A bacterial peptide effector undergoes proteolytic processing in plants and releases small peptides that alter plant morphology.
Abstract
Phytoplasmas are insect-borne plant pathogenic bacteria that alter host morphology. TENGU, a small peptide of 38 residues, is a virulence factor secreted by phytoplasmas that induces dwarfism and witches’ broom in the host plant. In this study, we demonstrate that plants process TENGU in order to generate small functional peptides. First, virus vector-mediated transient expression demonstrated that the amino-terminal 11 amino acids of TENGU are capable of causing symptom development in Nicotiana benthamiana plants. The deletion of the 11th residue significantly diminished the symptom-inducing activity of TENGU, suggesting that these 11 amino acids constitute a functional domain. Second, we found that TENGU undergoes proteolytic processing in vitro, generating peptides of 19 and 21 residues including the functional domain. Third, we observed similar processing of TENGU in planta, and an alanine substitution mutant of TENGU, for which processing was compromised, showed reduced symptom induction activity. All TENGU homologs from several phytoplasma strains possessed similar symptom induction activity and went through processing, which suggests that the processing of TENGU might be related to its function.
Phytoplasmas are small plant pathogenic bacteria that infect more than 700 plant species and cause significant damage to agricultural production. They reside intracellularly in plant phloem cells and are transmitted by phloem-feeding insects of the order Hemiptera, which mainly include leafhoppers (Christensen et al., 2005).
Phytoplasma-infected plants show various characteristic symptoms often accompanied by morphological deformation, including dwarfism, witches’ broom (proliferation of small branches resulting in a characteristic bushy look), purple top (purple coloration of leaves and stems), yellowing, virescence (green coloration of floral organs), phyllody (formation of leaf-like structures instead of floral organs), and proliferation (Christensen et al., 2005).
Recent studies have shown that several secreted phytoplasma proteins function as virulence factors to induce these morphological deformations (Hoshi et al., 2009; MacLean et al., 2011; Sugio et al., 2011). Among them, a small secreted peptide, TENGU, encoded by onion yellows phytoplasma mild strain (OY-M), was the first phytoplasma virulence factor identified that affects plant morphology (Hoshi et al., 2009). TENGU, a small peptide secreted from phytoplasma, is predicted to be translated as a 70-amino acid preprotein with a 32-amino acid signal peptide at its N terminus. The C-terminal 38 amino acids of TENGU preprotein are secreted into plant cytoplasm via the Sec system, a bacterial conserved protein translocation system on the cellular membrane, where the N-terminal signal peptide is cleaved. Transient or transgenic expression of this 38-amino acid peptide, the putative secreted region of TENGU in Nicotiana benthamiana and Arabidopsis (Arabidopsis thaliana), results in a short and bushy phenotype similar to the symptoms of phytoplasma-infected plants (Hoshi et al., 2009). However, the mechanism underlying this symptom development and the features of the active form of the TENGU peptide remain unclear.
Peptides have been found to play important roles in the regulation of plant development. In particular, secreted peptides act as signaling molecules mediating intercellular communications. A major group of such peptide signals, which includes many important peptides such as CLAVATA3 (CLV3; Fletcher et al., 1999), ROOT MERISTEM GROWTH FACTOR1 (Matsuzaki et al., 2010), and PHYTOSULFOKINE (PSK; Matsubayashi and Sakagami, 1996), is characterized by its small size (less than 20 amino acids) resulting from proteolytic processing (Matsubayashi, 2011).
In order to better understand the function of TENGU, we identified its functional domain. In doing so, we found that the N-terminal 11 amino acids of the secreted region of TENGU are sufficient for its function. Furthermore, the secreted region of TENGU is processed in vitro by plant extract, producing several C-terminally truncated peptides containing this functional domain. These peptides were not detected in the processing product of a TENGU mutant with reduced symptom induction activity, suggesting a relationship between TENGU processing and function.
RESULTS
The N-Terminal 11 Amino Acids Make Up the Functional Domain of TENGU
Transient expression of TENGU (38 amino acids) in N. benthamiana using a potato virus X (PVX) vector suppressed the elongation of the main stem. This led to an increase in total leaf number, presumably from enhancement of the outgrowth of lateral buds (Fig. 1A). To identify the functional domain of TENGU, which induces these morphological changes, we generated truncated mutants of TENGU and tested their symptom-inducing activities (Fig. 1B; Table I).
Figure 1.
Mapping the functional region of TENGU. A, Sequence of the 70-amino acid TENGU preprotein. The N-terminal 32 amino acids constitute the secretion signal sequence (underlined). N. benthamiana plants expressing the 38-amino acid TENGU mature protein (right) and plants inoculated with empty viral vector (left) are shown. B, Schematic diagram of TENGU truncation mutants. The secreted regions of TENGU and each truncation mutant are expressed with the engineered Met residue at their N termini (boldface). The amino acid sequence of the 38-amino acid secreted region of TENGU is shown at the top, and the symptom-inducing activity of each truncated mutant is indicated at the right. C, N. benthamiana plants expressing TENGU truncation mutants (N19, C19, N12, N11, and N10; right side of each panel) and the control plants inoculated with empty viral vector (left side of each panel). In A and C, photographs were taken at 21 dpi. In the bottom panels of A and C, stems are highlighted with red lines and leaf petioles with black lines. Bars = 5 cm.
Table I. Symptom-inducing activities of TENGU truncation mutants.
The number of plants used in a transient expression assay and the number of plants showing symptoms are shown.
| Parameter | Vector | TENGU | N19 | C19 | N12 | N11 | N10 | N8 |
|---|---|---|---|---|---|---|---|---|
| No. | 96 | 260 | 136 | 68 | 60 | 60 | 104 | 104 |
| Symptoma | 0 | 84 | 50 | 0 | 24 | 20 | 4 | 0 |
| Frequency (%) | 0 | 32.31b | 36.76b | 0 | 40.00b | 33.33b | 3.85 | 0 |
Plants with average internode length under 0.7 cm and with total leaf number of more than 15 were counted. bStatistical significance compared with the control vector, based on a χ2 test (P < 0.01).
Plants that expressed the N-terminal 19 amino acids of the secreted region of TENGU (hereafter referred to as N19) displayed a bushy phenotype characteristic of those expressing the TENGU 38-amino acid protein. However, plants that expressed the C-terminal 19 amino acids (C19) did not show any morphological changes (Fig. 1C), suggesting that the N-terminal domain of TENGU was responsible for symptom development. To further define the functional domain of TENGU, we analyzed the activities of four truncation mutants consisting of the N-terminal 12, 11, 10, and eight amino acids of TENGU (N12, N11, N10, and N8, respectively). As shown in Figure 1C, both N12 and N11 induced characteristic symptoms in N. benthamiana, whereas N10 and N8 did not.
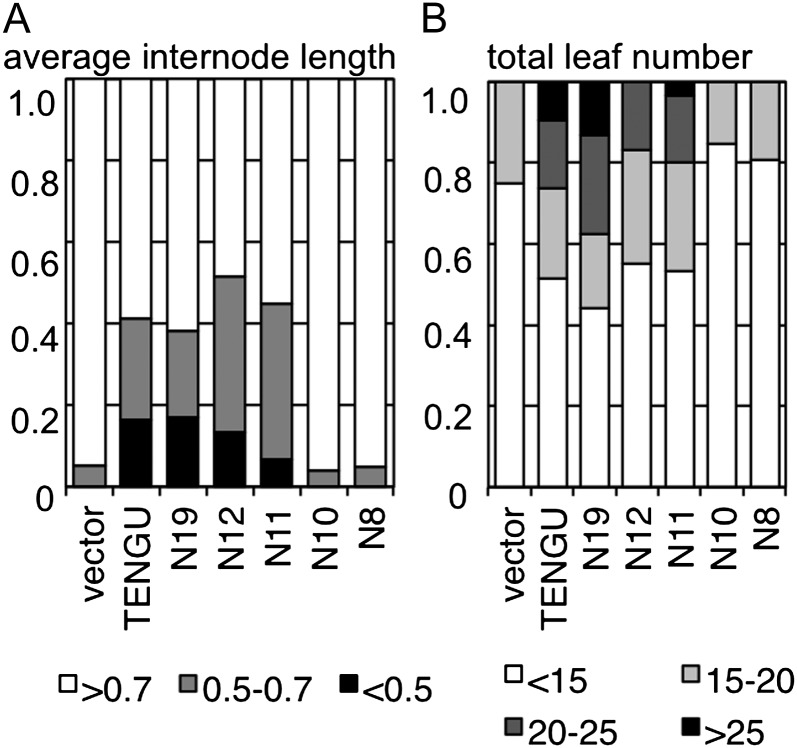
We quantified the morphological changes in plants expressing TENGU and its truncated mutants. Transient expression of the TENGU 38-amino acid protein induced symptoms including reduction in the average internode length and increase in the total leaf number of tested plants (Fig. 2). As judged from these two parameters, the activity of the C19 mutant was low, similar to the control vector that caused no symptoms, suggesting that the C-terminal region of TENGU is not involved in TENGU function. In addition, the activities of the N19, N12, and N11 mutants were comparable to those of full-length TENGU, whereas the activities of N10 and N8 mutants were similar to those of the negative control. These results suggest that the N-terminal 11 amino acids make up the functional domain of TENGU and are sufficient to cause symptom development. Moreover, we found that the deletion of the Leu residue at the 11th residue significantly diminished symptom-inducing activity, suggesting that this residue is important for TENGU function.
Figure 2.
Quantification of symptom-inducing activity of TENGU truncation mutants. A, Stacked bar graph showing the proportion of plants with an average internode length of less than 0.5 cm (black boxes), 0.5 to 0.7 cm (gray boxes), or more than 0.7 cm (white boxes). B, Stacked bar graph showing the proportion of plants with a total leaf number of more than 25 (black boxes), 20 to 25 (dark gray boxes), 15 to 20 (light gray boxes), and less than 15 (white boxes). The total number of plants is shown in Table I.
Generation of Small Fragments of TENGU by in Vitro Processing
In our transient expression assay of truncation mutants, we noted that the expression of N11 and N12, the N-terminal domain of TENGU, induced symptoms more consistently than full-length TENGU (Table I). Therefore, we hypothesized that the phytoplasma effector TENGU might be processed in plants. To test this hypothesis, we performed an in vitro processing assay according to previously reported methods with modifications (Ni and Clark, 2006).
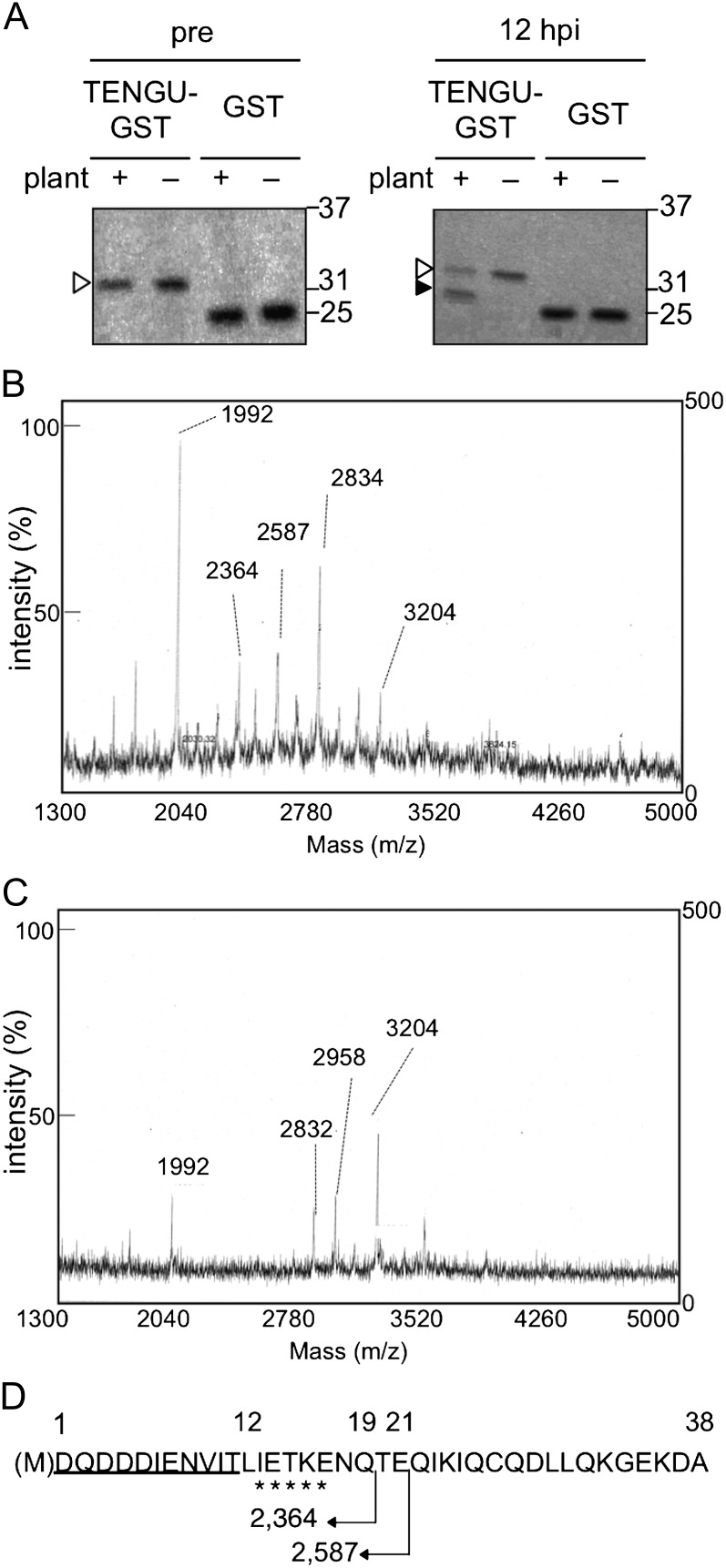
We constructed a recombinant protein of the secreted region of TENGU as a C-terminal fusion protein with glutathione-S-transferase (GST). We then incubated this fusion protein (TENGU-GST), as well as a GST protein control, in the presence of Arabidopsis plant extracts. The TENGU-GST protein was stable for up to 48 h when incubated with buffer used for protein extraction (Supplemental Fig. S1). After 12 h of incubation at room temperature with the plant extracts, a putative cleavage product of the TENGU-GST fusion protein was observed by gel electrophoresis (Fig. 3A), whereas no cleavage product was observed for the control GST protein. We purified this C-terminal cleavage product using Glutathione Sepharose beads and characterized it using N-terminal Edman sequencing (Supplemental Fig. S2). As a result, an amino acid sequence was obtained that corresponded to the 13th to the 17th residues of the secreted region of TENGU. This result suggests that TENGU can be processed between the 12th and 13th residues. To further characterize the N-terminal cleaved fragment, we performed a time-of-flight (TOF) mass spectrometry (MS) analysis of the unpurified TENGU-GST cleavage product. When we compared the MS spectrum of the TENGU-GST cleavage product (Fig. 3B) with that of the control solution wherein TENGU-GST was incubated with buffer (Fig. 3C), two peaks at mass-to-charge ratio 2,364 and 2,587 were specifically detected. The mass of these peaks matched the predicted mass of N-terminal 19- and 21-amino acid fragments of TENGU with engineered Met residue (N19 and N21, Fig. 3D). Together with the Edman sequencing, these results suggest that TENGU can be processed in vitro at several sites, generating a detectable level of cleaved peptides consisting of the N-terminal 19 and 21 residues of the secreted region of TENGU.
Figure 3.
In vitro processing of the recombinant TENGU-GST protein. A, TENGU-GST and GST protein were incubated with or without plant extract. Both proteins were analyzed by gel electrophoresis followed by Coomassie Brilliant Blue staining before (pre; left panel) and after 12 h of incubation (12 h post incubation [hpi]; right panel). White arrowheads indicate bands corresponding to full-length TENGU-GST, and the black arrowhead indicates that the band corresponds to the processing product. Molecular size markers (kD) are shown on the right. B, TOF-MS spectrum of TENGU-GST incubated with plant extract. C, TOF-MS spectrum of TENGU-GST incubated with buffer. m/z, Mass-to-charge ratio. D, Schematic representation of TENGU processing. Sequence obtained by Edman sequencing is marked by asterisks. The TENGU functional region (11 residues) is underlined. The predicted molecular mass (D) of the partial fragments of TENGU is indicated below the sequence.
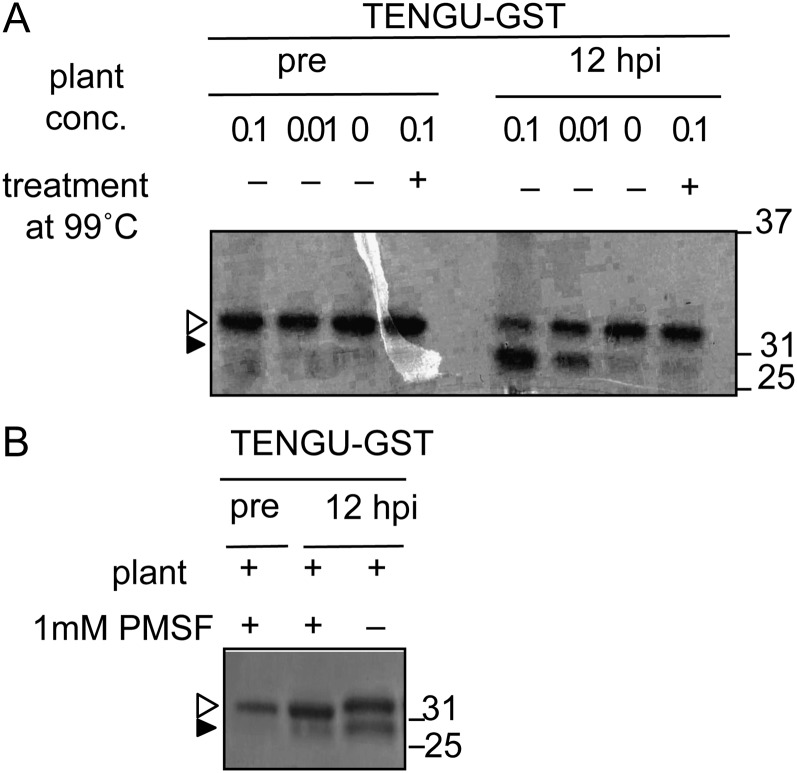
We further examined the characteristics of the in vitro processing of TENGU using plant extracts. We prepared Arabidopsis plant extracts at two different concentrations (100-fold and 10-fold dilution) and monitored them for in vitro TENGU processing activity (see “Materials and Methods”). As shown in Figure 4A, the 100-fold-diluted plant extract showed reduced processing activity compared with the 10-fold-diluted extract. Moreover, processing of TENGU-GST was inhibited by pretreatment of the plant extract at 99°C for 5 min. Therefore, TENGU processing was dependent on heat-labile host factor(s). Furthermore, the addition of a serine protease inhibitor, phenylmethylsulfonyl fluoride (PMSF), reduced the efficiency of TENGU processing, suggesting that a plant serine protease is involved in the in vitro processing (Fig. 4B).
Figure 4.
The dependence of TENGU in vitro processing on heat-labile host factor. A, Effects of heat treatment of the plant extract on TENGU-processing activity. Plant extract with and without treatment at 99°C for 5 min was incubated with TENGU-GST. The relative concentration of plant extract is indicated by 0.1, 0.01, and 0 (see “Materials and Methods”). B, Effects of the protease inhibitor on TENGU-GST processing. TENGU-GST protein and plant extract were incubated with or without 1 mm serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF). hpi, Hours post incubation.
TENGU Processing in Planta
The plant extract used in the in vitro processing assay was thought to contain a large amount of proteolytic enzymes, not all of which interact with TENGU under natural conditions. Hence, we further examined whether TENGU was processed in planta. We expressed myc-tagged GST-TENGU or myc-tagged GST under the control of the 35S promoter by agroinfiltration and examined the accumulation of both proteins by immunoblotting (Fig. 5). At 4 d post inoculation (dpi), both myc-GST-TENGU and myc-GST were detected as a single band at the expected molecular mass. However, at 5 dpi, an extra band was detected in the myc-GST-TENGU-expressing plant at a position corresponding to a processing product of myc-GST-TENGU. On the other hand, no extra band was detected in the plant expressing myc-GST, suggesting that TENGU was also processed in planta.
Figure 5.
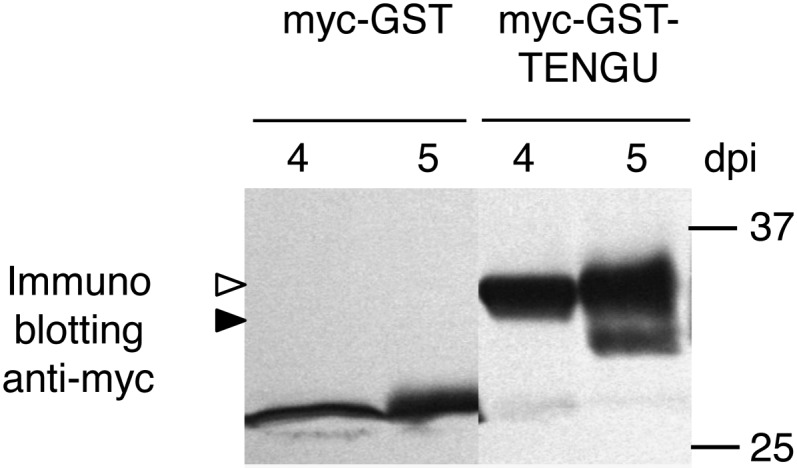
In planta processing of the TENGU fusion protein. myc-GST or myc-TENGU-GST protein was expressed in N. benthamiana by agroinfiltration. At 4 and 5 dpi, the expressed proteins were detected by immunoblotting using anti-myc antibody. The white arrowhead indicates the band corresponding to myc-GST-TENGU protein. A band of a putative processing product is marked with the black arrowhead.
Attenuation of TENGU Function by a Processing Site Mutation
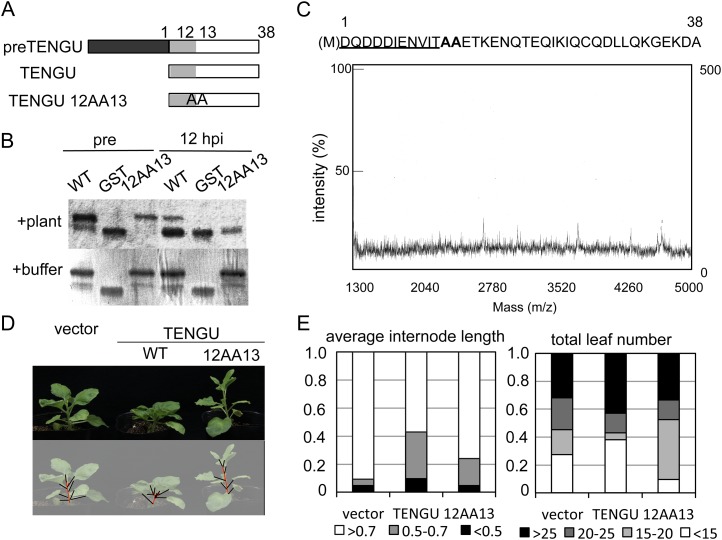
To examine the significance of TENGU processing in symptom-inducing activities, we produced an Ala substitution mutant, in which residues that flank one of the three processing sites, Leu-12 and Ile-13, were replaced by Ala (12AA13; Fig. 6A). We examined whether the mutant would be processed and whether it would induce TENGU-characteristic symptoms.
Figure 6.
Attenuation of TENGU function by a processing site mutation. A, Schematic representation of the TENGU-processing site mutant. The dark gray box indicates the secretion signal peptide. Light gray boxes indicate one of the N-terminal processing products predicted by in vitro processing analysis with symptom-inducing activities. White boxes indicate the C-terminal processing product. B, In vitro processing assay of the TENGU mutant. GST fusion proteins of wild-type TENGU (WT) and the 12AA13 TENGU mutant were incubated with plant extract or buffer and detected using gel electrophoresis followed by Coomassie Brilliant Blue staining. C, TOF-MS analysis of the 12AA13-GST-processing product. The sequence of the 12AA13 mutant is shown above the MS spectrum; the functional domain of TENGU is underlined; and mutated residues are shown in boldface. D, Symptom-inducing activities of TENGU and the 12AA13 mutant. E, Quantification of the symptom-inducing activity of the 12AA13 mutant.
We first performed an in vitro processing assay of the 12AA13 mutant fused to the N terminus of GST. After 12 h of incubation with plant extracts, the 12AA13-GST fusion protein appeared to be cleaved, as indicated by the emergence of a faster migrating band by SDS-PAGE (Fig. 6B). However, in the TOF-MS analysis of the 12AA13 mutant cleavage product, we observed fewer peaks with low intensity (Fig. 6C) compared with the wild-type TENGU (Fig. 3B). Importantly, fragments of the N19 and N21 amino acids that were detected in the cleavage product of wild-type TENGU were absent from the mutant cleavage product, suggesting that the proper accumulation of processed peptides was compromised by these Ala substitutions.
Next, we compared the symptom-inducing activities of the 12AA13 mutant with that of the TENGU wild type using a PVX-mediated transient expression assay. We found that transient expression of the 12AA13 mutant induced symptoms at a lower frequency than the wild type (Fig. 6, D and E; Table II).
Table II. Symptom-inducing activities of the Ala substitution mutant of TENGU.
The number of plants used in a transient expression assay and the number of plants showing symptoms are listed.
| Parameter | Vector | TENGU |
|
|---|---|---|---|
| Wild Type | 12AA13 | ||
| No. | 23 | 21 | 21 |
| Symptoma | 0 | 7 | 3 |
| Frequency (%) | 0 | 33.33 | 19.05 |
Plants with average internode length of less than 0.7 cm and with total leaf number of more than 15 were counted.
Processing of Functional TENGU Orthologs
To further test the biological significance of TENGU processing, we searched for tengu homologs and investigated whether they would also be processed. We first searched for tengu homologs using BLAST from three phytoplasma strains whose complete genomic sequences have been determined (aster yellows phytoplasma strain witches’ broom [AY-WB], Australian grapevine yellows, and apple proliferation; Bai et al., 2006; Kube et al., 2008; Tran-Nguyen et al., 2008). We identified only one tengu homolog (AYWB169) from AY-WB, which belonged to the same phylogenetic group as OY-M phytoplasma. Next, we cloned tengu homologs by PCR using primers that recognize the upstream and downstream genes of tengu. We obtained fragments containing tengu homologs from 11 phytoplasma strains in the aster yellows group (OY-W, onion yellows phytoplasma line W; PPT, potato purple top phytoplasma; ACLR, apricot chlorotic leaf role phytoplasma; KV, clover phyllody phytoplasma; PaWB, paulownia witches’ broom phytoplasma; sumac witches’ broom phytoplasma; GY, garlic yellows phytoplasma; MD, mulberry dwarf phytoplasma; WDWB, water dropwort witches’ broom phytoplasma; BamWB, bamboo witches’ broom phytoplasma; and PvWB, porcelain vine witches’ broom phytoplasma; Supplemental Fig. S3). We also attempted to amplify tengu-containing fragments from 11 phytoplasma strains belonging to other phylogenetic groups but were unsuccessful (data not shown).
The tengu homologs from the 11 phytoplasma strains were sequenced, and their amino acid sequences were analyzed with SignalP and SOSUI software. They contained similar features to TENGU from OY-M: a signal peptide, a signal cleavage site at their N termini, and a C-terminal 38-amino acid segment with no transmembrane domain. Therefore, the homologs are predicted to be secreted into plant cytoplasm (Fig. 7).
Figure 7.
Amino acid sequence alignment of TENGU homologs. Transmembrane regions are as predicted by the SOSUI program (underlined). Putative signal peptide cleavage sites are as predicted by the SignalP program (arrowhead). All TENGU homologs encode putative secretory peptides similar to TENGU, of which the C-terminal 38 residues are secreted to the host cytoplasm. Black boxes show identical residues. Gray boxes show similar residues. SWB, Sumac witches’ broom phytoplasma.
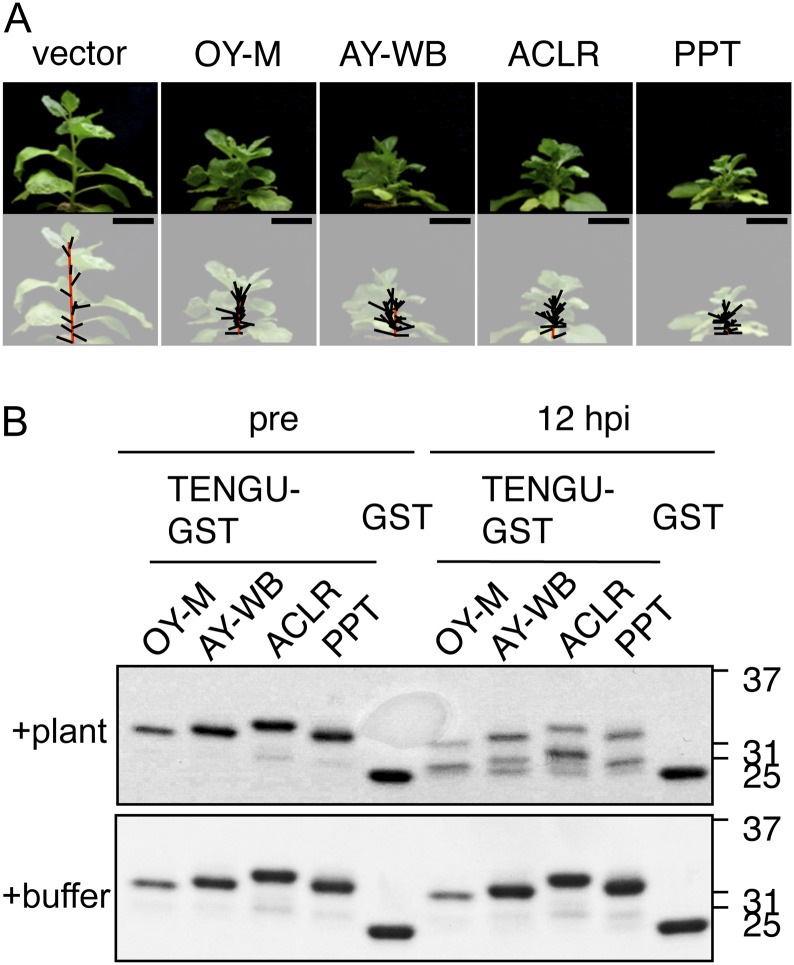
Although the amino acid sequence of TENGU was highly conserved among homologs, some amino acid polymorphisms were observed in the homologs from four phytoplasma strains (KV, PPT, AY-WB, and ACLR). Virus vector-mediated transient expression assays and in vitro processing assays demonstrated that all tengu homologs induced characteristic symptoms when transiently expressed in N. benthamiana (Fig. 8A) and that all tengu homologs were processed in vitro (Fig. 8B). These results suggested that these homologs are functional orthologs. The functional domain of TENGU, encompassing one to 11 amino acids of the secreted region, was consistently conserved among these homologs (Supplemental Fig. S4).
Figure 8.
Symptom-inducing activity and in vitro processing of tengu homologs. The secreted regions of the tengu homolog from KV strain and that from PPT strain were identical at the amino-acid level. A, Transient expression assay of tengu homologs. From left to right: empty viral vector (vector), a vector expressing TENGU from OY-M, and its homologs of AY-WB, ACLR, and PPT strains. All photographs were taken at 21 dpi. Bars = 5 cm. B, In vitro processing assay of TENGU homologs. Recombinant GST fusion proteins of TENGU and its homologs were incubated with plant extract (top panel) or buffer (bottom panel). Proteins were analyzed by gel electrophoresis before (pre) and after (12 h post incubation [hpi]).
DISCUSSION
TENGU Processing’s Potential Role in Symptom Induction
TENGU is a bacterial effector that is translated as a 70-amino acid preprotein, of which 38 C-terminal amino acids are released into plant phloem cells. Here, we found that the secreted region of TENGU was further processed by an unidentified plant host factor with protease activity, generating peptides of N19 and N21 residues containing 11-amino acid functional domains of TENGU. We suggest a scenario wherein TENGU is processed by a plant factor in order to generate functional peptides. The sequence of the 11-amino acid functional domain of TENGU was consistently conserved, and the presence of proteolytic processing was observed for all tested tengu orthologs. However, it is notable that the C-terminal region of TENGU, which is cleaved from the functional domain and is dispensable for symptom induction, is also conserved. This may suggest a conserved, but as of yet uncharacterized, function of the C-terminal region of TENGU.
The fact that the processed form of TENGU was found to have stronger symptom-inducing activity than the full-length TENGU (Table I) suggests that TENGU processing is important to symptom-inducing functions. Moreover, the significantly decreased accumulation of processed peptides from 12AA13, a TENGU mutant with reduced symptom-inducing activity, further strengthens the idea that small peptides generated through processing are functionally important. However, we cannot rule out the possibility that the substitution of the 12th and/or 13th residues attenuated TENGU function in a processing-independent manner, such as by compromising the proper folding or stability of the TENGU protein.
The Mechanism of TENGU Processing
Although the TENGU processing product was observed as a single dominant band by SDS-PAGE, we detected several distinct TENGU fragments upon TOF-MS analyses, suggesting that TENGU processing may be a complex process. Our in vitro processing experiments showed that TENGU recombinant protein could be processed at least at three sites: between Leu-12 and Ile-13, between Gln-19 and Thr-20, and between Glu-21 and Gln-22. The Ala substitution of Leu-12 and Ile-13 compromised the accumulation of peptides that were expected to be produced through processing at the latter two sites (Fig. 6C). It may seem unusual that the mutation of such distant residues influenced the processing rate. A possible explanation for this observation is that this substitution may affect the structure of the TENGU protein and interfere with the proper interaction between TENGU and host proteases. Alternatively, given that the recombinant GST fusion protein of the TENGU mutant seemed to be processed similarly to that of wild-type TENGU by SDS-PAGE, this substitution may reduce the stability of processed fragments.
We showed that TENGU processing is at least partly dependent on some heat-labile host plant factors, such as a serine protease(s). Interestingly, a precursor of Arabidopsis PSK and a precursor of peptide signal AtRALF23 are processed by subtilisin-like serine proteases named “AtSBT1.1” and “site 1 protease,” respectively. A loss of these proteases diminishes the processing of both precursor proteins (Srivastava et al., 2008, 2009). Identification of the TENGU-processing proteases would assist in analyzing the complex mechanisms of TENGU processing. In addition, such host protease(s) may be a candidate target for reducing symptom expression upon phytoplasma infection. Efforts are currently under way to purify the host protease(s) by tracing TENGU-processing activity.
TENGU’s Interference with Plant Development
To date, many plant bioactive peptides have been isolated and their functions have been documented. Similar to TENGU, which is released inside plant phloem cells and affects host development, some bioactive peptides, including ROTUNDIFOLIA4 (ROT4), are thought to act inside cells to regulate plant morphology. The peptide ROT4, of which the core functional region is the 30- to 34-amino acid central region, is involved in leaf development and is not thought to be processed (Narita et al., 2004; Ikeuchi et al., 2011). Although there is no known homology in the amino acid sequence, there might be a functional similarity between TENGU and these intracellular bioactive peptides.
In contrast to ROT4-type intracellular peptides, many bioactive peptides of plants are encoded as precursor proteins with a secretion signal peptide at their N termini and are secreted into the apoplast. They include important signaling molecules such as CLV3, which mediate non-cell-autonomous regulation of plant development (Clark, 2001; Hirakawa et al., 2008). Several common features are reported among these secreted peptides. Namely, their precursors undergo extensive processing such as glycosylation, Tyr sulfation, and proteolytic processing that release their mature peptides under 20 residues in length and allow them to function in vivo (Kondo et al., 2006; Matsuzaki et al., 2010). We found that TENGU is also processed, generating small peptides that contain its entire functional region. In addition, TENGU was previously found in phloem cells as well as other plant tissues that do not contain phytoplasma (Hoshi et al., 2009). This observation suggests that TENGU somehow moves within plants, as often reported for the effectors of plant pathogens (Khang et al., 2010; Rafiqi et al., 2010). This non-cell-autonomous feature of TENGU is different from ROT4, which acts locally (Ikeuchi et al., 2011). Thus, the mechanism of TENGU is in part similar to that of secreted peptides.
Recently, a striking homology was found between plant CLAVATA/ESR-related peptides (CLEs), which are encoded by multiple paralogs of CLV3 and regulate plant development (Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011), and small virulence factors of plant pathogenic nematodes. These nematode CLEs mimic host CLEs to support nematode infection (Wang et al., 2005; Guo et al., 2011). However, unlike nematode CLEs, TENGU does not show homology with any known proteins, including plant peptide signals. Therefore, TENGU seems to not simply mimic plant peptides but to induce characteristic morphological changes in host plants in an unknown but distinct manner. Deciphering the molecular mechanisms of TENGU function would provide new insight into the role of peptides in the regulation of plant growth.
MATERIALS AND METHODS
Phytoplasma Strains and Plant Materials
The phytoplasma strains used in this study are listed in Supplemental Table S1. Candidatus phytoplasma asteris OY (OY-M [AP006628] and OY-W) was maintained as described previously (Oshima et al., 2001). Samples of DNA from PPT, ACLR, and KV strains (Kakizawa et al., 2006) were kindly provided by Dr. A. Bertaccini (University of Bologna). Infected plants of PaWB, sumac witches’ broom phytoplasma, GY, and MD strains (Kakizawa et al., 2006) were kindly provided by Drs. T. Shiomi (National Institute for Agro-Environmental Sciences), T. Usugi (National Agricultural Research Center), N. Nishimura (Koibuchi College of Agriculture and Nutrition), and T. Tsuchizaki (Koibuchi College of Agriculture and Nutrition). Infected plants of BamWB, WDWB, and PvWB strains were kindly provided by Dr. H.-Y. Jung (Kyungpook National University). The Arabidopsis (Arabidopsis thaliana) and Nicotiana benthamiana plants were maintained in growth chambers with a 16-h-light/8-h-dark cycle at 23°C and 25°C, respectively.
Cloning of tengu Homologs
All primers used in this study are listed in Supplemental Table S2. Total DNA from healthy and phytoplasma-infected plants was extracted as described previously (Lee and Davis, 1986). Each tengu homolog was amplified from each phytoplasma-infected plant DNA using PCR with the primer pair PAM485F and PAM486R. The PCR-amplified fragments were sequenced directly, as described previously (Kakizawa et al., 2009).
Vector Construction
For viral vector-mediated expression of TENGU and its mutants, we used the binary PVX vector pCAMV. The vector pCAMV is a derivative of the binary vector pCAMBIA1301, which contains full-length PVX vector complementary DNA under the control of the 35S promoter derived from pP2C2S, and was kindly provided by Dr. David Baulcombe (Sainsbury Laboratory). A previously described pCAMV-tengu that expresses a 38-amino acid secreted region of TENGU with an N-terminal Met residue was used for the expression of TENGU (Hoshi et al., 2009). For TENGU expression, truncation mutants N19, C19, N12, N11, N10, and N8 and partial fragments of tengu were amplified from pCAMV-tengu and fused with an N-terminal Met residue by PCR using the following primers: for N19, PAM765-F and N19-R; for C19, C19-F and PAM765-R; for N12, N1211-F and N12-R; for N11, N1211-F and N11-R; for N10, N10-F and N10-R; and for N8, N8-F and N8-R. Similarly, the secreted regions of tengu homologs from ACLR and PPT phytoplasma strains were amplified from total DNA of phytoplasma-infected plants by PCR using the primers ACLR-F and ACLR-R or PPT-F and PPT-R, respectively. The secreted region of the tengu homolog from the AY-WB strain was generated by PCR using the primers AYWB-F and AYWB-R. All amplified fragments were cloned into the pCAMV vector between the SalI and SmaI sites, as described previously by Hoshi et al. (2009). To produce TENGU-GST recombinant protein, pET-tengu-gst was constructed using the following recombinant PCR technique. The tengu and gst genes were amplified by PCR with the primers pETtengu-F and TENGUgst-R or with the primers tenguGST-F and GSTecI-R from pCAMV-tengu or pGEX4T-1, respectively. The amplified fragments were mixed and combined by PCR using pETtengu-F and GSTecI-R primers, and the resultant chimeric gene, which encodes the secreted region of TENGU with an N-terminal Met residue and a C-terminal GST protein, was cloned into the pET30a vector between the NdeI and EcoRI sites.
The secreted regions of tengu homologs of the AY-WB, ACLR, and PPT strains were similarly combined with the gst gene using pETart-F and pETart-R primers instead of pETtengu-F and GSTecI-R. The amplified tengu homologs were ligated with linearized pET30a vector between the NdeI and EcoRI sites using the GENEART Seamless Cloning and Assembly kit (Life Technologies) according to the manufacturer’s instructions.
Amino acid substitution mutants at the processing site of TENGU (12AA13) were generated from pET-tengu-gst by PCR using primers 1213A-F or 1213E-F and GSTecI-R. The amplified fragments were cloned into pET30a between the NdeI and EcoRI sites. For the transient expression assay, both mutated tengu genes were amplified by PCR using primers PAM765-F and PAM765-R and cloned into pCAMV between the SalI and SmaI sites.
To construct 35S:myc-gst-tengu, the tengu gene was amplified with pGEXTENGU-F and pETTENGU-R primers and cloned into pGEX-4T-1 (GE Healthcare) between the BamHI and XhoI sites to generate pGEX-tengu carrying an N-terminal gst fusion gene of tengu. Subsequently, gst-tengu and gst as a control were amplified by PCR of pGEX-tengu or pGEX4T-1 using the primers GST-F and GSTTENGU-R or GST-F and GST-R, respectively, and then subcloned into pENTR (Life Technologies) between the SalI and EcoRI sites. Both genes were then introduced into the destination vector pEarleygate 203 (Earley et al., 2006) and fused in frame with an N-terminal cmyc tag using LR Clonase II Enzyme Mix (Life Technologies) according to the manufacturer’s instructions.
Transient Expression Assay
Agrobacterium tumefaciens EHA105 strains were transformed with each expression vector, and A. tumefaciens infiltration was performed as described previously (Senshu et al., 2011). Cultured bacterial cells were harvested and resuspended in infiltration buffer to a final optical density at 600 nm of 1.0. At 21 dpi, the length of the main stem, leaf number on the main stem, and total leaf number were measured for each plant. The average internode length of each tested plant was calculated as the length of the main stem divided by the leaf number of the main stem. Tested plants were categorized into three classes based on the average internode length (less than 0.5 cm, 0.5–0.7 cm, and more than 0.7 cm) or into four classes based on the total leaf number (less than 15, 15–20, 20–25, and more than 25).
In Vitro Processing Assay
A recombinant GST fusion protein of TENGU, TENGU amino acid substitution mutants, and tengu homologs were expressed in Escherichia coli BL21 cells. Expressed proteins were purified from cell culture using Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions. TENGU-GST fusion proteins, TENGU amino acid substitution mutants, tengu homologs, and GST were eluted with glutathione elution buffer (100 mm Tris, 20 mm glutathione, and 200 mm NaCl, pH 9.5). To prepare plant extracts, 2.0 g of Arabidopsis (whole-plant samples) was harvested 35 d after germination and ground in liquid nitrogen. Total protein was extracted with 4.0 mL of 20 mm Tris buffer (pH 8.0). The ground extract was centrifuged (10,000g for 5 min at 4°C), and the resulting supernatant was diluted 10-fold or 100-fold using 20 mm Tris buffer (pH 8.0) and used in the following in vitro processing assay. For the in vitro processing assay, 3.5 µg (10 µL) of TENGU-GST or its derivative protein was mixed with 90 µL of diluted plant extract and incubated for 12 h at room temperature. The concentrations of TENGU-GST and its derivative proteins were determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories).
TOF-MS Analyses
For TOF-MS analyses, 1,000 µL of in vitro processing reaction product was used. After 12 h of incubation, reaction products were mixed with 3 volumes of acetone and were frozen at −30°C for more than 3 h. Total proteins were collected by centrifugation at 10,000g for 15 min. The pellets were rinsed with 1,000 µL of acetone and then dried. Pellets were dissolved with 45 µL of 2% trifluoroacetic acid of which a 20-µL aliquot was purified with Ziptip (Millipore). Mass spectra were acquired by Voyager-DE STR (AB Sciex) using alpha-cyano-4-hydroxycinnamic acid matrix.
Immunoblotting
Immunoblotting was performed as described previously (Senshu et al., 2011). Briefly, N. benthamiana plants were inoculated with A. tumefaciens carrying each expression vector (35S:myc-GST-TENGU and 35S:myc-GST). Total protein from the infiltrated leaves was extracted with phosphate-buffered saline buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4OP·2H2O, and 1.8 mm KH2PO4, pH 7.4) containing complete mini protease inhibitor (Roche) at 4 or 5 dpi. Then, the protein was separated by SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and detected by anti-myc antibody clone 4A6 (1:5,000 dilution; Millipore).
Phylogenetic Analysis
A phylogenetic tree based on 16S ribosomal DNA was constructed as described previously using a 16S ribosomal DNA sequence of Acholeplasma laidlawii as an outgroup (Kakizawa et al., 2009). To construct the phylogenetic tree, the neighbor-joining method in MEGA version 3.1 was used. Any open reading frame similarities were analyzed using sequence interpretation tools (Institute of Medical Science, University of Tokyo) and the BLAST algorithm. Sequence alignments were performed using ClustalW version 1.8. The putative transmembrane domain was predicted using the SOSUI program version 1.11 (http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html), and the prediction of the signal sequence was performed using the SignalP program version 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB750352 to AB750362 (tengu homolog of GY, MD, SWD, OY-W, WDWB, PPT, BamWB, ACLR, PvWB, PaWB, and KV, respectively) and AB750363 (16S ribosomal RNA of GY).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Stability of the TENGU-GST protein.
Supplemental Figure S2. Characterization of the TENGU-GST processing product by Edman sequencing.
Supplemental Figure S3. Conservation of tengu homologs among phytoplasmas.
Supplemental Figure S4. Conservation of the 11-amino acid functional region among TENGU homologs.
Supplemental Table S1. Phytoplasma strains used in this study.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Dr. A. Bertaccini (University of Bologna) and Dr. H.-Y. Jung (Kyungpook National University) for providing plant materials. We also thank Dr. H. Tsukaya (the Graduate School of Science, the University of Tokyo) for technical advice. We thank Y. Hori and M. Kuroiwa of the Technology Advancement Center (the Graduate School of Agricultural and Life Sciences, the University of Tokyo) for the TOF-MS analysis and the Edman sequence analysis.
Glossary
- OY-M
onion yellows phytoplasma mild strain
- GST
glutathione-S-transferase
- TOF
time-of-flight
- MS
mass spectrometry
- dpi
days post inoculation
- PVX
potato virus X
- AY-WB
aster yellows phytoplasma strain witches’ broom
- OY-W
onion yellows phytoplasma line W
- PPT
potato purple top phytoplasma
- ACLR
apricot chlorotic leaf role phytoplasma
- KV
clover phyllody phytoplasma
- PaWB
paulownia witches’ broom phytoplasma
- GY
garlic yellows phytoplasma
- MD
mulberry dwarf phytoplasma
- WDWB
water dropwort witches’ broom phytoplasma
- BamWB
bamboo witches’ broom phytoplasma
- PvWB
porcelain vine witches’ broom phytoplasma
- CLEs
CLAVATA/ESR-related peptides
References
- Bai X, Zhang J, Ewing A, Miller SA, Jancso Radek A, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, et al. (2006) Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J Bacteriol 188: 3682–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen NM, Axelsen KB, Nicolaisen M, Schulz A. (2005) Phytoplasmas and their interactions with hosts. Trends Plant Sci 10: 526–535 [DOI] [PubMed] [Google Scholar]
- Clark SE. (2001) Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol 2: 276–284 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song KM, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Guo Y, Ni J, Denver R, Wang X, Clark SE. (2011) Mechanisms of molecular mimicry of plant CLE peptide ligands by the parasitic nematode Globodera rostochiensis. Plant Physiol 157: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S. (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci USA 106: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Yamaguchi T, Kazama T, Ito T, Horiguchi G, Tsukaya H. (2011) ROTUNDIFOLIA4 regulates cell proliferation along the body axis in Arabidopsis shoot. Plant Cell Physiol 52: 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizawa S, Oshima K, Ishii Y, Hoshi A, Maejima K, Jung HY, Yamaji Y, Namba S. (2009) Cloning of immunodominant membrane protein genes of phytoplasmas and their in planta expression. FEMS Microbiol Lett 293: 92–101 [DOI] [PubMed] [Google Scholar]
- Kakizawa S, Oshima K, Jung HY, Suzuki S, Nishigawa H, Arashida R, Miyata S, Ugaki M, Kishino H, Namba S. (2006) Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J Bacteriol 188: 3424–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park SY, Czymmek K, Kang S, Valent B. (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kube M, Schneider B, Kuhl H, Dandekar T, Heitmann K, Migdoll AM, Reinhardt R, Seemüller E. (2008) The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali.’ BMC Genomics 9: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IM, Davis RE. (1986) Prospects for in vitro culture of plant-pathogenic mycoplasmalike organisms. Annu Rev Phytopathol 24: 339–354 [Google Scholar]
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Tóth R, Nicolaisen M, Hogenhout SA. (2011) Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol 157: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011) Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol 52: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. (1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. (2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Mortier V, Herder GD, Whitford R, Velde WV, Rombauts S, D’haeseleer K, Holsters M, Goormachtig S. (2010) CLE peptide control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita NN, Moore S, Horiguchi G, Kubo M, Demura T, Fukuda H, Goodrich J, Tsukaya H (2004) Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J 38: 699–713 [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE. (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. (2009) Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 50: 67–77 [DOI] [PubMed] [Google Scholar]
- Oshima K, Shiomi T, Kuboyama T, Sawayanagi T, Nishigawa H, Kakizawa S, Miyata S, Ugaki M, Namba S (2001) Isolation and characterization of derivative lines of the onion yellows phytoplasma that do not cause stunting or phloem hyperplasia. Phytopathology 91: 1024–1029 [DOI] [PubMed] [Google Scholar]
- Rafiqi M, Gan PHP, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, Hardham AR, Dodds PN. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22: 2017–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. (2011) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact 24: 606–618 [DOI] [PubMed] [Google Scholar]
- Senshu H, Yamaji Y, Minato N, Shiraishi T, Maejima K, Hashimoto M, Miura C, Neriya Y, Namba S. (2011) A dual strategy for the suppression of host antiviral silencing: two distinct suppressors for viral replication and viral movement encoded by potato virus M. J Virol 85: 10269–10278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Liu J-X, Guo H, Yin Y, Howell SH. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu J-X, Howell SH. (2008) Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J 56: 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. (2011) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc Natl Acad Sci USA 108: E1254–E1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Nguyen LTT, Kube M, Schneider B, Reinhardt R, Gibb KS. (2008) Comparative genome analysis of “Candidatus Phytoplasma australiense” (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J Bacteriol 190: 3979–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Mitchum MG, Gao BL, Li CY, Diab H, Baum TJ, Hussey RS, Davis EL. (2005) A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol Plant Pathol 6: 187–191 [DOI] [PubMed] [Google Scholar]