The transcription factor SOC1 is regulated by two GATA transcription factors for the control of flowering while the GATAs are controlled by SOC1 to control greening and cold tolerance.
Abstract
The paralogous and functionally redundant GATA transcription factors GNC (for GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED) and GNL/CGA1 (for GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1) from Arabidopsis (Arabidopsis thaliana) promote greening and repress flowering downstream from the phytohormone gibberellin. The target genes of GNC and GNL with regard to flowering time control have not been identified as yet. Here, we show by genetic and molecular analysis that the two GATA factors act upstream from the flowering time regulator SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) to directly repress SOC1 expression and thereby repress flowering. Interestingly, this analysis inversely also reveals that the MADS box transcription factor SOC1 directly represses GNC and GNL expression to control cold tolerance and greening, two further physiological processes that are under the control of SOC1. In summary, these findings support the case of a cross-repressive interaction between the GATA factors GNC and GNL and the MADS box transcription factor SOC1 in flowering time control on the one side and greening and cold tolerance on the other that may be governed by the various signaling inputs that are integrated at the level of SOC1 expression.
Throughout evolution, plants have acquired the ability to adapt their growth and flowering to their environmental conditions to guarantee optimal reproductive success in their individual growth environments. Various signaling pathways are known that are integrated to control flowering time in response to light quality, daylength, temperature, and nutrient availability (Parcy, 2005; Franks et al., 2007; Izawa, 2007; Turck et al., 2008). In Arabidopsis (Arabidopsis thaliana), genetic studies have identified the transcription factors FLOWERING LOCUS T (FT), LEAFY, and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) as major integrators of flowering (Nilsson et al., 1998; Samach et al., 2000). FT protein was shown to act as the florigen that moves from the leaves in inductive conditions to the shoot apical meristem, where it promotes the transition from vegetative to reproductive meristem identity by forming a complex with the bZIP transcription factor FLOWERING LOCUS D (Corbesier et al., 2007; Mathieu et al., 2007; Taoka et al., 2011).
Under inductive long days, expression of the MADS box transcription factor SOC1 is essential for floral induction (Samach et al., 2000; Yoo et al., 2005). Under short days, SOC1 appears also to be the major integrator of flowering time stimulation in response to the phytohormone GA (Blázquez and Weigel, 1999; Moon et al., 2003). SOC1 expression is furthermore repressed by the MADS box transcription factor FLOWERING LOCUS C, which controls flowering in response to long cold periods (vernalization) and acts together with the MADS box transcription factor SHORT VEGETATIVE PHASE (Hartmann et al., 2000; Li et al., 2008; Tao et al., 2012). Additionally, SOC1 expression is controlled by an age-dependent regulatory system that involves SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors and their antagonistic microRNA regulator miRNA156 (Wang et al., 2009).
SOC1 expression and flowering are also repressed in response to short periods of cold that plants experience, such as during a cold spring season (Seo et al., 2009). During such cold periods, SOC1 expression is reduced and the concomitant delay in flowering correlates with an increase in the expression of cold-regulated genes such as C-REPEAT/DROUGHT-RESPONSIVE ELEMENT-BINDING FACTOR (CBF) and cold-response marker genes such as COLD-REGULATED15a (COR15a) and COR15b. Inversely, SOC1 overexpression results in the inverse regulation of cold-regulated genes, and this supports experimental data that indicate that SOC1 is a direct regulator of cold-regulated genes (Seo et al., 2009).
GAs have been implicated in a variety of growth responses in plants, including the control of flowering time (Hisamatsu and King, 2008; Itoh et al., 2008; Schwechheimer, 2011). GAs are perceived by the GIBBERELLIC ACID-INSENSITIVE DWARF1 GA receptors, which in turn bind to and induce the degradation of DELLA repressors such as GIBBERELLIC ACID INSENSITIVE (GAI) and REPRESSOR OF ga1-3 (RGA) from Arabidopsis (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Willige et al., 2007). DELLA proteins repress a broad range of different transcription factor activities, including that of the PHYTOCHROME INTERACTING FACTORS (PIFs), basic helix-loop-helix transcription factors that integrate GA as well as light signaling by interacting with DELLAs, and the phytochrome light receptors (de Lucas et al., 2008; Feng et al., 2008; Gallego-Bartolomé et al., 2010). Inversely, GA responses appear completely derepressed in mutants deficient in the function of the GlcNAc transferase SPINDLY (SPY; Jacobsen and Olszewski, 1993; Wilson and Somerville, 1995; Silverstone et al., 2007).
In long-day-grown wild-type plants, flowering is typically only moderately promoted by GA, because growth conditions in laboratory experiments are adjusted to guarantee a short generation time (Reeves and Coupland, 2001; Galvão et al., 2012). DELLA gene loss-of-function mutants with a constitutive GA response or spy mutants do not flower much faster than the wild type when grown under long days (Achard et al., 2003; Tseng et al., 2004). Based on the observation that the Landsberg erecta (Ler) allele of the GA biosynthesis mutant ga1-3 displays only a comparatively minor delay in flowering under long-day conditions, GA has for a long time been thought to be of minor importance for floral induction under long days. In contrast, ga1-3 mutants failed to flower even after 5 to 6 months in short-day conditions, and this has given rise to the long-standing hypothesis that, in Arabidopsis, GA is essential for flowering only under short days (Wilson et al., 1992; Reeves and Coupland, 2001). Several recent studies, however, report a strong flowering time delay also under long days in ga1 mutants and other Arabidopsis GA pathway mutants when examined in the Columbia ecotype (Willige et al., 2007; Hisamatsu and King, 2008; Galvão et al., 2012; Porri et al., 2012).
Plants use GAs also to control plant growth in response to abiotic stress such as cold, salt, and oxidative stress as well as biotic stress caused by pathogens (Achard et al., 2006, 2007, 2008a, 2008b; Navarro et al., 2008). The growth constraint resulting from the exposure to cold temperature correlates with and can be explained by a cold-induced stabilization of the DELLAs and results in increased cold tolerance in Arabidopsis seedlings (Achard et al., 2008a). The identities of the genes that confer cold tolerance downstream of the DELLAs have not been revealed as yet.
We have previously identified the two functionally paralogous GATA family transcription factors GNC (for GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED) and GNL/CGA1 (for GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1) from Arabidopsis as critical transcription targets downstream from GA, DELLAs, and PIFs (Bi et al., 2005; Naito et al., 2007; Richter et al., 2010). GNC and GNL expression is repressed in response to GA and increased in mutants with a block in GA signaling, such as ga1 and gid1abc. In agreement with the repressive activity of GAs on GNC and GNL expression, GNC and GNL overexpression plants, where GNC and GNL regulation is uncoupled from GA control, resemble ga1 or gid1abc mutants at the phenotypic and global gene expression levels. Similar to the GA pathway mutants, GNC and GNL overexpressors are dark-green dwarfs with a delay in germination and flowering (Richter et al., 2010). PIF3 is at least one member of the PIF family of transcription factors that controls GNC and GNL expression. As recently reported, the greening phenotype of gnc and gnl mutants may be explained by their role in directly and indirectly regulating the expression of chlorophyll biosynthetic genes and chloroplast development, growth, and division (Hudson et al., 2011; Chiang et al., 2012). GNC and GNL have also been identified as transcription repression targets of the floral homeotic genes APETALA3 and PISTILLATA (Mara and Irish, 2008). In the context of the role of GNC and GNL in regulating chlorophyll biosynthesis, it has been proposed that repression of the two GATAs is at least in part responsible for the nongreening of petals and stamens (Mara and Irish, 2008).
In this study, we show that GNL, as a representative GATA factor for the two functionally homologous GATAs GNC and GNL, represses flowering downstream from GA signaling and upstream from SOC1. We further show that GNL overexpression results in a decrease in SOC1 expression and that both GATAs can directly bind to the SOC1 promoter, suggesting that GNL may be a direct transcriptional repressor of SOC1 expression. Interestingly, we also find that GNC and GNL promote two other SOC1-dependent physiological responses downstream from SOC1, greening and cold tolerance, and that SOC1 can directly bind to the promoters of both GATAs. We thereby present a case for a cross-repressive interaction of GNC and GNL on the one side and SOC1 on the other in the control of flowering time, greening, and cold tolerance.
RESULTS
GNC and GNL Repress Flowering Downstream from GA Signaling
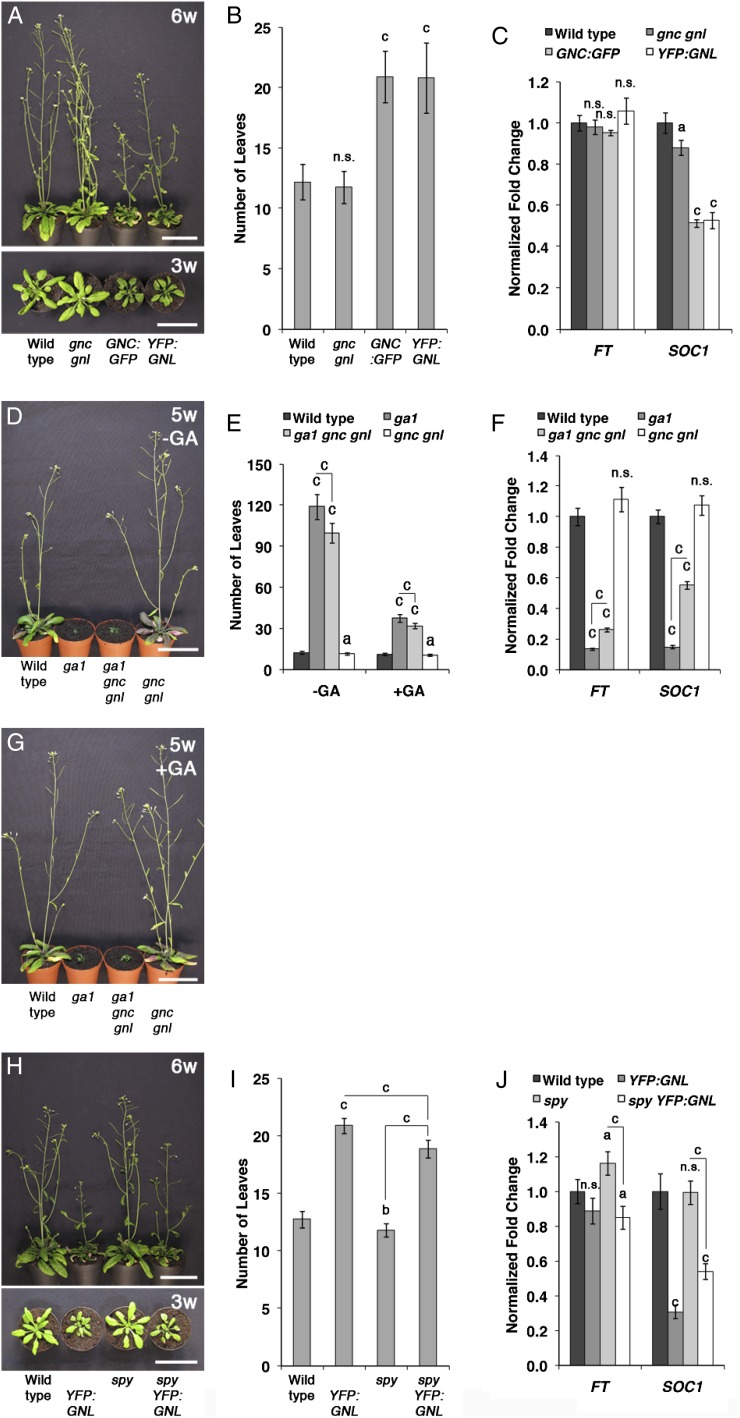
We have previously established that the GATA transcription factors GNC and GNL repress GA responses downstream from GA, DELLAs, and PIFs in Arabidopsis (Richter et al., 2010). The role for GNC and GNL in the control of flowering time is already suggested by a subtle acceleration in flowering in long-day-grown as well as short-day-grown gnc gnl mutants when compared with the wild type (Fig. 1, A and B; Supplemental Fig; S1; Richter et al., 2010). The repressive role of the two GATAs, however, is much more apparent in the GA-deficient ga1 background, where the loss of GNC and GNL in ga1 gnc gnl resulted in the partial suppression of the flowering time delay of the ga1 mutant (Fig. 1, D–F; Richter et al., 2010). Here, we found that ga1 gnc gnl mutants (99.7 ± 7.1 leaves; 79.8 ± 5.7 d) flower about 1 month earlier than ga1 (118.9 ± 9.1 leaves; 118.7 ± 10 d) when grown under long-day growth conditions. Since ga1 gnc gnl flowers still much later than the wild type (12.3 ± 1.1 leaves; 23.1 ± 1.1 d) or ga1 mutants after GA treatment (Fig. 1, D, E, and G), these findings suggest that other regulators in addition to GNC and GNL must repress flowering in the absence of GA. One the one side, such regulators could be functionally homologous GATA factors closely related to GNC and GNL or, on the other side, other unrelated proteins (Richter et al., 2010).
Figure 1.
GNC and GNL repress flowering downstream from the GA signaling pathway. A, D, G, and H, Representative photographs of Arabidopsis plants grown for 5 or 6 weeks (w) under long-day conditions. Plants shown in G were watered twice per week with 1 µm GA3. B, E, and I, Flowering time analysis (total rosette leaf number) of long-day-grown plants shown in A, D, G, and H. C, F, and J, Results of qRT-PCR analyses for the flowering time regulators FT and SOC1 performed with 10-d-old seedlings. Fold change was calculated relative to wild-type levels. Student’s t tests were performed in comparison with the wild type unless indicated otherwise: a = P ≤ 0.05, b = P ≤ 0.01, c = P ≤ 0.001; n.s., not significant. Bars = 5 cm.
In line with the proposed role of the two GATAs as repressors of flowering, we found that the overexpression of either GNC (GNC:GFP) or GNL (YELLOW FLUORESCENT PROTEIN [YFP]:GNL) results in a strong delay in flowering in the wild type (Fig. 1, A and B; Richter et al., 2010). In order to understand at which stage of flowering time regulation the GATA transgenes are active, we introduced YFP:GNL, which in our hands is the genetically more stable of the two GATA transgenes, into the spy and the rga gai DELLA gene loss-of-function backgrounds. Also in the early flowering spy mutant, which mimics the phenotype of a constitutive GA response mutant, we observed a strong delay in flowering in the presence of YFP:GNL (Fig. 1, H and I; Jacobsen and Olszewski, 1993). Flowering time was also strongly delayed in the rga gai loss-of-function mutant that normally flowers early, specifically in short-day conditions (Supplemental Fig. S2, A and B; Dill and Sun, 2001; Cheng et al., 2004). Since our previous analysis of GNC and GNL regulation had shown that the two GATAs are targets of PIF3, we also analyzed GNL overexpression in a PIF3:MYC overexpression line. Also in this background, flowering was strongly delayed, indicating that GNL represses flowering downstream from its transcriptional regulator PIF3 (Supplemental Fig. S2, C and D). In summary, these findings confirm the role of GNL as a repressor of flowering downstream from GA, DELLA, and PIF3. Since all our experiments at present suggest that GNC and GNL are functionally redundant, we speculate that these observations also hold true for GNC.
SOC1 Is an Essential Flowering Time Integrator Downstream from GNC and GNL
Different flowering time pathways converge on the regulation of the two positive flowering time regulators FT and SOC1. To understand the contribution of FT and SOC1 expression to the flowering time phenotypes of the genotypes described above, we performed quantitative real-time (qRT)-PCR using 10-d-old seedlings to assess their transcript abundance in the presence of the GATA overexpression constructs (Fig. 1C). While the abundance of FT is not significantly altered by the presence or absence of the GATAs, the abundance of SOC1 is strongly repressed when either GNC or GNL is overexpressed. Furthermore, the reduced transcript abundance of SOC1 also correlates with the late-flowering phenotype of the spy YFP:GNL and PIF3:MYC YFP:GNL genotypes (Fig. 1J; Supplemental Fig. S2D). Inversely, SOC1 transcript levels are increased in ga1 gnc gnl when compared with ga1 (Fig. 1F).
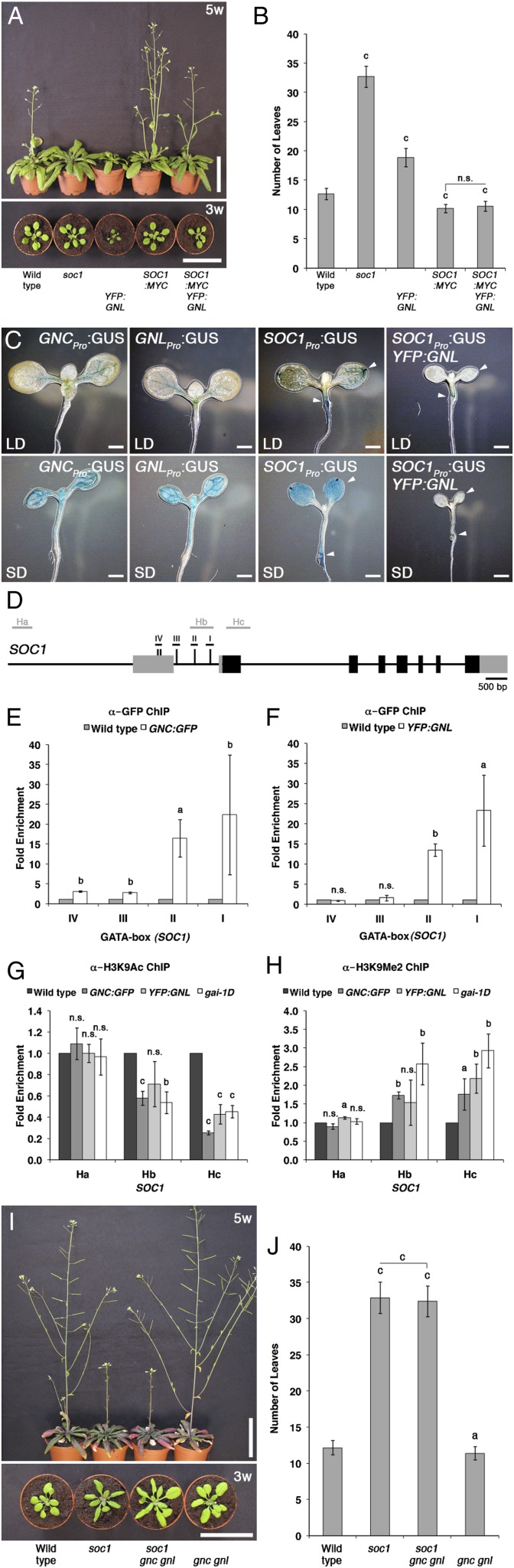
Since these findings invited the hypothesis that GNC and GNL may be regulators of SOC1 expression, we became interested in examining the relationship between SOC1 and the GATAs in more detail. To this end, we introduced the YFP:GNL overexpression transgene into a SOC1:MYC overexpression background. Flowering is strongly delayed in soc1 loss-of-function mutants (32.7 ± 2.8 leaves) and slightly accelerated in a SOC1:MYC overexpression line (10.2 ± 0.7 leaves) when compared with the wild type (12.7 ± 1.0 leaves; Fig. 2, A and B). Interestingly, and unlike what we had observed when we introduced YFP:GNL into the GA pathway mutants (Fig. 1), YFP:GNL overexpression does not delay flowering in the SOC1:MYC background, where SOC1 expression is under the control of the 35S cauliflower mosaic virus promoter and thus uncoupled from its native transcriptional regulation (10.6 ± 0.9 leaves; Fig. 2, A and B). This suggests that SOC1:MYC promotes flowering downstream from GNL. Using promoter-GUS lines for SOC1 (SOC1Pro:GUS), GNC (GNCPro:GUS), and GNL (GNLPro:GUS), we could show in subsequent experiments that the three genes are coexpressed in the leaves of 10-d-old seedlings and that the overexpression of YFP:GNL represses SOC1:GUS expression in this tissue (Fig. 2C).
Figure 2.
GNC and GNL act upstream of SOC1. A, Representative photographs of 3- and 5-week-old (w) Arabidopsis plants. B, Flowering time analysis (total rosette leaf number) of long-day-grown plants shown in A. C, GUS expression in the first pair of true leaves in 10-d-old long-day-grown (LD) and short-day-grown (SD) Arabidopsis seedlings expressing GNCPro:GUS, GNLPro:GUS, and SOC1Pro:GUS in the wild type and the YFP:GNL overexpression background. Arrows indicate expression differences between the GUS transgenes. Note that YFP:GNL overexpression results in a dwarfed seedling phenotype (Richter et al., 2010). D, Schematic representation of the SOC1 genomic locus. Black boxes, exons; gray boxes, untranslated regions; underlined roman numbers, promoter regions containing GATA boxes (vertical lines); gray lines, predicted nucleosome-binding regions. E and F, Fold enrichment (GNC:GFP/wild type [E] and YFP:GNL/wild type [F]) of promoter fragment amplification after ChIP-PCR with anti-GFP. G and H, Fold enrichment after ChIP with anti-H3K9Ac and anti-H3K9me2 and promoter fragment amplification by PCR when compared with the wild type. I, Representative photographs of 6-week-old Arabidopsis plants. J, Flowering time analysis (total rosette leaf number) of long-day-grown plants shown in I. Student’s t tests were performed in comparison with the wild type unless indicated otherwise: a = P ≤ 0.05, b = P ≤ 0.01, c = P ≤ 0.001; n.s., not significant. Bars = 5 cm.
Since we could identify various GATA boxes in the promoter of SOC1, we reasoned that GNL and possibly also GNC may regulate SOC1 expression directly (Fig. 2D). To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) for GNC:GFP and YFP:GNL and, indeed, detected a strong binding of both GATA factors to two GATA boxes (boxes I and II) in the first intron and exon of the SOC1 promoter (Fig. 2, E and F). In support of a model where the decrease in SOC1 transcript abundance in the GNC:GFP or YFP:GNL overexpressors is the result of transcriptional repression, we further detected decreased abundance of K9-acetylated histone 3 (H3K9Ac), a marker for active chromatin, and increased abundance of K9-dimethylated histone 3 (H3K9me2), a marker for inactive chromatin, at two promoter regions, Hb and Hc, that are proximal to the ATG start codon of SOC1 (Fig. 2, G and H). At the same time, binding of the two histone H3 variants to a more remote site, Ha, was unaltered (Fig. 2, G and H). A similar H3-variant chromatin-binding preference was observed when we tested H3K9Ac and H3K9me2 binding to the SOC1 promoter in the GA-insensitive gai-1D mutant, where GNC and GNL expression is increased as a consequence of DELLA protein stabilization (Fig. 2, G and H; Supplemental Fig. S3; Richter et al., 2010). Since this analysis may suffer from the criticism that the observed promoter binding is the result of an off-target amplification of the overexpressed YFP:GNL, we established also a GNLPro:GNL:HA transgene that complemented the gnc gnl mutant phenotype (Supplemental Fig. S4, A and B). We then tested binding of the hemagglutinin (HA)-tagged GNL protein to promoter boxes IV (negative control) and I (positive control) and could in both cases confirm the negative (IV) as well as the positive (I) binding of GNL:HA to the predicted target sites in the SOC1 promoter (Supplemental Fig. S4C).
Since our experiments indicated that GNC and GNL are direct transcriptional repressors upstream from SOC1, we also expected that the late-flowering phenotype of soc1, unlike the late-flowering phenotype of the ga1 mutant, is not suppressed in a soc1 gnc gnl triple mutant. Indeed, we found no significant difference in the late flowering of soc1 gnc gnl (32.4 ± 2.1 leaves) when compared with the soc1 single mutant (32.9 ± 2.2 leaves). Therefore, we concluded that SOC1 is critical for flowering time control downstream from GNC and GNL (Fig. 2, I and J). In line with a role of GA upstream of SOC1 in the control of flowering, we also noted that the flowering time delay of the soc1 mutant cannot be suppressed by GA treatments that are sufficient to accelerate flowering in the wild type, suggesting that SOC1 is essential to promote flowering downstream from GA (Supplemental Fig. S5, A and B). In summary, these findings suggest that GNC and GNL are direct repressors of SOC1 and that the GNC- and GNL-dependent repression of SOC1 transcript abundance correlates with activating and repressive chromatin changes at the SOC1 promoter and with the late-flowering phenotype of GNC and GNL overexpressors. Thus, SOC1 is the essential flowering time integrator downstream from GNC and GNL.
GNC and GNL Promote Greening Downstream from SOC1
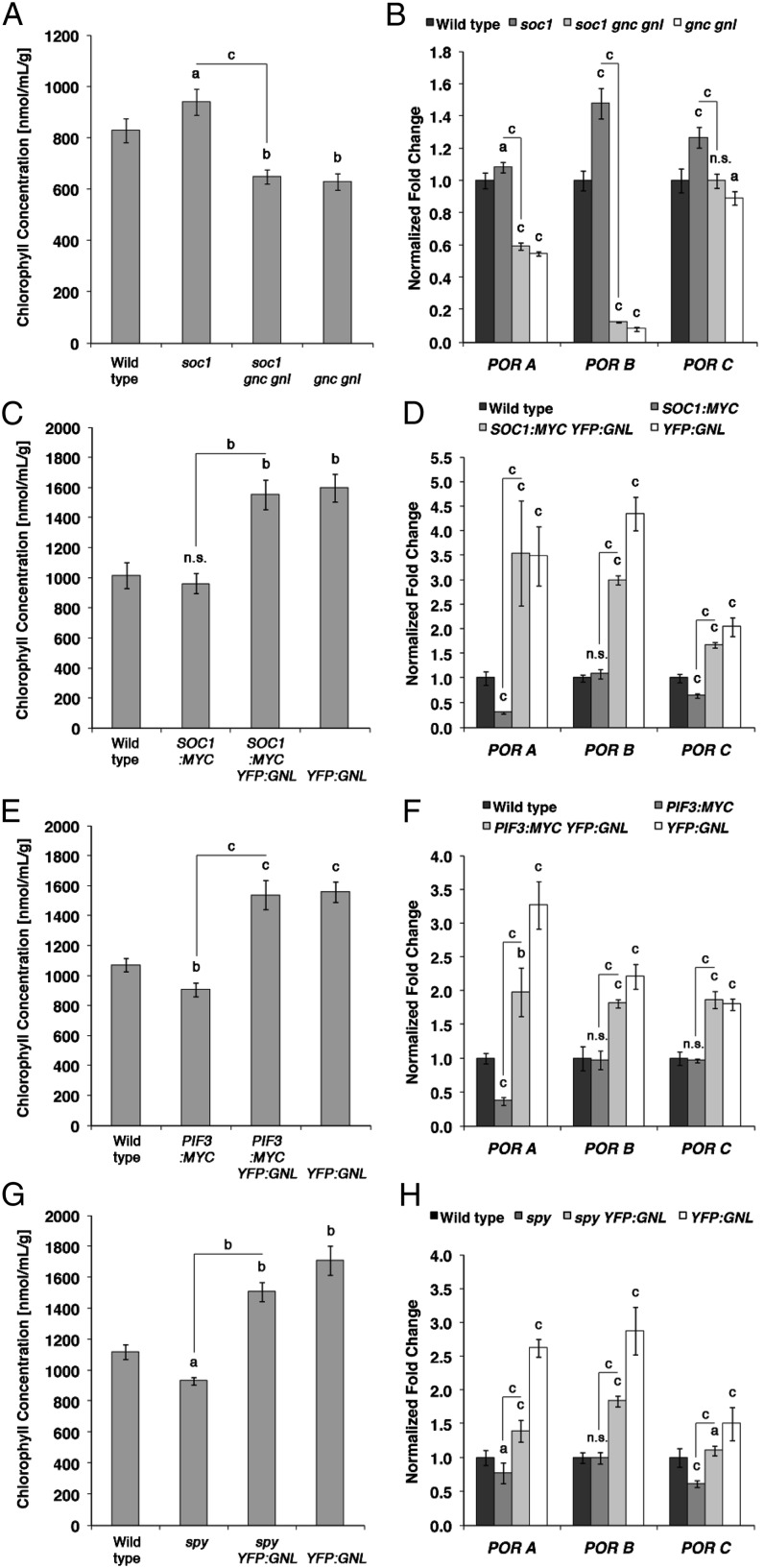
A reduction in chlorophyll content had been the original phenotype associated with the gnc mutant in Arabidopsis (Bi et al., 2005). Both GNC and GNL have recently been characterized as positive regulators of chlorophyll biosynthesis and chloroplast division (Richter et al., 2010; Hudson et al., 2011; Chiang et al., 2012). gnc gnl loss-of-function mutants are light green; conversely, GNC and GNL overexpression lines show increased greening and chlorophyll accumulation. Interestingly, we observed that the soc1 gnc gnl triple mutant is visibly less green and contains less chlorophyll than the dark-green soc1 single mutant (Fig. 3A; Supplemental Fig. S6). When compared with soc1 and the wild type, this reduction in chlorophyll accumulation of the soc1 gnc gnl triple mutant also correlated with a decrease in the transcript abundance of the three PROTOCHLOROPHYLLIDE OXYDOREDUCTASE (POR) genes PORA, PORB, and PORC, the gene products of which control a critical step in chlorophyll biosynthesis (Fig. 3B; Thomas, 1997). While our analysis of flowering time control had suggested that GNC and GNL function upstream of SOC1, the greening phenotype suggested a role of GNC and GNL downstream from SOC1 in the control of chlorophyll biosynthesis. This antagonism in the genetic interaction between the GATAs and SOC1 with regard to greening and flowering time control, respectively, was further supported by the greening phenotype of the YFP:GNL SOC1:MYC background. While we had observed that the YFP:GNL overexpressor cannot delay flowering in the early-flowering SOC1:MYC background, we found that the overexpression of YFP:GNL resulted in a stronger chlorophyll accumulation as well as increased POR transcript levels in SOC1:MYC (Fig. 3, C and D). In this regard, the YFP:GNL overexpressor when introduced into SOC1:MYC behaved in a similar manner as it did in the PIF3:MYC or spy background, and this finding suggested that YFP:GNL is downstream of SOC1 in the control of greening (Fig. 3, E–H). Furthermore, while GA treatments had not been sufficient to promote flowering in soc1 (Supplemental Fig. S5, A and B), indicating that SOC1 acts downstream of GA signaling in the control of flowering, we found that GA treatments of soc1 are sufficient to suppress the soc1 mutant’s greening phenotype, clearly demonstrating that SOC1 is upstream of GA in the control of greening (Supplemental Fig. S5C). In summary, these findings suggested a role for YFP:GNL downstream from the GA pathway and SOC1 in the control of chlorophyll biosynthesis and greening.
Figure 3.
GNC and GNL promote greening downstream from SOC1. A, C, E, and G, Chlorophyll concentration. B, D, F, and H, qRT-PCR analyses of POR gene expression from 10-d-old seedlings as shown in A, C, E, and G. Student’s t tests were performed in comparison with the wild type unless indicated otherwise: a = P ≤ 0.05, b = P ≤ 0.01, c = P ≤ 0.001; n.s., not significant.
GNC and GNL Promote Cold Tolerance Downstream from SOC1
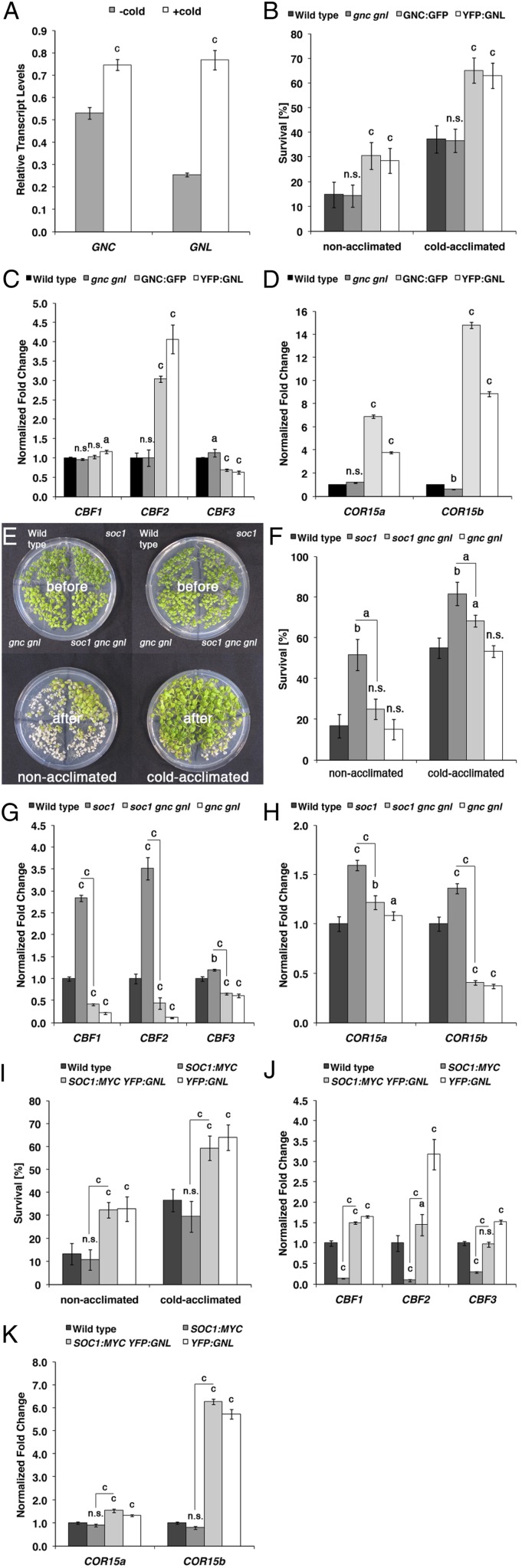
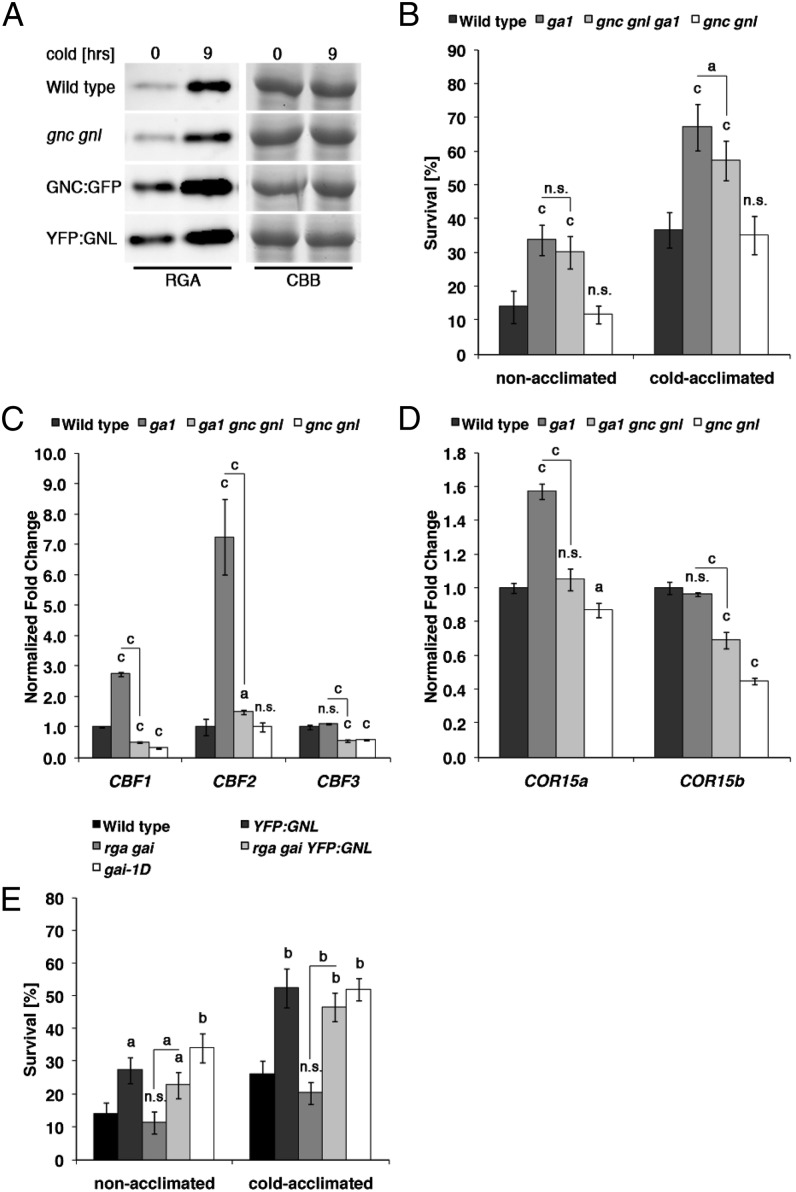
Besides its prominent role in the regulation of flowering time, SOC1 was previously also shown to interfere with cold tolerance (Seo et al., 2009). Interestingly, a qRT-PCR analysis of mock- and cold-treated 10-d-old seedlings revealed that GNC and GNL are regulated by cold temperatures, pointing at a putative role of the two genes in cold response (Fig. 4A). Indeed, when we tested cold tolerance in nonacclimated and cold-acclimated gnc gnl loss-of-function mutants and the GNC:GFP or YFP:GNL overexpression lines, we found that seedling survival in the GNC or GNL overexpressors is approximately twice as high as in the wild type, regardless of prior acclimation (Fig. 4B). At the molecular level, the increased survival correlated with an increase in the expression of CBF2 and the two cold-response marker genes COR15a and COR15b (Fig. 4, C and D; Medina et al., 1999).
Figure 4.
GNC and GNL promote cold tolerance downstream from SOC1. A, qRT-PCR analyses of GNC and GNL expression of 10-d-old seedlings in response to a 9-h cold treatment (4°C). B, F, and I, Seedling survival of nonacclimated and cold-acclimated 10-d-old seedlings after cold treatment. C, D, G, H, J, and K, qRT-PCR analyses of CBF (C, G, and J) and COR15 (D, H, and K) gene expression in 10-d-old seedlings. E, Representative photograph of nonacclimated and cold-acclimated 10-d-old seedlings before (top panels) and after (bottom panels) cold treatment. Student’s t tests were performed in comparison with the wild type unless indicated otherwise: a = P ≤ 0.05, b = P ≤ 0.01, c = P ≤ 0.001; n.s., not significant.
As we will show later, we also detected strongly increased GNC and GNL transcript levels in the soc1 mutant (Fig. 6A). Since increased cold tolerance is also a phenotype of soc1 mutants (Seo et al., 2009), we reasoned that the elevated GNC and GNL transcript levels in soc1 may be causative for the cold tolerance phenotype of this mutant. In support of this hypothesis, we found that the increased cold tolerance phenotype of soc1 is strongly suppressed in the soc1 gnc gnl triple mutant (Fig. 4, E and F). This genetic suppression of the soc1 cold tolerance phenotype was mirrored by a decrease in the abundance of the three CBF genes as well as that of COR15a and COR15b in soc1 gnc gnl when compared with the soc1 single mutant (Fig. 4, G and H). Along the same lines, and in contrast to the dominant effect of SOC1:MYC overexpression over YFP:GNL overexpression with regard to the control of flowering, we observed that the overexpression of YFP:GNL is dominant over the overexpression of SOC1:MYC with regard to the cold tolerance of nonacclimated and cold-acclimated seedlings, CBF expression, as well as COR expression (Fig. 4, I–K). Taken together, these findings suggest that the cold tolerance phenotype of soc1 is at least partially mediated by the increase in GNC and GNL transcript abundance and that GNC and GNL function downstream from SOC1 in the control of cold tolerance.
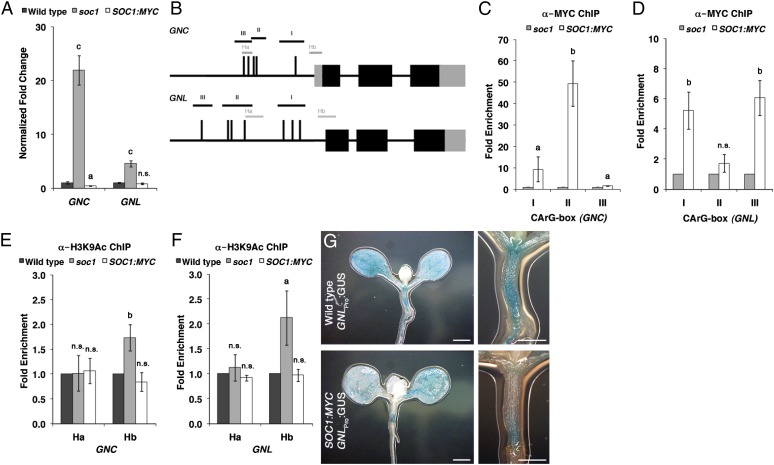
Figure 6.
SOC1 is a direct repressor of GNC and GNL transcription. A, qRT-PCR analysis of GNC and GNL transcript abundance in soc1 and SOC1:MYC. B, Schematic representation of the GNC and GNL genomic loci. Black boxes, exons; gray boxes, untranslated regions; underlined roman numbers, promoter regions containing CArG boxes (vertical lines); gray lines, predicted nucleosome-binding regions Ha and Hb. Note that there is no known 5′ untranslated region for GNL. C and D, Fold enrichment (SOC1:MYC/soc1) after PCR amplifications of GNC (C) and GNL (D) promoter fragments following ChIP with anti-MYC. E and F, Fold enrichment after ChIP with anti-H3K9Ac and PCR amplification of GNC (E) and GNL (F) promoter fragments in the wild type, soc1, and SOC1:MYC. G, Representative GUS staining of 10-d-old seedlings expressing GNL:GUS in the wild type and the SOC1:MYC background. Student’s t tests were performed in comparison with the wild type unless indicated otherwise: a = P ≤ 0.05, b = P ≤ 0.01, c = P ≤ 0.001; n.s., not significant. Scale bars = 0.5 mm.
It was previously established that cold-induced growth arrest and cold tolerance in Arabidopsis are promoted by DELLA repressors and that the exposure to cold temperatures correlates with an increase in DELLA protein abundance (Achard et al., 2008a). In line with a cold-induced stabilization of the DELLAs, we also detected increased levels of the DELLA protein RGA in 10-d-old cold-treated wild-type and gnc gnl seedlings (Fig. 5A). Interestingly, however, we also found that RGA levels are increased already at ambient temperature in GNC and GNL overexpressors of the same age (Fig. 5A). Thus, the GNC and GNL overexpressors mimic the DELLA accumulation phenotype of cold-treated wild-type seedlings. Since we had previously established that DELLA protein abundance correlates with GNC and GNL transcript abundance (Richter et al., 2010), we reasoned that the increased transcript abundance of the GATAs may be the molecular cause for the cold tolerance phenotype of ga1. However, when we compared the cold tolerance phenotype of ga1 gnc gnl with that of ga1, we detected only a mild, and in our conditions only in cold-acclimated conditions statistically significant, suppression of the cold tolerance phenotype of ga1 when examining ga1 gnc gnl (Fig. 5B). Surprisingly, the molecular analysis performed in parallel revealed a strong reduction in CBF and COR15 gene expression when compared with ga1 that we would have expected to correlate with a strong decrease in cold tolerance (Fig. 5, C and D). The absence of a strong suppression of the ga1 cold tolerance phenotype and the concomitant presence of a molecular suppression of CBF and COR gene expression suggests that the accumulation of GNC and GNL transcript alone is not causative for the cold tolerance observed in the GA-deficient ga1 mutant, that other genes downstream from GA and DELLA promote cold tolerance in the absence of GA, and that the reduction of CBF and COR transcript abundance is not sufficient to predict cold tolerance in a ga1 mutant background.
Figure 5.
Analysis of GNC- and GNL-mediated cold tolerance in GA pathway mutants. A, Immunoblot of RGA protein abundance in 10-d-old Arabidopsis seedlings grown before and after a 9-h cold treatment (4°C). CBB, Coomassie Brilliant Blue as a loading control. B and E, Seedling survival of nonacclimated and cold-acclimated 10-d-old seedlings after cold treatment. C and D, qRT-PCR analyses of CBF (C) and COR15 (D) gene expression in 10-d-old seedlings. Student’s t tests were performed in comparison with the wild type unless indicated otherwise: a = P ≤ 0.05, b = P ≤ 0.01, c = P ≤ 0.001; n.s., not significant.
When we subsequently explored the relationship between YFP:GNL and the rga gai loss-of-function mutant with regard to cold tolerance, we found that YFP:GNL expression can induce cold tolerance also in this background, indicating that cold tolerance is promoted by GNL downstream from GA and DELLAs and, thus, that accumulation of the DELLAs as observed in YFP:GNL is not causative for promoting cold tolerance (Fig. 5E). In summary, these findings suggest that GNC and GNL as well as other factors are required to confer cold tolerance in the absence of GA. In this respect, the partial suppression of the cold tolerance phenotype of ga1 in ga1 gnc gnl is reminiscent of the partial suppression of the flowering time defect of ga1 in ga1 gnc gnl. As proposed above for the control of flowering time in the ga1 gnc gnl mutant, the cold response phenotype of ga1 also may be regulated by other GATA transcription factors or by unrelated regulators in addition to GNC and GNL.
SOC1 Represses GNC and GNL Transcription
The suppression of the soc1 greening and cold tolerance phenotypes in soc1 gnc gnl suggested a regulation of GNC and GNL gene expression by SOC1. Indeed, and as already mentioned before, we detected strongly increased GNC and GNL transcript levels in the soc1 mutant when compared with the wild type (Fig. 6A). Since this may be the consequence of a direct repressive activity of SOC1 on the promoters of the two GATA genes, we tested the binding of SOC1:MYC to CArG boxes in the GNC and GNL promoters using ChIP analysis. Indeed, we found binding of SOC1 to two promoter regions that span four CArG boxes in each gene promoter (boxes I and II in GNC and boxes I and III in GNL; Fig. 6, B–D). In both promoters, we further found increased binding of the open chromatin marker H3K9Ac in the soc1 mutant at a nucleosome-binding site proximal to their ATG start codons (Hb), while binding to a distal site (Ha) was unaffected (Fig. 6, E and F). Although the effect of SOC1:MYC overexpression on the transcript abundance of GNC and GNL was not strong when we examined whole seedlings using qRT-PCR, the repressive action of SOC1 on GNL became apparent when we examined GNLPro:GUS expression in the wild type and the SOC1:MYC background. Here, a strong reduction of GNLPro:GUS expression was detectable (e.g. in the upper region of the hypocotyl and the shoot meristem of long-day-grown seedlings; Fig. 6G). Taken together, these observations suggest that SOC1 is a direct transcriptional repressor of GNC and GNL transcription. There is thus a cross-repressive interaction between SOC1 and the two GATA factor genes. Their respective gene products can reciprocally repress their transcription and thereby promote or repress SOC1-dependent flowering or GNC- and GNL-dependent greening and cold tolerance, respectively.
DISCUSSION AND CONCLUSION
In this study, we analyze the role of the functionally paralogous GATA factors GNC and GNL in flowering time control, greening, and cold tolerance. This analysis was stimulated by our observations that GNC and GNL repress flowering in the GA-deficient ga1 mutant, on the one side, and that overexpression of the GATA factors is sufficient to delay flowering in the wild type, on the other (Fig. 1, D and E; Richter et al., 2010). Furthermore, we found here that overexpression of GNL, as a representative for the two GATAs, can delay flowering in a range of early-flowering GA pathway mutants. These findings are thus in line with a role of the two GATAs as repressors of flowering downstream from the GA signaling pathway. Our further observations that GNC and GNL repress flowering in wild-type plants only to a minor extent, under long-day as well as short-day conditions, and that GNC and GNL only partially repress flowering in ga1 suggest that other regulators in addition to GNC and GNL repress flowering in the absence of the two GATAs. Several possibilities can be envisioned that can serve to explain this phenotype. First, we know from our previous analyses and the analysis of others that the gnl allele (SALK_003995) used in our study has reduced GNL expression but is not a null allele (Mara and Irish, 2008). Unfortunately, we and others have been unable in repeated attempts to reisolate a presumed gnl null allele (SALK_021362; Bi et al., 2005; Mara and Irish, 2008). Thus, the residual flowering time repression in ga1 gnc gnl could potentially be the result of repression by residual GNL. Second, it may be that other related GATA factors repress flowering in addition to GNC and GNL. Arabidopsis has at least four GATA factors that are closely related to GNC and GNL and that may act redundantly in the repression of flowering in the absence of ga1 (Richter et al., 2010). Third, it may be that other unrelated proteins repress flowering in addition to GNC and GNL in the absence of GA. In this regard, it is interesting that the protein SPL9 was recently identified as a DELLA-controlled flowering time regulator in Arabidopsis and that GA was recently described to promote the expression of the SPL genes SPL3, SPL4, and SPL5 in a SOC1-dependent manner (Wu et al., 2009; Jung et al., 2012; Yu et al., 2012). Thus, DELLA repression of SPL gene expression and SPL protein activity could potentially delay flowering in ga1 in parallel to GNC and GNL.
That the suppression of the phenotypes caused by the GA deficiency of ga1 must be genetically complex is already suggested by the results of several ga1 suppressor screens that had been conducted in the past. In summary, only three genetic loci were identified as genetic suppressors of ga1, RGA, SPY, as well as a gain-of-function allele of the F-box protein subunit SLEEPY1 that promotes DELLA protein degradation (Wilson and Somerville, 1995; Silverstone et al., 1997; Fu et al., 2004). In fact, although it is known to date that all five Arabidopsis DELLA proteins contribute to the strong phenotype of the ga1 mutant, only the suppression of ga1 by the RGA loss-of-function allele significantly suppressed the ga1 mutant (Silverstone et al., 1997; Cheng et al., 2004). In turn, the founding member of the DELLA gene family, GAI, had been identified based on the gain-of-function phenotype of the gai-1 mutant, which, as we understand now, encodes for a GA-insensitive stabilized variant of GAI, and the reversion from this gain-of-function phenotype in the intragenic gai-t6 suppressor mutation (Peng et al., 1997). Here, we see some parallels between the suppression of ga1 by the loss of multiple DELLA genes that ultimately fully suppressed the ga1 phenotype and the partial suppression of ga1 in ga1 gnc gnl that could potentially be enhanced by the loss of other GATA factors or other regulators. Along the same lines, we see parallels in the genetics of the DELLA and the GNC or GNL loss-of-function mutants. In both cases, the repressive function is not obvious in the wild-type background but only apparent in GA deficiency or when protein content is increased due to a stabilization of the DELLA protein or due to an overexpression of the GATA factor.
While GNL overexpression delays flowering in the wild type, we further found that GNL overexpression cannot delay flowering when SOC1 is overexpressed; its expression is thus uncoupled from its normal transcriptional control in the wild type. Since the strong delay in flowering in our GNL overexpression studies correlated with the repression of SOC1, we hypothesized and subsequently tested successfully by ChIP-PCR that GNC and GNL directly bind to SOC1 to mediate its transcriptional repression. Using a complementing GNL promoter-driven GNL transgene, we could further substantiate our findings of SOC1 as a direct target of the GATAs and thus eliminate the possible criticism that the binding observed with ChIP-PCR using GNC:GFP and YFP:GNL is an artifactual off-target binding and amplification resulting from their overexpression. Taken together, our data strongly support a model whereby GNC and GNL are transcriptional repressors of SOC1.
Interestingly, our genetic interaction studies revealed an inverse relationship between SOC1 and the GATAs in the control of greening and cold tolerance. The regulatory role of SOC1 on the expression of GNC and GNL became apparent due to the visible suppression of the enhanced greening phenotype of soc1 in a soc1 gnc gnl mutant. Since SOC1 has been implicated in the control of cold tolerance, a finding that was initially based on the observation that cold-regulated genes are expressed in soc1 mutants even at ambient temperature, we also tested the contribution of GNC and GNL to the enhanced cold tolerance of the soc1 mutant as well as the cold tolerance phenotype of the late-flowering GNC and GNL overexpression lines. Here, we observed a suppression of the increased cold tolerance of soc1 in the absence of GNC and GNL as well as an increase in cold tolerance when the two GATAs are overexpressed. It was proposed that the antagonistic regulation of cold tolerance and flowering time by SOC1 may be relevant during short periods of cold temperature as they can be experienced by plants during cold periods in spring (Seo et al., 2009). Two studies have recently reported the genome-wide identification of direct SOC1 targets using ChIP (Immink et al., 2012; Tao et al., 2012). Interestingly, neither analysis has resulted in the identification of GNC or GNL as a direct SOC1 target. Since the different approaches employed make use of different parameters for data analysis, the differences in the results with regard to GNC and GNL promoter binding may simply be due to differences in the data analysis and stringency in data analysis.
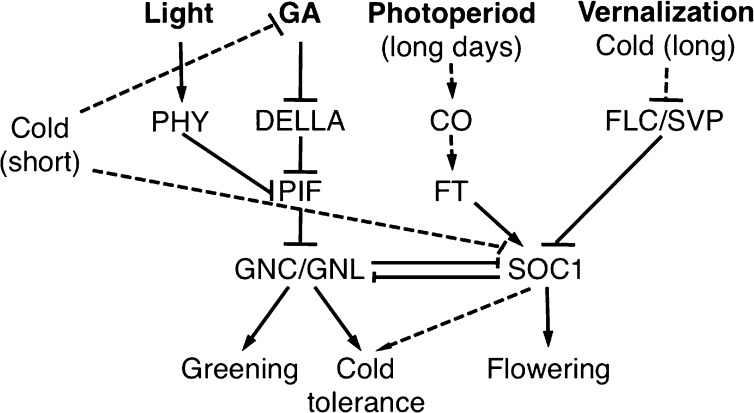
Besides positioning the GATA factors and SOC1 within their respective pathways, our observation of a mutual repression of the GATAs and SOC1 with regard to flowering time control, on the one side, and greening and cold tolerance, on the other, is particularly intriguing. In Arabidopsis, vernalization, temperature, photoperiod, as well as light quality and also GA biosynthesis are integrated at the level of SOC1 expression to ultimately promote flowering (Fig. 7). Therefore, it is interesting to speculate that, at least in Arabidopsis, the repression of the pathways for cold tolerance and greening following SOC1 activation and the transition to reproductive growth may ensure that sufficient resources can be allocated to flowering and thereby guarantee the plant’s reproductive success (Fig. 7). Inversely, activation of GNC and GNL expression during unfavorable growth conditions, such as cold stress, that will lead to increased DELLA protein levels in the Arabidopsis wild type, would result in increased GNC and GNL expression and shift the balance toward the cold tolerance-promoting GNC and GNL pathway (Fig. 7).
Figure 7.
Model of the interactions of the GATAs GNC and GNL and the MADS box transcription factor SOC1 in the control of greening, cold tolerance, and flowering with their respective regulators pathways. CO, CONSTANS; FLC, FLOWERING LOCUS C; PHY, phytochrome; SVP, SHORT VEGETATIVE PHASE.
MATERIALS AND METHODS
Biological Material
The following mutants and transgenic lines were used in this study: ga1 (Salk_109115; Willige et al., 2007); gai-1D (Peng et al., 1997); gnc (SALK_001778), gnl (SALK_003995), GNC:GFP (35S:GNC:GFP), GNCPro:GUS, YFP:GNL (35S:YFP:GNL), and GNLPro:GUS (Richter et al., 2010); PIF3:MYC (Clack et al., 2009); rga-24 gai-t6 (King et al., 2001); soc1-2, SOC1:MYC(9x), and SOC1Pro:GUS (Liu et al., 2008); and spy-3 (Jacobsen and Olszewski, 1993). With the exception of rga-24 gai-t6 and gai-1D (Ler), all mutants and transgenic lines are in the Arabidopsis (Arabidopsis thaliana) ecotype Columbia. YFP:GNL was introduced into the different genetic backgrounds by genetic crosses from a stably expressing YFP:GNL line in the Columbia or Ler background.
GNLPro:GNL
HA was obtained by insertion of a genomic fragment obtained by PCR amplification into the Gateway system-compatible cloning vector pEarleyGate 301 (Earley et al., 2006). The resulting T-DNA construct was directly transformed into gnc gnl mutants using the floral dip transformation method (Clough and Bent, 1998). For a list of relevant primers, see Supplemental Table S1.
Physiological Experiments
For flowering time analyses, plants were randomly arranged and grown in 150 μmol m−2 s−1 white light in MobyLux GroBanks (CLF Plant Climatics) under long-day (16 h of light/8 h of dark, 21°C/18°C) or short-day (8 h of light/16 h of dark, 21°C/18°C) conditions. The time of bolting was scored from at least 18 plants by counting the number of rosette leaves (Richter et al., 2010). For GA treatments, plants were watered twice per week. Cold tolerance experiments were performed as described previously (Achard et al., 2008a) with the following modification: 14-d-old seedlings that had or had not been cold acclimated for 2 d at 4°C were transferred to −20°C and kept at this temperature until the agar or soil temperature reached −6°C. Subsequently, seedlings were transferred to ambient temperature for an additional 4 d before seedling survival was quantified. For chlorophyll quantification, chlorophyll was extracted and quantified from 10-d-old seedlings (three independent replicates) as described previously (Inskeep and Bloom, 1985).
qRT-PCR
Total RNA for qRT-PCR was isolated with a NucleoSpin RNA plant kit from 10-d-old whole seedlings (Macherey-Nagel). DNA was removed by an on-column treatment with rDNase (Macherey-Nagel), and 2 µg of total RNA was subsequently reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Fermentas) using an oligo(dT) primer. The complementary DNA equivalent of 60 to 80 ng of total RNA was used in a 10-μL PCR in a CFX96 Real-Time System Cycler (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad) in a 40-cycle two-step amplification protocol (10 s at 95°C, 25 s at 60°C). Relevant primers are listed in Supplemental Table S1.
GUS Staining
GUS staining was performed according to previously published methods (Dohmann et al., 2008).
Immunoblots and ChIP
Immunoblotting was performed with the SuperSignal Femto West substrate as described previously (Thermo Fisher Scientific) using the anti-RGA antibody (Willige et al., 2007; Richter et al., 2010). Immunoblots were imaged using a LAS-4000 Mini image analyzer (FUJIFILM). ChIP and the subsequent quantitative PCRs were performed as described by others using antibodies against H3K9Ac (ab10812; Abcam) and H3K9me2 (ab1220; Abcam), with the GFP Vector Fusion Aid kit (Axorra) for GNC:GFP and YFP:GNL, anti-c-Myc agarose (Sigma) for SOC1:MYC, and an anti-HA antibody (3F10; Roche) together with A/Gplus-agarose beads (Santa Cruz Biotechnology) for GNL:HA (Fode and Gatz, 2009; Oh et al., 2009).
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: CBF1 (AT4G25490), CBF2 (AT4G25470), CBF3 (AT4G25480), COR15A (AT2G42540), COR15B (AT2G42530), FT (AT1G65480), GA1 (AT4G02780), GAI (AT1G14920), GID1A (AT3G05120), GID1B (AT3G63010), GID1C (AT5G27320), GNC (AT5G56860), GNL/CGA1 (AT4G26150), PIF3 (AT1G09530), PORA (AT5G54190), PORB (AT4G27440), PORC (AT1G03630), RGA (AT2G01570), SOC1 (AT2G45660), and SPY (AT3G11540).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Contributions of GNC and GNL to the repression of flowering in short-day conditions.
Supplemental Figure S2. GNC and GNL repress flowering downstream from DELLAs and PIF3.
Supplemental Figure S3. GNC and GNL transcript levels are increased in gai-1D when compared with the wild type.
Supplemental Figure S4. Transgenic GNL expressed from its own promoter binds to the SOC1 promoter.
Supplemental Figure S5. Differential GA sensitivity of soc1 mutants with regard to flowering time but not with regard to chlorophyll accumulation.
Supplemental Figure S6. Suppression of the soc1 chlorophyll accumulation phenotype in soc1 gnc gnl.
Supplemental Table S1. List of primers used in this study.
Acknowledgments
We thank Björn Willige and Ulrich Lutz for critical comments on the manuscript and Hao Yu for providing the SOC1:MYC and SOC1Pro:GUS transgenic lines.
Glossary
- Ler
Landsberg erecta
- qRT
quantitative real-time
- ChIP
chromatin immunoprecipitation
- H3K9Ac
K9-acetylated histone 3
- H3K9me2
K9-dimethylated histone 3
- HA
hemagglutinin
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP. (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. (2008a) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P. (2008b) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S. (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Weigel D. (1999) Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol 120: 1025–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE. (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA. (2009) Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T. (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EM, Levesque MP, De Veylder L, Reichardt I, Jürgens G, Schmid M, Schwechheimer C. (2008) The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135: 2013–2022 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode B, Gatz C. (2009) Chromatin immunoprecipitation experiments to investigate in vivo binding of Arabidopsis transcription factors to target sequences. Methods Mol Biol 479: 261–272 [DOI] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. (2007) Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci USA 104: 1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. (2010) Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Galvão VC, Horrer D, Küttner F, Schmid M. (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hisamatsu T, King RW. (2008) The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J Exp Bot 59: 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi YM, Rothstein SJ. (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk AD, Schmid M, et al. (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR. (1985) Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physiol 77: 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Matsuoka M. (2008) Molecular biology of gibberellins signaling in higher plants. Int Rev Cell Mol Biol 268: 191–221 [DOI] [PubMed] [Google Scholar]
- Izawa T. (2007) Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot 58: 3091–3097 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM. (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69: 577–588 [DOI] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP. (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Mara CD, Irish VF. (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Pérez-Alonso M, Salinas J. (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T. (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Nilsson O, Lee I, Blázquez MA, Weigel D. (1998) Flowering-time genes modulate the response to LEAFY activity. Genetics 150: 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F. (2005) Flowering: a time for integration. Int J Dev Biol 49: 585–593 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Coupland G. (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126: 1085–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. (2011) Gibberellin signaling in plants: the extended version. Front Plant Sci 2: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I. (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martínez EC, Sun TP. (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Thomas H. (1997) Chlorophyll: a symptom and a regulator of plastid development. New Phytol 136: 163–181 [Google Scholar]
- Tseng TS, Salomé PA, McClung CR, Olszewski NE. (2004) SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Somerville CR. (1995) Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol 108: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]