completely eliminated the spontaneous cell death phenotype in the sscd1 mutant. Furthermore, we found that treatment of Arabidopsis wild-type seedlings with succinylacetone, an abnormal metabolite caused by loss of FAH in the Tyr degradation pathway (Lindblad et al., 1977), is able to mimic the sscd1 cell death phenotype. These results demonstrate that disruption of FAH leads to cell death in Arabidopsis and suggest that the Tyr degradation pathway is essential for plant survival under SD.
RESULTS
Isolation and Characterization of the sscd1 Mutant
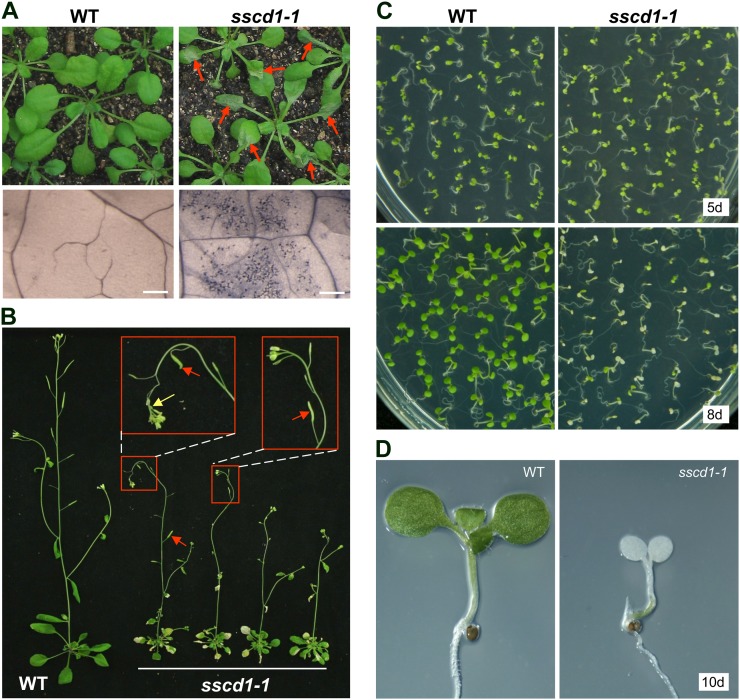
Approximately 100,000 M2 seeds from the ethyl methanesulfonate-mutagenized Arabidopsis population were first grown under LD for about 2 weeks and then transferred to SD to screen for mutants with spontaneous cell death phenotypes. One mutant, referred to as sscd1-1, exhibited obvious symptoms of wilting leaves a few days after transfer to SD (Fig. 1A, top panels). Further trypan blue staining, which assays for cell death (Dietrich et al., 1994; Bowling et al., 1997), confirmed that cell death occurred in the wilted leaves of sscd1-1 (Fig. 1A, bottom panels). The sscd1-1 mutant was crossed to wild-type Columbia (Col-0), and the F2 populations segregated for sscd1-1 and wild-type phenotypes in a ratio of 1:3, indicating that sscd1-1 resulted from the mutation of a single recessive Mendelian locus.
Figure 1.
The sscd1-1 mutant exhibits a cell death phenotype under SD. A, Wild-type (WT) and sscd1-1 seedlings grown in soil under LD for 2 weeks and then under SD for 4 d (top panels). Trypan blue staining of wild-type and wilted sscd1-1 leaves is shown in the bottom panels. The red arrows indicate some of the wilted leaves. Bars = 250 µm. B, Wild-type and sscd1-1 plants grown in soil under LD for 2 weeks and then under SD for 3 weeks. The red arrows indicate some fertile siliques, and the yellow arrow indicates wilting inflorescences. C, Wild-type and sscd1-1 seedlings grown on MS under SD for 5 d (5d; top panels) or 8 d (8d; bottom panels). D, Wild-type and bleached sscd1-1 seedlings grown on MS under SD for 10 d (10d).
When germinated and grown under LD for about 2 weeks and then transferred to SD, the sscd1-1 mutant plants exhibited obvious cell death symptoms but were able to grow and produce new leaves, inflorescences, and occasional partially fertilized siliques (Fig. 1, A and B). However, when germinated and grown under SD, most of sscd1-1 seedlings became bleached and then died (Fig. 1, C, bottom panels, and D) within 10 d, although the seedling phenotype of sscd1-1 was similar to the wild type at the early stage within 5 d (Fig. 1C, top panels).
Map-Based Cloning of the SSCD1 Gene
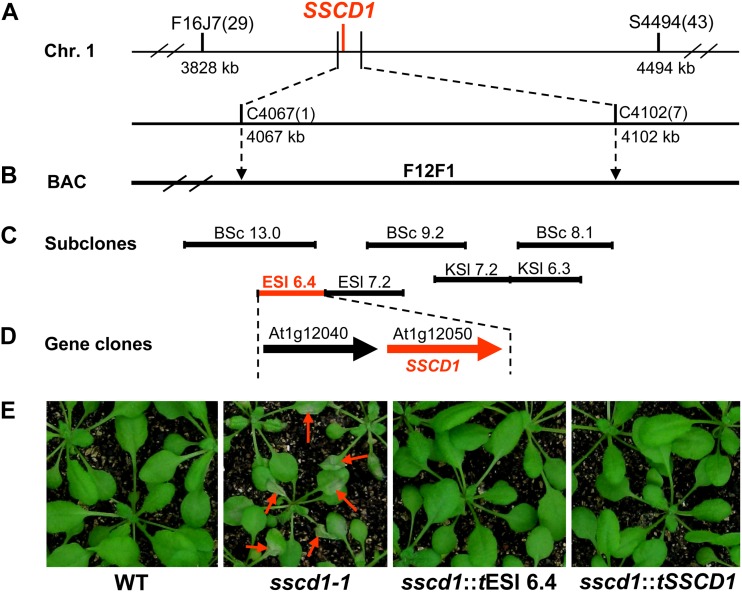
Based on the linkage analysis among molecular markers and the sscd1 phenotype of approximately 2,500 seedlings from the F2 population of the cross between sscd1-1 and wild-type Landsberg erecta, the SSCD1 gene was localized on chromosome I between two simple sequence-length polymorphic markers (F16J7 and S4494; Fig. 2A). Further mapping placed SSCD1 between two cleaved-amplified polymorphic sequence (CAPS) markers (C4067 and C4102), which are included in the bacterial artificial chromosome (BAC) clone F12F1 (Fig. 2B).
Figure 2.
Mapping of the SSCD1 gene. The SSCD1 locus was mapped to an approximately 35-kb interval between the CAPS markers C4067 and C4102 on chromosome I (A), which is included in a single BAC F12F1 (B). Complementation assays with subcloned fragments of F12F1 (C) and the genes (D) showed that the phenotypes of sscd1-1 transgenic for the subclone ESl 6.4 (sscd1::tESl 6.4) and At1g12050 (sscd1::tSSCD1) were restored to the wild type (E). The positions shown below the markers indicate the locations of the Arabidopsis Genome Initiative map on the chromosome. The numbers in parentheses indicate the number of recombinant plants. Restriction endonucleases used to generate subclones (BSc, BamHI + SacI; KSl, KpnI + SalI; ESl, EcoRI + SalI) are shown. The fragment and gene marked in red can complement sscd1-1. The red arrows in E indicate wilted leaves.
The SSCD1 gene was finally localized onto an approximately 6-kb fragment (ESl 6.4) by functional complementation with the subcloned fragments of F12F1 (Fig. 2, C and E). The ESl 6.4 fragment contains two genes, At1g12040 and At1g12050 (Fig. 2D). At1g12050 could fully rescue the cell death phenotype in the sscd1-1 mutant background (Fig. 2E), which was referred to as the SSCD1 gene.
The corresponding sequence of the SSCD1 gene from sscd1-1 deviated from that of the wild type by a single nucleotide substitution, G to A, at position +1,030 relative to its translation start codon (Fig. 3A). The G1030A nucleotide substitution is predicted to convert codon 157 (W) into a translation stop codon (Fig. 3B). Sequence analysis reveals that the SSCD1 gene encodes a putative polypeptide of 421 amino acids (Fig. 3B) with 53% identity to the human FAH, an enzyme involved in Tyr degradation (Lindblad et al., 1977). The SSCD1 protein is predicted to contain a domain of unknown function, DUF1969, and a FAH domain using the Plant Proteomics Database (http://ppdb.tc.cornell.edu; Sun et al., 2009; Fig. 3C).
Figure 3.
Characterization of the sscd1-1 mutation. A, Structure of the SSCD1 gene and the mutated base in the sscd1-1 mutant. Black boxes and uppercase letters represent exons. Thick lines and lowercase letters represent introns. White boxes represent untranslated regions. The numbers indicate the locations of bases relative to the translation start codon (ATG). TGA is the stop codon. B, The coding sequence of the SSCD1 gene and the corresponding amino acid sequence. A single base change from g to a at the 470th base in the sscd1-1 mutant results in the conversion of the 157th codon (tgg) for W into a stop codon (tag). The coding sequence is shown in lowercase letters (top row), and the corresponding deduced amino acid sequence is shown in single-letter code (bottom row). The asterisk indicates the translation stop. C, The SSCD1 protein is predicted to contain a domain of unknown function, DUF1969, and a FAH domain (top) that is defective in the sscd1-1 mutant (bottom). The numbers above the columns indicate the locations of the amino acids. [See online article for color version of this figure.]
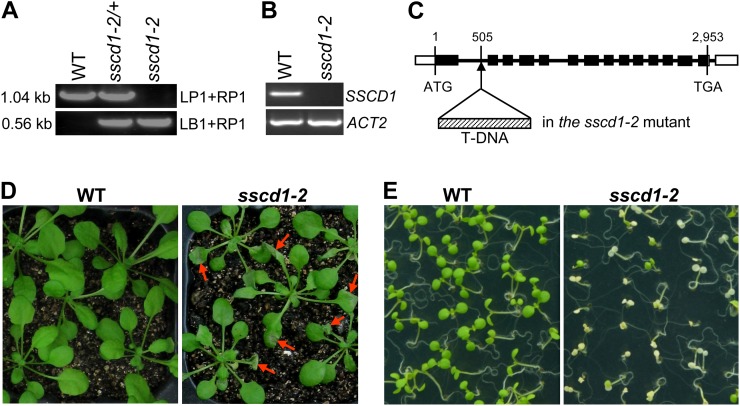
In addition to the sscd1-1 mutant, we identified another mutant (SAIL_128_B11; Fig. 4A), referred to as the sscd1-2 mutant, with the transferred DNA (T-DNA) insertion into the first intron at +505 bp relative to the translation start codon in the SSCD1 gene (Fig. 4C). Reverse transcription (RT)-PCR analysis showed that expression of the SSCD1 gene was completely abolished in sscd1-2 (Fig. 4B). Similar to sscd1-1, the sscd1-2 mutant also exhibited cell death phenotypes under SD (Fig. 4, D and E), which further verified that disruption of FAH leads to cell death phenotypes in Arabidopsis.
Figure 4.
A T-DNA insertion line in the SSCD1 gene shows a cell death phenotype. A, PCR-amplified products from the wild type (WT), heterozygote (sscd1-2/+), and homozygote (sscd1-2) using two specific primer pairs, LP1 + RP1 and LB1 + RP1 (see “Materials and Methods”). The sizes of the PCR products are indicated. B, RT-PCR analysis of SSCD1 expression in the wild type and the sscd1-2 mutant. ACT2 served as a control. C, Location of the T-DNA insert in the SSCD1 gene in the sscd1-2 mutant. Black boxes and thick lines represent exons and introns, respectively. White boxes represent untranslated regions. The numbers indicate the locations of bases relative to the translation start codon (ATG). TGA is the stop codon. D, Wild-type and sscd1-2 seedlings grown in soil under LD for 2 weeks and then under SD for 4 d. The red arrows indicate some of the wilted leaves. E, Wild-type and sscd1-2 seedlings grown on MS under SD for 8 d.
Cell Death in sscd1 Was Eliminated by Knockout of the HGO Gene
In humans, loss of fumarylacetoacetate (FAA) causes HT1, a lethal disorder (St-Louis and Tanguay, 1997), which probably results from an accumulation of toxic Tyr degradation intermediates such as maleylacetoacetate (MAA) and FAA (Kvittingen, 1986; Jorquera and Tanguay, 2001). To investigate whether the cell death in the sscd1 mutant might also result from an accumulation of the intermediates MAA and FAA, we sought to interrupt the production of these intermediates in the sscd1 mutant by the inactivation of HGO essential for the generation of MAA and FAA.
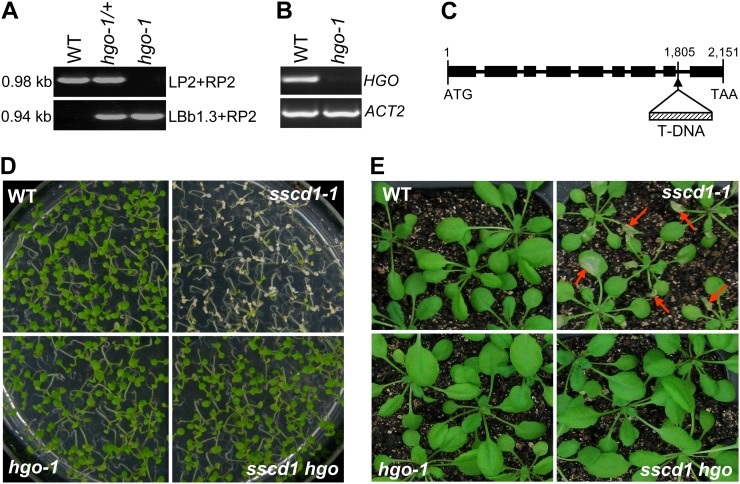
Arabidopsis contains a single gene (At5g54080) encoding a putative HGO, which shows strong amino acid identity (57%) to the human HGO (Fernández-Cañón and Peñalva, 1995a; Dixon and Edwards, 2006). We identified a T-DNA insertion mutant (SALK_027807; Alonso et al., 2003) in At5g54080, referred to as the hgo-1 mutant, where HGO expression was completely abolished (Fig. 5, A–C). We generated the sscd1 hgo double mutant through genetic crossing of sscd1-1 with hgo-1. Phenotypic analysis showed that the sscd1 hgo double mutant did not display the cell death phenotypes under SD (Fig. 5, D and E). These results demonstrated that the hgo-1 mutation, which interrupts the production of Tyr degradation intermediates MAA and FAA, could completely suppress cell death in the sscd1 mutant, suggesting that the accumulation of these intermediates causes the cell death phenotype in the sscd1 mutant.
Figure 5.
The hgo-1 mutant completely suppresses the cell death phenotype of the sscd1-1 mutant. A, PCR-amplified products from the wild type (WT), heterozygote (hgo-1/+), and homozygote (hgo-1) using two specific primer pairs, LP2 + RP2 and LBb1.3 + RP2 (see “Materials and Methods”). The sizes of the PCR products are indicated. B, RT-PCR analysis of HGO expression in the wild type and the hgo-1 mutant. C, Structure of the HGO gene (from the translation start codon to the stop codon) and the location of the T-DNA insertion in the hgo-1 mutant. Black boxes and thick lines represent exons and introns, respectively. The numbers indicate the locations of bases relative to the start codon (ATG). TAA is the stop codon. D, Wild-type, sscd1-1, hgo-1, and sscd1 hgo seedlings grown on MS under SD for 9 d. E, Wild-type, sscd1-1, hgo-1, and sscd1 hgo seedlings grown in soil under LD for 2 weeks and then under SD for 6 d. The red arrows indicate some of the wilted leaves.
Succinylacetone Induces Cell Death in Arabidopsis
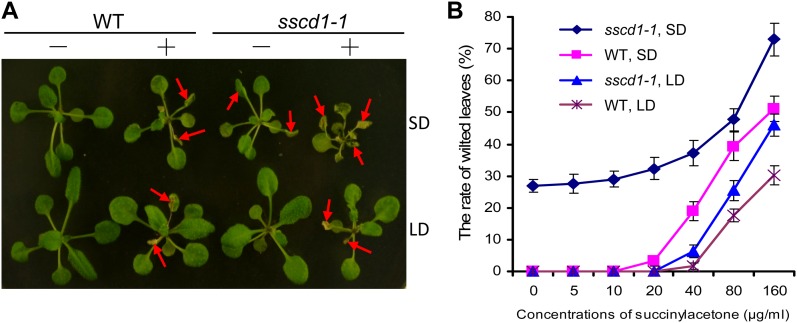
Once MAA and FAA accumulate, resulting from the loss of FAH, both of them are able to undergo spontaneous reduction to succinylacetoacetate followed by spontaneous nonenzymatic decarboxylation to succinylacetone (Lindblad et al., 1977). In both human HT1 and mouse fah mutants, the metabolic intermediates (MAA, FAA, succinylacetoacetate, and succinylacetone) were highly accumulated, which are toxic to cells and tissues (Lindblad et al., 1977; Grompe et al., 1993; Aponte et al., 2001; Jorquera and Tanguay, 2001). Due to technical difficulties, it is so far not possible to detect these metabolic intermediates in entire worms (Fisher et al., 2008) and in Arabidopsis plants (data not shown). To verify that the cell death phenotype in the sscd1 mutant might also have resulted from the accumulation of succinylacetoacetate and succinylacetone, we followed the approach used in worms (Fisher et al., 2008) and treated Arabidopsis seedlings with succinylacetone, the unique product that is commercially available among these metabolic intermediates. We found that treatment of wild-type seedlings with succinylacetone mimicked the sscd1 phenotypes: the treated wild-type seedlings exhibited wilted leaves and slow-growth symptoms under LD (Fig. 6A); these symptoms were more severe under SD (Fig. 6). Moreover, we found that the sscd1 mutant was more sensitive to succinylacetone than the wild type under LD (Fig. 6). Application of succinylacetone was able to significantly exacerbate the cell death of the sscd1 mutant under SD (Fig. 6). In addition, we found that the toxicity of succinylacetone to Arabidopsis seedlings is dose dependent (Fig. 6B). For example, treatment with 5 and 10 μg mL−1 succinylacetone was unable to cause leaf wilting in wild-type seedlings under SD (Fig. 6B); however, severe symptoms (approximately 50% wilted leaves) developed in wild-type seedlings when they were treated with 160 μg mL−1 succinylacetone (Fig. 6B). Taken together, these results demonstrated that the accumulation of the metabolic intermediates, including succinylacetone, causes the cell death phenotype in the sscd1 mutant.
Figure 6.
Treatment of Arabidopsis seedlings with succinylacetone causes cell death. A, Wild-type (WT) and sscd1-1 seedlings grown under LD for 12 d were treated without (−) or with (+) 160 µg mL−1 succinylacetone and grown for an addition 5 d under SD or LD. The red arrows indicate some of the wilted leaves. B, Rate of wilted leaves in the indicated seedlings. Error bars represent sd (n > 60). The experiment was repeated three times.
DISCUSSION
In plants, Tyr can be converted into 4-hydroxyphenylpyruvate by Tyr aminotransferase and then transformed into homogentisate by 4-hydroxyphenylpyruvate dioxygenase (Löffelhardt and Kindl, 1979; Fiedler et al., 1982; Lopukhina et al., 2001). Homogentisate is the precursor of tocopherol biosynthesis in plants and also can be produced from other sources such as the shikimate pathway (Fiedler et al., 1982; Arango and Heise, 1998; Collakova and DellaPenna, 2003). The Arabidopsis complementary DNAs encoding the HGO, MAAI, and FAH homologs were expressed in Escherichia coli and able to degrade homogentisate into fumarate and acetoacetate in the in vitro assays (Dixon and Edwards, 2006), indicating that the Tyr degradation pathway may exist in plants. However, the role of the Tyr degradation pathway in plants is still unknown. In this study, we found that disruption of FAH, an enzyme catalyzing FAA into fumarate and acetoacetate in the final step of the Tyr degradation pathway (Lindblad et al., 1977), leads to spontaneous cell death in Arabidopsis. Our results reveal the importance of the Tyr degradation pathway in plants and suggest that the Tyr degradation pathway is essential for plant survival under SD.
In animals, Tyr degradation is an essential pathway (Lindblad et al., 1977), and blockage of it results in metabolic disorder diseases (Lindblad et al., 1977; Ruppert et al., 1992; Grompe et al., 1993). Deficiency of FAH in animals causes lethality (Grompe et al., 1993; St-Louis and Tanguay, 1997), which is most likely due to the accumulation of the Tyr degradation intermediates MAA and FAA (Kvittingen, 1986; Jorquera and Tanguay, 2001). The HGO, an enzyme upstream of FAH in the Tyr degradation pathway, is essential for the catalysis of homogentisate into MAA and FAA (Lindblad et al., 1977). Inactivation of HGO in fungi, mice, and worms, therefore, was able to rescue the lethality associated with FAH mutations (Fernández-Cañón and Peñalva, 1995b; Manning et al., 1999; Fisher et al., 2008). Through generation of the double mutant sscd1 hgo, we also found that disruption of the Arabidopsis putative HGO is able to completely eliminate the spontaneous cell death in the sscd1 mutant (Fig. 5). These results suggest that the cell death in the sscd1 mutant could, directly or indirectly, result from the accumulation of MAA and FAA. Together with the observations from fungi, mice, and worms (Fernández-Cañón and Peñalva, 1995b; Manning et al., 1999; Fisher et al., 2008), our results suggest that the Tyr degradation pathway may be a universal and conserved pathway.
The accumulation of MAA and FAA, resulting from the loss of FAH, would lead to abnormal metabolism of these compounds, both of which are reduced to succinylacetoacetate that is then converted to succinylacetone (Lindblad et al., 1977). Studies on human HT1 and its animal models have shown that the accumulation of MAA, FAA, succinylacetoacetate, and succinylacetone causes direct tissue damage (Lindblad et al., 1977; Grompe et al., 1993; Sun et al., 2000; Aponte et al., 2001; Jorquera and Tanguay, 2001; Bergeron et al., 2006). In this study, we found that treatment of Arabidopsis wild-type seedlings with succinylacetone is able to mimic the sscd1 cell death phenotype (Fig. 6A) in a dosage-dependent manner (Fig. 6B), suggesting that accumulation of the metabolic intermediates, including succinylacetone, may lead to the cell death phenotype in the sscd1 mutant.
The sscd1 mutant seedlings do not display a cell death phenotype under LD (data not shown). Compared with the wild type, sscd1 mutant seedlings, under LD, are more sensitive to exogenous succinylacetone in a dosage-dependent manner (Fig. 6). Consistent with the cell death phenotypes in the sscd1 mutant under SD but not under LD (Figs. 1 and 4; data not shown), we found that, under SD but not under LD, the sscd1 mutant accumulated higher levels of the Arabidopsis zeta class glutathione transferase1, which encodes the Arabidopsis putative MAAI (Dixon and Edwards, 2006) responsible for isomerizing MAA to FAA (Dixon et al., 2000; Chen et al., 2003; data not shown). Together with the data shown in Figures 5 and 6, these data indicate that higher levels of Tyr degradation intermediates might accumulate to cause cell death in the sscd1 mutant under SD but not under LD. It is possible that LD may suppress the accumulation of Tyr degradation intermediates.
We propose a possible mechanistic explanation for the relationships among cell death, photoperiod, and the Tyr degradation pathway. In the sscd1 mutant under SD, disruption of FAH leads to the accumulation of Tyr degradation intermediates, including MAA, FAA, succinylacetoacetate, and succinylacetone, which are toxic to plant cells and cause cell death. However, LD may suppress the accumulation of these Tyr degradation intermediates through yet unidentified mechanisms. The accumulation level of these intermediates could be very lower under LD, which is insufficient to damage cells and tissues. When the biosynthesis of these intermediates is abolished through the disruption of HGO, the cell death phenotype disappears under SD in the sscd1 hgo double mutant.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines SAIL_128_B11 and SALK_027807 (Alonso et al., 2003) were ordered from the Arabidopsis Biological Resource Center.
Seeds were surface sterilized, plated on plant growth medium (Murashige and Skoog medium supplemented with 1% Suc [MS]), chilled at 4°C for 2 d, and then transferred to a growth chamber under LD (16 h of light/8 h of dark) or SD (8 h of light/16 h of dark) at 150 μmol m−2 s−1 and 22°C. Similar conditions were followed for soil-grown plants.
Mutant Screening
Approximate 30,000 seeds of Arabidopsis Col-0 were mutagenized with 0.3% ethyl methanesulfonate following routine procedures. About 70% of mutagenized seeds (approximately 20,000 seeds) were able to grow in soil and produce M2 seeds. M2 seeds were routinely plated on MS, grown under LD for 2 weeks, and then moved to SD for screening of mutants with the wilting leaf phenotype.
Trypan Blue Staining
Trypan blue staining for the detection of cell death was performed according to Bowling et al. (1997). The leaves were boiled for approximately 2 min in a lactic acid-phenol-trypan blue solution (2.5 mg mL−1 trypan blue, 25% [w/v] lactic acid, 23% water-saturated phenol, and 25% glycerol) and then stained overnight. Next, samples were decolorized with a chloral hydrate solution (2.5 g mL−1) for 3 d. After multiple exchanges of chloral hydrate solution, samples were equilibrated for several hours in 70% glycerol and examined with JSZ8 series zoom stereo microscope with a Nikon digital sight DS-Fi1.
Molecular Markers
The SCCD1 gene was identified using the map-based cloning approach (Xie et al., 1998). The CAPS markers C4067 and C4102 showed a DNA polymorphism between Col-0 and Landsberg erecta when SspI and NcoI were used, respectively, to digest the PCR fragment amplified with their corresponding primers (C4067, 5′-AACACTAGTGCCGTCACGTG-3′ and 5′-GCTGTCTAGATTCAATGTATC-3′; C4102, 5′-ATCATGTAGAAGAAGGGTATC-3′ and 5′-GAAGGATACAATTTCAAAGAAC-3′). The simple sequence-length polymorphic markers F16J7 and S4494 showed a polymorphism between Col-0 and Landsberg erecta when the PCR fragment was amplified with their corresponding primers (F16J7, 5′-TGATGTTGAGATCTGTGTGCAG-3′ and 5′-GTGTCTTGATACGCGTCGAT-3′; S4494, 5′-AATCCAATTAACAGTAGGCA-3′ and 5′-AGGTAATCATAACACAAATAATGT-3′).
Complementation Test
The BAC F12F1 (AC002131) was digested with the restriction enzymes indicated in Figure 2. Each digested fragment was recovered and cloned into pCambia1301 vector.
A 3,175-bp genomic fragment of the SSCD1 gene was amplified by FastPfu DNA polymerase (TransGen) from the wild-type (Col-0) plants using a forward primer (5′-GTACCCGGGATGGCGTTGCTGAAGTCTTT-3′) located at the SSCD1 start codon with a SmaI site added and a reverse primer (5′-CGAGAGCTCACTTATTGTTAATGGGTTGG-3′) corresponding to 3′ of the stop codon with the addition of a SacI site and then cloned into the pROK2 vector under the control of a 35S promoter.
All constructs were mobilized into Agrobacterium tumefaciens by electroporation and then introduced into the sscd1-1 mutant by the floral dip method of in planta A. tumefaciens-mediated transformation (Clough and Bent, 1998).
The sscd1-1 plants transgenic for an approximately 6.4-kb insert of F12F1 and the SSCD1 gene were referred to as sscd1::tESl 6.4 and sscd1::tSSCD1, respectively.
Characterization of T-DNA Insertion Lines
The specific primers for identifying the sscd1-2 mutant from the SAIL_128_B11 line were LP1 (5′-CATCATCTCAAAAACGGAAGC-3′), RP1 (5′-TGCATCAAACAGCAATAAACG-3′), and LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′), and the specific primers for identifying the hgo-1 mutant from the SALK_027807 line were LP2 (5′-CCTCCTCGATGGTTGGTTGC-3′), RP2 (5′-GTCGGTAGCTCGGTGTTTGT-3′), and LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′).
RT-PCR Analysis
Two-week-old seedlings grown in MS were harvested. RT-PCR analysis was performed following routine procedures. The SSCD1 gene was amplified with primers 5′-CCTCGTCCTGCCGTCGCTAT-3′ and 5′-CTTGTGGATGGCCCTGACCT-3′, and the HGO gene was amplified with primers 5′-CGGTGAACTCTTTACTGCTA-3′ and 5′-ATCTAAACCAACACCGTTAT-3′. The ACT2 gene was used as the internal control and amplified with primers 5′-TTCCGCTCTTTCTTTCCAAGCTCA-3′ and 5′-AAGAGGCATCAATTCGATCACTCA-3′. The program of PCR was as follows: 95°C for 2 min; 25 cycles of 94°C for 30 s, 51°C to 55°C for 30 s, and 72°C for 1 min; and then 72°C for 10 min.
Generation of the sscd1 hgo Double Mutant
The sscd1-1 homozygous plants were crossed to the hgo-1 mutant. Then, hgo-1 homozygous plants were identified from the F2 progeny by PCR amplification with primers (LP2 + RP2 and LBb1.3 + RP2), among which the sscd1-1 homozygous plants were further identified through sequencing of the PCR products amplified with the SSCD1-specific primers (5′-CCTCGTCCTGCCGTCGCTAT-3′ and 5′-CTTGTGGATGGCCCTGACCT-3′).
Treatment of Arabidopsis Seedlings with Succinylacetone
Twelve-day-old seedlings grown under LD were sprayed with different concentrations of succinylacetone. Succinylacetone treatment was carried out once per day for 4 d. The rate of seedlings with wilted leaves was counted at day 5, and the seedlings were photographed at day 6. The experiment was repeated three times.
Sequence data from this article can be found in the GenBank data library under accession number NM_101077.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for distributing the T-DNA insertion lines and the BAC clone and Dr. Guo-Liang Wang for helpful suggestions.
Glossary
- PCD
programmed cell death
- LD
long days
- SD
short days
- HGO
homogentisate dioxygenase
- MAAI
maleylacetoacetate isomerase
- FAH
fumarylacetoacetate hydrolase
- HT1
hereditary tyrosinemia type I
- Col-0
Columbia
- CAPS
cleaved-amplified polymorphic sequence
- BAC
bacterial artificial chromosome
- T-DNA
transfer DNA
- RT
reverse transcription
- MAA
maleylacetoacetate
- FAA
fumarylacetoacetate
- MS
Murashige and Skoog medium supplemented with 1% Suc
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aponte JL, Sega GA, Hauser LJ, Dhar MS, Withrow CM, Carpenter DA, Rinchik EM, Culiat CT, Johnson DK. (2001) Point mutations in the murine fumarylacetoacetate hydrolase gene: animal models for the human genetic disorder hereditary tyrosinemia type 1. Proc Natl Acad Sci USA 98: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango Y, Heise KP. (1998) Tocopherol synthesis from homogentisate in Capsicum annuum L. (yellow pepper) chromoplast membranes: evidence for tocopherol cyclase. Biochem J 336: 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron A, Jorquera R, Orejuela D, Tanguay RM. (2006) Involvement of endoplasmic reticulum stress in hereditary tyrosinemia type I. J Biol Chem 281: 5329–5334 [DOI] [PubMed] [Google Scholar]
- Berkey R, Bendigeri D, Xiao S. (2012) Sphingolipids and plant defense/disease: the “death” connection and beyond. Front Plant Sci 3: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jørgensen LB, Brown RE, Mundy J. (2002) Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kawarasaki Y, Nakano H, Yamane T. (2003) Cloning and in vitro and in vivo expression of plant glutathione S-transferase zeta class genes. J Biosci Bioeng 95: 594–600 [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D. (2003) Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol 131: 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. (1996) Death don’t have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. (1994) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Cole DJ, Edwards R. (2000) Characterisation of a zeta class glutathione transferase from Arabidopsis thaliana with a putative role in tyrosine catabolism. Arch Biochem Biophys 384: 407–412 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. (2006) Enzymes of tyrosine catabolism in Arabidopsis thaliana. Plant Sci 171: 360–366 [DOI] [PubMed] [Google Scholar]
- Donahue JL, Alford SR, Torabinejad J, Kerwin RE, Nourbakhsh A, Ray WK, Hernick M, Huang X, Lyons BM, Hein PP, et al (2010) The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 22: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X. (2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA 104: 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Peñalva MA. (1995a) Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J Biol Chem 270: 21199–21205 [DOI] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Peñalva MA. (1995b) Fungal metabolic model for human type I hereditary tyrosinaemia. Proc Natl Acad Sci USA 92: 9132–9136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler E, Soll J, Schultz G. (1982) The formation of homogentisate in the biosynthesis of tocopherol and plastoquinone in spinach chloroplasts. Planta 155: 511–515 [DOI] [PubMed] [Google Scholar]
- Fisher AL, Page KE, Lithgow GJ, Nash L. (2008) The Caenorhabditis elegans K10C2.4 gene encodes a member of the fumarylacetoacetate hydrolase family: a Caenorhabditis elegans model of type I tyrosinemia. J Biol Chem 283: 9127–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS. (1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89: 25–31 [DOI] [PubMed] [Google Scholar]
- Greenberg JT. (1996) Programmed cell death: a way of life for plants. Proc Natl Acad Sci USA 93: 12094–12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG, Soriano P. (1993) Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev 7: 2298–2307 [DOI] [PubMed] [Google Scholar]
- Hu G, Yalpani N, Briggs SP, Johal GS. (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Okamoto H, Iwasaki Y, Asahi T. (2001) A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J 27: 89–99 [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Tanaka H, Nakai M, Asahi T. (2003) Deletion of a chaperonin 60 β gene leads to cell death in the Arabidopsis lesion initiation 1 mutant. Plant Cell Physiol 44: 255–261 [DOI] [PubMed] [Google Scholar]
- Jorquera R, Tanguay RM. (2001) Fumarylacetoacetate, the metabolite accumulating in hereditary tyrosinemia, activates the ERK pathway and induces mitotic abnormalities and genomic instability. Hum Mol Genet 10: 1741–1752 [DOI] [PubMed] [Google Scholar]
- Kvittingen EA. (1986) Hereditary tyrosinemia type I: an overview. Scand J Clin Lab Invest Suppl 184: 27–34 [PubMed] [Google Scholar]
- Lam E. (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT. (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17: 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad B, Lindstedt S, Steen G. (1977) On the enzymic defects in hereditary tyrosinemia. Proc Natl Acad Sci USA 74: 4641–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin RA, Zakeri Z. (2004) Apoptosis, autophagy, and more. Int J Biochem Cell Biol 36: 2405–2419 [DOI] [PubMed] [Google Scholar]
- Löffelhardt W, Kindl H. (1979) Conversion of 4-hydroxyphenylpyruvic acid into homogentisic acid at the thylakoid membrane of Lemna gibba. FEBS Lett 104: 332–334 [DOI] [PubMed] [Google Scholar]
- Lopukhina A, Dettenberg M, Weiler EW, Holländer-Czytko H. (2001) Cloning and characterization of a coronatine-regulated tyrosine aminotransferase from Arabidopsis. Plant Physiol 126: 1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, Balagué C, Roby D. (2004) Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16: 2217–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. (2001) The Arabidopsis accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Al-Dhalimy M, Finegold M, Grompe M. (1999) In vivo suppressor mutations correct a murine model of hereditary tyrosinemia type I. Proc Natl Acad Sci USA 96: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M, Bollich C, Uecker F. (1983) Spontaneous occurrence of the Sekiguchi lesion in two American rice lines: its induction, inheritance, and utilization. Phytopathology 73: 603–606 [Google Scholar]
- Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, Henry Y, Soubigou-Taconnat L, Lelarge-Trouverie C, Saindrenan P, et al (2009) Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL. (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ 4: 671–683 [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert S, Kelsey G, Schedl A, Schmid E, Thies E, Schütz G. (1992) Deficiency of an enzyme of tyrosine metabolism underlies altered gene expression in newborn liver of lethal albino mice. Genes Dev 6: 1430–1443 [DOI] [PubMed] [Google Scholar]
- St-Louis M, Tanguay RM. (1997) Mutations in the fumarylacetoacetate hydrolase gene causing hereditary tyrosinemia type I: overview. Hum Mutat 9: 291–299 [DOI] [PubMed] [Google Scholar]
- Sun MS, Hattori S, Kubo S, Awata H, Matsuda I, Endo F. (2000) A mouse model of renal tubular injury of tyrosinemia type 1: development of de Toni Fanconi syndrome and apoptosis of renal tubular cells in Fah/Hpd double mutant mice. J Am Soc Nephrol 11: 291–300 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P, Feussner K, Werner S, Lerche J, Iven T, Heilmann I, Riezman H, Feussner I. (2011) Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol 192: 841–854 [DOI] [PubMed] [Google Scholar]
- Wang W, Yang X, Tangchaiburana S, Ndeh R, Markham JE, Tsegaye Y, Dunn TM, Wang GL, Bellizzi M, Parsons JF, et al. (2008) An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. (1993) The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Gen Genet 239: 122–128 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]