The analysis of a mutant in the main enzyme responsible for cyanide detoxification, the mitochondrial β-cyanoalanine synthase, uncovers a new signaling role for cyanide in the plant response to pathogens.
Abstract
Cyanide is produced concomitantly with ethylene biosynthesis. Arabidopsis (Arabidopsis thaliana) detoxifies cyanide primarily through the enzyme β-cyanoalanine synthase, mainly by the mitochondrial CYS-C1. CYS-C1 loss of function is not toxic for the plant and leads to an increased level of cyanide in cys-c1 mutants as well as a root hairless phenotype. The classification of genes differentially expressed in cys-c1 and wild-type plants reveals that the high endogenous cyanide content of the cys-c1 mutant is correlated with the biotic stress response. Cyanide accumulation and CYS-C1 gene expression are negatively correlated during compatible and incompatible plant-bacteria interactions. In addition, cys-c1 plants present an increased susceptibility to the necrotrophic fungus Botrytis cinerea and an increased tolerance to the biotrophic Pseudomonas syringae pv tomato DC3000 bacterium and Beet curly top virus. The cys-c1 mutation produces a reduction in respiration rate in leaves, an accumulation of reactive oxygen species, and an induction of the alternative oxidase AOX1a and pathogenesis-related PR1 expression. We hypothesize that cyanide, which is transiently accumulated during avirulent bacterial infection and constitutively accumulated in the cys-c1 mutant, uncouples the respiratory electron chain dependent on the cytochrome c oxidase, and this uncoupling induces the alternative oxidase activity and the accumulation of reactive oxygen species, which act by stimulating the salicylic acid-dependent signaling pathway of the plant immune system.
The gaseous hormone ethylene is known to regulate multiple physiological and developmental processes in plants, such as seedling emergence, leaf and flower senescence, climacteric fruit ripening, and organ abscission. Ethylene is also involved in the response of plants to abiotic and biotic stresses (Wang et al., 2002; Broekaert et al., 2006; van Loon et al., 2006). Enhanced ethylene production is an early, active response of plants to the perception of pathogen attack and is associated with the induction of defense reactions. During ethylene biosynthesis, S-adenosyl-l-Met is converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase. ACC is finally oxidized by ACC oxidase to form ethylene, carbon dioxide, and cyanide (Hartley et al., 1998; Wang et al., 2002). Hydrogen cyanide is a colorless and highly volatile liquid. The anion cyanide is toxic and renders the cells of an organism unable to use oxygen, primarily through the chelation of divalent and trivalent metal ions in the prosthetic groups of several metalloenzymes, including copper/zinc superoxide dismutase, catalase, nitrate and nitrite reductase, nitrogenase, peroxidases, and the mitochondrial cytochrome c oxidase (Isom and Way, 1984; Donato et al., 2007).
Cyanide must be rapidly detoxified and metabolized by the plant to keep the concentration below toxic levels. Plants detoxify cyanide primarily through the enzyme β-cyanoalanine synthase (CAS), for which considerable levels of activity are constitutively found in many plant species. Rhodanese and mercaptopyruvate sulfurtransferase activities also make minor contributions to the cyanide detoxification process (Miller and Conn, 1980). CAS is a pyridoxal phosphate-dependent enzyme that converts Cys and cyanide to hydrogen sulfide and β-cyanoalanine, which is later converted to Asn, Asp, and ammonia by NIT4 class nitrilases (Piotrowski, 2008). Arabidopsis (Arabidopsis thaliana) plants carry the mitochondrial CAS CYS-C1 (At3g61440; Watanabe et al., 2008), which belongs to the family of β-substituted Ala synthase enzymes. The family also includes the three major O-acetyl-serine(thiol)lyase enzymes OAS-A1 (At4g14880), OAS-B (At2g43750), and OAS-C (At3g59760; Watanabe et al., 2008), the l-Cys desulfhydrase DES1 (At5g28030; Álvarez et al., 2010), the S-sulfocysteine synthase CS26 (At3g03630; Bermúdez et al., 2010), and the functionally unknown cytosolic isoforms CYS-D1 (At3g04940) and CYS-D2 (At5g28020). Mutations in CYS-C1 result in plants that accumulate cyanide and that display abnormal root hair (García et al., 2010), suggesting that cyanide has a signaling role in root development. The lack of the mitochondrial O-acetyl-serine(thiol)lyase isoform OAS-C, which is necessary to detoxify the sulfide released by the CAS activity, causes an accumulation of sulfide and cyanide and a root phenotype similar to the cys-c1 loss-of-function mutant (Álvarez et al., 2012b).
Several authors have suggested that cyanide could act as a regulator of other metabolic processes in addition to performing the described role in plant root development (Siegien and Bogatek, 2006). It has been observed that this molecule is released during seed germination and that exogenously applied hydrogen cyanide breaks seed dormancy in several plants (Cohn and Hughes, 1986; Fol et al., 1989; Bogatek et al., 1991; Bethke et al., 2006). The role of cyanide as a regulatory molecule is not restricted to plants, and it has been demonstrated that cyanide is generated in leukocytes from Gly via a peroxidase (Stelmaszyńska, 1986) as well as in the central nervous system, where it has been hypothesized to act as a neuromodulator (Gunasekar et al., 2000; Cipollone and Visca, 2007). Cyanide production can be stimulated by opiates and decreased by treatment with muscarinic receptor agonists (Borowitz et al., 1997; Gunasekar et al., 2004).
Despite the variety of known functions for cyanide in different organisms, the role of cyanide production in plants seems to have been unevaluated to date. In cyanogenic plants, cyanide is produced during the degradation of cyanogenic lipids and from the catabolism of cyanogenic glycosides (Poulton, 1990). Cyanide and cyanogenic compounds play an important role in plant defense against herbivores (Zagrobelny et al., 2008). In noncyanogenic plants, cyanide is a coproduct of ethylene biosynthesis. The molecule is also produced during the biosynthesis of camalexin, a phytoalexin formed in Arabidopsis plants upon infection by a large variety of microorganisms, including bacteria, fungi, and oomycetes (Glawischnig, 2007). During camalexin biosynthesis, the Trp-derived intermediate indole-3-acetonitrile is conjugated with Cys and serves as a substrate for the cytochrome P450 enzyme CYP71B15. This enzyme catalyzes the formation of the thiazoline ring as well as the release of cyanide and subsequent oxidative decarboxylation of dihydrocamalexic acid to camalexin (Glawischnig, 2007; Böttcher et al., 2009). Since both cyanide sources, camalexin and ethylene, are produced after pathogen attack, cyanide should be produced at significant levels during the plant response to pathogens. It has been shown that exogenous cyanide can enhance the resistance of tobacco (Nicotiana tabacum) and Arabidopsis leaves to Tobacco mosaic virus and Turnip vein clearing virus, respectively (Chivasa and Carr, 1998; Wong et al., 2002). Recently, it has been demonstrated that exogenously applied cyanide increases the resistance of young rice (Oryza sativa) plants to blast fungus infection, suggesting that cyanide rather than ethylene contributes to plant resistance (Seo et al., 2011).
This work aims to further investigate the role of endogenously produced cyanide in the plant immune response by analyzing the behavior of Arabidopsis knockout mutants of the mitochondrial CAS CYS-C1 and the regulation of CYS-C1 in response to pathogen attack.
RESULTS
The cys-c1 Mutant Transcriptome Shows a High Correlation with Biotic Stresses
The loss of function of the CYS-C1 enzyme has previously been characterized in root tissues, but its function in leaves has not been studied to date (García et al., 2010). Phenotypic analysis of the cys-c1 null mutant shows no obvious alterations in the aerial parts of the plant whether grown in long- or short-day photoperiods. To analyze the effect of the loss of function of the CYS-C1 enzyme at the molecular level, we performed a comparative transcriptomic analysis of leaves of cys-c1 and wild-type plants grown under identical long-day conditions on Murashige and Skoog (MS) medium for 14 d. Total RNA was prepared and analyzed using the Affymetrix Arabidopsis ATH1 GeneChip array. Three biological replicates were performed for each genotype. Restricting the analysis to the genes whose expression was changed at least 2-fold as a threshold and at a significance level of P < 0.05, we identified 51 genes that exhibited alterations in transcription level. Among them, 31 genes were up-regulated in the cys-c1 mutant plant compared with the wild-type plant, and 20 genes were down-regulated (Microarray Gene Expression Omnibus database accession no. GSE19242; Supplemental Table S1). To detect physiologically relevant patterns, the genes with altered expression were assigned to functional categories based on classification by the Bio-Array Resource for Arabidopsis Functional Genomics (Toufighi et al., 2005). The resulting group lists revealed that a high proportion of both up- and down-regulated genes in the cys-c1 mutant were associated with the plant’s responses to biotic and abiotic stress and signaling (Supplemental Fig. S1). The induction of selected genes such as WRKY33 (encoding a WRKY transcription factor), ERF6 (encoding an ethylene response transcription factor), CYP81F2 (encoding a cytochrome P450 involved in glucosinolate biosynthesis), and GSTU24 (coding for a putative glutathione-S-transferase) was confirmed by real-time reverse transcription (RT)-PCR, thus validating the data obtained by the array (Supplemental Fig. S2)
A meta-analysis of the cys-c1 transcript profile data was performed by comparison with the available Affymetrix Arabidopsis ATH1 GeneChip array databases and the analytical tools of Genevestigator (Hruz et al., 2008). Biclustering and hierarchical clustering analysis of the up- and down-regulated genes in cys-c1 showed that 80% were coregulated with genes that were deregulated in wild-type seeds of the ecotype Columbia (Col-0) after treatment with 0.1% oxygen for 6 d (GSE14420; Christianson et al., 2009; Supplemental Fig. S3; Supplemental Table S2). In comparing microarray data for the gene subset categorized as biotic, 54% of the genes identified overlapped with those already shown to be affected by fungal pathogens or altered in Pseudomonas syringae pv tomato (Pst)-infected Arabidopsis plants or elicitor-treated plants (Supplemental Figs. S4 and S5; Supplemental Table S2). Among the genes identified in these groups are several transcription factors related to the biotic defense response, such as WRKY18, WRKY33, WRKY40, and the gene coding for FLG22-INDUCED RECEPTOR-LIKE KINASE1 (AT2G19190). No correlation was found with ACC treatments or mutants in ethylene signaling (Supplemental Fig. S6).
In the light of this analysis, it is interesting to speculate that cyanide plays a role in signaling and defense against pathogen infection in leaf tissues. We aimed then to investigate this hypothesis further.
Cyanide Accumulates during the Infection of Arabidopsis Plants with Botrytis cinerea
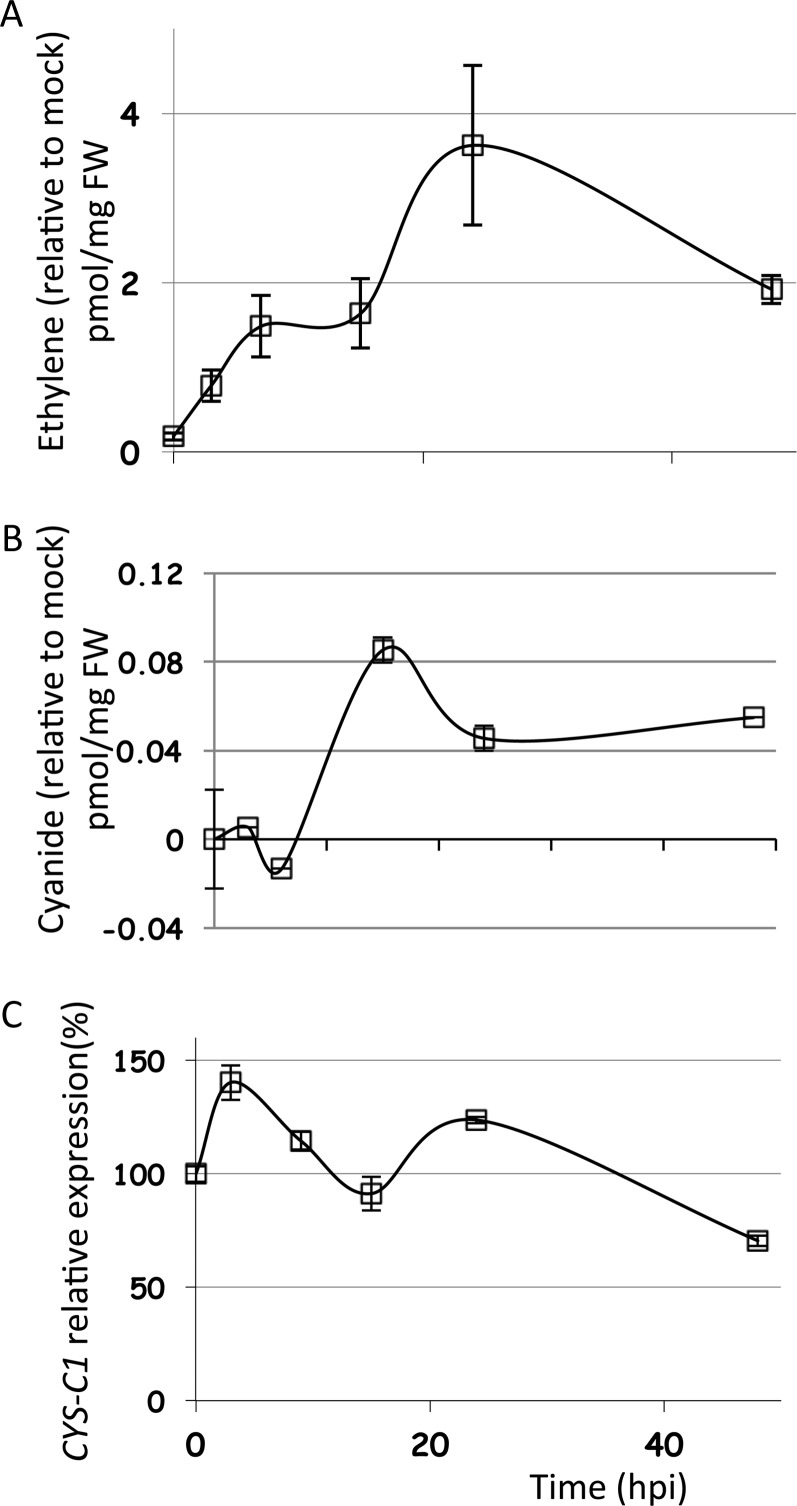
B. cinerea is a necrotrophic pathogen that causes gray mold diseases in many crop plants, resulting in significant crop losses. B. cinerea and other necrotrophic pathogens promote and benefit from host cell death during pathogenesis, as dead cells and necrotic tissues provide a base for saprophytic growth from which B. cinerea further colonizes healthy tissue (AbuQamar et al., 2006). When plants are infected by B. cinerea, they produce high levels of ethylene (Cristescu et al., 2002; Han et al., 2010). Figure 1A shows that the ethylene production increases rapidly in Arabidopsis when challenged with B. cinerea, reaching a maximum level at 24 h post infection (hpi). We investigated the accumulation of the cyanide coproduced during the B. cinerea-Arabidopsis interaction as well as the regulation of the CYS-C1 gene under these conditions. At the beginning of the interaction, the level of cyanide dropped transiently at 9 hpi and then started accumulating, reaching a maximum of 190% of the basal level at 15 hpi (Fig. 1B); accordingly, CYS-C1 expression shows a waving curve with expression peaks at 3 and 24 hpi and a valley at 15 hpi, this last level coinciding with the higher level of cyanide (Fig. 1C).
Figure 1.
Time course of the accumulation of ethylene and cyanide and the regulation of CYS-C1 transcript during the Arabidopsis-B. cinerea interaction. A and B, Ethylene (A) and cyanide (B) were measured in leaf extracts of wild-type plants grown for 6 to 7 weeks and infected with a spore suspension of B. cinerea. The results presented are expressed as means ± sd of a representative experiment in which 12 to 14 leaves from infected plants were pooled and three independent extractions were made from the pooled material. The experiment was repeated three times, with similar results obtained each time. FW, Fresh weight. C, The expression level of CYS-C1 was analyzed by real-time RT-PCR and referred to the UBQ10 internal control. The data correspond to means ± sd of three independent analyses using material grown in different batches at different times. For each analysis, five to six plants were pooled, and three independent RNA extractions were made from the pooled material. Two experimental replicates were made for each sample. The data were normalized against the data obtained from plants treated with a mock solution. Nonnormalized data are shown in Supplemental Figures S7A, S8A, and S9A.
Cyanide Accumulation and CYS-C1 Gene Expression Are Negatively Correlated during Compatible and Incompatible Plant-Bacteria Interactions
The bacterial pathogen P. syringae is a hemibiotrophic pathogen that produces bacterial specks in a wide range of plant species. In the early stages of compatible infections, host cell death does not occur. Later stages of infection, however, are associated with host tissue chlorosis and necrosis (Glazebrook, 2005).
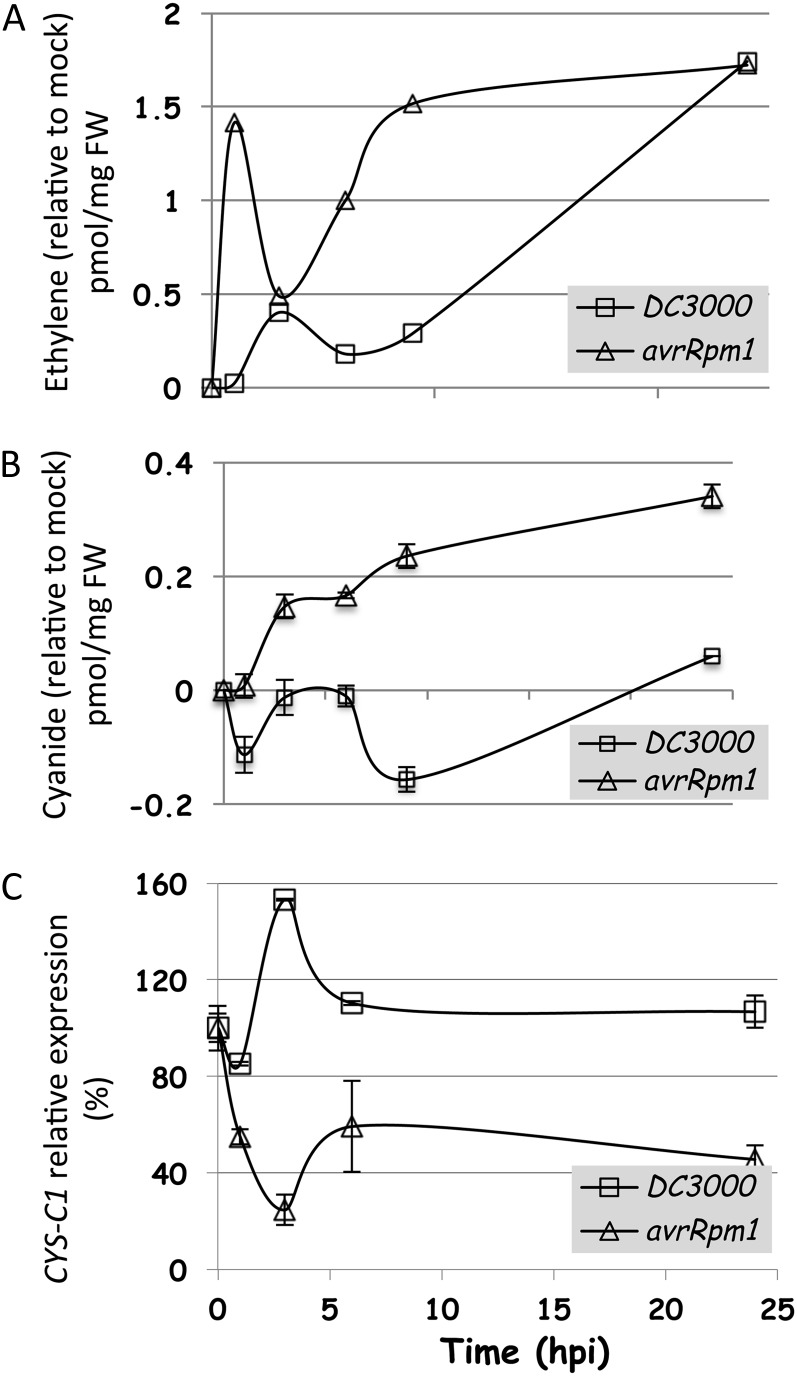
Besides the nonhost resistance, plants have the capacity to recognize pathogen-associated molecular patterns by surface pattern-recognition receptors and to induce a response leading to a basal or pathogen-associated molecular pattern-triggered immunity (PTI; Jones and Dangl, 2006). Some pathogens have evolved to avoid recognition by delivering effectors that suppress PTI, and this results in a compatible plant-pathogen interaction. For their defense, plants have also evolved RESISTANCE genes that encode receptors recognizing specific pathogen effectors, resulting in effector-triggered immunity (ETI; Jones and Dangl, 2006). In addition to the PTI response, Pst DC3000 can elicit an ETI reaction in Arabidopsis when expressing the type III effector AvrRpm1 (Bent et al., 1994; Mindrinos et al., 1994; Grant et al., 1995). When tobacco plants are infected by P. syringae, they produce ethylene. The production is monophasic if the bacteria do not elicit a hypersensitive response (HR) and produce a disease and biphasic if the bacteria induce a HR and do not subsequently produce a disease (Mur et al., 2008). Moreover, transcriptomic data suggest that genes encoding ethylene biosynthetic enzymes were up-regulated in Arabidopsis following challenge with avirulent bacteria (Mur et al., 2008). To investigate this response further, the production of ethylene was monitored during compatible and incompatible interactions. Arabidopsis plants were infected with a virulent Pst DC3000 or an avirulent Pst DC3000 avrRpm1 strain. Samples were taken at 1, 3, 6, 9, and 24 hpi. Ethylene was accumulated in the early stages of both interactions, although the accumulation occurred earlier in the incompatible interaction than in the compatible interaction. A second rise occurred at 9 hpi of the avirulent interaction (Fig. 2A). The infection with Pst DC3000 induced ethylene accumulation only at the very late stages of the interaction (24 hpi). These data are in agreement with the results already published for the tobacco-P. syringae interaction (Mur et al., 2008).
Figure 2.
Time course of the accumulation of ethylene and cyanide and the regulation of CYS-C1 transcript during the Arabidopsis-P. syringae interactions. A and B, Ethylene (A) and cyanide (B) were measured in leaf extracts of wild-type plants grown for 6 to 7 weeks and infected with a bacterial suspension of either Pst DC3000 or Pst DC3000 avrRpm1 as described in “Materials and Methods.” The results presented are expressed as means ± sd of a representative experiment in which 12 to 14 leaves from infected plants were pooled and three independent extractions were made from the pooled material. The experiment was repeated three times, with similar results obtained each time. FW, Fresh weight. C, The expression level of CYS-C1 was analyzed by real-time RT-PCR and referred to the UBQ10 internal control. The data correspond to means ± sd of three independent analyses using material grown in different batches at different times. For each analysis, five to six plants were pooled, and three independent RNA extractions were made from the pooled material. Moreover, two experimental replicates were made for each sample. The data were normalized against the data obtained from plants treated with a mock solution. Nonnormalized data are shown in Supplemental Figures S7B, S8B, and S9B.
We also determined the kinetics of the accumulation of cyanide in the same samples. Interestingly, cyanide accumulated at different rates in the two Arabidopsis-Pst interactions, being detoxified preferentially during the compatible interaction (Fig. 2B). In fact, during ETI, cyanide started accumulating at 3 hpi, and its level did not decrease significantly during the infection. In contrast, during the PTI, cyanide content decreased at 1 hpi, increased to the basal level at 3 and 6 hpi, then decreased and started increasing again after 9 hpi, to reach the basal level of 24 hpi. Accordingly, the transcription of CYS-C1 was induced during the compatible interaction and was repressed during the ETI, with the curve showing an opposite peak at 3 hpi (Fig. 2C).
Mitochondrial Cyanide Differentially Affects the Response to Necrotrophic and Biotrophic Pathogens, and This Effect Is Reversed with Hydroxocobalamin Treatment
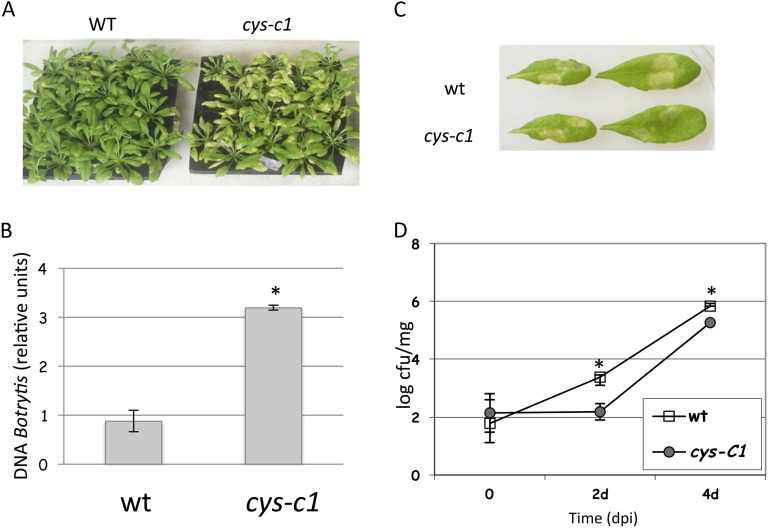
Nonlethal concentrations of cyanide can enhance the resistance of plants to fungi (Seo et al., 2011). cys-c1 mutant plants have been shown to accumulate more cyanide in both root and leaf tissues and to exhibit less ethylene accumulation than wild-type plants (García et al., 2010). To investigate the possible role of mitochondrial cyanide in plant defense against pathogens, cys-c1 mutant plants defective in the mitochondrial CAS (García et al., 2010) were challenged by a necrotrophic compatible pathogen (B. cinerea) and a hemibiotrophic compatible pathogen (Pst DC3000). When challenged with the fungus, cys-c1 showed more severe symptoms than wild-type plants and accumulated six times more B. cinerea DNA (Fig. 3, A and B). Conversely, the cys-c1 mutant exhibited a higher tolerance to the infection by Pst DC3000 than the Col-0 wild type, as it showed less severe symptoms than wild-type plants and accumulated 12-fold less Pst DC3000 colony-forming units (cfu) mg−1 fresh weight at 2 d post infection (dpi) than Col-0; the difference was 6-fold at 4 dpi (Fig. 3, C and D). However, the susceptibility to an avirulent strain of Pst DC3000 is not affected by the cys-c1 mutation (Supplemental Fig. S10).
Figure 3.
Responses of the cys-c1 mutant to pathogen infection. A and B, Susceptibility of the wild type (wt) and the cys-c1 mutant to B. cinerea infection. A, Wild-type and cys-c1 mutant plants after 5 d of B. cinerea infection. B, Quantification of fungus growth was performed by real-time PCR amplification of the B. cinerea creA gene, which was normalized against the Arabidopsis UBQ10 gene. DNA was isolated from leaves 5 d after spore inoculation of 6- to 7-week-old wild-type and mutant plants grown in parallel. The data correspond to means ± sd of at least three independent analyses made from material grown in different batches at different times. For each analysis, 20 infected plants were pooled, and six independent DNA extractions were made from the pooled material. Moreover, two experimental replicates were made from each sample. C and D, Susceptibility of the wild type and the cys-c1 mutant to infection with virulent Pst DC3000 bacteria. C, Wild-type and cys-c1 mutant leaves after 3 d of Pst DC3000 infection. cys-c1 leaves show less severe symptoms than the wild type. D, The cfu were counted at 0, 2, and 4 dpi of 6- to 7-week-old wild-type and mutant plants grown in parallel. At total of 12 to 14 leaves were pooled for each analysis, in which three independent counts were made from the pooled material and two experimental replicates were made from each sample. The data correspond to means ± sd of one representative experiment. *P < 0.05. The experiment was performed three times with material grown in different batches at different times; similar results were obtained for each iteration.
To confirm that the observed phenotype of the cys-c1 mutant plants was indeed due to the disruption of the CYS-C1 gene, complementation analysis was performed using the full-length CYS-C1 genomic fragment including its promoter region (Pcys-c1). cys-c1 plants transformed with the Pcys-c1-CYS-C1 fragment displayed pathogen sensitivity similar to that of the wild type (Supplemental Fig. S11).
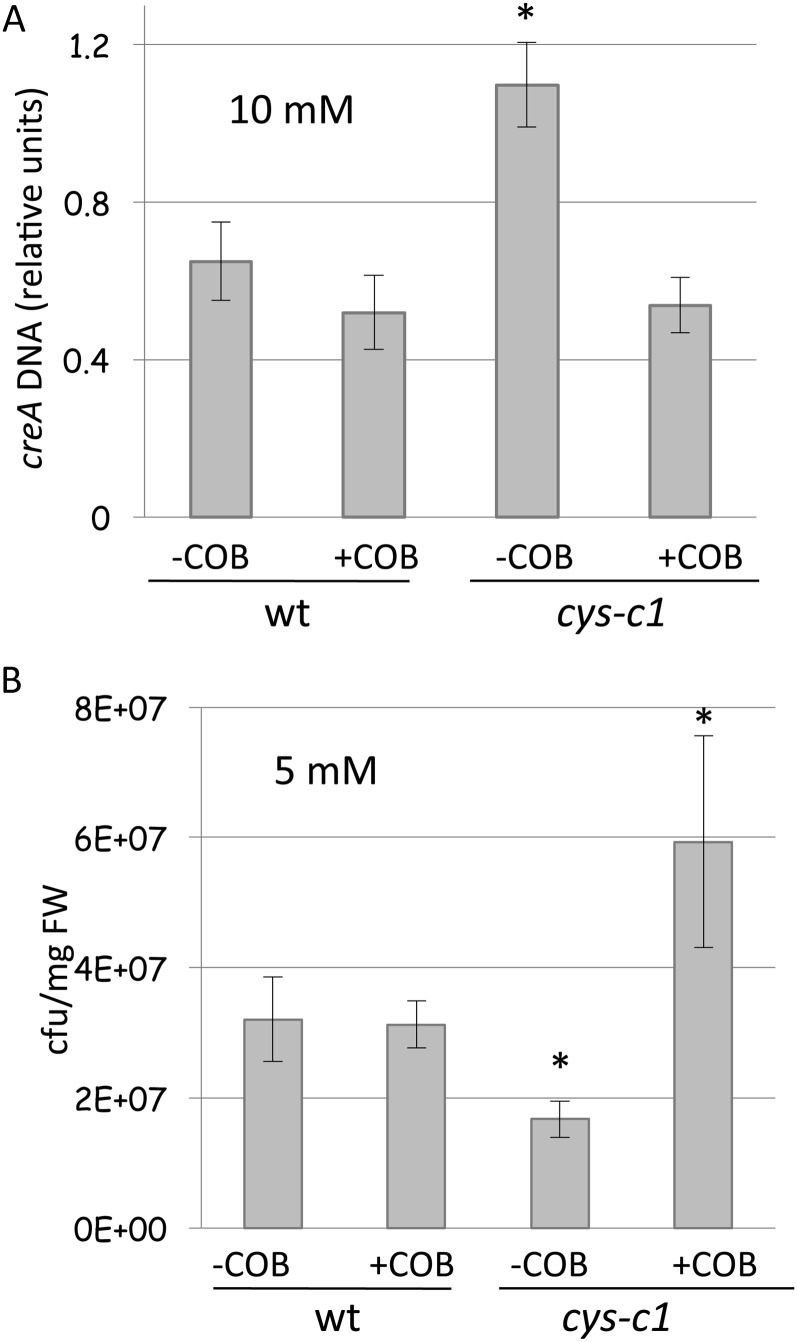
Hydroxocobalamin is a natural form of vitamin B12 that is commonly used as an antidote for severe acute cyanide poisoning in humans (Borron et al., 2007; Hall et al., 2007). Hydroxocobalamin can penetrate cells and act at an intracellular level to bind cyanide and form nontoxic cyanocobalamin, which is excreted in the urine (Astier and Baud, 1996). In plants, hydroxocobalamin has been used to antagonize the effect of cyanide in roots, reverting the root hairless phenotype in cys-c1 lines to that of wild-type plants (García et al., 2010). The addition of 10 mm hydroxocobalamin at the time of infection with B. cinerea reverted the sensitivity phenotype exhibited by the cys-c1 mutant, decreasing the accumulation of B. cinerea DNA in infected cys-c1 leaves to wild-type levels (Fig. 4A). Moreover, this effect was dose dependent, as the treatment with 5 mm hydroxocobalamin partially reverted the susceptibility of the cys-c1 mutant to B. cinerea to levels similar to those of wild-type plants (Supplemental Fig. S12). Similarly, treatment with hydroxocobalamin altered the phenotype of resistance to Pst DC3000 exhibited by the cys-c1 mutant, as bacteria were able to develop even better in cys-c1 plants treated with the antidote than in wild-type plants in either the presence or absence of hydroxocobalamin (Fig. 4B). To exclude the possibility that the hydroxocobalamin directly affected pathogen growth, we performed growth tests of Pst DC3000 in solid culture Luria-Bertani (LB) medium in the absence and presence of 5 mm hydroxocobalamin. No differences were observed in either of the two conditions (Supplemental Fig. S13). Therefore, the possibility of a direct effect of hydroxocobalamin in the pathogen’s growth rather than rescuing the cys-c1 phenotype is excluded.
Figure 4.
Hydroxocobalamin effect on plant susceptibility to pathogens. Wild-type (wt) and cys-c1 mutant plants were infected with B. cinerea (A) or Pst DC3000 (B), as indicated in “Materials and Methods.” Pathogens were collected in suspensions containing (+COB) or not containing (−COB) hydroxocobalamin at the concentration indicated and used to perform the susceptibility assays. Quantification of fungus growth was performed by real-time PCR amplification of the B. cinerea creA gene, which was normalized against the Arabidopsis UBQ10 gene. DNA was isolated from leaves 5 d after spore inoculation of 6- to 7-week-old wild-type and mutant plants grown in parallel. The cfu were counted at 3 dpi, with 12 to 14 leaves pooled for each analysis. Three independent determinations were made from the pooled material, and two experimental replicates were made from each sample. The data correspond to means ± sd of one representative experiment. *P < 0.05. FW, Fresh weight.
Mitochondrial Cyanide Is Correlated with Plant Resistance to Viral Pathogens
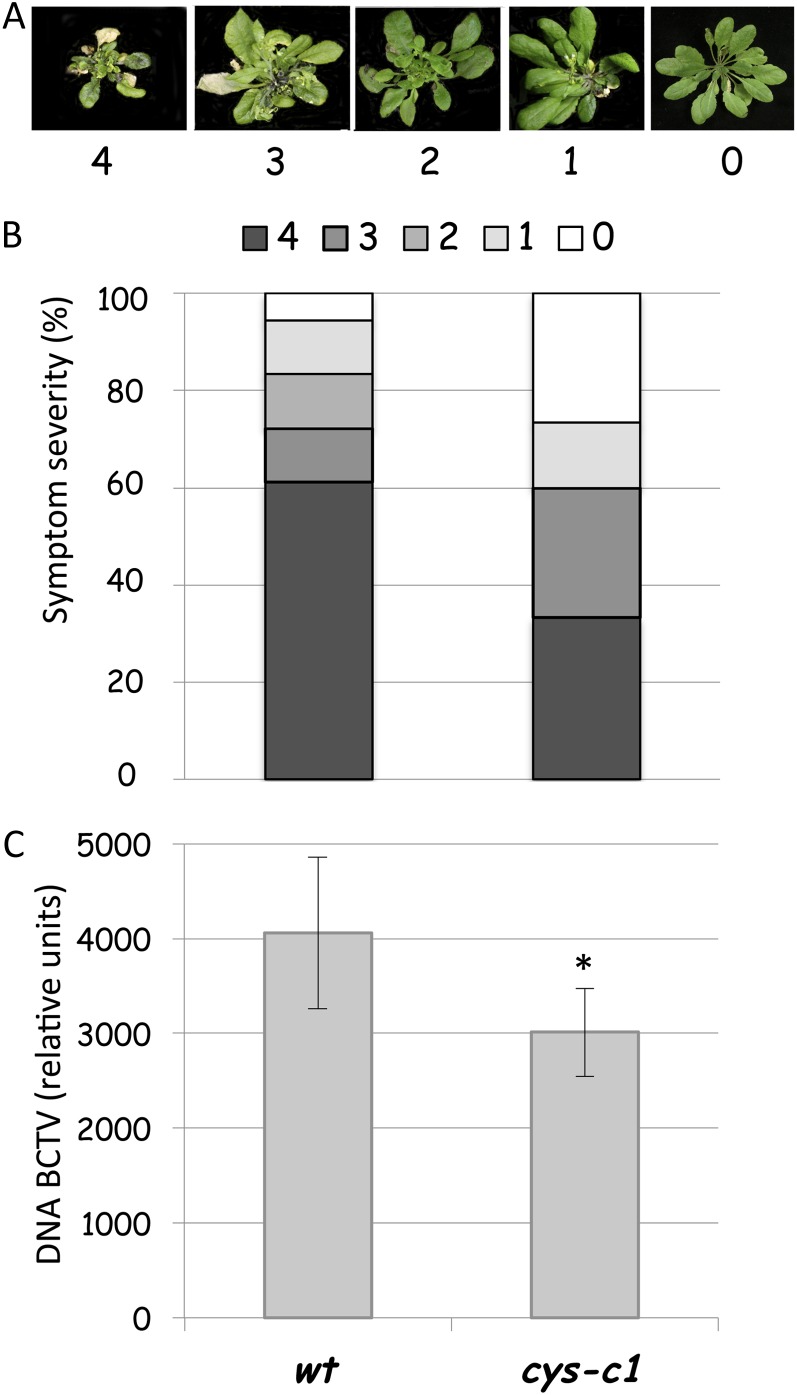
Nonlethal concentrations of cyanide can enhance the resistance of plants to viral infection (Chivasa and Carr, 1998; Wong et al., 2002). Members of the geminivirus family are plant viruses with circular, single-stranded DNA genomes (Rojas et al., 2005) that infect a wide range of plant species and that cause extensive losses in crops. To determine whether mitochondrial cyanide accumulation is involved in the cyanide-related resistance to viruses, wild-type and cys-c1 mutant plants were challenged with the geminivirus Beet curly top virus (BCTV). When infected with the virus, cys-c1 plants exhibited symptoms less severe than those of respective wild-type plants (Fig. 5, A and B). Plants showing no symptoms (cataloged as 0 by the severity index described by Baliji et al. [2007]) constituted 26.6% in the case of the cys-c1 mutant and 5.5% in the case of the wild-type plants. Moreover, the sum of plants showing the category 0 (asymptomatic) plus 1 (mild symptoms) was 40% for the cys-c1 mutant and only 16.6% for the wild-type plants. On the other hand, 33.3% of the cys-c1 mutant and 61.1% of the wild-type plants showed the most severe symptoms, exhibiting almost no plant growth (categorized as 4 in the severity index). When viral DNA present in infected plants was quantified by real-time PCR, the results clearly showed that the cys-c1-infected plants accumulated less viral DNA than did wild-type plants (Fig. 5C). These results indicate that endogenously produced cyanide can protect plants from virus attack just as exogenously applied cyanide does.
Figure 5.
Response of the cys-c1 mutant to virus. A, Example of the severity index described in “Materials and Methods” and Baliji et al. (2007): 0, no symptoms; 1 to 4, increasing severity of symptoms. B, Susceptibility of the wild type (wt) and the cys-c1 mutant to BCTV infection. Whole 6- to 7-week-old plants of each genotype were agroinoculated, and the symptom severity was recorded at 28 dpi. C, Quantification of virus growth was performed in the same plants at 28 dpi by real-time PCR amplification of the viral DNA, which was normalized against the Arabidopsis UBQ10 gene. The data correspond to means ± sd of three independent analyses made from material grown in different batches at different times. For each analysis, at least 10 infected plants were pooled, and six independent DNA extractions were made from the pooled material. Two experimental replicates were performed from each sample. *P < 0.05. [See online article for color version of this figure.]
The cys-c1 Mutation Produces a Reduction in Respiration Rate in Leaves and an Induction of Alternative Oxidase and PR1 Expression
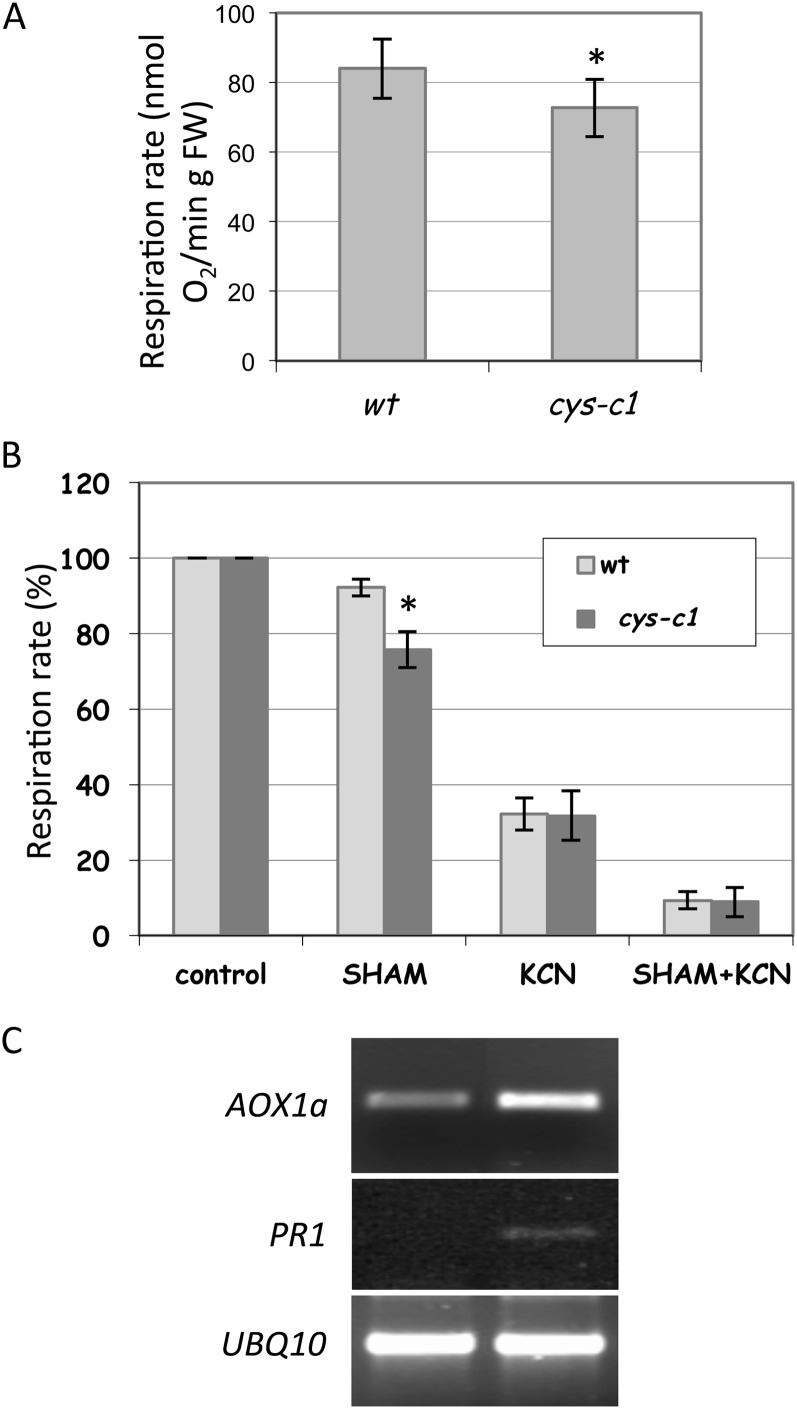
Cyanide binds to the heme iron of the mitochondrial cytochrome c oxidase, thereby blocking the cytochrome respiration pathway and the utilization of oxygen in cellular functions (Donato et al., 2007). In higher plants, an alternative cyanide-resistant respiratory pathway is catalyzed by the alternative oxidase (AOX), which is located in the mitochondrial inner membrane and acts as a terminal oxidase in the mitochondrial electron transport chain. AOX branches from the main respiratory chain at the level of the ubiquinone pool and catalyzes the four-electron reduction of oxygen to water, releasing the energy as heat (Millenaar and Lambers, 2003). Much work has revealed that the genes encoding AOX, AOX protein, and the alternative respiratory pathway are frequently induced during plant-pathogen interactions (Hanqing et al., 2010). The cys-c1 mutant displays a reduction of root (García et al., 2010) and leaf (Fig. 6A) respiration rates. The addition of salicylhydroxamic acid (SHAM), an inhibitor of the AOX pathway, affects the respiration rate of wild-type and mutant plants differently, as it decreases the respiration rate of wild-type leaves only about 8% but alters the respiration rate of the cys-c1 mutant leaves by about 24% (Fig. 6B). In both wild-type and mutant plants, the addition of potassium cyanide (KCN) reduces the oxygen uptake drastically to about 30% of the maximum respiration rate. The addition of KCN plus SHAM reduces oxygen uptake to levels lower than 10% (Fig. 6B). The increase of the AOX pathway in the cys-c1 mutant correlates with an increase in transcript abundance of the AOX1a gene (Fig. 6C), which is induced by an alteration of the cytochrome respiration pathway (Albury et al., 2009; García et al., 2010). In addition, the expression of PR1, a pathogenesis-related protein induced by the salicylic acid-dependent pathway (An and Mou, 2011), is induced in cys-c1 plants in the absence of stress (Fig. 6C), suggesting that endogenously produced cyanide can modulate this pathway in Arabidopsis plants.
Figure 6.
Respiration rates (A and B) and AOX1a and PR1 expression levels (C) in leaves of wild-type (wt) and cys-c1 mutant plants. Cyanide-independent and AOX respiration were determined in the presence of 0.5 mm KCN or 4 mm SHAM, respectively. The transcription level of the AOX gene AOX1a, PR1, and the control UBQ10 was determined by RT-PCR in leaves of noninfected 6- to 7-week-old plants. The data correspond to means ± sd of at least three independent analyses made from material grown in different batches at different times. *P < 0.05. FW, Fresh weight.
The cys-c1 Mutant Accumulates Reactive Oxygen Species But Does Not Show Programmed Cell Death Lesions
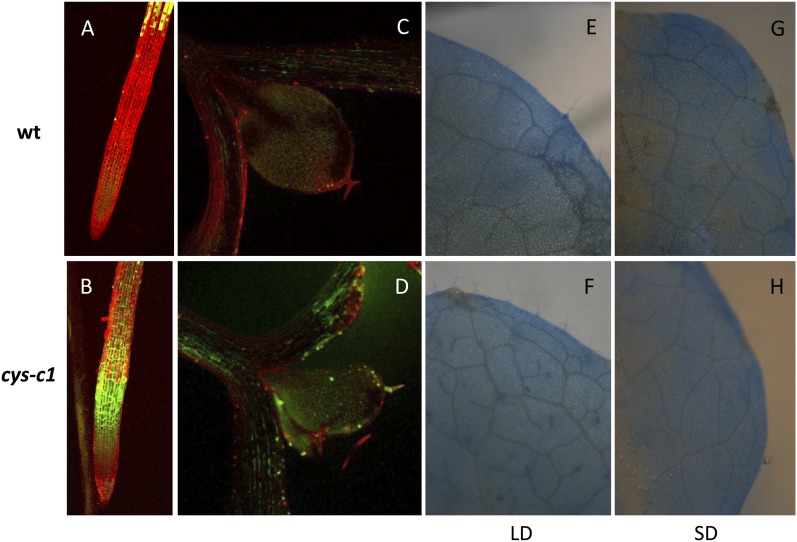
One of the earliest responses to pathogen infection is the production of reactive oxygen species (ROS; Lamb and Dixon, 1997), which together with nitric oxide and salicylic acid can promote the HR (Delledonne et al., 1998; Álvarez, 2000) and lead to the activation of systemic acquired resistance, a broad-spectrum form of disease resistance (Vlot et al., 2008). Since a reduction of the respiration rate can produce an accumulation of ROS, we compared the accumulation of ROS in cys-c1 and wild-type seedlings grown under control conditions (Fig. 7). Imaging of ROS in vivo in plant tissues by confocal laser microscopy is a very useful technique (Schneider et al., 1998). We observed a fluorescence emission resulting from the oxidation of the nonfluorescent 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) to a highly fluorescent product; this signal reflects significant production of hydrogen peroxide (H2O2). In roots, this fluorescence was higher in cys-c1 specimens than in wild-type samples (Fig. 7, A and B). Although chlorophyll autofluorescence interferes with the H2O2 detection in green tissues, we were able to observe a higher fluorescence in cys-c1 than in wild-type cotyledons (Fig. 7, C and D).
Figure 7.
Accumulation of H2O2 and lesion formation in the wild type (wt) and the cys-c1 mutant. A to D, H2O2 was detected by H2DCFDA staining in root (A and B) and cotyledons (C and D) from 5-d-old wild-type and cys-c1 mutant plants cultured in MS medium. E to H, Lactophenol trypan blue was used to stain spontaneous cell death lesions. Detached leaves of plants grown in soil for 3 weeks in long-day conditions (LD; E and F) or 6 to 7 weeks in short-day conditions (SD; G and H) were used for the assay. All the experiments were repeated at least three times, with similar results obtained each time.
Because H2O2 is a signaling intermediate molecule in programmed cell death, we stained the leaves of plants grown in long-day and short-day conditions with lactophenol trypan blue. We did not observe lesions characteristic of spontaneous cell death in the leaves of the cys-c1 mutant (Fig. 7, E–H).
DISCUSSION
Plants synthesize ethylene in response to different environmental stimuli, including pathogen attack (Wang et al., 2002; Pandey and Somssich, 2009). The role of ethylene in defense signaling in plants has been studied extensively, but its involvement remains controversial. Treatment with ethylene increases or decreases resistance to pathogens depending on the plant-pathogen interaction, and the use of mutants defective in ethylene signaling indicates a limited or different role of ethylene in the resistance to some biotrophic and necrotrophic pathogens, including fungi, bacteria, and viruses (Pieterse et al., 1998; Brading et al., 2000; Broekaert et al., 2006; Iwai et al., 2006). Cyanide is produced concomitantly with ethylene biosynthesis. In this work, however, we show different patterns of ethylene and cyanide accumulation during infection of Arabidopsis with both the fungus B. cinerea and the virulent and avirulent P. syringae. In addition, we show that the lack of mitochondrial CAS of Arabidopsis, which leads to an accumulation of cyanide in plant tissues (García et al., 2010), results in an altered response to plant pathogens. The response is completely dependent on cyanide, as demonstrated by genetic and chemical complementation. All these data suggest that cyanide also acts in the regulation of the plant immune responses. Furthermore, the transcriptional regulation of the CYS-C1 gene during the three plant-pathogen interactions analyzed allows a differential accumulation of cyanide in each interaction, suggesting that CYS-C1 is involved in the signaling pathway, leading to resistance or sensitivity depending on the type of pathogen.
The classification of genes differentially expressed in cys-c1 and wild-type plants reveals that the high endogenous cyanide content of the cys-c1 mutant is correlated with the biotic stress response. More specifically, the cyanide accumulation is correlated with the induction of genes encoding proteins involved in the plant signaling pathway. Among the induced genes in the cys-c1 mutant, three WRKY transcription factors, WRKY18, WRKY33, and WRKY40, are involved in the modulation of host defenses toward phytopathogens (Pandey and Somssich, 2009). WRKY33 in particular was shown to be required for resistance to the necrotrophs Alternaria brassicicola and B. cinerea (Zheng et al., 2006), while WRKY18 and WRKY40 appear to be necessary for the resistance to P. syringae (Xu et al., 2006). Often, WRKY factors interact both physically and functionally in a complex pattern of overlapping or antagonistic roles. For instance, the mutation of either WRKY18 or WRKY40 does not affect the susceptibility of plants to either necrotrophic or biotrophic pathogens. Double wrky18wrky40 mutants, however, are more susceptible to P. syringae and more resistant to B. cinerea than wild-type plants (Xu et al., 2006). The simultaneous activation of WRKY18, WRKY33, and WRKY40 in the cys-c1 mutant, then, does not necessarily lead to an additive effect for the expression of each WRKY factor separately. In fact, we have found that cyanide accumulation correlates with an increased susceptibility to a necrotrophic pathogen, and this association would probably be due to a deleterious but nonlethal effect of cyanide itself. An intriguing increase of the tolerance to biotrophic pathogens is observed concurrently. This increased tolerance is applicable to both a bacteria and a virus and occurs together with the induction of the pathogenesis-related PR1 mRNA in the absence of pathogens. Both susceptibility to the necrotrophic fungus and resistance to the biotrophic bacteria are reversed by treatment with the antidote hydroxocobalamin, demonstrating that the effect observed is specifically related to cyanide. Although ROS are more abundant in the cys-c1 mutant than in wild-type plants, no PCD lesions or microlesions are observed in the mutant, which demonstrates that cyanide does not induce a lesion-mimic phenotype that could be responsible for the resistance to Pst DC3000 (Lorrain et al., 2003)
To discriminate between distinctive pathogens and to activate appropriate responses, plants use phytohormones for signaling. In general, responses against biotrophic pathogens include a signaling cascade dependent on salicylic acid, while necrotrophic organisms induce signaling pathways dependent on ethylene and jasmonic acid (Pieterse et al., 2009). Interactions between the different signaling pathways have been demonstrated, indicating a complex network of hormone cross talk (Koornneef and Pieterse, 2008; Spoel and Dong, 2008; Leon-Reyes et al., 2010). Exogenously applied cyanide mimics salicylic acid-induced resistance of tobacco, Arabidopsis, and tomato (Solanum lycopersicum) plants to viruses (Chivasa and Carr, 1998; Wong et al., 2002). More recently, this treatment has been shown to confer resistance of rice to the biotrophic fungus Magnaporthe oryzae, and it has been suggested that cyanide increases during the HR (Seo et al., 2011). We have demonstrated that Arabidopsis plants accumulate more cyanide when they are infected with an avirulent strain of Pst DC3000 than when they are challenged with the virulent strain, suggesting that this molecule has a role in the establishment of the HR. The repression of CYS-C1 expression during the avirulent interaction and its activation during the virulent interaction further support this hypothesis. Finally, the resistance of the cys-c1 mutant to biotrophic pathogens indicates that cyanide mimics or induces the salicylic acid signaling pathway in Arabidopsis plants.
Interestingly, cys-c1 mutant leaves exhibit a reduced respiration rate that is more sensitive to the alternative pathway inhibitor SHAM and an enhanced expression of the AOX1a gene, showing that the alternative respiration pathway is activated in the mutant plants. AOX allows flexibility of plant respiratory metabolism, especially under environmental stresses (Vanlerberghe and McIntosh, 1997; Mackenzie and McIntosh, 1999), and it is induced by many adverse conditions (Hanqing et al., 2010). The induction of AOX1a in the cys-c1 mutant in nonstressed conditions could prepare it to better respond to a pathogen attack, probably by inducing a signal transduction dependent on ROS that culminates in the induction of defense proteins such as PR1 and other proteins related to pathogenesis. It has been suggested that tobacco and tomato cyanide-induced resistance to virus is mediated by AOX, which would contribute to the signal transduction pathway leading to resistance (Chivasa and Carr, 1998; Fu et al., 2010). Strikingly, when overexpressing either the native AOX or a version of AOX mutated at its active site, tobacco mosaic virus vectors increase systemic movements in Nicotiana benthamiana (Murphy et al., 2004).
In summary, our results suggest that cyanide, a low-Mr and highly hydrophilic molecule, acts as a signal in plants. Nitric oxide and oxygen peroxide are also low-Mr molecules that are toxic at high concentrations but that exhibit a signaling role at low concentrations; their roles have been extensively demonstrated and accepted (Delledonne et al., 1998; Laloi et al., 2004). In our model, cyanide that is produced in the cys-c1 mutant uncouples the respiratory electron chain dependent on the cytochrome c oxidase, and this uncoupling induces the AOX activity and the accumulation of ROS, which act by stimulating the salicylic acid-dependent signaling pathway of the plant immune system.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type ecotype Col-0 and the SALK_022479 mutant were used in this work. The plants were grown in soil with a photoperiod of 8 h of white light (120 µE m−2 s−1) at 20°C/16 h of dark at 18°C. Plants were cultivated for 6 to 7 weeks. For some experiments, the plants were cultivated in solid MS medium in petri dishes supplemented with 1% Suc with a photoperiod of 16 h of white light (120 µE m−2 s−1) at 20°C/8 h of dark at 18°C.
To generate the cys-c1 complementation line (cys-c1::Pcys-c1-CYS-C1), a 2,949-bp genomic fragment containing the full-length coding sequence of CYS-C1 plus the intergenic region between CYS-C1 and its contiguous PIP1 gene (At3g61430) was obtained by PCR amplification using the proofreading Platinum Pfx DNA polymerase (Invitrogen) and the primers C1GW-F and C1GW-R (Supplemental Table S3). The fragment was cloned into the pENTR/D-TOPO vector (Invitrogen) and transferred into the pMDC99 vector (Curtis and Grossniklaus, 2003) using the Gateway system (Invitrogen) according to the manufacturer’s instructions. The final construct, Pcys-c1-CYS-C1, was generated by transformation into Agrobacterium tumefaciens and then introduced into cys-c1 null plants using the floral dip method (Clough and Bent, 1998).
Respiration Measurements in Leaves
Wild-type and mutant plants were grown for 6 to 7 weeks in soil. Approximately 50 mg of leaf tissues was cut and transferred into air-tight cuvettes containing 20 mm HEPES (pH 7.2) and CaCl2, and oxygen uptake was measured as a decrease of oxygen concentration in the dark using a Clark-type electrode. Cyanide-resistant oxygen uptake was measured in the presence of 0.5 mm KCN. The component of the change due to the AOX pathway was determined by measurement in the presence of 4 mm of the inhibitor SHAM.
Bacterial Pathogen Infections
The bacterial strains used in this study were Pseudomonas syringae pv tomato DC3000 and Pst DC3000 bearing a plasmid containing the avrRpm1 avirulence gene (Grant et al., 1995). For treatment of the plants, bacterial cultures were collected from plates in 10 mm MgCl2, and their concentrations were adjusted to 5 × 106 bacteria mL−1 (optical density at 600 nm = 0.01; Pst DC3000 avrRpm1) or 2.5 × 106 bacteria mL−1 (optical density at 600 nm = 0.005; Pst DC3000). Sterile 10 mm MgCl2 was used as a mock solution. The bacterial suspension or the mock solution was then pressure infiltrated into the abaxial side of the leaves of 6- to 7-week-old plants using a syringe without a needle. Wild-type, mutant, and complemented plants were grown at the same time using the same conditions (Swanson et al., 1988).
Bacteria and Growth Tests
Pst DC3000 avrRpm1 bacteria were collected from LB plates supplemented with rifampicin (50 µg mL−1) in 10 mm MgCl2, and their concentration was adjusted to 5 × 106 bacteria mL−1 (optical density at 600 nm = 0.01). To determine whether hydroxocobalamin affects bacterial viability, growth tests were performed as described previously (Álvarez et al., 2012a) by supplementing the growth medium with 5 mm hydroxocobalamin instead of 0.5 mm Cys. Six series of 1:10 dilutions were performed. In all, 10 µL of the resulting suspensions was plated, grown for 48 h at 28°C, and subsequently photographed (Supplemental Fig. S13).
In Planta Growth of Virulent or Avirulent Pst DC3000
The protocol for measuring the growth of bacteria was adapted from (Tornero and Dangl, 2001). Wild-type, mutant, and complemented plants were grown for 6 to 7 weeks at the same time and in the same conditions and inoculated with bacterial pathogens as described above. One hour after the inoculation, the samples for day 0 were taken. To determine bacterial growth, 100 mg of leaves was ground in 500 µL of 10 mm MgCl2 and gently vortexed. In all, 20 µL from each sample was added to the wells of a microtiter plate containing 180 µL of 10 mm MgCl2, and serial 10-fold dilutions were plated on petri dishes containing 50 mg mL−1 rifampicin. The plates were incubated at 30°C, and the number of colonies was counted 30 h later. The number of cfu mg−1 fresh weight was determined by the formula cfu mg−1 fresh weight = k(N × 10d−1)/(weight of the tissue), where N is the number of colonies counted in the dilution number d and the constant k (500 in our case) represents the number of cfu present in the sample per colony appearing in the first dilution (Tornero and Dangl, 2001).
Fungal Infections
The Botrytis cinerea strain ME4 was grown in a solid strawberry broth for 12 d, and spore suspensions were prepared at a concentration of 5 × 105 spores mL−1 in 12 g L−1 potato dextrose broth. Six- to 7-week-old wild-type, mutant, and complemented plants grown at the same time and in the same conditions were pulverized with a Preval sprayer with spore suspension. Approximately 2 mL of spore suspension per plant was used. The plants were covered with a transparent film to maintain 100% humidity. The samples were collected for PCR analysis after 5 d.
Quantification of B. cinerea DNA Accumulation in Infected Plants
DNA from infected plants was quantified by real-time PCR according to a previous study (Calo et al., 2006). DNA from the B. cinerea creA gene (Tudzynski et al., 2000) was amplified using the oligonucleotides creABOT-F and creABOT-R (Supplemental Table S3). As an internal standard to normalize the real-time PCR Arabidopsis UBQ10 DNA was amplified using the oligonucleotides qUBQ10F and qUBQ10R (Supplemental Table S3). Relative quantifications were performed by subtracting the cycle threshold (CT) value of UBQ10 from the CT value of creA (ΔCT). The relative B. cinerea DNA was calculated as 2−ΔCT.
Geminivirus Infection Assays
Infection of Arabidopsis plants with Beet curly top virus was performed by whole plant agroinoculation as described (Briddon et al., 1989; Lozano-Durán et al., 2011). Inoculated plants were scored for the appearance of symptoms typical of a BCTV infection on systemically infected tissue. Symptom severity was evaluated at 28 dpi according to the severity index described by Baliji et al. (2007), where 0 represents symptomless plants and 1 to 4 represent plants showing increasing symptom severity. The infection assay was performed in triplicate.
Quantification of BCTV DNA Accumulation in Infected Plants
Total DNA of infected plants was extracted at 28 dpi using the DNeasy Plant Mini Kit (Qiagen) and digested with DpnI to differentiate between viral DNA originating from a replication in planta, which is not methylated, and viral DNA originating from replication in the inoculum of A. tumefaciens, which is methylated. Viral DNA accumulation was quantified by real-time PCR using the primers BCTV-F and BCTV-R (Supplemental Table S2). As an internal standard to normalize the real-time PCR, Arabidopsis UBQ10 DNA was amplified using the oligonucleotides qUBQ10F and qUBQ10R (Supplemental Table S3). Relative quantifications were performed by subtracting the CT value of UBQ10 from the CT value of BCTV (ΔCT). The relative BCTV DNA was calculated as 2−ΔCT.
H2O2 Detection
For the fluorimetric detection of H2O2, 5-d-old seedlings were incubated with 10 µm H2DCFDA (Molecular Probes) for 5 min in the presence of 10 µm propidium iodide (López-Martín et al., 2008). Samples were observed using a Leica TCS SP2 spectral confocal microscope with excitation of 488 nm and an emission range of 500 to 550 nm for fluorescein detection and 600 to 650 nm for propidium iodide detection.
Cell Death Staining
Trypan blue staining for dead cells in leaves was performed as described previously (Carol and Dolan, 2006) by incubating the leaves in a lactic acid-phenol-trypan blue solution (2.5 mg mL−1 trypan blue, 25% [w/v] lactic acid, 23% phenol, and 25% glycerol), heating them over boiling water for 1 min, and finally destaining them using a 2.5 g mL−1 chloral hydrate solution before photographing the leaves.
Ethylene Determination by Gas Chromatography
A total of 100 to 300 mg of infected leaves was collected, weighted, placed inside a 12-mL vial, and finally sealed. The amount of ethylene produced and released to the gas phase during 24 h was determined by gas chromatography by injecting 1 mL of the head space onto a GC2010 apparatus equipped with an activated alumina column and a flame ionization detector. The oven and the detector temperatures were isothermally maintained at 80°C and 150°C, respectively. The results are expressed as means ± sd from at least five replicate samples, and the experiment was repeated three times from independent samples.
Cyanide Determination by HPLC
A total of 100 mg of plant tissue was homogenized using a mortar and pestle with liquid nitrogen and resuspended in cold borate-phosphate extraction buffer (2 mL g−1 fresh weight) containing 27 mm sodium borate and 47 mm potassium phosphate, pH 8.0. Homogenates were centrifuged at 15,000g for 15 min at 4°C. Extracted cyanide was subsequently quantified by reverse-phase HPLC after derivatization with 2,3-naphthalenedialdehyde to form a 1-cyano-2-alkyl-benz[f]isoindole derivative by previously described methods (Lin et al., 2005; García et al., 2010).
RNA Isolation and Semiquantitative RT-PCR
Total RNA was extracted from Arabidopsis leaves using the RNeasy Plant Mini Kit (Qiagen) and reverse transcribed using an oligo(dT) primer and the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer’s instructions. AOX1a and PR1 expression was determined by semiquantitative PCR using an aliquot of the complementary DNA (cDNA) and the oligonucleotides shown in Supplemental Table S3. The constitutively expressed UBQ10 gene was used as a control. The PCR conditions were as follows: a denaturation cycle of 2 min at 94°C; 30 amplification cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; and an extension cycle of 5 min at 72°C.
Real-Time RT-PCR
Quantitative real-time RT-PCR was used to validate microarray data and to analyze the expression of the CYS-C1 gene. First-strand cDNA was synthesized as described above. Gene-specific primers for each gene were designed using the Vector NTI Advance 10 software (Invitrogen; Supplemental Table S3). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad), and the signals were detected on an iCYCLER (Bio-Rad) according to the manufacturer’s instructions. The cycling profile consisted of 95°C for 10 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. A melt curve from 60°C to 90°C was run following the PCR cycling. The expression level of each gene was normalized to that of the constitutive UBQ10 gene by subtracting the CT value of UBQ10 from the CT value of the gene (ΔCT). The fold change was calculated as 2−(ΔCT mutant − ΔCT wild type).
RNA Extraction and Microarray Hybridization
For microarray studies of the cys-c1 mutant, plants were grown on MS plates supplemented with 1% Suc under a photoperiod of 16 h of white light (120 µE m−2 s−1) at 20°C/8 h of dark at 18°C. Leaves of 15-d-old plants were used for total RNA isolation with TRIzol reagent (Invitrogen) and cleaning with the RNeasy Plant Mini Kit (Qiagen). The resulting material was used to synthesize biotinylated complementary RNA (cRNA) for hybridization to Arabidopsis ATH1 arrays (Affymetrix) using the 3′ Amplification One-Cycle Target Labeling Kit. Briefly, 4 mg of RNA was reverse transcribed to produce first-strand cDNA using a (dT)24 primer with a T7 RNA polymerase promoter site added to the 3′ end. After second-strand synthesis, in vitro transcription was performed using T7 RNA polymerase and biotinylated nucleotides to produce biotin-labeled cRNA. The cRNA preparations (15 μg) were fragmented into fragments of 35 to 200 bp at 95°C for 35 min. These fragmented cRNAs were hybridized to the Arabidopsis ATH1 microarrays at 45°C for 16 h. Each microarray was washed and stained in the Affymetrix Fluidics Station 400 following standard protocols. Microarrays were scanned using the Affymetrix GeneChip Scanner 3000.
Microarray Data Analysis
Microarray analysis was performed using the affylmGUI R package (Wettenhall et al., 2006). The Robust Multiarray Analysis algorithm was used for background correction, normalization, and summarizing expression levels (Irizarry et al., 2003). Differential expression analysis was performed using Bayes t statistics and the linear models for microarray data (Limma), which are included in the affylmGUI package. P values were corrected for multiple testing using the false discovery rate method (Benjamini and Hochberg, 1995; Reiner et al., 2003). Cutoff values of 2-fold change and P < 0.05 were adopted to discriminate the expression of genes that were differentially expressed in the mutant plant with respect to the wild type. Gene classification into functional groups was determined using the Bio-Array Resource for Arabidopsis Functional Genomics (Toufighi et al., 2005) and MapMan software (http://gabi.rzpd.de/projects/MapMan/). The microarray data for the cys-c1 mutant were meta-analyzed using Genevestigator (Hruz et al., 2008).
Statistical Analysis
For all the experiments shown, at least three independent samples were analyzed (for details, see the figure legends). An ANOVA statistical analysis of data was performed using the program OriginPro 7.5 (OriginLab).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CYS-C1 (At3g61440) and CYS-C1 T-DNA mutant (SALK_022479). The microarray Gene Expression Omnibus accession number is GSE19242.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the cys-c1 transcriptome.
Supplemental Figure S2. Relative expression levels of selected genes in the cys-c1 mutant plants.
Supplemental Figure S3. Graphic display of the hierarchical clustering of cys-c1 up- or down-regulated genes in response to hypoxia, performed with Genevestigator (Hruz et al., 2008).
Supplemental Figure S4. Graphic display of the meta-profile analysis of cys-c1 up- or down-regulated genes in response to biotic stresses, performed with Genevestigator (Hruz et al., 2008).
Supplemental Figure S5. Graphic display of the hierarchical clustering of cys-c1 up- or down-regulated genes in response to elicitors and pathogens, performed with Genevestigator (Hruz et al., 2008).
Supplemental Figure S6. Graphic display of the hierarchical clustering of cys-c1 up- or down-regulated genes in response to ACC treatment and in the etr1-1 mutant, performed with Genevestigator (Hruz et al., 2008).
Supplemental Figure S7. Time course of the accumulation of ethylene during the Arabidopsis-B. cinerea interaction (A) and the Arabidopsis-P. syringae interactions (B).
Supplemental Figure S8. Time course of the accumulation of cyanide during the Arabidopsis-B. cinerea interaction (A) and the Arabidopsis-P. syringae interactions (B).
Supplemental Figure S9. Time course of the expression of CYS-C1 during the Arabidopsis-B. cinerea interaction (A) and the Arabidopsis-P. syringae interactions (B).
Supplemental Figure S10. Susceptibility of the wild type and the cys-c1 mutant to infection with avirulent Pst DC3000 avrRpm1 bacteria.
Supplemental Figure S11. Genetic complementation of the pathogen-associated phenotype of the cys-c1 mutant.
Supplemental Figure S12. Dose-dependent effect of hydroxocobalamin on plant susceptibility to B. cinerea.
Supplemental Figure S13. Growth tests of Pst DC3000 bacteria grown in LB medium supplemented with rifampicin and 5 mm hydroxocobalamin (COB 5 mm) or with rifampicin alone (−COB).
Supplemental Table S1. List of differentially regulated genes in leaves of the cys-c1 mutant compared with the wild type.
Supplemental Table S2. Pathogen- and hypoxia-regulated genes in the cys-c1 mutant.
Supplemental Table S3. Oligonucleotide sequences used in this work.
Acknowledgments
We acknowledge Dr. Olga Del Pozo for providing the bacterial strains used in this work.
Glossary
- ACC
1-aminocyclopropane-1-carboxylic acid
- CAS
β-cyanoalanine synthase
- MS
Murashige and Skoog
- RT
reverse transcription
- Col-0
Columbia
- hpi
hours post infection
- PTI
pathogen-associated molecular pattern-triggered immunity
- ETI
effector-triggered immunity
- Pst
Pseudomonas syringae pv tomato
- HR
hypersensitive response
- dpi
days post infection
- cfu
colony-forming units
- LB
Luria-Bertani
- BCTV
Beet curly top virus
- qPCR
quantitative PCR
- AOX
alternative oxidase
- SHAM
salicylhydroxamic acid
- ROS
reactive oxygen species
- H2DCFDA
2′,7′-dichlorodihydrofluorescein diacetate
- H2O2
hydrogen peroxide
- CT
cycle threshold
- cDNA
complementary DNA
- cRNA
copy RNA
- cRNA
complementary RNA
References
- AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T. (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48: 28–44 [DOI] [PubMed] [Google Scholar]
- Albury MS, Elliott C, Moore AL. (2009) Towards a structural elucidation of the alternative oxidase in plants. Physiol Plant 137: 316–327 [DOI] [PubMed] [Google Scholar]
- Álvarez C, Bermúdez MA, Romero LC, Gotor C, García I. (2012a) Cysteine homeostasis plays an essential role in plant immunity. New Phytol 193: 165–177 [DOI] [PubMed] [Google Scholar]
- Álvarez C, Calo L, Romero LC, García I, Gotor C. (2010) An O-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol 152: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, García I, Romero LC, Gotor C. (2012b) Mitochondrial sulfide detoxification requires a functional isoform O-acetylserine(thiol)lyase C in Arabidopsis thaliana. Mol Plant 5: 1217–1226 [DOI] [PubMed] [Google Scholar]
- Álvarez ME. (2000) Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol Biol 44: 429–442 [DOI] [PubMed] [Google Scholar]
- An C, Mou Z. (2011) Salicylic acid and its function in plant immunity. J Integr Plant Biol 53: 412–428 [DOI] [PubMed] [Google Scholar]
- Astier A, Baud FJ. (1996) Complexation of intracellular cyanide by hydroxocobalamin using a human cellular model. Hum Exp Toxicol 15: 19–25 [DOI] [PubMed] [Google Scholar]
- Baliji S, Sunter J, Sunter G. (2007) Transcriptional analysis of complementary sense genes in Spinach curly top virus and functional role of C2 in pathogenesis. Mol Plant Microbe Interact 20: 194–206 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Bermúdez MA, Páez-Ochoa MA, Gotor C, Romero LC. (2010) ArabidopsisS-sulfocysteine synthase activity is essential for chloroplast function and long-day light-dependent redox control. Plant Cell 22: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Reinöhl V, Jones RL. (2006) Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223: 805–812 [DOI] [PubMed] [Google Scholar]
- Bogatek R, Dziewanowska K, Lewak S. (1991) Hydrogen-cyanide and embryonal dormancy in apple seeds. Physiol Plant 83: 417–421 [Google Scholar]
- Borowitz JL, Gunasekar PG, Isom GE. (1997) Hydrogen cyanide generation by mu-opiate receptor activation: possible neuromodulatory role of endogenous cyanide. Brain Res 768: 294–300 [DOI] [PubMed] [Google Scholar]
- Borron SW, Baud FJ, Mégarbane B, Bismuth C. (2007) Hydroxocobalamin for severe acute cyanide poisoning by ingestion or inhalation. Am J Emerg Med 25: 551–558 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glawischnig E. (2009) The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brading PA, Hammond-Kosack KE, Parr A, Jones JD. (2000) Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J 23: 305–318 [DOI] [PubMed] [Google Scholar]
- Briddon RW, Watts J, Markham PG, Stanley J. (1989) The coat protein of Beet curly top virus is essential for infectivity. Virology 172: 628–633 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Delauré SL, De Bolle MF, Cammue BP. (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Calo L, García I, Gotor C, Romero LC. (2006) Leaf hairs influence phytopathogenic fungus infection and confer an increased resistance when expressing a Trichoderma alpha-1,3-glucanase. J Exp Bot 57: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Dolan L. (2006) The role of reactive oxygen species in cell growth: lessons from root hairs. J Exp Bot 57: 1829–1834 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Carr JP. (1998) Cyanide restores N gene-mediated resistance to tobacco mosaic virus in transgenic tobacco expressing salicylic acid hydroxylase. Plant Cell 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollone R, Visca P. (2007) Is there evidence that cyanide can act as a neuromodulator? IUBMB Life 59: 187–189 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Hughes JA. (1986) Seed dormancy in red rice. V. Response to azide, hydroxylamine, and cyanide. Plant Physiol 80: 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu SM, De Martinis D, Te Lintel Hekkert S, Parker DH, Harren FJ. (2002) Ethylene production by Botrytis cinerea in vitro and in tomatoes. Appl Environ Microbiol 68: 5342–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Donato DB, Nichols O, Possingham H, Moore M, Ricci PF, Noller BN. (2007) A critical review of the effects of gold cyanide-bearing tailings solutions on wildlife. Environ Int 33: 974–984 [DOI] [PubMed] [Google Scholar]
- Fol M, Frachisse JM, Petel G, Gendraud M. (1989) Effect of cyanide on the membrane-potential of Jerusalem artichoke (Helianthus tuberosus L) tuber parenchyma: different responses in relation to dormancy. C R Acad Sci III Sci Vie 309: 551–556 [Google Scholar]
- Fu LJ, Shi K, Gu M, Zhou YH, Dong DK, Liang WS, Song FM, Yu JQ. (2010) Systemic induction and role of mitochondrial alternative oxidase and nitric oxide in a compatible tomato-Tobacco mosaic virus interaction. Mol Plant Microbe Interact 23: 39–48 [DOI] [PubMed] [Google Scholar]
- García I, Castellano JM, Vioque B, Solano R, Gotor C, Romero LC. (2010) Mitochondrial β-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22: 3268–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E. (2007) Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Gunasekar PG, Borowitz JL, Turek JJ, Van Horn DA, Isom GE. (2000) Endogenous generation of cyanide in neuronal tissue: involvement of a peroxidase system. J Neurosci Res 61: 570–575 [DOI] [PubMed] [Google Scholar]
- Gunasekar PG, Prabhakaran K, Li L, Zhang L, Isom GE, Borowitz JL. (2004) Receptor mechanisms mediating cyanide generation in PC12 cells and rat brain. Neurosci Res 49: 13–18 [DOI] [PubMed] [Google Scholar]
- Hall AH, Dart R, Bogdan G. (2007) Sodium thiosulfate or hydroxocobalamin for the empiric treatment of cyanide poisoning? Ann Emerg Med 49: 806–813 [DOI] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S. (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Hanqing F, Kun S, Mingquan L, Hongyu L, Xin L, Yan L, Yifeng W. (2010) The expression, function and regulation of mitochondrial alternative oxidase under biotic stresses. Mol Plant Pathol 11: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SA, Kato K, Salter KJ, Kozlowski RZ. (1998) Functional evidence for a novel suramin-insensitive pyrimidine receptor in rat small pulmonary arteries. Circ Res 83: 940–946 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Isom GE, Way JL. (1984) Effects of oxygen on the antagonism of cyanide intoxication: cytochrome oxidase, in vitro. Toxicol Appl Pharmacol 74: 57–62 [DOI] [PubMed] [Google Scholar]
- Iwai T, Miyasaka A, Seo S, Ohashi Y. (2006) Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol 142: 1202–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A. (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Van der Does D, De Lange ES, Delker C, Wasternack C, Van Wees SC, Ritsema T, Pieterse CM. (2010) Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232: 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Wong BK, Burgey CS, Gibson CR, Singh R. (2005) In vitro metabolism of a thrombin inhibitor and quantitation of metabolically generated cyanide. J Pharm Biomed Anal 39: 1014–1020 [DOI] [PubMed] [Google Scholar]
- López-Martín MC, Becana M, Romero LC, Gotor C. (2008) Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Plant Physiol 147: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D. (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R, Rosas-Díaz T, Gusmaroli G, Luna AP, Taconnat L, Deng XW, Bejarano ER. (2011) Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23: 1014–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L. (1999) Higher plant mitochondria. Plant Cell 11: 571–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar FF, Lambers H. (2003) The alternative oxidase: in vivo regulation and function. Plant Biol 5: 2–15 [Google Scholar]
- Miller JM, Conn EE. (1980) Metabolism of hydrogen cyanide by higher plants. Plant Physiol 65: 1199–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mur LA, Laarhoven LJ, Harren FJ, Hall MA, Smith AR. (2008) Nitric oxide interacts with salicylate to regulate biphasic ethylene production during the hypersensitive response. Plant Physiol 148: 1537–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AM, Gilliland A, York CJ, Hyman B, Carr JP. (2004) High-level expression of alternative oxidase protein sequences enhances the spread of viral vectors in resistant and susceptible plants. J Gen Virol 85: 3777–3786 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M. (2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69: 2655–2667 [DOI] [PubMed] [Google Scholar]
- Poulton JE. (1990) Cyanogenesis in plants. Plant Physiol 94: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43: 361–394 [DOI] [PubMed] [Google Scholar]
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K. (1998) The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev 12: 2013–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Mitsuhara I, Feng J, Iwai T, Hasegawa M, Ohashi Y. (2011) Cyanide, a coproduct of plant hormone ethylene biosynthesis, contributes to the resistance of rice to blast fungus. Plant Physiol 155: 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegien I, Bogatek R. (2006) Cyanide action in plants: from toxic to regulatory. Acta Physiol Plant 28: 483–497 [Google Scholar]
- Spoel SH, Dong X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Stelmaszyńska T. (1986) Formation of HCN and its chlorination to ClCN by stimulated human neutrophils. 2. Oxidation of thiocyanate as a source of HCN. Int J Biochem 18: 1107–1114 [DOI] [PubMed] [Google Scholar]
- Swanson J, Kearney B, Dahlbeck D, Staskawicz BJ. (1998) Cloned avirulence gene of Xanthomonas campestris pv. vesicatoria complements spontaneous race change mutant. Mol Plant Microbe Interact 1: 5–9 [Google Scholar]
- Tornero P, Dangl JL. (2001) A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J 28: 475–481 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tudzynski B, Liu S, Kelly JM. (2000) Carbon catabolite repression in plant pathogenic fungi: isolation and characterization of the Gibberella fujikuroi and Botrytis cinerea creA genes. FEMS Microbiol Lett 184: 9–15 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703–734 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Geraats BP, Linthorst HJ. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park SW. (2008) Systemic acquired resistance: the elusive signal(s). Curr Opin Plant Biol 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kusano M, Oikawa A, Fukushima A, Noji M, Saito K. (2008) Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol 146: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22: 897–899 [DOI] [PubMed] [Google Scholar]
- Wong CE, Carson RA, Carr JP. (2002) Chemically induced virus resistance in Arabidopsis thaliana is independent of pathogenesis-related protein expression and the NPR1 gene. Mol Plant Microbe Interact 15: 75–81 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrobelny M, Bak S, Møller BL. (2008) Cyanogenesis in plants and arthropods. Phytochemistry 69: 1457–1468 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]