Arabidopsis responds to simultaneous water stress and nematode infection by activating a unique program of gene expression that is distinct from the response to individual stresses.
Abstract
In field conditions, plants may experience numerous environmental stresses at any one time. Research suggests that the plant response to multiple stresses is different from that for individual stresses, producing nonadditive effects. In particular, the molecular signaling pathways controlling biotic and abiotic stress responses may interact and antagonize one another. The transcriptome response of Arabidopsis (Arabidopsis thaliana) to concurrent water deficit (abiotic stress) and infection with the plant-parasitic nematode Heterodera schachtii (biotic stress) was analyzed by microarray. A unique program of gene expression was activated in response to a combination of water deficit and nematode stress, with 50 specifically multiple-stress-regulated genes. Candidate genes with potential roles in controlling the response to multiple stresses were selected and functionally characterized. RAPID ALKALINIZATION FACTOR-LIKE8 (AtRALFL8) was induced in roots by joint stresses but conferred susceptibility to drought stress and nematode infection when overexpressed. Constitutively expressing plants had stunted root systems and extended root hairs. Plants may produce signal peptides such as AtRALFL8 to induce cell wall remodeling in response to multiple stresses. The methionine homeostasis gene METHIONINE GAMMA LYASE (AtMGL) was up-regulated by dual stress in leaves, conferring resistance to nematodes when overexpressed. It may regulate methionine metabolism under conditions of multiple stresses. AZELAIC ACID INDUCED1 (AZI1), involved in defense priming in systemic plant immunity, was down-regulated in leaves by joint stress and conferred drought susceptibility when overexpressed, potentially as part of abscisic acid-induced repression of pathogen response genes. The results highlight the complex nature of multiple stress responses and confirm the importance of studying plant stress factors in combination.
Plants are adapted to respond to diverse environmental stress conditions, activating specific molecular and physiological changes to minimize damage. In field conditions, plants may be exposed to a variety of concurrent stresses (Mittler and Blumwald, 2010). Despite this, the majority of laboratory studies on plant stress factors have analyzed each stress in isolation. There is increasing evidence that the response to multiple environmental stresses is distinct from that for individual stresses, and not merely additive (Atkinson and Urwin, 2012). This has been demonstrated in transcriptome studies on plants subjected to multiple abiotic stresses. In both tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana), a combination of drought and heat stress induces a novel program of gene expression, activating transcripts that are not induced by either stress individually (Rizhsky et al., 2002, 2004). Similarly, microarray analysis has revealed that exposure to multiple biotic stresses (two species of herbivorous insect) elicits a transcriptional response that is distinct from each individual response (Voelckel and Baldwin, 2004). It has therefore been proposed that each stress combination should be studied as an entirely new stress (Mittler and Blumwald, 2010). Understanding such mechanisms will be crucial for the future development of broad-spectrum stress-tolerant crops.
The response of plants to simultaneous biotic and abiotic stresses is of particular interest, as the signaling pathways of individual stress responses interact and antagonize one another, a process controlled principally by hormones (Anderson et al., 2004; Asselbergh et al., 2008b; Atkinson and Urwin, 2012). Abscisic acid (ABA) is produced primarily in response to abiotic stresses and induces a range of downstream processes resulting in stress tolerance. By contrast, the response to biotic stresses is defined by antagonism between the hormones jasmonic acid, salicylic acid, and ethylene. However, ABA can also act as a global regulator of stress responses by dominantly suppressing biotic stress signaling pathways. This may facilitate fine-tuning of plant stress responses to focus on the most severe threat and can have both positive and negative effects on defense against pathogens (Anderson et al., 2004; Yasuda et al., 2008; Ton et al., 2009). For example, treatment with ABA has been shown to repress systemic acquired resistance to pathogens and prevent the accumulation of defense compounds such as lignins and phenylpropanoids (Mohr and Cahill, 2007; Kusajima et al., 2010). Drought stress or ABA treatment can actually increase the susceptibility of Arabidopsis to an avirulent strain of Pseudomonas syringae, while ABA treatment in tomato (Solanum lycopersicum) increases susceptibility to the pathogens Botrytis cinerea and Erwinia chrysanthemi (Audenaert et al., 2002; Mohr and Cahill, 2003; Asselbergh et al., 2008a). ABA-deficient mutants of both Arabidopsis and tomato show increased resistance to pathogens (Audenaert et al., 2002; Mohr and Cahill, 2003; Anderson et al., 2004). By contrast, ABA signaling is necessary for defense against some fungi and oomycetes, and ABA-induced stomatal closure can help prevent microbial invasion (Ton and Mauch-Mani, 2004; Melotto et al., 2006; Adie et al., 2007). Downstream regulation of multiple stress signaling pathways is achieved by a complex network of interacting components (Atkinson and Urwin, 2012). Transcription factors and mitogen-activated protein kinases may be particularly important in defining signal specificity, as they frequently control a wide range of downstream events and many are induced by more than one stress (Fujita et al., 2006; Zhang et al., 2006).
Water deficit and infection with plant-parasitic nematodes represent two environmental stresses with interacting effects under field conditions. Nematode infection can exacerbate the effects of water stress on plants, as their parasitism of roots severely disrupts plant water relations (Bird, 1974; Haverkort et al., 1991; Smit and Vamerali, 1998). The cyst nematode Heterodera sacchari increases drought-related losses in upland rice (Oryza sativa) by contributing to reduced leaf water potential, stomatal conductance, and leaf dry weight (Audebert et al., 2000). The potato cyst nematode Globodera pallida causes root retardation in potato (Solanum tuberosum), which in turn has the effect of reducing drought tolerance (Smit and Vamerali, 1998). Water stress and infection with nematodes both negatively affect growth in potatoes but, in combination, produce nonadditive effects (Fasan and Haverkort, 1991; Haverkort et al., 1991).
Transcriptome analysis and functional characterization of individual genes involved in the multiple stress response can provide further opportunities for fully understanding the complex interactions that regulate multiple stress responses. In this work, the transcriptome response of Arabidopsis plants to combined biotic and abiotic stress was analyzed using Affymetrix ATH1 whole genome arrays. Water stress was imposed through a short dehydration treatment, while biotic stress comprised infection with the beet cyst nematode Heterodera schachtii. Candidate genes with potential roles in controlling stress interaction were selected and their function characterized through the analysis of overexpression lines and loss-of-function mutants under control and stress conditions. The interaction with plant hormones was then investigated through the use of hormone signaling mutants.
RESULTS
Transcriptional Profiling of Arabidopsis Plants Subjected to Combined Water Deficit and Nematode Stress
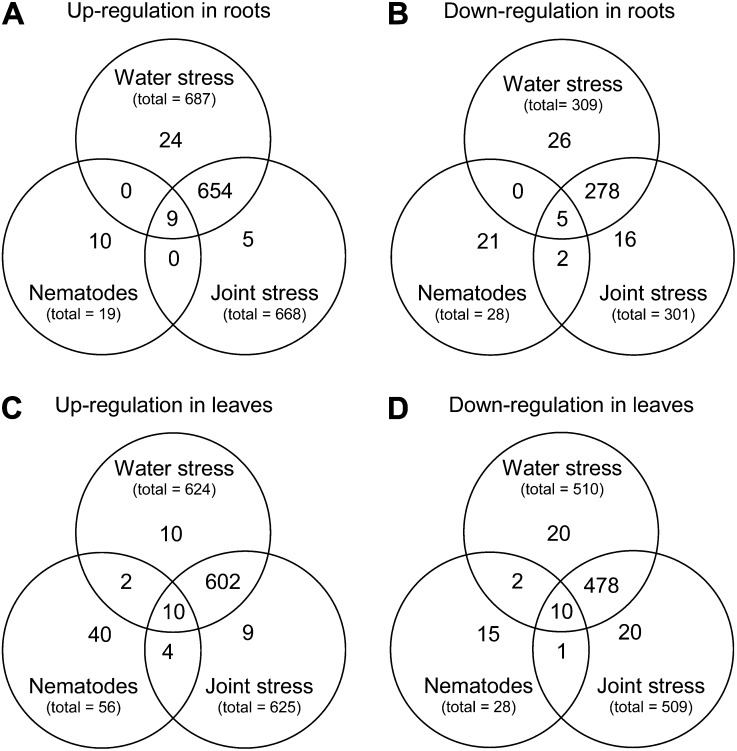
Microarrays (ATH1 chips; Affymetrix) were used to determine changes in Arabidopsis gene transcript levels as a result of exposure to water deficit, nematode infection, or the two stresses in combination. Root and leaf tissue were examined separately. Genes were considered differentially regulated if their expression was significantly different from the control treatment (P < 0.05). Each individual or joint stress treatment induced a unique set of differentially expressed genes (Fig. 1; Supplemental Table S1). The number of genes differentially regulated in response to water stress, 996 in roots and 1,134 in leaves, was far greater than that induced by nematode stress, 47 in roots and 84 in leaves. Around one-quarter of the nematode-induced genes were also regulated by water stress and joint stress, suggesting a more generalized role in stress response. In response to a combination of biotic and abiotic stress, a total of 969 transcripts were found to be significantly different from the control treatment in roots (668 induced and 301 repressed), while 1,134 were differentially regulated in leaves (625 induced and 509 repressed). The overall pattern of gene induction was highly similar to that observed for water stress, as 96% of transcripts regulated by joint stress were also regulated by water stress alone. By contrast, only 2% of joint stress-regulated transcripts were regulated by nematode stress alone. In addition to these overlapping transcript changes, the joint stress treatment induced a set of specific changes that were not differentially regulated by either of the single stresses, revealing that a novel and unique program of gene expression is activated in response to the combined stresses. Quantitative reverse transcription (qRT)-PCR verification of expression for 12 genes selected to encompass a range of fold changes correlated highly (R2 = 0.729) with the results of the microarray (Supplemental Table S2). All of the genes showed the same direction of fold change using both systems; however, almost all genes showed a greater magnitude of fold change when measured by qRT-PCR.
Figure 1.
Venn diagrams showing overlap between genes differentially regulated by water stress, nematode stress, or the two in combination. Genes up- and down-regulated in roots (A and B, respectively) and leaves (C and D, respectively) are shown separately. Genes are shown whose expression levels differed significantly from control arrays where P < 0.05. Overlapping sections represent genes that were up- or down-regulated by more than one stress treatment. Data shown represent three biological replicates.
The aim of this study was to identify genes differentially regulated between individual and dual stress treatments. These were genes specifically activated or repressed by the addition of a second stress and which therefore stood out as potential candidates for orchestrating the multiple stress response. As the response to water deficit and to joint stress was transcriptionally similar, genes with a different pattern of expression under the two conditions were of particular interest. Student’s t tests were used to identify these ‘interaction’ genes that were differentially regulated by joint stress compared with individual stress, as opposed to comparison with unstressed plants (Supplemental Table S3). This list included any genes that were differentially regulated by both individual and joint stress but to a significantly greater or lesser extent when the stresses occurred together. This system allowed the exclusion of any genes with a very similar pattern of transcription under individual water stress and joint stress, but which only reached the significance threshold in one comparison. Of the interaction genes, several categories of gene function were highly prominent (Supplemental Table S4). These included genes with both functional and regulatory roles. In roots, 23 up-regulated genes had functions in cell wall modification, including extensins, pectinesterases, polygalacturonases, and xyloglucan transferases. Carbohydrate metabolism genes were abundant among the up- and down-regulated interaction genes in roots and leaves, particularly glycosyl and glycoside hydrolases. Sixteen interaction genes with disease resistance annotations, including those with Leu rich repeat domains, were down-regulated in leaf tissue, while four were down-regulated in roots. Several genes encoding proteins with oxidoreductase functions were repressed in leaves. Signal transduction and regulation genes, such as those encoding transcription factors, protein kinases, and Rapid Alkalinization Factor (RALF)-like signal molecules, were also prevalent among the interaction genes. A total of 32 and 27 transcription factors were up-regulated in leaves and roots, respectively, while 22 and 28 were repressed in those tissues. The largest group of differentially regulated transcription factors belonged to the MYB family, while also abundant were those from the no apical meristem (NAM) family, as well as the APETALA2 (AP2), zinc finger (C2H2 type), basic helix-loop-helix, and Dof-type families. Supplemental Figure S1 shows the functional Gene Ontology categories of the interaction genes.
Genes were selected for further analysis from overrepresented families of transcription factors, those strongly regulated by hormones (as determined using data from Genevestigator; Supplemental Fig. S2), or those with the largest fold change in expression between individual and joint stress. The role of the candidate genes in stress response mechanisms was then investigated through loss-of-function and constitutive overexpression mutants. The findings obtained from the study of three genes are reported here, namely RAPID ALKALINIZATION FACTOR-LIKE8 (AtRALFL8), METHIONINE GAMMA LYASE (AtMGL), and AZELAIC ACID INDUCED1 (AZI1). These were selected as genes likely to play prominent roles in governing the response to multiple stresses.
Expression of Candidate Genes during Stress Treatments
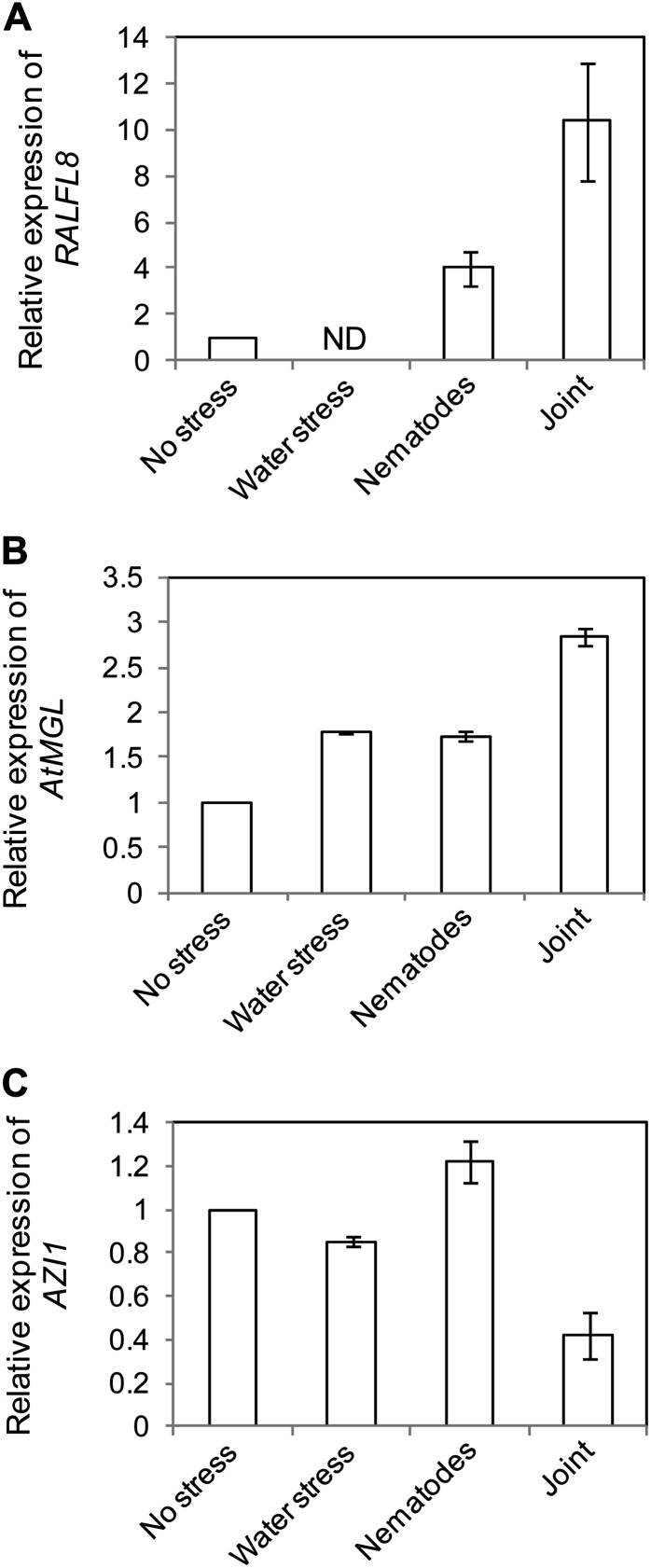
The genes AtRALFL8 (At1g61563) and AtMGL (At1g64660) were among the ‘interaction’ genes identified in the microarray study that were significantly up-regulated due to simultaneous water deficit and nematode stress in root tissue (Supplemental Table S3). AtRALFL8 encodes a short signaling peptide with similarity to tobacco RALF (Olsen et al., 2002). Three other RALFL genes (AtRALFL23, AtRALFL33, and AtRALFL34) were also induced by the stresses in combination. AtMGL encodes a Met γ-lyase involved in cellular Met homeostasis and Ile synthesis (Rébeillé et al., 2006). AZI1 (At4g12470) is an example of a gene down-regulated in leaf tissue by the stresses in combination (Supplemental Table S3). This gene belongs to a family of lipid transfer proteins and is important for defense priming in systemic plant immunity (Jung et al., 2009). Differential regulation of the genes was confirmed by qRT-PCR (Fig. 2), and in each case, the expression differential was greater than that shown by microarray, a commonly observed effect likely to arise from the differing sensitivities of the two technologies (Clarke and Zhu, 2006).
Figure 2.
Relative expression of candidate genes in leaves of plants under differing stress treatments. The expression levels of AtRALFL8 (A), AtMGL (B), and AZI1 (C) are shown relative to the unstressed samples, as analyzed by qRT-PCR. RNA was pooled from 40 plants. Error bars represent se of the means of three technical replicates. ND, Not detected.
Phenotype of Loss-of-Function and Constitutive Expression Lines under Control Conditions
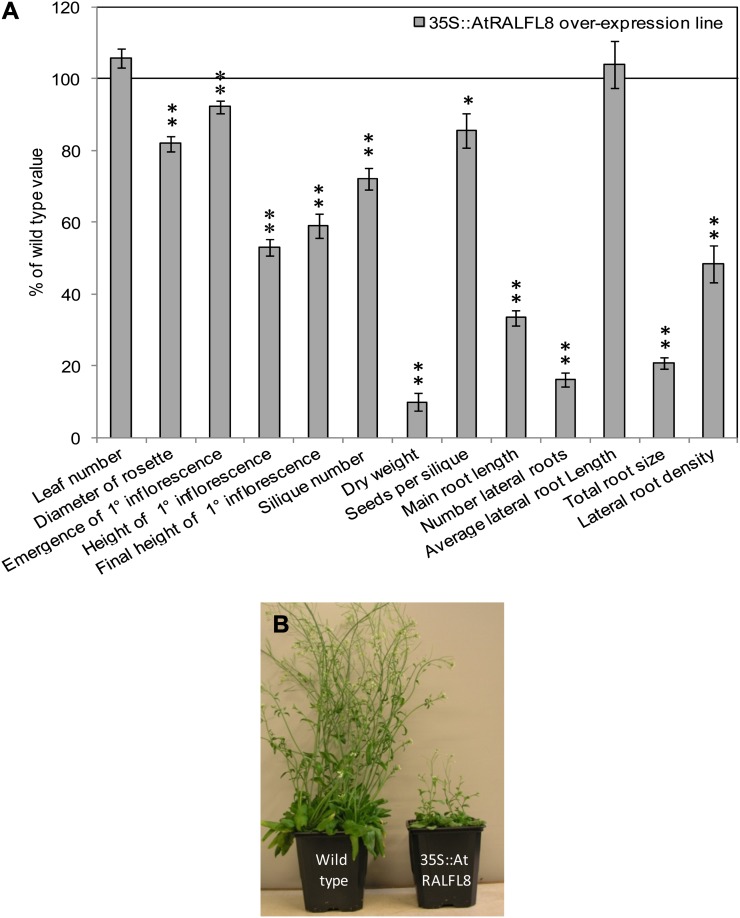
Constitutive overexpression lines were created for each gene using the Cauliflower mosaic virus 35S promoter and their relative levels of expression quantified. The most highly overexpressing line for each gene was used for subsequent experiments. The selected 35S::AZI1 line produced 12,500 times the wild-type level of AZI1 transcript, the 35S::AtMGL line had 1,590-fold increased expression, and the 35S::AtRALFL8 line 21,000-fold increased expression. Transfer DNA insertion lines were obtained for AtMGL and AZI1, although none was available for AtRALFL8. The effect of altered AtRALFL8, AtMGL, and AZI1 expression was investigated by studying the growth characteristics of mutants and overexpression lines under control conditions. Figures 3 to 5 show a range of phenotypic measurements for each mutant compared with wild-type plants. 35S::AtRALFL8 plants had a severely stunted phenotype, as characterized by a root system that was only 20% of the wild-type size (Fig. 3). Main root length, number of lateral roots, total root size, and lateral root density were significantly reduced. The rosette diameter was smaller than the wild type, while biomass accumulation was also greatly reduced, giving plants whose aerial parts weighed only 50 mg (dry weight) after 35 d compared with the wild-type weight of 500 mg. The inflorescence was significantly shorter than the wild type, even when fully mature, and developed fewer siliques, which, in turn, contained fewer seeds.
Figure 3.
Phenotype of AtRALFL8 overexpression line under control conditions. 35S::AtRALFL8 plants were grown in soil to measure the phenotype of aerial parts of the plants and on tissue culture plates to analyze the root systems. A, Phenotypic measurements are shown as a percentage of the value obtained for wild-type plants grown in parallel. The line at 100% represents the wild-type value. Asterisks show a significant difference from the wild type (n = 16; **P < 0.01, *P < 0.05). B, Photograph shows wild-type and transgenic plants 35 d after sowing. [See online article for color version of this figure.]
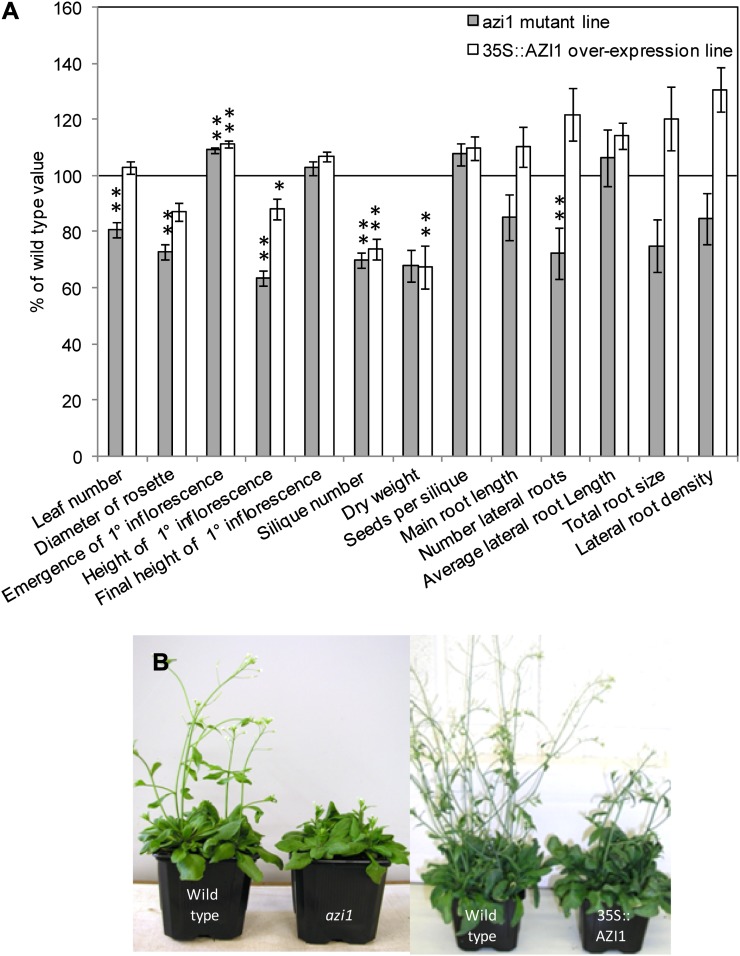
Figure 5.
Phenotype of AZI1 mutant and overexpression lines under control conditions. Mutant azi1 and 35S::AZI1 plants were grown in soil to measure the phenotype of aerial parts of the plants and on tissue culture plates to analyze the root systems. A, Phenotypic measurements are shown as a percentage of the value obtained for wild-type plants grown in parallel. The line at 100% represents the wild-type value. Asterisks show a significant difference from the wild type (n = 16; **P < 0.01, *P < 0.05). B, Photographs show wild-type and mutant/overexpressing plants 35 d after sowing. [See online article for color version of this figure.]
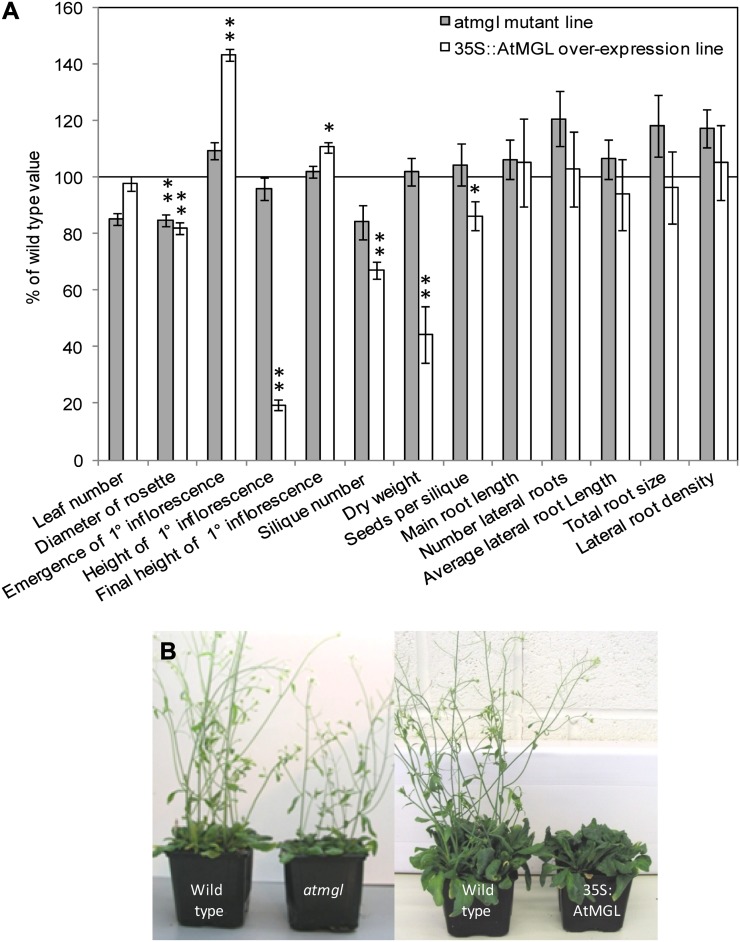
The only visible effect of AtMGL gene inactivation was a 15% smaller rosette diameter at 16 d (Fig. 4). However, the overexpression of this gene caused several phenotypic differences in development in aerial plant parts. 35S::AtMGL rosettes were slightly reduced in diameter and had a very low rate of biomass accumulation, with a dry weight only 44% of the wild-type value. Seed yield was also significantly reduced, although the final inflorescence height was slightly greater than the wild type.
Figure 4.
Phenotype of AtMGL mutant and overexpression lines under control conditions. Mutant atmgl and 35S::AtMGL plants were grown in soil to measure the phenotype of aerial parts of the plants and on tissue culture plates to analyze the root systems. A, Phenotypic measurements are shown as a percentage of the value obtained for wild-type plants grown in parallel. The line at 100% represents the wild-type value. Asterisks show a significant difference from the wild type (n = 16; **P < 0.01, *P < 0.05). B, Photographs show wild-type and mutant/overexpressing plants 35 d after sowing. [See online article for color version of this figure.]
Both the azi1 mutant and the 35S::AZI1 constitutive expression line exhibited slow growth in the aerial parts of the plant (Fig. 5). The azi1 mutant had a more severe phenotype, with reduced leaf number, rosette diameter, height of primary inflorescence, silique number, and dry weight, as well as fewer lateral roots. The 35S::AZI1 plants had fewer siliques and a lower dry weight than the wild type, although no difference could be found between the root systems and those of control plants. Primary inflorescences in both the mutant and overexpression lines emerged later than the wild type, but seed yield was not affected. To confirm that the observed phenotypes resulted directly from overexpression of the transgene, two additional lines per construct were analyzed for their growth characteristics under control conditions, yielding similar results (Supplemental Fig. S3).
Stress Tolerance Phenotype of Loss-of-Function and Constitutive Expression Lines
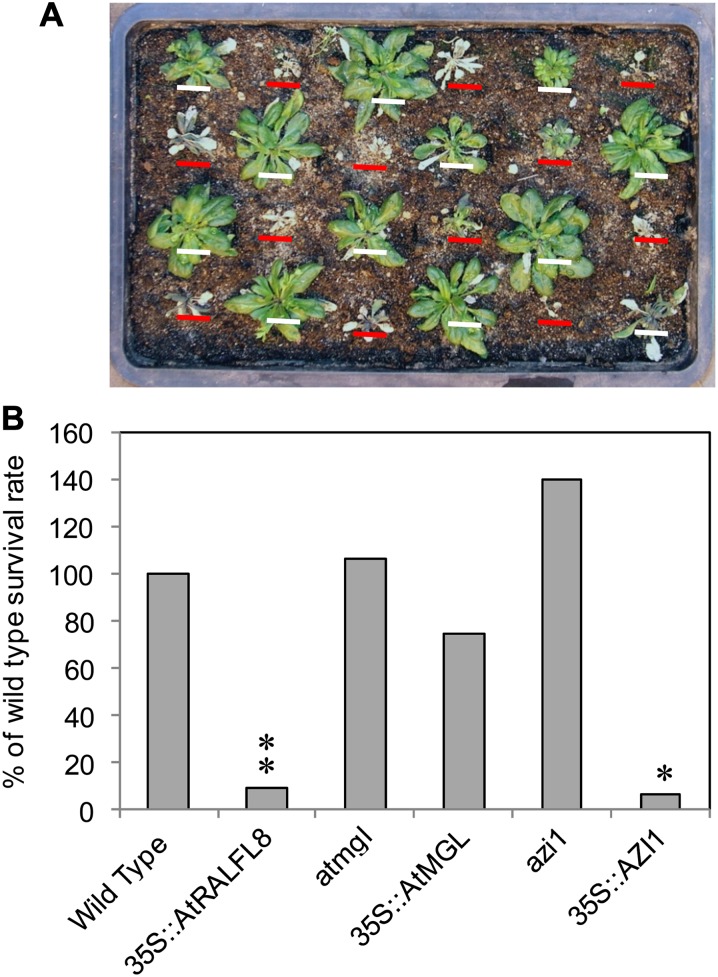
Drought tolerance of Arabidopsis mutant and overexpression lines was assessed by measuring survival rate after a prolonged period without irrigation. 35S::AtRALFL8 plants had a significantly lower survival rate than control plants, with a recovery rate that was only 9% of the wild-type value (Fig. 6, A and B). 35S::AZI1 plants also exhibited increased susceptibility to drought stress, showing a recovery rate of 6.7% of the wild-type value. The drought tolerance of 35S::AtMGL, atmgl, and azi1 plants was not affected by the altered gene expression.
Figure 6.
Survival rate of mutant and overexpression lines after drought stress. Irrigation was withheld from 12 21-d-old seedlings of each genotype until soil moisture content dropped to less than 3% (approximately 2 weeks). Plants were rewatered and scored for survival after a further week. A, Wild-type (white bars) and 35S::AtRALFL8 plants (red bars) 1 week after rewatering. B, Survival rate of each genotype compared with wild-type plants (shown as 100%). Asterisks show a significant difference from the wild type (**P < 0.01, *P < 0.05) according to χ2 tests.
The susceptibility of loss-of-function and overexpression lines to infection with the plant-parasitic nematode H. schachtii was assessed. The number of successful parasitism events on each genotype was compared with that on wild-type plants. 35S::AtRALFL8 plants supported 2.6 times more nematodes than wild-type plants (P < 0.05). By contrast, significantly fewer nematodes were observed on 35S::AtMGL plants, which had only 24% the infection rate of wild-type plants (P < 0.05). The genotypes atmgl, azi1, and 35S::AZI1 showed no altered susceptibility to H. schachtii (Fig. 7).
Figure 7.
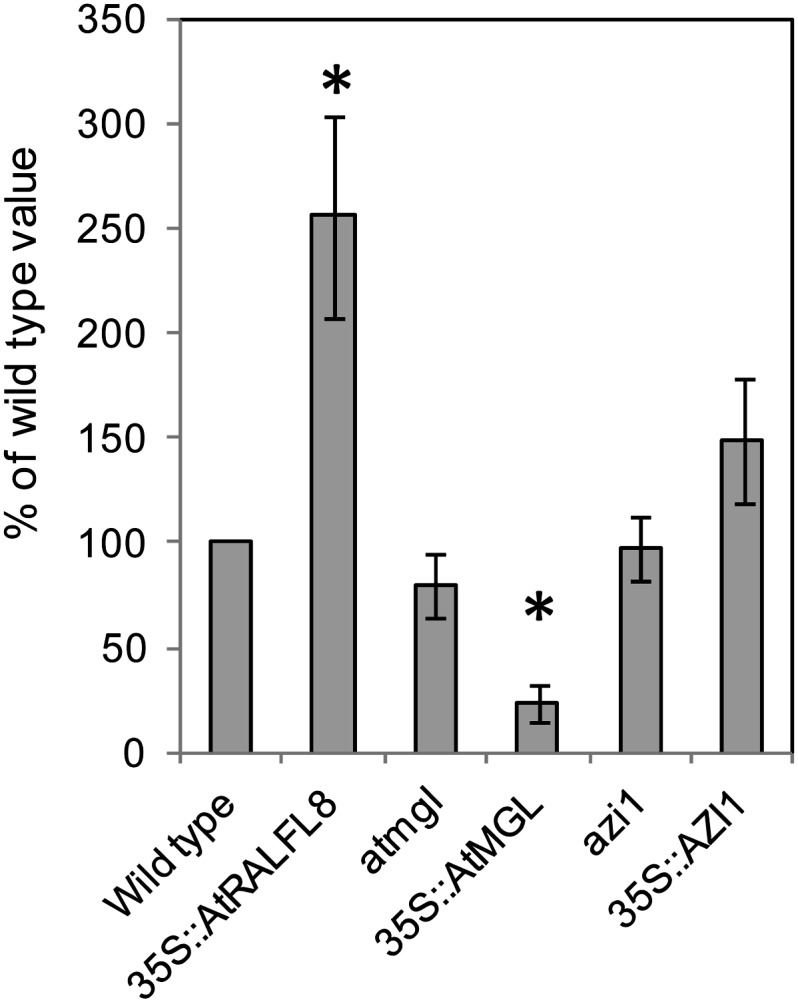
Nematode susceptibility assays. Mutant and overexpression lines for each candidate gene were exposed to 100 J2 H. schachtii nematodes per plant. Nematodes were allowed to develop for 10 d, then roots were stained and the number of established third- and fourth-stage juvenile nematodes (J3 and J4) counted per plant. The mean number of nematodes per plant for each genotype is expressed as a percentage of the wild-type value (100%). Asterisks show a significant difference from the wild type (n = 10–12; *P < 0.05).
Expression of Candidate Genes in Hormone Signaling Mutants
To investigate the relationship of candidate genes with known hormone signaling pathways, the expression levels of AtRALFL8, AtMGL, and AZI1 were analyzed in Arabidopsis lines deficient in ethylene, ABA, or salicylic acid signaling pathways using qRT-PCR. There were significant differences in expression of each gene in one or more mutants compared with the level in wild-type plants (Fig. 8). Expression of AtRALFL8 was significantly higher in ethylene-insensitive ein3-1 plants, with 2.5 times the level of wild-type transcript (P < 0.05; Fig. 8A). Expression of AtRALFL8 was similar in the constitutive ethylene response mutant constitutive triple response1 (ctr1) and in ecotype Columbia (Col-0) plants. AtMGL was expressed 3.8-fold higher in ABA-insensitive abi2-1 mutant plants, which are deficient in ABA signal transduction, than in the control Landsberg erecta plants (P < 0.01; Fig. 8B). Altered expression of AZI1 was also observed in hormone signaling mutants (Fig. 8C). In the ctr1 mutant, AZI1 expression was 13 times the wild-type value (P < 0.01), while in the ein3-1 mutant, the expression was lower than, although not significantly different from, the control. AZI1 expression was increased 6-fold in the abi2-1 mutant (P < 0.01; Fig. 8C). None of the three genes had altered expression in the abi4-1 mutant or in the salicylic acid-deficient transgenic line expressing salicylate hydroxylase (NahG).
Figure 8.
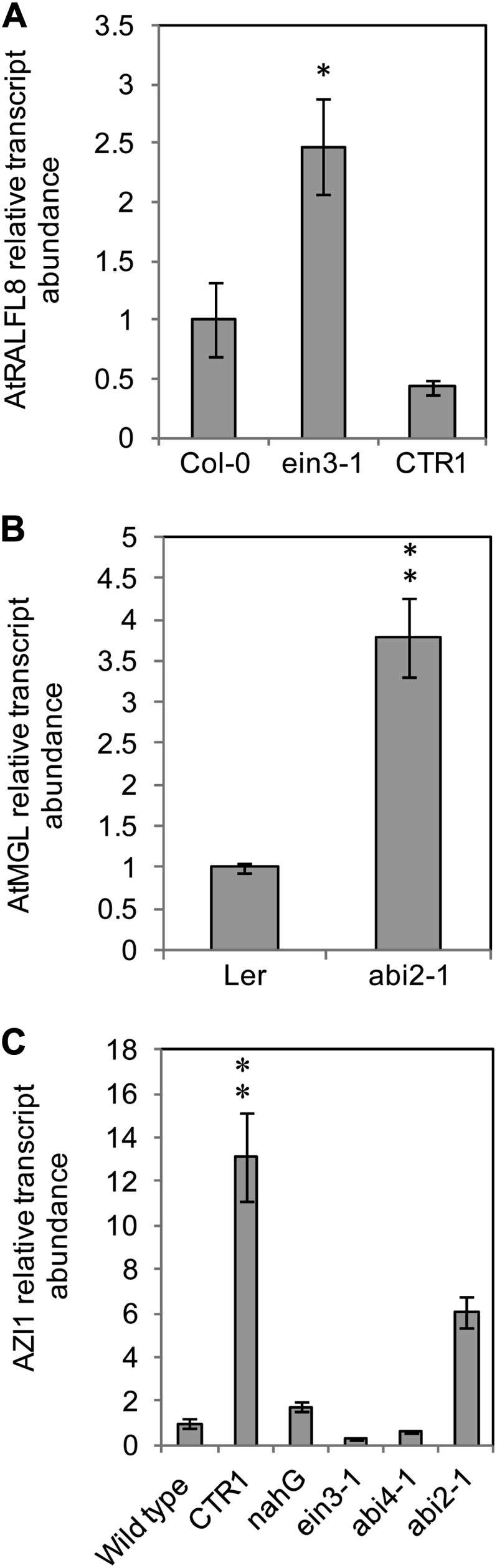
Expression levels of candidate genes in hormone signaling mutants. The relative transcript abundance of candidate genes was analyzed in Arabidopsis hormone signaling mutants using qRT-PCR. AtRALFL8 (A), AtMGL (B), and AZI1 (C). Values are the average of three biological replicates of four plants each. Asterisks show significant differences in candidate gene expression level compared with the wild-type value (**P < 0.01, *P < 0.05).
Investigation of 35S::AtRALFL8 Phenotype
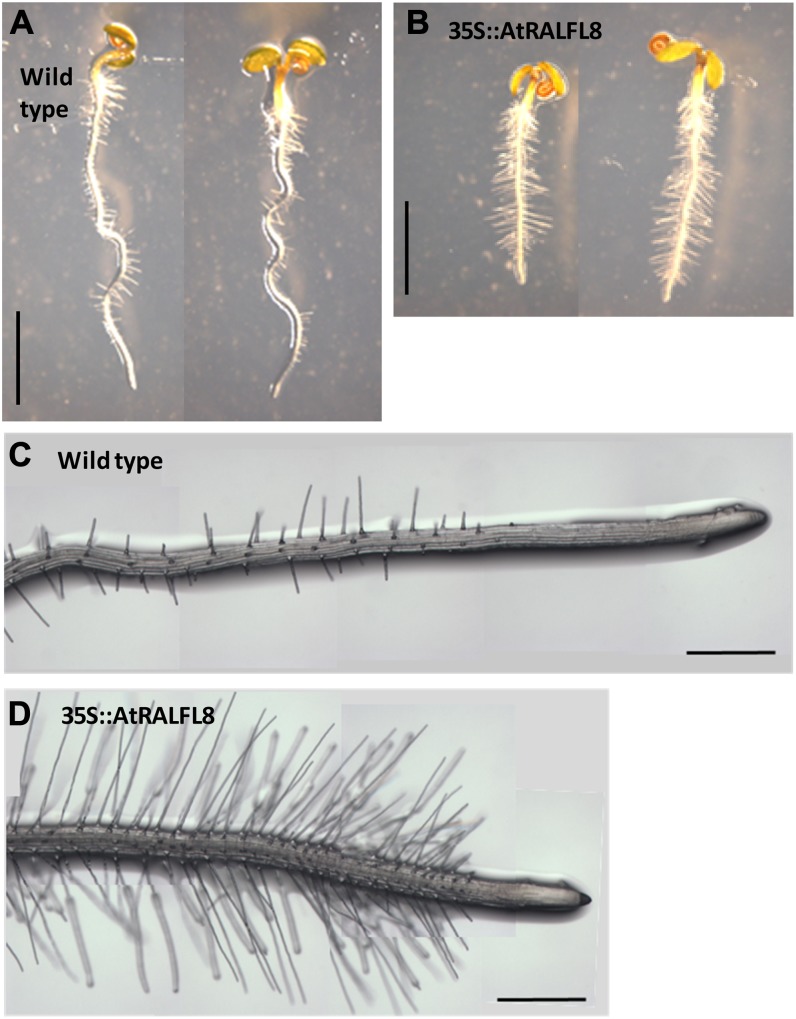
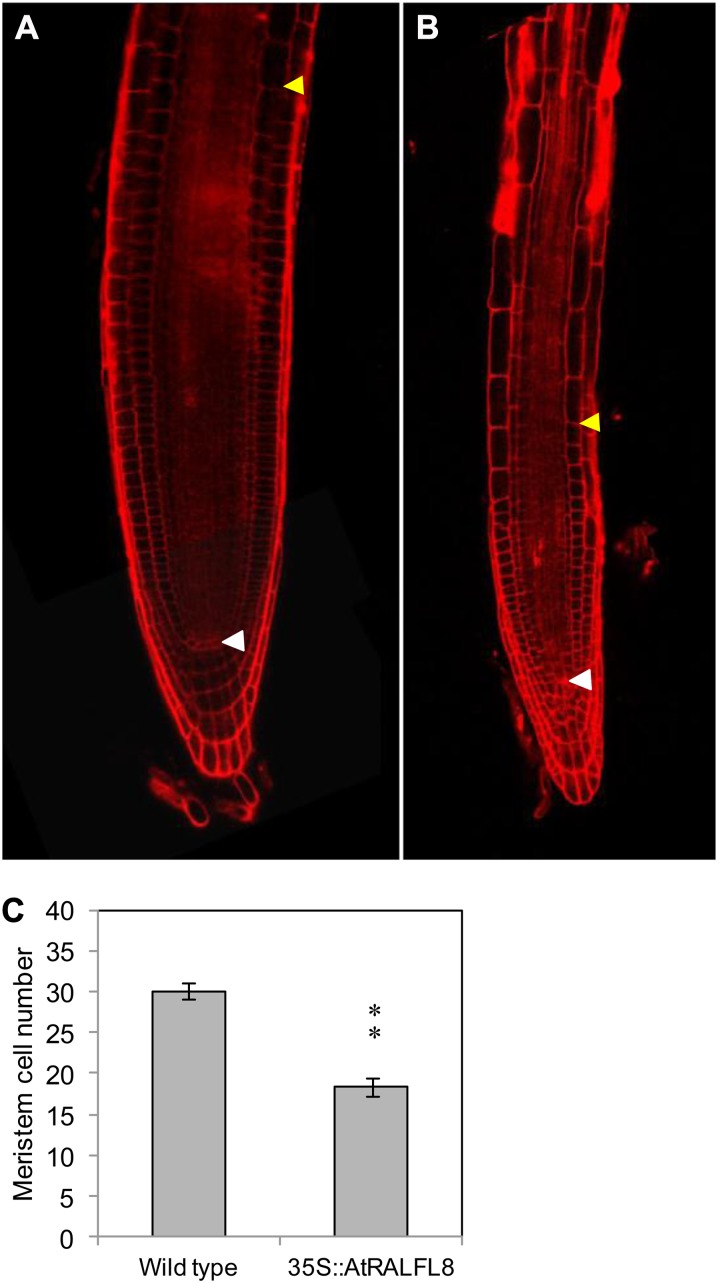
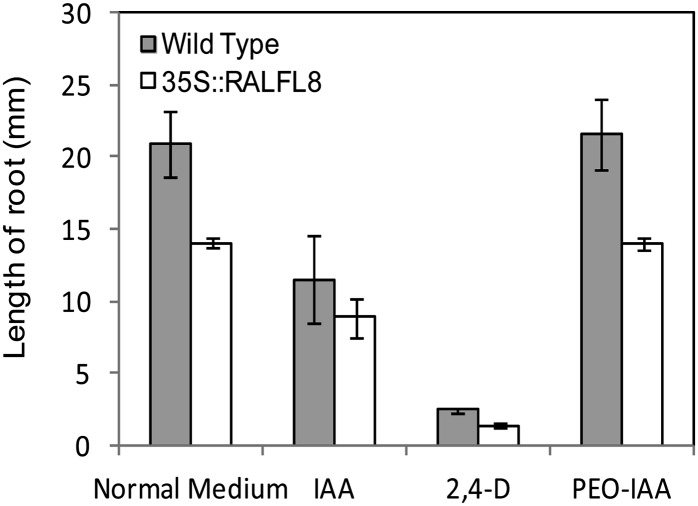
In addition to the severely stunted root system of 35S::AtRALFL8 (Fig. 3A), roots had longer and more numerous root hairs than the wild type (Fig. 9) and a shorter meristem, as determined by the number of cortical cells between the quiescent center and the transition zone of elongation differentiation (Fig. 10). This phenotype suggests disruption in auxin or ethylene signaling (Knox et al., 2003; Moubayidin et al., 2010) and was observed in four independent 35S::AtRALFL8 transgenic lines, thus eliminating the possibility of it being a secondary effect of the transgene insertion (data not shown). On close observation, the root epidermal cells were seen to maintain a normal pattern of hair cells and nonhair cells, although the epidermal cells appeared shorter. To determine whether this phenotype was related to a disruption in auxin signaling, wild-type and 35S::AtRALFL8 plants were grown on media containing auxin indole-3-acetic acid (IAA), synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-d), and antiauxin α-(phenylethyl-2-one; PEO)-IAA and the primary root lengths measured (Fig. 11). The 35S::AtRALFL8 plants responded to each compound in a similar manner to wild-type plants, producing a slightly shorter root on IAA, a very short root on 2,4-d, and a normal length root on PEO-IAA. 35S::AtRALFL8 homozygous plants were crossed with the auxin-insensitive mutant auxin resistant3 (axr3-1). This mutant carries a gain-of-function mutation in AXR3, which prevents its auxin-mediated degradation, thus causing auxin insensitivity and giving a phenotype with no root hairs, reduced root elongation, and agravitropism. The resulting 35S::AtRALFL8/axr3-1 heterozygotes were indistinguishable from the homozygous axr3-1 plants (data not shown), suggesting that the dominant axr3-1 mutation overrides the effect of overexpressing AtRALFL8 from an ectopic promoter. The spatial pattern of auxin response in 35S::AtRALFL8 plants was assessed using the auxin-responsive marker DR5rev::GFP; however, no overall changes were observed in the pattern of DR5::GFP expression (data not shown). Coexpression analysis using the Arabidopsis Coexpression Data Mining Tool (http://www.arabidopsis.leeds.ac.uk/act) revealed that the expression of AtRALFL8 is extremely highly correlated with pectin methylesterase genes (r < 0.99). Expression of three pectin methylesterase genes that are coexpressed with AtRALFL8 (At2g47040, At1g69940, and At3g62170) was analyzed in the 35S::AtRALFL8 overexpression line. No difference in expression was detected between the transgenic line and the wild-type plants (data not shown), suggesting that an increase in AtRALFL8 expression does not affect pectin methylesterases at the transcriptional level.
Figure 9.
Root hair phenotype of 35S::AtRALFL8 plants. Wild-type (A and C) and 35S::AtRALFL8 (B and D) seedlings were grown on one-half-strength MS media and photographed after 4 d of growth. Long, dense root hairs are clearly visible in the overexpression line. Bars = 2.5 mm (A and B) and 500 μm (C and D). [See online article for color version of this figure.]
Figure 10.
Reduced meristem size in 35S::AtRALFL8 roots. Cortical meristematic cells between the quiescent center (white arrows) and the transition zone (yellow arrow) were counted in wild-type (A) and 35S::AtRALFL8 (B) plants 10 d post germination. Red indicates cell walls as stained by propidium iodide. C, The meristem was significantly shorter in the 35S::AtRALFL8 plants (n = 5; **P < 0.01).
Figure 11.
Effect of auxin on root length in 35S::AtRALFL8 overexpression line. Wild-type and 35S::AtRALFL8 seedlings were grown on different media and the root length measured after 7 d (n = 8). IAA is a natural auxin, 2,4-d is a synthetic auxin, and PEO-IAA is an antiauxin agent.
DISCUSSION
The effect of two or more concurrent environmental stresses can be far more detrimental to plants than an individual stress, leading to severe agricultural losses (Craufurd and Peacock, 1993; Savin and Nicolas, 1996; Mittler, 2006). The ability of plants to recognize and respond to specific stress combinations may be extremely important, particularly when those individual stresses would elicit conflicting responses. However, the combination of abiotic and biotic stress factors on the plant whole-genome transcriptome is not well documented. Previous studies have aimed to identify genes important in multiple stress tolerance by comparing lists of genes induced by each stress individually (Seki et al., 2002; Swindell, 2006; Kilian et al., 2007; Kant et al., 2008). With our current knowledge of how stress responses interact, this type of research is no longer considered sufficient for understanding multiple stress responses (Mittler and Blumwald, 2010). Here, the transcriptome response of Arabidopsis following a combination of water deficit and nematode stress treatments displayed a distinct program of gene expression, supporting the theory that plant responses to stress are highly specialized and unique to the exact set of environmental conditions encountered (Mittler, 2006; Yasuda et al., 2008; Ton et al., 2009). Similar results have been observed in studies of two different abiotic stresses (Rizhsky et al., 2004). Furthermore, a recent study by Rasmussen et al. (2013) has also confirmed that the response of Arabidopsis to dual stress is very different to and cannot be predicted from the individual responses by comparing single or combined cold, heat, high-light, salt, and flagellin treatments.
Repression of Pathogen Defense Pathways in Response to Concurrent Biotic and Abiotic Stress
When water deficit and nematode stress were applied to plants in combination, the resulting gene expression profile resembled that of the plant under water deficit alone more closely than under nematode stress alone. The impact of water deficit may be more profound than nematode stress, as water deficit causes rapid physiological changes throughout the plant, giving widespread and cellular osmotic imbalance and turgor loss (Chaves et al., 2003). By contrast, plant-parasitic nematodes have evolved mechanisms by which to minimize damage to plant tissues and thus evade standard plant defense systems (Wubben et al., 2008). Therefore, when the stresses occur together, the plant response may prioritize the potentially more damaging abiotic stress. Antagonistic cross talk between biotic and abiotic signaling pathways may also play a role in creating the observed response to the stresses in combination. ABA, although primarily responsible for orchestrating plant response to abiotic stress, also has a prominent role in pathogen and disease resistance (Asselbergh et al., 2008b; Yasuda et al., 2008; Ton et al., 2009). There is evidence that ABA produced during abiotic stress suppresses defense pathways, including systemic acquired resistance (SAR), which is known to be induced by nematode invasion (Wubben et al., 2008; Yasuda et al., 2008). In addition, the down-regulation of genes with disease resistance annotations in this study, including many containing Leu-rich repeat domains, supports an active suppression of pathogen response pathways as a result of concurrent biotic and abiotic stresses.
AZI1 is a pathogen response gene that was down-regulated when water deficit and nematode stress were imposed in combination. AZI1 is a lipid transfer protein that is locally induced following pathogen infection and important for the establishment of systemic immunity priming in distal tissues. It acts to mobilize lipids as signaling molecules (Parker, 2009) in a process that is dependent on salicylic acid and the mobile metabolite azelaic acid (Jung et al., 2009). AZI1 also plays a role in abiotic stress response and its overexpression can lead to improved freezing tolerance in Arabidopsis (Xu et al., 2011). Analysis of overexpression and mutant azi1 lines in this study allowed further characterization of this gene. AZI1 overexpression conferred drought stress susceptibility. The chemical induction of SAR can negatively influence the production of ABA and the activation of ABA-responsive genes (Yasuda et al., 2008). Therefore, the observed drought susceptibility may have been due to an overactivation of the SAR priming system, which led to an inhibition of ABA-induced drought response genes. In addition, the expression of AZI1 was 6-fold higher in the ABA-insensitive mutant abi2-1 but no different from the wild type in abi4-1. Thus, ABA may negatively regulate AZI1-mediated systemic immunity priming downstream of the ABA signal transduction gene ABI2, but not dependent on the ABA-responsive AP2 transcription factor ABI4. Ethylene is likely to be a positive regulator of the systemic priming system, however, as shown by the increased expression of AZI1 in ctr1 mutants. The overexpression of AZI1 conferred no resistance to nematode infection. Although a local salicylic acid response is important for resistance to cyst nematodes (Wubben et al., 2008), these results suggest that azelaic acid-associated priming does not influence susceptibility to cyst nematode infection. When biotic and abiotic stresses are encountered simultaneously, AZI1 and the systemic immunity pathway may be specifically repressed in an ABA-responsive manner to focus resources on the abiotic stress response.
Signal Transduction and Regulation
Many of the interaction genes identified in this study were regulatory genes such as transcription factors and small signal molecules. The large number of MYB transcription factors identified among the interaction genes contributes to increasing evidence that these factors are central in controlling cross talk and specificity between different stress signaling pathways (Rizhsky et al., 2004; Mattana et al., 2005; Vannini et al., 2007; Abuqamar et al., 2009; Dubos et al., 2010; Atkinson and Urwin, 2012). MYBs regulate stress-related production of secondary metabolites in the phenylpropanoid pathway such as anthocyanins and lignin, as well as cell wall biosynthesis (Jin et al., 2000; Patzlaff et al., 2003; Wuyts et al., 2006; Dubos et al., 2010), and may make excellent candidates for the improvement of broad-spectrum stress tolerance (Jin et al., 2000; Vannini et al., 2004). In addition, the action of NAM and AP2 transcription factors in straddling both biotic and abiotic stress signaling pathways is further supported (Nakashima et al., 2007; Xu et al., 2011).
The expression of four small RALF-like signaling molecules was induced by the combination of water deficit and nematode stress (AtRALFL8, AtRALFL23, AtRALFL33, and AtRALFL34). These short signaling peptides show similarity to tobacco RALF (Pearce et al., 2001). They are growth regulators that can inhibit cell elongation in roots and pollen tubes (Wu et al., 2007; Matos et al., 2008; Srivastava et al., 2009; Covey et al., 2010). RALFLs are expressed in widely differing locations throughout the plant (Supplemental Fig. S4; Olsen et al., 2002; Hruz et al., 2008), although they have a high degree of sequence similarity. Their identification in this study suggests a mechanism whereby plants induce such signal molecules to reduce growth during severe stress.
Cell Wall Modification
Cell wall modifications can provide an increased physical barrier against potential pathogens, while improving tolerance to drought and oxidative stress and maintaining turgor during osmotic stress (Piro et al., 2003; Pelloux et al., 2007; An et al., 2008; Leucci et al., 2008). The observed up-regulation of cell wall modification proteins in response to combined nematode and dehydration stress may therefore be a general defensive mechanism activated to confer broad-spectrum tolerance against multiple stresses. AtRALFL8 may play a role in cell wall modification, as it is coexpressed extremely highly with genes encoding cell wall pectinesterases. These are crucial for cell wall remodeling in a variety of growth, reproductive, and defense processes, and their transcript level varies in response to biotic and abiotic stresses, including nematode infection (Pelloux et al., 2007; An et al., 2008). Different pectinesterases are specifically active at varying pH levels, and their function can be modulated by alkalinization (Feijó et al., 1999; Pelloux et al., 2007). AtRALFL8 is myristoylated at its N terminus and may therefore act as a signaling molecule that binds to the cell membrane and causes alkalinization of the cell wall, a similar mechanism to that proposed for SlPRALF in tomato (Boisson et al., 2003; Covey et al., 2010). Root surface pH varies along the root tip with distance from the meristem, being lowest at the zone of cell elongation (Staal et al., 2011). Therefore, the constitutive expression of an alkalinization factor could directly inhibit the expansion of cells in this zone. Alkalinization of the apoplast may have also been responsible for the abnormally long and numerous root hairs of the AtRALFL8 overexpression line, as changes in pH are crucial during rapid cell elongation processes, including the growth of pollen tubes, root epidermal cells, and root hair growth (Bibikova et al., 1998; Wu et al., 2007). Changes in cell wall modification proteins may also account for the hypersusceptibility of the AtRALFL8 overexpression line to drought stress as well as parasitism by the nematode H. schachtii.
Root alkalinization by RALF-like signal molecules may be a point of interaction between the hormones auxin and ethylene during multiple stress responses. There is evidence that auxin acts in concert with ethylene to mediate the alkalinization of the root surface during growth inhibition (Staal et al., 2011). This, combined with the shortened meristem in the AtRALFL8 overexpression line and the increase in AtRALFL8 expression in the ethylene signaling mutant ein3-1, suggests that AtRALFL8 may be involved in modulating events in the auxin response pathway (Knox et al., 2003; Moubayidin et al., 2010). AtRALFL8 induction during simultaneous biotic and abiotic stress is an illustration of the complex mechanisms by which plants act through different hormone signaling pathways to produce cellular changes that protect them from stress.
Cellular and Metabolic Responses to Multiple Stresses
The production and homeostasis of amino acids is intrinsic to many aspects of both biotic and abiotic stress responses. Pro and Ile accumulate as osmoprotectants during drought and osmotic stress and can act as scavengers of reactive oxygen species, regulators of pH, or substrates for the synthesis of stress-related proteins (Nambara et al., 1998; Joshi et al., 2010). Ile also has the crucial role of combining with jasmonic acid to make the active defense and pathogen response hormone JA-Ile (Nambara et al., 1998; Joshi and Jander, 2009). The up-regulation of AtMGL, a Met homeostasis gene, in joint-stressed plants in this study suggested that a shift in the metabolism of this amino acid may be important in the response to concurrent biotic and abiotic stresses (Rébeillé et al., 2006; Goyer et al., 2007; Joshi and Jander, 2009). Most cellular Met is converted to S-adenosyl-Met for use in essential plant processes such as DNA replication and methylation and the synthesis of ethylene, cell walls, chlorophyll, and secondary metabolites. However, AtMGL catabolizes Met in an alternative pathway, leading to the synthesis of Ile. Previously, AtMGL has been induced in response to P. syringae and Phytophthora parasitica (Genevestigator) and in response to a combination of heat and drought stress (Rizhsky et al., 2004). Drought-stressed atmgl plants accumulate less Ile than wild-type plants (Joshi and Jander, 2009). Altering amino acid homeostasis by the channeling of Met into the Ile pathway may be an adaptive strategy, providing protection from severe or multiple stresses, albeit with a growth penalty. The increase of AtMGL transcript in the ABA-resistant mutant abi2-1 suggests that this process is negatively regulated by ABA.
The analysis of atmgl mutants revealed a phenotype that was little different from wild-type plants under control or drought-stressed conditions, a result also described by Joshi and Jander (2009). These observations are likely to result from redundancy with threonine deaminase, an alternative Ile biosynthesis mechanism (Joshi et al., 2010). The overexpression of AtMGL, by contrast, severely affected the growth of aerial parts of the plant under normal conditions. As 80% of Met is normally directed into S-adenosyl-Met, the overactivity of AtMGL would convert excess Met into the alternative pathway, depleting the pool used as methyl donors for essential plant processes and resulting in the reduced growth phenotype. 35S::AtMGL plants were less susceptible to infection with the nematode H. schachtii. This may be attributed to a number of factors. Plant-parasitic nematodes are net consumers that depend on amino acids from their hosts. A high sink strength in the feeding cells of H. schachtii leads to an enriched Met concentration as well as an increase in transcription of Met scavenging genes (Szakasits et al., 2009; Hofmann et al., 2010). The depletion of available Met by the overexpression of AtMGL may therefore inhibit nematode protein synthesis, preventing their development. In addition, the increased production of Ile due to AtMGL overexpression may boost levels of JA-Ile, creating a heightened defense mechanism that could respond more effectively to the nematode invasion.
CONCLUSION
The study of simultaneous biotic and abiotic stresses highlights the complex mechanisms by which plants tailor their response to precise environmental conditions. A new pattern of transcription was observed that differed from that of either stress individually, which, in addition, supports the hypothesis that the response to a potentially more damaging abiotic stress can override the response to biotic stress. The importance of various processes such as cell wall remodeling, Met homeostasis, and immune system priming in the response to multiple stresses has been demonstrated through the functional analysis of individual genes. While the results reported here underline the importance of studying plant stress factors in combination, to fully understand how plants respond to multiple stresses in field conditions, further work will be required.
MATERIALS AND METHODS
Plant and Nematode Material, Growth Conditions, and Stress Treatments for Microarray Experiment
Arabidopsis (Arabidopsis thaliana Col-0) seeds were sterilized by soaking in 95% (v/v) ethanol for 2 min and 10% (v/v) bleach for 5 min, followed by five washes in sterile distilled water. Seeds were stratified at 4°C for 48 h, then germinated and grown in square petri dishes on solid media containing one-half-strength Murashige and Skoog (MS) salts with vitamins (Duchefa), 1% (w/v) Suc (Sigma), and 1% (w/v) plant agar (Duchefa). Four seeds were sown per 10-cm plate, and plates were held at an angle of approximately 70° in a growth cabinet (Sanyo MLR) at 20°C with a light intensity of 140 μmol m–2 s–1 and under 16-h/8-h light/dark cycles. Heterodera schachtii cysts were extracted from soil, then sterilized and hatched as described in Urwin et al. (1997). Overnight hatches of second-stage juveniles (J2s) were sterilized in 0.5 mg ml–1 hexadecyltrimethylammonium bromide and 0.1% (v/v) chlorhexidine digluconate for 30 min and washed twice in sterile tap water. After 18 d of growth, plants were divided into four treatment groups, control, water deficit, nematode infection, and combined stress. Plants from the nematode-infected and combined stress treatment groups were each challenged with 175 H. schachtii J2s. All other plants were mock inoculated with sterile tap water. Ten days later, dehydration treatment was imposed on the water deficit and combined stress treatment groups. Control and nematode-infected plants were removed from the agar and then immediately replaced and returned to the growth cabinet for 45 min. Dehydration-treated plants were removed from the medium and placed in a clean flow of air for 15 min, as described by Kilian et al. (2007), during which time plants lost 10% of their fresh weight. Plants were then placed back on the agar and returned to growth cabinets for 30 min before harvesting tissue from all four treatment groups. Aerial tissues were separated from roots, and both were frozen in liquid nitrogen. Forty plants were used per treatment group. The entire experiment was carried out twice more on different occasions, giving three biological replicates.
RNA Isolation, Complementary DNA Synthesis, and Affymetrix GeneChip Analysis
Plant tissue from each complete biological replicate was divided into five pools of eight plants per treatment and RNA isolated from each pool separately using the RNeasy Plant Mini Kit (Qiagen). Equal amounts of RNA were then combined into one sample per treatment group and replicate. A 2100 Expert Bioanalyzer (Agilent Technologies) was used to confirm the quality of all RNA samples before microarray analysis. Twenty-four arrays were used in total, representing four treatments, two tissue types, and three replicates. Hybridization of biotin-labeled RNA to Affymetrix ATH1 GeneChip arrays and array scanning were performed by the Nottingham Arabidopsis Stock Centre transcriptomics service (Craigon et al., 2004) following the standard Affymetrix protocol. Normalization and analysis of differential expression was carried out using GeneSpring GX10 (Agilent Technologies). Baseline preprocessing, normalization, and summarization were carried out using the Robust Multiarray Average summarization algorithm, as described by Irizarry et al. (2003). Genes were considered differentially regulated if their normalized expression value was significantly different from the control (P < 0.05). One-way ANOVA with Benjamini Hochberg multiple testing correction (false discovery rate of 0.05) was used to identify genes differentially regulated between treatment groups. Further Student’s t test analyses were used to detect ‘interaction’ genes regulated by joint stress compared with individual water deficit stress. From these lists, several genes of interest were selected for further study. Data has been deposited in the public repository NASCArrays and is accessible at http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl with the reference number NASCARRAYS-489.
The expression changes in a subset of 12 genes were verified using qRT-PCR. Complementary DNA was generated from 250 ng total RNA per sample using BioScript MMLV reverse transcriptase (Bioline) and qRT-PCR carried out on the resulting complementary DNA using Brilliant II SYBR Green Master Mix (Agilent Technologies). qRT-PCR conditions consisted of 93°C for 3 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Fluorescence data were collected at the end of the annealing phase. Differences in transcript level were determined using MxPro software (Stratagene), with ACTIN2 as a normalizing gene. All primer pairs had an amplification efficiency of 90% to 110%, and R2 correlation coefficients for standard curves ranged between 0.94 and 1.00. See Supplemental Table S5 for details of primers used in qRT-PCR analysis.
Generating Overexpression Lines
Coding regions of the genes of interest were cloned into the plant transformation binary vector pBI121 from which the GUS gene had been removed and replaced by a KpnI restriction site. The gene coding sequences were placed under the control of a Cauliflower mosaic virus 35S promoter with the neomycin phosphotransferaseII (nptII) gene for kanamycin resistance as the selectable marker. The constructs were introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform Arabidopsis plants using the floral dip method. Transgenic lines were validated by PCR and the increase in target gene expression quantified using qRT-PCR as described earlier. Primer sequences are provided in Supplemental Table S5.
Transfer DNA Insertion Mutants
Insertion mutants were obtained for AtMGL (SALK_074592C) and AZI1 (SALK_085727C) from the Nottingham Arabidopsis Stock Centre. Homozygosity of the lines was confirmed by PCR, using primers annealing either to the left border region of the transfer DNA and a 3′ flanking sequence to amplify the insertion or to 5′ and 3′ flanking regions to amplify the wild-type allele.
Phenotypic Analysis of Transgenic Plants
Seeds of each transgenic overexpression and mutant Arabidopsis line, together with wild-type Col-0 plants, were sown onto trays of compost (Sinclair Potting and Growing Medium, East Riding Horticulture) with a depth of 5 cm and grown in a growth chamber (Sanyo) at 20°C under 16-h/8-h light/dark cycles (200 μmol m–2 s–1, 60% humidity). Approximately 14 d after sowing, seedlings were removed from trays and repotted into 9-cm pots for phenotypic analysis. For overexpression lines, analysis was carried out on the most highly expressing line per construct and two additional independent transgenic lines with lower expression. Phenotypic measurements were recorded at various stages throughout plant development under control conditions, including rosette diameter and leaf number 16 d after sowing, timing of primary inflorescence emergence, height of primary inflorescence 35 d after sowing, final height of primary inflorescence, silique number on primary inflorescence 40 d after sowing, and seed number per silique (n = 16). Dry weight of aerial plant material was also determined 35 d after sowing (n = 4). For root system analysis, plants were grown on one-half-strength MS medium as described previously. Fourteen days after sowing, the plates were scanned using a Hewlett Packard ScanJet 5370C, and root parameters were measured from the digital images using Image-Pro Plus software version 7.0 (MediaCybernetics; n = 9). Phenotypic differences between plant genotypes were analyzed using ANOVA or the Kruskal-Wallis H test.
Stress Treatments of Transgenic Plants
For drought tolerance assays, plants were grown in 25- × 40- × 5-cm trays on compost mixed with sand and loam soil at a ratio of 2:1:1 to facilitate drainage. Plants were transplanted into these trays 2 weeks after germinating. Twelve transgenic plants were alternated with 12 wild type per tray. Trays were then watered to field capacity for 1 week, and then irrigation was ceased until the soil moisture level dropped to 3% to 4% (2 weeks on average). The plants were then rewatered to field capacity for 1 week and scored for survival. For nematode susceptibility assays, plants were grown in sterile conditions on one-half-strength MS medium as described previously. Eighteen days after sowing, each plant was infected with 100 J2 H. schachtii nematodes. Nematodes were allowed to develop for 10 d, the root systems stained in acid fuchsin (Fuller et al., 2007), and the number of established third- and fourth-stage juvenile nematodes (J3 and J4) per plant counted.
Analysis of Gene Expression in Hormone Signaling Mutants
The Arabidopsis hormone signaling mutants abi2-1 (ecotype Landsberg erecta), abi4-1 (Col-0), ein3-1 (Col-0), NahG (Col-0), and ctr1 (Col-0), together with the corresponding wild-type ecotypes, were germinated on one-half-strength MS medium as described previously. ABA (3 µm) was included in the media of abi2-1 and abi4-1 plants, and the media for ein3-1 plants contained 1-aminocyclopropane-1-carboxylic-acid (1 µm). Fourteen-day-old seedlings were planted out into compost/sand/loam. After a further 14 d, leaf tissue was harvested from three pools of four plants for each genotype. RNA was isolated from each pool and analyzed by qRT-PCR as detailed previously, using primers for each of the genes of interest. Differences in gene expression were analyzed using ANOVA or the Kruskal-Wallis H test.
Analysis of 35S::AtRALFL8 Root Phenotype
To analyze auxin responses, plants were grown on unsupplemented Arabidopsis thaliana salts media (Knox et al., 2003) or that containing 0.1 μm IAA, 0.1 μm 2,4-d, or 1 μm PEO-IAA. The plates were held in an upright position and root length measured 7 d post germination. Expression levels of the pectin methylesterase genes At2g47040, At1g69940, and At3g62170 were analyzed in the leaves of 14-d-old 35S::AtRALFL8 plants by qRT-PCR. Primer sequences are provided in Supplemental Table S5. Root tips were stained with propidium iodide, and confocal microscopy was carried out using an LSM Meta 510 microscope (Zeiss). The spatial pattern of auxin response was visualized by crossing 35S::RALFL8 plants with those containing the DR5rev::GFP auxin responsive promoter (Friml et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_104837/At1g61563 (AtRALFL8); NM_105141/At1g64660 (AtMGL); and NM_117317/At4g12470 (AZI1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Functional Gene Ontology categories of interaction genes.
Supplemental Figure S2. Figure compiled from Genevestigator, showing the expression pattern of candidate genes in response to various hormone treatments.
Supplemental Figure S3. Comparative phenotypes of additional AtRALFL8, AtMGL, and AZI1 overexpression lines under control conditions.
Supplemental Figure S4. Figure compiled from Genevestigator, showing the expression pattern of RALFL genes in different plant tissues.
Supplemental Table S1. Lists of genes differentially regulated by each individual stress treatment.
Supplemental Table S2. Expression data for genes used in qRT-PCR validation of microarray experiment.
Supplemental Table S3. Interaction genes identified by comparison of joint stress with individual stress treatment.
Supplemental Table S4. Lists of interaction genes in highly represented functional groups.
Supplemental Table S5. Sequences of all primers used in the study.
Acknowledgments
We thank Hanma Zhang and Stefan Kepinski (University of Leeds) for providing the Arabidopsis hormone signaling lines used in this study.
Glossary
- ABA
abscisic acid
- Col-0
ecotype Columbia
- SAR
systemic acquired resistance
- MS
Murashige and Skoog
- IAA
indole-3-acetic acid
- PEO
α-(phenylethyl-2-one)
- 2,4-d
2,4-dichlorophenoxyacetic acid
- qRT
quantitative reverse transcription
References
- Abuqamar S, Luo HL, Laluk K, Mickelbart MV, Mengiste T. (2009) Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J 58: 347–360 [DOI] [PubMed] [Google Scholar]
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK. (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228: 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Achuo AE, Höfte M, Van Gijsegem F. (2008a) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol Plant Pathol 9: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Höfte M. (2008b) Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact 21: 709–719 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Audebert A, Coyne DL, Dingkuhn M, Plowright RA. (2000) The influence of cyst nematodes (Heterodera sacchari) and drought on water relations and growth of upland rice in Cote d’Ivoire. Plant Soil 220: 235–242 [Google Scholar]
- Audenaert K, De Meyer GB, Höfte MM. (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Jacob T, Dahse I, Gilroy S. (1998) Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125: 2925–2934 [DOI] [PubMed] [Google Scholar]
- Bird AF. (1974) Plant response to root-knot nematode. Annu Rev Phytopathol 12: 69–85 [Google Scholar]
- Boisson B, Giglione C, Meinnel T. (2003) Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J Biol Chem 278: 43418–43429 [DOI] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS. (2003) Understanding plant responses to drought - from genes to the whole plant. Funct Plant Biol 30: 239–264 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Zhu T. (2006) Microarray analysis of the transcriptome as a stepping stone towards understanding biological systems: practical considerations and perspectives. Plant J 45: 630–650 [DOI] [PubMed] [Google Scholar]
- Covey PA, Subbaiah CC, Parsons RL, Pearce G, Lay FT, Anderson MA, Ryan CA, Bedinger PA. (2010) A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol 153: 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. (2004) NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res 32: D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craufurd PQ, Peacock JM. (1993) Effect of heat and drought stress on sorghum (Sorghum bicolor). 2. Grain-yield. Exp Agric 29: 77–86 [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Fasan T, Haverkort AJ. (1991) The influence of cyst nematodes and drought on potato growth. 1. Effects on plant-growth under semi-controlled conditions. Neth J Plant Pathol 97: 151–161 [Google Scholar]
- Feijó JA, Sainhas J, Hackett GR, Kunkel JG, Hepler PK. (1999) Growing pollen tubes possess a constitutive alkaline band in the clear zone and a growth-dependent acidic tip. J Cell Biol 144: 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Fuller VL, Lilley CJ, Atkinson HJ, Urwin PE. (2007) Differential gene expression in Arabidopsis following infection by plant-parasitic nematodes Meloidogyne incognita and Heterodera schachtii. Mol Plant Pathol 8: 595–609 [DOI] [PubMed] [Google Scholar]
- Goyer A, Collakova E, Shachar-Hill Y, Hanson AD. (2007) Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. Plant Cell Physiol 48: 232–242 [DOI] [PubMed] [Google Scholar]
- Haverkort AJ, Fasan T, Vandewaart M. (1991) The influence of cyst nematodes and drought on potato growth. 2 Effects on plants water relations under semi-controlled conditions. Neth J Plant Pathol 97: 162–170 [Google Scholar]
- Hofmann J, El Ashry AelN, Anwar S, Erban A, Kopka J, Grundler F. (2010) Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. Plant J 62: 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jin HL, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Jander G. (2009) Arabidopsis methionine γ-lyase is regulated according to isoleucine biosynthesis needs but plays a subordinate role to threonine deaminase. Plant Physiol 151: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Joung JG, Fei ZJ, Jander G. (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39: 933–947 [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kant P, Gordon M, Kant S, Zolla G, Davydov O, Heimer YM, Chalifa-Caspi V, Shaked R, Barak S. (2008) Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ 31: 697–714 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777 [DOI] [PubMed] [Google Scholar]
- Kusajima M, Yasuda M, Kawashima A, Nojiri H, Yamane H, Nakajima M, Akutsu K, Nakashita H. (2010) Suppressive effect of abscisic acid on systemic acquired resistance in tobacco plants. J Gen Plant Pathol 76: 161–167 [Google Scholar]
- Leucci MR, Lenucci MS, Piro G, Dalessandro G. (2008) Water stress and cell wall polysaccharides in the apical root zone of wheat cultivars varying in drought tolerance. J Plant Physiol 165: 1168–1180 [DOI] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS. (2008) A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett 582: 3343–3347 [DOI] [PubMed] [Google Scholar]
- Mattana M, Biazzi E, Consonni R, Locatelli F, Vannini C, Provera S, Coraggio I. (2005) Overexpression of Osmyb4 enhances compatible solute accumulation and increases stress tolerance of Arabidopsis thaliana. Physiol Plant 125: 212–223 [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E. (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30: 461–469 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. (2007) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct Integr Genomics 7: 181–191 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LSP, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51: 617–630 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S. (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Mundy J, Skriver K. (2002) Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biol 2: 441–451 [PubMed] [Google Scholar]
- Parker JE. (2009) The quest for long-distance signals in plant systemic immunity. Sci Signal 2: pe31. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, et al. (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36: 743–754 [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA., Jr (2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Piro G, Leucci MR, Waldron K, Dalessandro G. (2003) Exposure to water stress causes changes in the biosynthesis of cell wall polysaccharides in roots of wheat cultivars varying in drought tolerance. Plant Sci 165: 559–569 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S. (2006) Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103: 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang HJ, Mittler R. (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang HJ, Shuman J, Shulaev V, Davletova S, Mittler R. (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savin R, Nicolas ME. (1996) Effects of short periods of drought and high temperature on grain growth and starch accumulation of two malting barley cultivars. Aust J Plant Physiol 23: 201–210 [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Smit AL, Vamerali T. (1998) The influence of potato cyst nematodes (Globodera pallida) and drought on rooting dynamics of potato (Solanum tuberosum L.). Eur J Agron 9: 137–146 [Google Scholar]
- Srivastava R, Liu J-X, Guo H, Yin Y, Howell SH. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Staal M, De Cnodder T, Simon D, Vandenbussche F, Van der Straeten D, Verbelen JP, Elzenga T, Vissenberg K. (2011) Apoplastic alkalinization is instrumental for the inhibition of cell elongation in the Arabidopsis root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid. Plant Physiol 155: 2049–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. (2006) The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics 174: 1811–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FMW, Bohlmann H. (2009) The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J 57: 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B. (2004) β-Amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38: 119–130 [DOI] [PubMed] [Google Scholar]
- Urwin PE, Lilley CJ, McPherson MJ, Atkinson HJ. (1997) Resistance to both cyst and root-knot nematodes conferred by transgenic Arabidopsis expressing a modified plant cystatin. Plant J 12: 455–461 [DOI] [PubMed] [Google Scholar]
- Vannini C, Campa M, Iriti M, Genga A, Faoro F, Carravieri S, Rotino GL, Rossoni M, Spinardi A, Bracale M. (2007) Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci 173: 231–239 [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37: 115–127 [DOI] [PubMed] [Google Scholar]
- Voelckel C, Baldwin IT. (2004) Herbivore-induced plant vaccination. Part II. Array-studies reveal the transience of herbivore-specific transcriptional imprints and a distinct imprint from stress combinations. Plant J 38: 650–663 [DOI] [PubMed] [Google Scholar]
- Wu J, Kurten EL, Monshausen G, Hummel GM, Gilroy S, Baldwin IT. (2007) NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J 52: 877–890 [DOI] [PubMed] [Google Scholar]
- Wubben MJE, Jin J, Baum TJ. (2008) Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol Plant Microbe Interact 21: 424–432 [DOI] [PubMed] [Google Scholar]
- Wuyts N, Lognay G, Swennen R, De Waele D. (2006) Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J Exp Bot 57: 2825–2835 [DOI] [PubMed] [Google Scholar]
- Xu ZY, Zhang X, Schläppi M, Xu ZQ. (2011) Cold-inducible expression of AZI1 and its function in improvement of freezing tolerance of Arabidopsis thaliana and Saccharomyces cerevisiae. J Plant Physiol 168: 1576–1587 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Liu Y, Yang T, Zhang L, Xu S, Xue L, An L. (2006) Diverse signals converge at MAPK cascades in plant. Plant Physiol Biochem 44: 274–283 [DOI] [PubMed] [Google Scholar]