Analysis of symbiotic nodulation indicates low auxin activity and auxin hypersensitivity during nodule initiation, and regulatory feedback with auxin and cytokinin during nodule development.
Abstract
Symbiotic root nodules in leguminous plants result from interaction between the plant and nitrogen-fixing rhizobia bacteria. There are two major types of legume nodules, determinate and indeterminate. Determinate nodules do not have a persistent meristem, while indeterminate nodules have a persistent meristem. Auxin is thought to play a role in the development of both these types of nodules. However, inhibition of rootward auxin transport at the site of nodule initiation is crucial for the development of indeterminate nodules but not determinate nodules. Using the synthetic auxin-responsive DR5 promoter in soybean (Glycine max), we show that there is relatively low auxin activity during determinate nodule initiation and that it is restricted to the nodule periphery subsequently during development. To examine if and what role auxin plays in determinate nodule development, we generated soybean composite plants with altered sensitivity to auxin. We overexpressed microRNA393 to silence the auxin receptor gene family, and these roots were hyposensitive to auxin. These roots nodulated normally, suggesting that only minimal/reduced auxin signaling is required for determinate nodule development. We overexpressed microRNA160 to silence a set of repressor auxin response factor transcription factors, and these roots were hypersensitive to auxin. These roots were not impaired in epidermal responses to rhizobia but had significantly reduced nodule primordium formation, suggesting that auxin hypersensitivity inhibits nodule development. These roots were also hyposensitive to cytokinin and had attenuated expression of key nodulation-associated transcription factors known to be regulated by cytokinin. We propose a regulatory feedback loop involving auxin and cytokinin during nodulation.
Auxin plays a crucial role in the initiation and development of a number of plant organs (for review, see Vanneste and Friml, 2009). Auxin is perceived by a set of TRANSPORT INHIBITOR RESPONSE1(TIR1)-like F-box proteins that form part of an SCF-type ubiquitin ligase complex (Ruegger et al., 1998; Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Molecular and biochemical characterization of TIR1 in Arabidopsis (Arabidopsis thaliana) revealed that it directly binds auxin and aids in the degradation of auxin/indole-3-acetic acid (Aux/IAA) repressor proteins (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Aux/IAAs are short-lived nuclear proteins encoded in general by primary/early auxin response genes (Ulmasov et al., 1997). Aux/IAA proteins form protein complexes with auxin response factor (ARF) transcriptional activators and repress the expression of auxin-responsive genes (Reed, 2001; Tiwari et al., 2001, 2004). Dissociation of ARF-Aux/IAA protein complexes by TIR1 in an auxin-dependent manner results in auxin-responsive gene expression (Gray et al., 1999, 2001). The ARF gene family consists of both transcriptional activators and repressors that bind with specificity (at auxin response elements) to promoters of primary/early auxin response genes. Repressor ARFs are capable of repressing gene expression with or without auxin (Ulmasov et al., 1999; Tiwari et al., 2003). It has been hypothesized recently that they are more likely to act independently of Aux/IAA proteins (Guilfoyle and Hagen, 2012). Presumably, repressor ARFs act independently of TIR1 as well, and silencing their expression or degradation of repressor ARF proteins would result in auxin-responsive gene expression. There is clear genetic evidence for the role of specific auxin receptors, Aux/IAAs and ARFs (for review, see Vanneste and Friml, 2009), as well as complex interaction networks among them (Vernoux et al., 2011) in governing plant development.
The role of auxin in root nodule development in leguminous plants is not completely understood. Legume nodules are specialized root lateral organs that result from symbiotic interaction between the plant and a compatible group of nitrogen-fixing soil bacteria collectively termed rhizobia (for review, see Murray, 2011). Symbiosis begins with a chemical exchange between partners, which is the first step that determines host specificity. Legume roots release specific flavonoid compounds that are recognized by compatible rhizobia species; in turn, they synthesize and release lipochitooligosaccharide “Nod” signals. The perception of Nod signals and the colonization of root hairs by rhizobia cells lead to root hair deformation and curling to entrap rhizobia. Subsequent invagination of the root hair cells forms specialized structures termed infection threads. Simultaneously, cell divisions occur in cortex and pericycle cell layers in preparation to form nodule primordia. Infection threads transport rhizobia into the developing nodules (formed by cortex cell divisions), where they differentiate into bacteroids and fix nitrogen. Ultimately, the two cell division foci merge and symplastic and vascular connections are formed, enabling the transport of nutrients to and from the mature nodules. There are two major types of nodules formed in legume roots: indeterminate and determinate (for review, see Hirsch, 1992; Sprent, 2007). Indeterminate nodules are oblong and characterized by the presence of a persistent meristem. Examples of plants that form indeterminate nodules include temperate legumes such as pea (Pisum sativum), Medicago truncatula, and white clover (Trifolium repens). In contrast, determinate nodules are spherical and lack a persistent meristem. There is no sustained cell division during determinate nodule development; cell expansion rather than cell division results in nodule growth. Examples of plants producing determinate nodules include tropical legumes such as soybean (Glycine max), common bean (Phaseolus vulgaris), and Lotus japonicus. Additionally, indeterminate nodules arise from inner cortical cell layers whereas determinate nodules arise from outer/mid cortical cell layers. A number of different plant hormones, including auxin and cytokinin, are hypothesized or shown to play a role in the initiation and development of both these types of nodules (for review see Hirsch and Fang, 1994; Ferguson and Mathesius, 2003; Ding and Oldroyd, 2009; Suzaki et al., 2013).
The roles of auxin in nodule initiation and development were hypothesized initially based on the effect of exogenous application of auxin or auxin transport inhibitors (Allen et al., 1953; Hirsch et al., 1989), subsequently based on the expression of reporter gene constructs (e.g. GH3:GUS; Mathesius et al., 1998; Boot et al., 1999; Pacios-Bras et al., 2003; Takanashi et al., 2011), and more recently demonstrated through the modulation of auxin transport using reverse-genetic tools (Subramanian et al., 2006; Wasson et al., 2006; Zhang et al., 2009). It was discovered very early that auxin transport inhibitors induce nodule-like structures in legume roots (Allen et al., 1953), including the induction of certain early nodulin genes (Hirsch et al., 1989), suggesting that auxin might initiate nodule development. Indeed, the expression of an auxin-responsive marker (GH3:GUS) was observed during nodule development in both determinate (L. japonicus [Pacios-Bras et al., 2003; Takanashi et al., 2011]) and indeterminate (white clover [Mathesius et al., 1998]) nodule-producing legumes. However, the expression pattern was clearly distinct in the roots and other nodule tissues between the two types of nodules. For example, GH3:GUS expression was reduced in the vasculature rootward to the inoculation site but increased in cortex cells around the site of rhizobial inoculation in white clover, an indeterminate nodule-forming legume (Mathesius et al., 1998). Subsequently, GH3:GUS expression was detected in the dividing primordium cells but moved to the peripheral cells (presumably those forming the nodule vasculature) during differentiation. In contrast, GH3:GUS expression in the root vasculature was not reduced in response to rhizobial inoculation in L. japonicus, a determinate nodule-forming legume. Nevertheless, GUS expression was detected initially in the dividing cells, later in peripheral cells in nodule primordia, and in the nodule vasculature in mature nodules (Pacios-Bras et al., 2003; Takanashi et al., 2011). The GH3:GUS expression pattern in the root vasculature and measurement of auxin transport suggested that inhibition of rootward auxin transport might precede the initiation of indeterminate nodules but not determinate nodules (Mathesius et al., 1998; Boot et al., 1999). We and others obtained clear genetic evidence for the role of auxin transport inhibition in nodule development by silencing the biosynthesis of flavonoids, a group of phenolic compounds that act as endogenous auxin transport inhibitors (Subramanian et al., 2006; Wasson et al., 2006; Zhang et al., 2009). These data conclusively demonstrated that inhibition of auxin transport in the roots at the site of nodule initiation is crucial for the development of indeterminate nodules but not for determinate nodules. Other studies showed that components of the auxin transport machinery (PIN-FORMED2 and LIKE AUX1) might play a role in the development of indeterminate nodules as well (de Billy et al., 2001; Huo et al., 2006). We concluded that the requirements of auxin distribution during primordium initiation/development are different between these two types of nodules (Subramanian et al., 2007).
Another line of evidence pointing toward a role for auxin in nodule development is the coordinate expression of microRNAs (miRNAs) regulating auxin signaling during this process in soybean (Subramanian et al., 2008) and other legumes (for review, see Simon et al., 2009; Bazin et al., 2012). miRNAs are small noncoding RNAs that regulate the expression of protein-coding genes primarily through posttranscriptional mechanisms and are known to play a vital role in plant development (for review, see Chen, 2009). The expression of several auxin signaling components is regulated by miRNAs. For example, miR393 regulates members of the auxin receptor TIR1/AFB gene family (Parry et al., 2009; Vidal et al., 2010; Si-Ammour et al., 2011), miR160 regulates members of the ARF10/ARF16/ARF17 repressor ARF family (Mallory et al., 2005; Wang et al., 2005), miR167 regulates the ARF8 family (Wu et al., 2006), and miR390 regulates the ARF3 family (Fahlgren et al., 2006; Marin et al., 2010). In soybean roots, the expression of miR393, which regulates the TIR1/AFB auxin receptor gene family, was transiently up-regulated, whereas miR160, which regulates a group of repressor ARFs, was down-regulated upon Bradyrhizobium japonicum inoculation. More recently, detailed analysis of the specific and sensitive auxin-inducible marker, DR5:GFP-NLS, in L. japonicus showed that auxin induction during nodule primordium development might occur downstream of cytokinin perception (Suzaki et al., 2012). While all of these data strongly suggested that auxin signaling is regulated during nodule development, it was not known if and what role auxin plays during the development of symbiotic nodules, specifically determinate nodules. We sought to address this long-standing question by (1) monitoring the activity of the auxin-specific synthetic promoter DR5 during soybean nodule development and (2) examining nodule development in soybean roots with altered auxin sensitivity.
RESULTS
Auxin-Inducible Gene Expression during Root Nodule Initiation in Soybean
We examined auxin-inducible gene expression during nodule development in soybean transgenic composite plants (Collier et al., 2005) using a marker gene construct where the synthetic auxin-responsive DR5 promoter (Ulmasov et al., 1997) drove the expression of the fluorescent protein tandem dimer Tomato (tdT; Campbell et al., 2002). To distinguish autofluorescence observed on the soybean root surface when using confocal microscopy, we examined DR5:tdT together with GFP driven by the constitutive superubiquitin promoter. The yellowish red tdT + GFP signal can be clearly distinguished from the bright red autofluorescence on the root surface. DR5:tdT expression was detected in the root meristem and columella cells of the root cap (Fig. 1A) and lateral root (LR) primordia (Fig. 1B). Treatment of the roots with exogenous auxin (1 μm 2,4-dichlorophenoxyacetic acid [2,4-D] for 24 h) resulted in an increased tdT expression domain at the root tips accompanied by a noticeable “swelling” of the root tips (Supplemental Fig. S1, A and B). The expression pattern and auxin responsiveness were consistent with what has been reported in Arabidopsis and other species (Ni et al., 2001; Ilina et al., 2012), indicating that the construct was suitable for monitoring auxin-responsive gene expression in soybean composite plant roots.
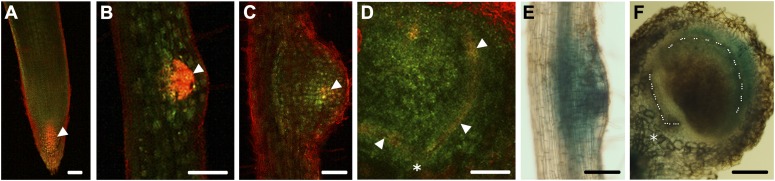
Figure 1.
Expression of the auxin-inducible marker gene constructs, DR5:tdT and DR5:GUS, in soybean roots. A to D, Expression of DR5:tdT and sUbi:GFP in root tips (A), lateral root primordia (B), emerging nodules (C), and mature nodules (D). A to C are confocal optical sections of whole mounts, and D is a confocal image of a hand-sectioned mature nodule. Cells expressing both DR5:tdT and sUbi:GFP appear yellow/reddish yellow (arrowheads), and root surface autofluorescence appears bright red. E and F, Expression of DR5:GUS in nodule primordia (E) and mature nodules (F). The image in F is that of a hand-sectioned mature nodule. Asterisks in D and F indicate the point of contact between the root and the nodule. Dotted lines in F mark the nodule vasculature. Bars = 100 μm (A–D) and 200 μm (E and F).
We inoculated these plants with B. japonicum cells and examined DR5:tdT expression at 3, 7, and 14 d post inoculation (dpi). Surprisingly, we observed a much lower expression of DR5:tdT in the majority of nodule primordia and emerging nodules (Fig. 1C). DR5:tdT expression was localized primarily to the nodule “apex.” The level of DR5:tdT expression in nodule primordia was so low that it could not be detected reliably by regular fluorescence microscopy (Supplemental Fig. S1E) but only by confocal microscopy. In contrast, DR5:tdT expression in the root tips and LR primordia was readily detectable by regular fluorescence microscopy (Supplemental Fig. S1, C and D). In transverse sections of mature nodules, DR5:tdT expression was detected closer to the nodule periphery primarily along the vasculature and was clearly absent from the central infection zone (Fig. 1D).
We also examined DR5:GUS expression during nodule development, due to the relatively longer half-life and higher sensitivity of GUS. Consistent with results from confocal microscopy, we detected the expression of DR5:GUS in the majority of nodule primordia (Fig. 1E), with intense staining closer to the nodule apex. In transverse sections of mature nodules, DR5:GUS expression was limited to the nodule periphery and not detected in the infection zone (Fig. 1F). Together, these observations suggested that (1) there is auxin activity during nodule initiation and development in soybean, but the level of auxin activity in nodule primordia is much lower compared with that in LR primordia; (2) there is no or minimal auxin activity in the nodule infection zone during subsequent nodule development; and (3) the majority of the auxin activity is localized around the vasculature in mature nodules.
Manipulating Auxin Sensitivity Using miR393 and miR160
To examine the role of auxin in symbiotic nodule initiation in soybean, we sought to manipulate auxin signaling in transgenic composite plant roots by overexpressing miRNAs against the auxin receptor and a set of repressor ARF genes. We reasoned that this would help overcome functional redundancy usually associated with components of auxin signaling in classical genetic mutants (see “Discussion”). Second, we have previously shown that the levels of these miRNAs are influenced by B. japonicum inoculation in soybean (Subramanian et al., 2008). We overexpressed miR393 to silence the TIR1/AFB auxin receptor family and to obtain auxin-hyposensitive plants. Overexpression of miR393 precursor using the cassava vein mosaic virus (CsVMV) CVP2 promoter (a kind gift from Dr. Claude Fauquet) resulted in increased levels of mature miR393 (Supplemental Fig. S2A) and a corresponding decrease in two of the three validated targets of this miRNA (Supplemental Fig. S2B). We overexpressed miR160 to silence a set of repressor ARFs belonging to the ARF10/ARF16/ARF17 family and to obtain auxin-hypersensitive plants. Overexpression of miR160 precursor also resulted in increased levels of the corresponding mature miRNA (Supplemental Fig. S2C) and a decrease in the levels of all nine validated targets (Supplemental Fig. S2D).
We assayed physiological responses to auxin in the roots of these plants by treating them with 0, 0.2, or 1.0 μm of the synthetic auxin analog 2,4-D and examining root growth and the initiation of LRs after 1 week. In the absence of exogenous auxin, we observed no statistically significant difference between the vector control and miRNA-overexpressing lines in either the rate of root growth (Supplemental Table S1A) or LR density (Supplemental Table S1B). When treated with auxin, the elongation of vector control roots was significantly inhibited in response to both levels of auxin treatment (Supplemental Table S1A) in a dose-dependent manner (Fig. 2A). However, inhibition of root growth in miR393-overexpressing (miR393ox) plants was significantly lower than that of vector control at both levels of auxin treatment (Fig. 2A; Supplemental Table S1A), suggesting that these roots were hyposensitive to auxin. In contrast, there was significantly enhanced inhibition of root growth in miR160-overexpressing (miR160ox) plants relative to vector control plants at both levels of auxin treatment (Fig. 2A; Supplemental Table S1A). This suggested that miR160ox roots were hypersensitive to auxin. We also examined the increase in LR density (both primordia and emerged LRs) in response to auxin treatment in miR393ox and miR160ox roots. Vector control roots had a significant increase in LR density in a dose-dependent manner in response to auxin (Fig. 2B; Supplemental Table S1B). Similar to the response observed for inhibition of root elongation, miR393ox roots displayed auxin hyposensitivity (reduced LR density versus vector control; Fig. 2B; Supplemental Table S1B), whereas miR160ox roots displayed auxin hypersensitivity (significantly higher LR density versus vector control; Fig. 2B; Supplemental Table S1B).
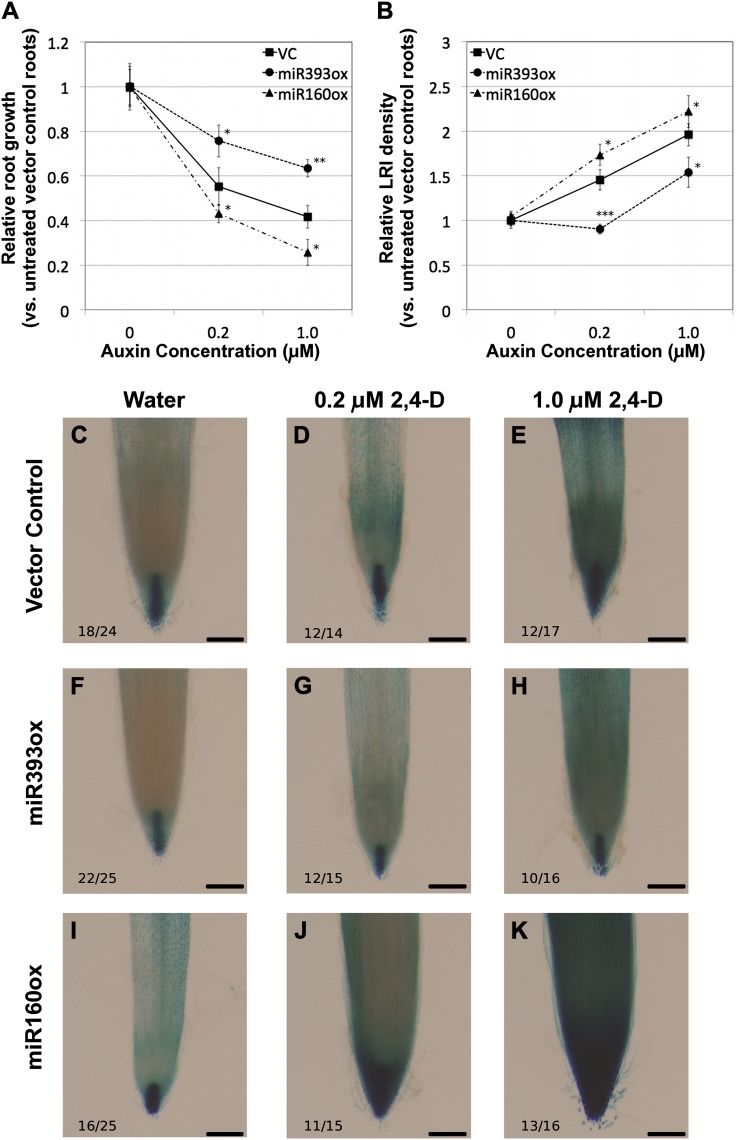
Figure 2.
Physiological and molecular responses of miR393- and miR160-overexpressing soybean composite plant roots to exogenous auxin. A and B, Relative root elongation (A) and lateral root density (B) in response to treatment with 0, 0.2, or 1.0 μm 2,4-D compared with untreated vector control (VC) roots. Data presented are averages ± se from at least 20 roots per data point. Asterisks indicate the level of statistically significant difference, if any, compared with vector control roots (*P < 0.05, **P < 0.01, ***P < 0.001). C to K, DR5:GUS expression in root tips of untreated (C, F, and I) and auxin-treated (D, G, and J, 0.2 μm; E, H, and K, 1.0 μm) vector control (C–E), miR393ox (F–H), and miR160ox (I–K) roots. The number of independent transgenic roots showing the representative staining pattern out of the number of roots examined is indicated in each panel. Bars = 200 μm.
In addition to the above physiological responses, we also assayed molecular responses to auxin in miRNA-overexpressing roots by examining the expression of DR5:GUS, the synthetic auxin-responsive marker. In untreated vector control roots, expression of DR5:GUS (“auxin maximum”) was observed primarily in the root cap columella and the root meristem (Fig. 2C), consistent with our DR5:tdT results. Treatment with 0.2 or 1.0 μm 2,4-D for 12 h induced DR5:GUS expression in a dose-dependent manner, indicated by an expanded expression domain and darker staining (Fig. 2, D and E). DR5:GUS expression in untreated miR393ox roots was similar or slightly reduced compared with that in vector control roots (Fig. 2D). However, auxin treatment resulted in a notably reduced induction of DR5:GUS expression (Fig. 2, G and H) compared with vector control roots, indicating that these roots were insensitive/hyposensitive to auxin. In contrast, DR5:GUS expression was clearly induced in miR160ox roots in response to auxin, with an expanded domain of expression and darker staining in the root tips (Fig. 2, J and K), compared with vector control roots. This suggested that these roots were hypersensitive to auxin.
We also examined the expression of seven endogenous auxin-responsive marker genes in miRNA-overexpressing roots by reverse transcription-quantitative PCR (Table I). Of these, five were clearly induced by auxin in vector control roots. For example, a 40-fold induction of GH3 expression was observed in response to treatment with 1 μm 2,4-D for 6 h in vector control roots (Table I). Auxin induction of GH3 was significantly attenuated in miR393ox roots (Table I). On the other hand, in miR160ox roots, there was a significantly higher induction of GH3 in response to auxin (Table I). Similarly, four of the six Aux/IAA genes examined were strongly induced by auxin treatment (GmIAA1, IAA8, IAA13, and IAA20). Similar to GH3, auxin induction of all four genes was significantly attenuated in miR393ox roots compared with the vector control (Table I). Auxin induction of three of these genes was significantly enhanced in miR160ox roots (the exception being GmIAA13) compared with the vector control (Table I). In summary, at least four of the five auxin-inducible marker genes had an attenuated response in miR393ox roots and an enhanced response in miR160ox roots. All the above results strongly indicate that miR393ox resulted in auxin hyposensitivity while miR160ox resulted in auxin hypersensitivity. We used these roots to examine the effect of altered auxin sensitivity on symbiotic nodule development.
Table I. Expression of selected marker genes in response to 1.0 μm 2,4-D in vector control, miR393ox, and miR160ox roots.
Data shown are fold change in gene expression relative to untreated (0 μm 2,4-D) controls. Original gene expression values were normalized to that of actin and further confirmed using two additional housekeeping genes. Numbers in parentheses indicate the range of possible fold change values based on sd between replicates (two independent experiments). Asterisks indicate the level of statistically significant difference, if any, compared with vector control roots (Student’s t test: *P < 0.05, **P < 0.01).
| Marker Gene | Expression (Fold Change Compared with Untreated Controls) |
||
|---|---|---|---|
| Vector Control | miR393ox | miR160ox | |
| GmGH3 (Glyma05g21680) | 42.81 (40.04–45.78) | 30.07* (27.81–32.51) | 115.09** (106.59–124.28) |
| IAA1 (Glyma10g32330) | 4.62 (4.22–5.07) | 3.06** (2.56–3.66) | 8.11** (7.16–9.19) |
| IAA8 (Glyma09g33630) | 5.50 (5.04–6.01) | 3.85* (3.12–4.74) | 6.31* (5.44–7.31) |
| IAA9 (Glyma01g24100) | 1.94 (1.68–2.22) | 1.62 (1.52–1.73) | 1.56 (1.41–1.72) |
| IAA13 (Glyma17g12080.2) | 231.25 (169.32–315.85) | 74.03** (59.50–92.11) | 97.46* (75.11–126.45) |
| IAA14 (Glyma10g32340) | 1.35 (1.23–1.49) | 1.17 (1.04–1.32) | 1.73 (1.56–1.93) |
| IAA20 (Glyma19g40970) | 6.23 (5.27–7.37) | 2.68* (2.39–2.99)* | 9.43* (8.55–10.39) |
Auxin Hypersensitivity in the Roots or Nodule Primordia Inhibits Nodulation
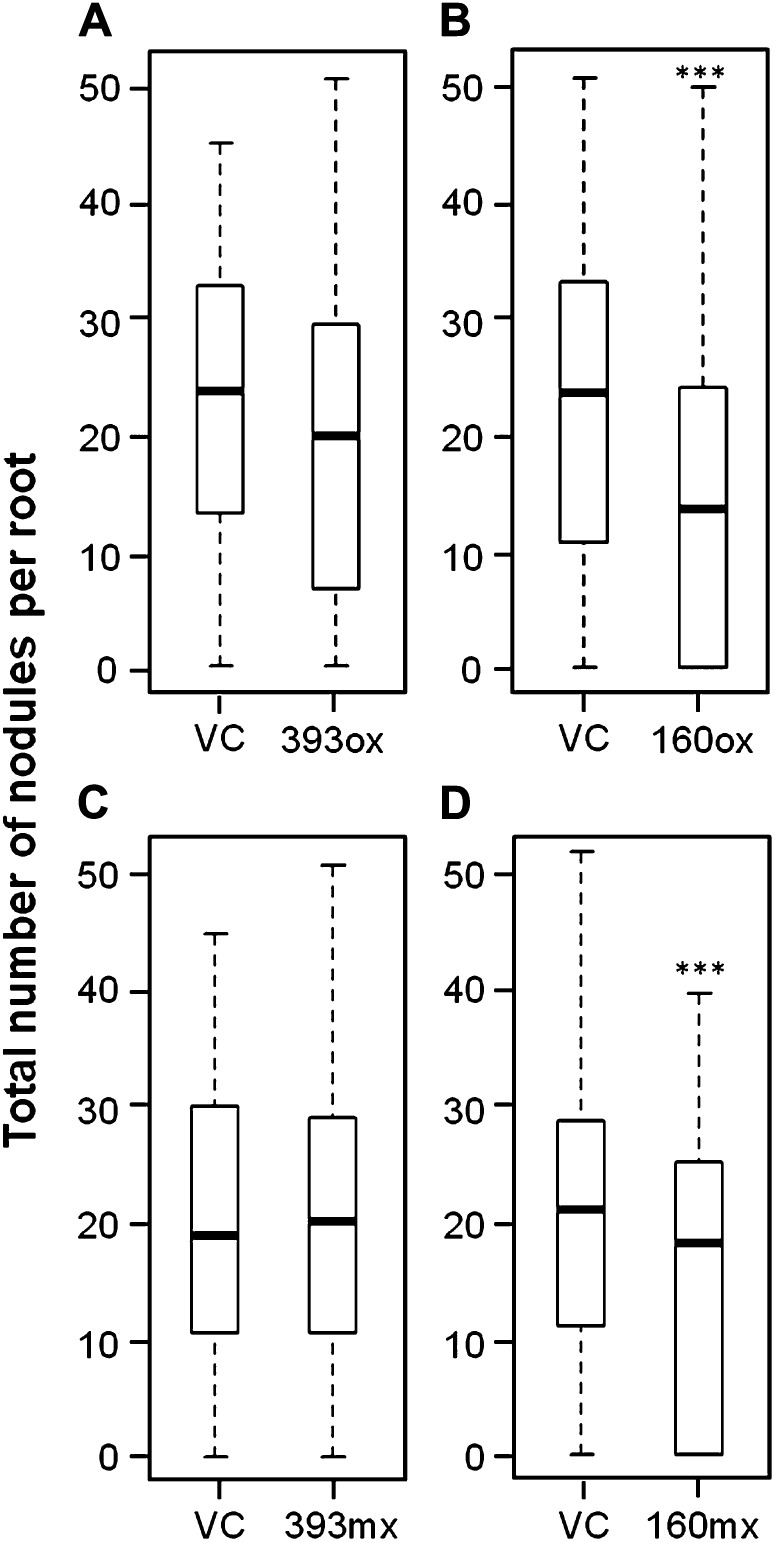
miR393ox and miR160ox composite plants were inoculated with B. japonicum cells, and the extent of nodulation was examined 14 dpi. Vector control roots had an average of 20.2 ± 1.8 nodules per root. The number of nodules on miR393ox roots (16.3 ± 2.2) did not significantly differ from that of vector control roots (Poisson distribution analysis), suggesting that auxin hyposensitivity did not significantly influence nodulation in soybean (Fig. 3A). In contrast, miR160ox roots had significantly fewer nodules (19.7 ± 1.7 in vector control versus 9.6 ± 1.4 in miR160ox roots), suggesting that auxin hypersensitivity inhibited nodulation in soybean (Fig. 3B). However, this experiment did not answer if this inhibition is a direct effect of auxin sensitivity on nodule tissues or a pleiotropic effect of altered auxin sensitivity in the root system. To address this question, we “misexpressed” these miRNAs using the nodule-specific soybean EARLY NODULIN40 (ENOD40) promoter (Yang et al., 1993). When examined in composite plant roots using an ENOD40:tdT construct, soybean ENOD40 was not expressed or was expressed at very low levels in uninoculated roots (including root tips and LRs) but was highly expressed in nodule primordia, emerging nodules, and mature nodules in soybean (Supplemental Fig. S3). Therefore, altered auxin sensitivity in roots misexpressing these miRNAs would be limited to these target tissues. Composite plants misexpressing miR393 (miR393mx) nodulated as efficiently as vector control plants (14.9 ± 2.2 versus 17.4 ± 2.6 nodules per root, respectively; Fig. 3C), consistent with results from miR393ox plants. Interestingly, miR160mx plants had significantly fewer nodules compared with vector control roots (15.8 ± 2.4 versus 7.7 ± 1.0; Fig. 3D), as observed in miR160ox plants. This suggested that auxin hypersensitivity during and at the sites of nodule primordia formation was sufficient to inhibit nodulation in soybean.
Figure 3.
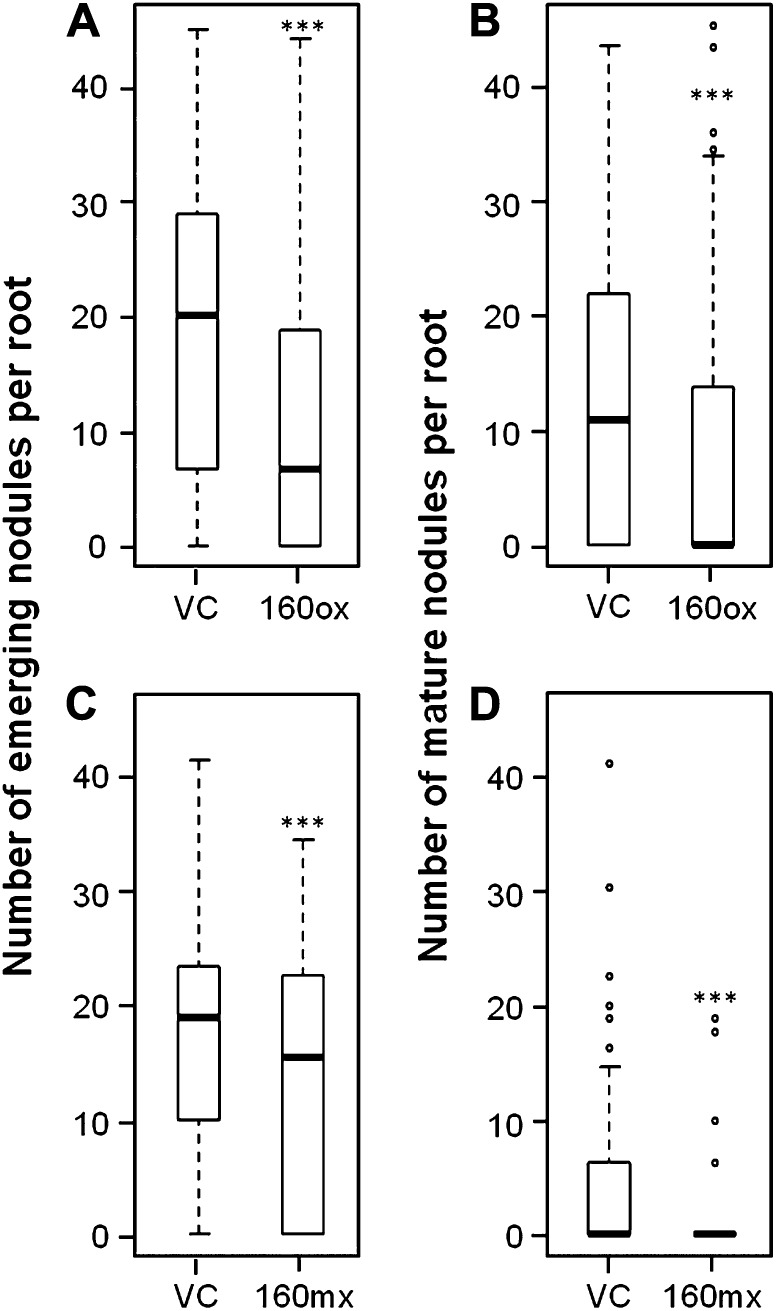
Nodulation in soybean composite plant roots overexpressing or misexpressing (using the ENOD40 promoter) miR393 or miR160. Box plots show the distribution of nodule numbers per root in miR393ox (A), miR160ox (B), miR393mx (C), and miR160mx (D) roots compared with the respective vector controls (VC). Data shown are from at least 80 roots for each construct. Boxes indicate data within the first and third quartiles, and the thick black line indicates the median. Asterisks indicate the level of statistically significant difference, if any, compared with the respective vector control roots (***P < 0.001).
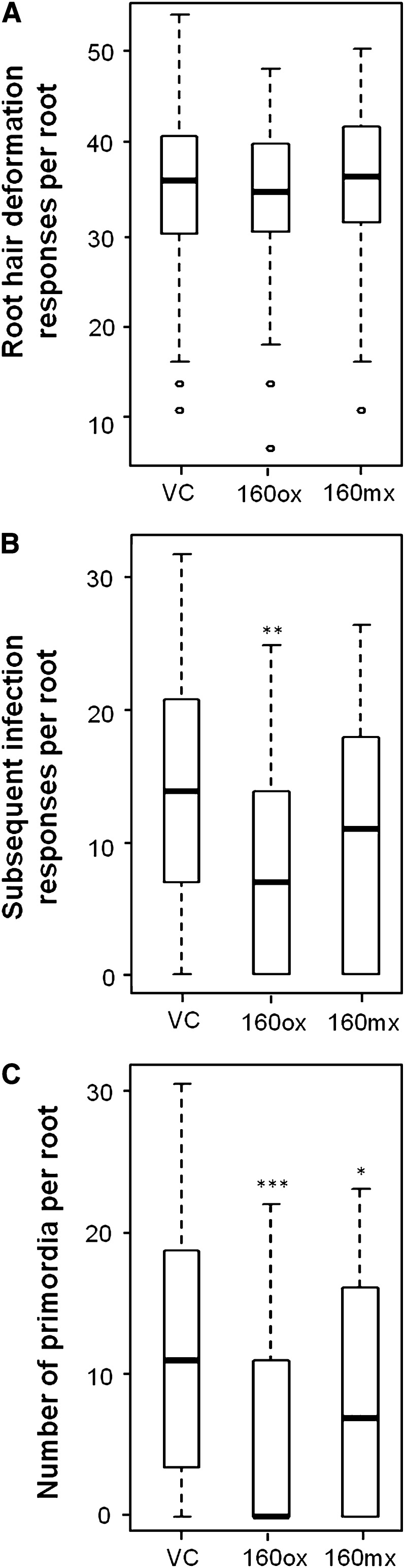
Auxin Hypersensitivity Did Not Affect Root Hair Responses to Rhizobia But Affected Nodule Primordium Formation
We closely examined nodule development in miR160ox and miR160mx roots to identify which stage(s) of nodule development is influenced by auxin hypersensitivity. We used B. japonicum expressing an nptII:GUS construct (a kind gift from Dr. Gary Stacey) to follow root hair colonization, infection thread development, and nodule primordium formation. In vector control plants, there were 45 ± 7 infections per root that showed root hair deformation/curling responses and/or infection thread formation limited to the epidermal cells 5 dpi (Fig. 4A; Supplemental Fig. S4, A and B). There was no significant difference in the number of infections showing these responses in miR160ox (39 ± 5; P = 0.52) or miR160mx (48 ± 7; P = 0.72) plants (Fig. 4A), suggesting that auxin hypersensitivity did not affect root hair colonization or deformation/curling responses in soybean. In addition to these infections, there were on average 5 ± 1 infections per root in vector control plants where the infection thread had proceeded farther into the cortex cells or had reached dividing primordia at 5 dpi (Fig. 4B; Supplemental Fig. S4, C and D). miR160ox roots had significantly fewer infections (2 ± 1) that showed this response (P = 0.002; Fig. 4B), but there was no significant difference between vector control and miR160mx (4 ± 1) roots (P = 0.1; Fig. 4B). This observation suggested that auxin hypersensitivity in the entire root system (resulting from miR160ox) inhibited infection thread growth in the cortex while auxin hypersensitivity in nodule-associated cells (resulting from miR160mx) did not affect this process.
Figure 4.
Early symbiotic responses in roots overexpressing or misexpressing miR160. Box plots show the distribution of root hair deformation or curling responses limited to the epidermis at 5 dpi (A), infection thread growth into the cortex at 5 dpi (B), and nodule primordium initiation responses at 8 dpi (C) in vector control (VC), miR160ox, and miR160mx roots. Data shown are from at least 40 roots for each construct. Boxes indicate data within the first and third quartiles, and the thick black line indicates the median. Potential outliers, if any, are depicted as circles. Asterisks indicate the level of statistically significant difference, if any, compared with the respective vector control roots (*P < 0.05, **P < 0.01, ***P < 0.001).
We also counted the number of nodule primordia in these roots 8 dpi. Vector control roots had 3.9 ± 0.8 primordia per root (Fig. 4C). Both miR160ox (1.3 ± 0.3) and miR160mx (2.2 ± 0.4) plants had significantly fewer nodule primordia (Fig. 4C), but with different levels of severity indicated by statistical significance (P = 0.0002 and 0.02, respectively). We conclude that auxin hypersensitivity in the entire root system (resulting from miR160ox) caused a severe inhibition of nodule primordium formation while auxin hypersensitivity in nodule-associated cells (resulting from miR160mx) was sufficient to cause a moderate inhibition of nodule primordium formation.
Next, we examined nodule development and maturation in miR160ox and miR160mx plants by classifying the number of nodules at 14 dpi into emerging and mature nodules (see “Materials and Methods”). Consistent with a reduction in the number of infection threads reaching the cortex and a severe reduction in the number of nodule primordia, miR160ox roots had a severe reduction in the number of emerging nodules compared with the vector control roots (12.4 ± 1.1 versus 5.9 ± 0.9; Fig. 5A). Interestingly, miR160mx roots also had a severe reduction in the number of emerging nodules (13.9 ± 2.0 versus 7.2 ± 1.0; Fig. 5C), even though these roots were unaffected in the number of infections reaching the cortex and had only a moderate reduction in the number of nodule primordia. This observation suggested that auxin hypersensitivity in nodule tissues might inhibit the development of nodule primordia into emerging nodules. Consistent with the severe reduction in emerging nodules, both miR160ox and miR160mx roots had severe reductions in the number of mature nodules as well (Fig. 5, B and D). Together, our results suggest that auxin hypersensitivity resulting from ectopic expression of miR160 inhibits not only the formation of nodule primordia but also their subsequent development in soybean.
Figure 5.
Nodule development in roots overexpressing or misexpressing miR160. Box plots show the distribution of emerging (A and C) and mature (B and D) nodules per root in miR160ox (A and B) and miR160mx (C and D) roots compared with the respective vector control (VC) roots. Data shown are from at least 90 roots for each construct. Boxes indicate data within the first and third quartiles, and the thick black line indicates the median. Potential outliers, if any, are depicted as circles. Asterisks indicate the level of statistically significant difference, if any, compared with the respective vector control roots (*P < 0.05, **P < 0.01, ***P < 0.001).
Roots Overexpressing miR160 Are Hyposensitive to Cytokinin
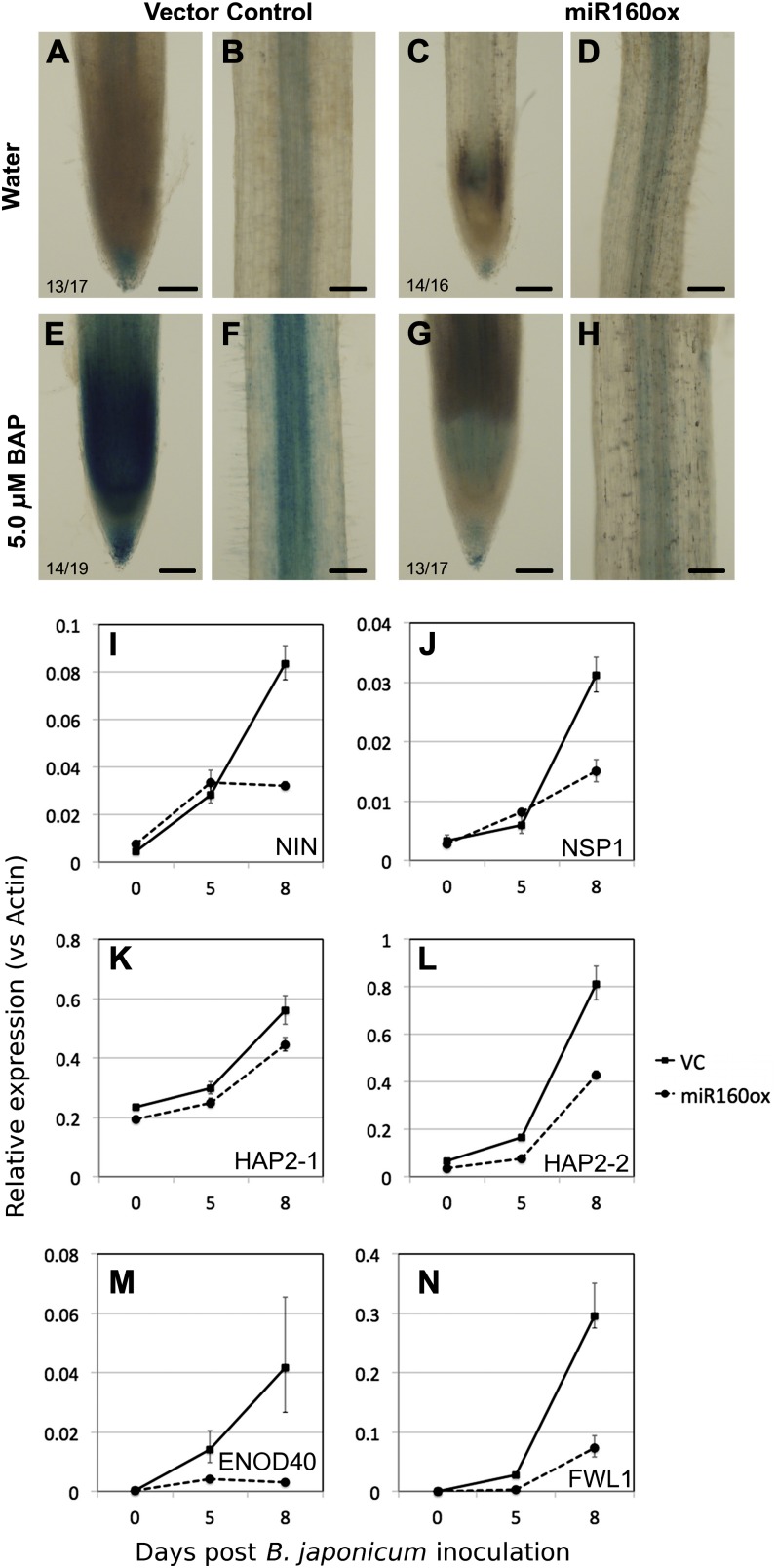
Our results suggested that hypersensitivity to auxin inhibits nodule primordium formation, and it is known that cytokinin promotes this process (see “Discussion”). Since auxin and cytokinin are known to act antagonistically during a number of different plant developmental processes, we examined cytokinin sensitivity in roots overexpressing miR160. First, we examined the expression of ARABIDOPSIS RESPONSE REGULATOR5 (ARR5):GUS, which has been used as a marker for cytokinin activity in legumes (Lohar et al., 2004). In untreated vector control roots, AtARR5:GUS expression was detected primarily in the root cap (Fig. 6A), vasculature (Fig. 6B), and at the base of LR primordia of mature root regions (data not shown). There was no obvious difference in AtARR5:GUS expression between untreated vector control and miR160ox roots (Fig. 6, C and D). When roots were treated with 5 μm 6-benzylaminopurine (BAP) for 6 h, there was a clear induction of AtARR5:GUS expression in the root tip and vasculature of vector control roots (Fig. 6, E and F). In contrast, a very weak induction was observed in miR160ox roots (Fig. 6, G and H). This suggested that these roots were indeed hyposensitive to cytokinin.
Figure 6.
A to H, Expression of cytokinin-responsive gene expression in miR160ox roots. Expression is shown for AtARR5:GUS in root tips (A, C, E, and G) and vasculature (B, D, F, and H) of untreated (A–D) and cytokinin (5 μm BAP)-treated (E–H) vector control (A, B, E, and F) and miR160ox (C, D, G, and H) roots. The number of independent transgenic roots showing the representative staining pattern out of the number of roots examined is indicated in each panel. I to N, Expression of cytokinin-dependent nodulation genes, NIN (I), NSP1 (J), HAP2-1 (K), HAP2-2 (L), ENOD40 (M), and FWL1 (N), along a time course of B. japonicum inoculation in vector control (VC) and miR160ox roots. Data presented are averages ± sd of three replicates.
Next, we examined the expression of soybean orthologs of Arabidopsis ARR5 and ARR9 in response to cytokinin. Some of these type A RESPONSE REGULATOR genes were shown to regulate nodule development in M. truncatula (Op den Camp et al., 2011). Based on BLASTx searches and previously published information, we identified four potential soybean orthologs each for AtARR5 and AtARR9. All eight genes were significantly induced by 5 μm BAP as early as 30 min in both vector control roots and miR160ox roots (Table II). However, the level of cytokinin induction was significantly attenuated in miR160ox roots compared with vector control roots for six of these RESPONSE REGULATOR genes (Table II). Together, these observations suggested that miR160ox roots were hyposensitive to cytokinin.
Table II. Expression of selected marker genes in response to 5.0 μm BAP in vector control and miR160ox roots.
Data shown are fold change in gene expression relative to untreated (0 μm BAP) controls. Original gene expression values were normalized to that of actin and further confirmed using two additional housekeeping genes. Numbers in parentheses indicate the range of possible fold change values based on sd between replicates (two independent experiments). Asterisks indicate the level of statistically significant difference, if any, compared with vector control roots (Student’s t test; *P < 0.05, **P < 0.01).
| Marker Gene | Expression (Fold Change Compared with Untreated Controls) |
|
|---|---|---|
| Vector Control | miR160ox | |
| GmRR5-1 (Glyma04g34820) | 1.69 (1.28–2.23) | 4.02** (3.28–4.91) |
| GmRR5-2 (Glyma06g19870) | 8.01 (5.96–10.76) | 3.07** (2.25–4.19) |
| GmRR5-3 (Glyma05g01730) | 9.60 (8.21–11.23) | 3.72** (2.93–4.23) |
| GmRR5-4 (Glyma17g10170) | 4.53 (3.88–5.29) | 3.52* (2.94–4.22) |
| GmRR9-1 (Glyma04g29250.2) | 7.64 (5.68–10.27) | 4.50** (2.62–7.71) |
| GmRR9-2 (Glyma11g21650) | 11.50 (9.93–13.32) | 4.24** (4.00-4.49) |
| GmRR9-3 (Glyma13g26770) | 4.39 (3.18–6.04) | 3.82* (3.05–4.78) |
| GmRR9-4a (Glyma15g37770) | 11.99 (3.06–46.81) | 9.13 (3.75–22.16) |
Due to very low expression levels, this gene had poor reproducibility between replicate samples.
Finally, we sought to examine the influence of miR160ox on nodulation pathway genes dependent on cytokinin perception/activity. In L. japonicus and soybean, the expression of NODULE INCEPTION (NIN), NODULE SIGNALING PATHWAY1 (NSP1), HAPLESS2 (HAP2)-1, and HAP2-2 is induced during nodule development (Heckmann et al., 2011; Hayashi et al., 2012). NIN and HAP2 are directly induced by cytokinin treatment, and all these genes act downstream of cytokinin perception during nodule development in L. japonicus. We examined the expression of soybean orthologs of these genes along a time course of B. japonicum inoculation in vector control and miR160ox roots. The expression of NIN and NSP1 increased moderately at 5 dpi and very highly at 8 dpi in response to B. japonicum inoculation (Fig. 6, E and F). In miR160ox roots, the expression of NIN and NSP1 also increased to moderate levels at 5 dpi, but their expression was much lower than in vector control roots at 8 dpi (Fig. 6, E and F). Between the two HAP2 genes we examined, HAP2-2 was induced at much higher levels compared with HAP2-1 in response to B. japonicum inoculation in vector control roots (Fig. 6, G and H). The expression of these genes was lower in miR160ox roots compared with vector control roots along the entire time course (Fig. 6, G and H). However, the induction of HAP2-2 was affected to a larger extent in miR160ox roots compared with HAP2-1. We also examined the expression of two nodulation-inducible marker genes, ENOD40 (Yang et al., 1993) and FRUIGHT WEIGHT2.2-LIKE1 (FWL1) (Libault et al., 2010), at 0, 5, and 8 dpi in vector control and miR160ox roots. The expression of both ENOD40 and FWL1 was very low or undetectable at 0 dpi but significantly increased at 5 and 8 dpi in vector control roots (Fig. 6, M and N). Consistent with the reduced expression of NIN and NSP1 and reduced nodule primordia initiation, miR160ox roots had significantly lower expression of these genes in response to B. japonicum inoculation (Fig. 6, M and N).
Our data clearly show that overexpression of miR160 resulted in hypersensitivity to auxin and hyposensitivity to cytokinin. Consistently, the expression of cytokinin-dependent nodulation genes was also reduced in miR160ox roots in response to B. japonicum inoculation. These results suggest that hypersensitivity to auxin and hyposensitivity to cytokinin results in impaired nodule development in soybean.
DISCUSSION
There Is Relatively Low Auxin Activity during Determinate Nodule Initiation
Using the auxin-inducible marker gene constructs, DR5:tdT and DR5:GUS, we identified that there is low or transient auxin activity during soybean nodule initiation and development. Previous studies have also identified the expression of a different auxin-responsive marker gene construct (GH3:GUS) during the initiation of both determinate and indeterminate nodules (Mathesius et al., 1998; Pacios-Bras et al., 2003; Takanashi et al., 2011). However, subsequent sustained GH3:GUS expression in nodule primordia is detected only in indeterminate nodules, since determinate nodules do not have a persistent nodule meristem. Recently, Suzaki et al. (2012) reported that the expression of DR5:GFP-NLS (where the marker gene gets localized to the nucleus, offering increased detectability) was observed in cortex cells dividing to form nodule primordia in L. japonicus, a determinate nodule-forming legume. Subsequently, DR5:NLS-GFP expression was limited to the periphery of nodule primordia and was absent in the rhizobium-colonized zone, consistent with our results. Therefore, the use of markers with enhanced detectability (e.g. GFP-NLS) or stability (e.g. GUS) detected auxin-inducible gene expression during nodule initiation, suggesting that it was low or transient. The observation that DR5:tdT was readily detectable in LR primordia and emerging LRs, but not in nodule primordia or nodules, suggested that the level of auxin activity during determinate nodule initiation and development is much lower than that during LR initiation. In addition, it appears that an initial auxin maximum occurs during nodule primordium initiation and subsequently diminishes from the primordium into the nodule periphery during nodule maturation (Suzaki et al., 2012; this study). We conclude that the requirement of auxin activity changes in a spatiotemporal manner during determinate nodule development.
We overexpressed or misexpressed specific miRNAs to modulate auxin sensitivity in soybean roots. We reasoned that the use of miRNAs would overcome the pitfalls of functional redundancy often associated with genetic mutants, especially in auxin signaling. For example, while single order mutants of ARF10, ARF16, or ARF17 show no obvious phenotypes in Arabidopsis, triple order mutants or miR160-overexpressing lines (where all three of the ARFs are silenced) show obvious defects in plant development (Mallory et al., 2005; Wang et al., 2005). Similarly, multiorder mutants show distinct and severe phenotypes compared with the single order mutants tir1 and afb in Arabidopsis (Parry et al., 2009). Phylogenetic analysis of the soybean TIR1/AFB (TribeMCL00611 in legumeIP) and ARF10/ARF16/ARF17 (TribeMCL00296 in legumeIP) families also suggested similar functional redundancy among family members. Indeed, overexpression of miR393 resulted in auxin hyposensitivity in soybean roots, as has previously been reported in Arabidopsis (Parry et al., 2009). However, the level of reduction in auxin sensitivity in miR393ox Arabidopsis plants is not as severe as observed in the multiorder mutants tir and afb, likely due to the partial resistance of some of the target genes to miR393-mediated cleavage (Parry et al., 2009). Unfortunately, soybean mutants with genetic lesions in TIR/AFB genes are not available for such a comparison with miR393ox roots. Nevertheless, reduced sensitivity to auxin in miR393ox soybean roots resulted in reduced LR initiation but did not affect nodule formation. Due to the expression of DR5:tdT along the vasculature in mature nodules, we also examined miR393ox nodules with a light microscope for any defects in vascular development. We observed no obvious defects in vascular development in miR393ox nodules. This suggested that only minimal TIR1/AFB activity is required for proper nodule formation and development in soybean (and likely other determinate nodule-forming legumes as well). This is consistent with the reduced level of auxin-responsive gene expression observed during determinate nodule formation. Alternatively, TIR1/AFB-independent auxin signaling mechanisms (e.g. AUXIN-BINDING PROTEIN1; Jones et al., 1998; Braun et al., 2008) might govern determinate nodule formation and development.
Auxin Hypersensitivity Inhibits Determinate Nodule Development
Overexpression of miR160 resulted in a clear auxin hypersensitivity in soybean roots. Similar observations have been made in Arabidopsis as well. For example, miR160ox Arabidopsis plants had increased LR density (Wang et al., 2005), and proteins encoded by ARF10, ARF16, and ARF17 (targets of miR160) have been proposed to encode repressor ARF proteins (Mallory et al., 2005). The presence of Proline- and Serine-rich middle regions suggested that soybean ARF10/ARF16/ARF17 family members might act as repressors as well (Supplemental Fig. S5). Indeed, a mutation in miR160 leading to increased expression of ARF10, ARF16, and ARF17 resulted in auxin-resistant phenotypes in Arabidopsis (Liu et al., 2010). In addition, miR160ox resulted in consumption of the root cap in Arabidopsis (Wang et al., 2005). Surprisingly, we observed intact root caps in soybean miR160ox roots when examined normally (Fig. 2) or through columella starch staining (data not shown). It is possible that this difference is due to the different promoters used to drive the overexpression of miR160 in Arabidopsis and soybean.
Results from miR160mx roots suggested that auxin hypersensitivity in nodule tissues causes only a moderate inhibition of primordium formation but a severe inhibition of subsequent nodule development in soybean. What role does auxin play in nodule development? Signaling events during nodule development occur in two distinct phases, the first one in the epidermis in response to Nod factor perception and the second one in cortex cells following the activation of cytokinin signaling (Oldroyd et al., 2011). Auxin hypersensitivity does not appear to affect epidermal responses (Fig. 4A) but clearly affects the second phase of events in the cortex influencing infection thread growth, primordium formation, and subsequent development (Fig. 4, B and C). Does auxin regulate infection thread growth? We think this might be an indirect effect. Infection thread growth in the cortex is thought to be determined by cells dividing to form nodule primordia (Murray et al., 2007; Oldroyd and Downie, 2008). Therefore, it is likely that the lack of nodule primordia caused the arrest of infection thread growth in the cortex of miR160ox soybean roots. Consistent with this observation, a moderate reduction in primordium initiation did not affect infection thread growth in the cortex of miR160mx roots. We cannot exclude the possibility that auxin directly influenced the rhizobial infection process similar to its effect on infection by bacterial pathogens in Arabidopsis (Navarro et al., 2006; Cui et al., 2013). However, while auxin sensitivity promotes susceptibility to bacterial pathogens, we observed inhibition of rhizobial infection threads, suggesting that these mechanisms are not likely to be conserved.
What effect does auxin have on nodule initiation in the cortex? There is very low auxin-responsive gene expression during nodule initiation, and it appears specifically in the apex region of nodule primordia (Fig. 1C). We examined nodule initials (cell division foci) in vector control and miR160ox roots using roots expressing the DR5:GUS marker (Supplemental Fig. S6). The number of nodule initials per root was not significantly different between vector control and miR160ox roots at 3 dpi (2.7 ± 0.9 versus 2.0 ± 0.6, respectively; Poisson distribution analysis; P = 0.2). We conclude that auxin hypersensitivity did not affect nodule initial cell divisions. During postinitiation stages of nodule development, the majority of auxin activity gets restricted to the nodule periphery, and there is no or minimal auxin activity in the infection zone, at least in determinate nodule-forming legumes (Fig. 1D; Suzaki et al., 2012). Interestingly, ENOD40 and DR5 have minimal overlap between their expression domains (Fig. 1D; Supplemental Fig. S3F). Therefore, it is likely that there was sustained auxin activity in the nodule initials/infection zone of miR160ox and miR160mx roots, and this might have inhibited nodule development. We attempted to test this hypothesis by monitoring DR5:GUS expression during postinitiation stages of nodule development in miR160ox roots. Significantly reduced primordium formation in these roots precluded such an experiment. It appears that the suppression of auxin concentration/sensitivity in specific nodule cell types, and/or the maintenance of a correct window of auxin concentration/sensitivity in specific cell types, is crucial for proper development of the determinate nodule primordium.
It is currently not clear if auxin hypersensitivity inhibits indeterminate nodule formation. Auxin-sensitive genotypes of Medicago spp. nodulated faster and/or had more nodules (Kondorosi et al., 1993). Similarly, autoregulation-defective supernodulating mutants of M. truncatula display an increase in shoot-to-root long-distance auxin transport (van Noorden et al., 2007) compared with wild-type plants. These results suggested that auxin promotes nodule formation in these species. On the other hand, an apparent auxin resistance resulting from the silencing of CELL DIVISION CYCLE16, a cell cycle component, resulted in increased nodulation and a reduced number of LRs in M. truncatula (Kuppusamy et al., 2009). Interestingly, auxin appears to play a crucial role during the infection process in actinorhizal nodules. The expression of auxin influx carriers and the accumulation of auxin have been observed in Frankia spp.-infected cells during nodule formation in Casuarina glauca (Péret et al., 2007; Perrine-Walker et al., 2010).
Auxin-Cytokinin Regulation of Nodule Development
Auxin and cytokinin antagonize each other in a number of different plant development processes, including meristem/primordium development (e.g. shoot apical meristem, root meristem, LR primordia, and leaf primordia; for review, see Su et al., 2011). Do auxin and cytokinin play opposite roles in nodule initiation as well? It is known that cytokinin promotes nodule primordium formation and development. For example, the cytokinin-responsive marker, AtARR5:GUS, is specifically induced in cortex cells, dividing to form nodule primordia in L. japonicus (Lohar et al., 2004). Similarly, cytokinin insensitivity caused an inability to initiate nodule primordia in both L. japonicus (a determinate nodule-forming legume; Murray et al., 2007) and M. truncatula (an indeterminate nodule-forming legume; Plet et al., 2011). In clear agreement with these observations, gain-of-function mutations in L. japonicus HISTIDINE KINASE1 (LHK1) resulted in spontaneous nodule formation even in the absence of rhizobia (Tirichine et al., 2007). We show that miR160ox resulted in hypersensitivity to auxin, hyposensitivity to cytokinin, and a reduction in nodule primordium formation. Interestingly, loss of cytokinin sensitivity in L. japonicus hit1-1/lhk1 mutant plants resulted in nodulation phenotypes similar to that of soybean miR160ox roots. While infection threads formed normally, nodule primordium initiation was significantly reduced in these mutants (Murray et al., 2007). Similarly, both miR160ox roots as well as lhk1 mutants had an attenuated expression of NIN and ENOD40 in response to rhizobial inoculation. Together, these results suggest that reduced nodulation in auxin-hypersensitive miR160ox roots is due to the suppression of cytokinin activity. Is a balance between auxin and cytokinin (biosynthesis and/or signaling) crucial for proper nodule primordium development? Indeed, such a hypothesis was proposed recently by Oldroyd et al. (2011). However, such interactions are likely to be cell type and developmental stage specific. For example, it was discovered very early that the addition of both auxin and cytokinin was necessary to initiate cell divisions opposite xylem poles in isolated pea cortex cell explants (Libbenga et al., 1973). Recent results indicate that auxin activity in nodule initial cells is under the control of both cytokinin and NIN in L. japonicus. In addition, auxin-inducible gene expression was observed during cortex cell divisions as well as in the “infection zone” of uncolonized spontaneous nodules in the cytokinin gain-of-function snf2 mutants (Suzaki et al., 2012). Computer models (Deinum et al., 2012) as well as genetic evidence (Plet et al., 2011) also suggested that inhibition of auxin efflux by cytokinin is the most likely mechanism that leads to an auxin maximum during nodule initiation. These observations suggested that cytokinin might govern the localized accumulation of auxin during nodule primordium initiation. Our results suggest that enhanced auxin sensitivity (potentially uncoupling its regulation by cytokinin) results in the inhibition of nodule formation. In addition, despite the observation that ENOD40 acts downstream of NIN during nodule development (Grønlund et al., 2005), we observed moderate reduction of primordium development in ENOD40:miR160 roots. We hypothesize that a feedback loop involving cytokinin, NIN, miR160, and auxin (Supplemental Fig. S7) governs proper nodule formation and development.
MATERIALS AND METHODS
Plant Material and DNA Vectors
Three different pCAMGFP-promoter:Gateway (GW) vectors were generated by cloning the respective promoters DR5 (Ulmasov et al., 1997), CsVMV (described by Graham et al., 2007), or GmENOD40 (Yang et al., 1993) in front of an attR1-ccdB-attR2 GW cassette into pCAM-sUbi:GFP (described by Subramanian et al., 2005). pCAMGFP-DR5:tdT was generated through an LR Clonase (Invitrogen) reaction between pDONR/Zeo-tdT (a kind gift from Dr. Gary Stacey) and pCAMGFP-DR5:GW. Precursors of miR393 (miRBaseID MI0007216) and miR160 (miRBaseID MI0001774) were cloned by PCR and subsequently by TOPO-TA cloning into pCR8/GW (Invitrogen). Overexpression and misexpression constructs were generated using pCAMGFP-CsVMV:GW and pCAMGFP-Enod40:GW vectors, respectively, as the destination vector and pCR8/GW/miR393 and pCR8/GW/miR160 as the entry vector in an LR Clonase (Invitrogen) reaction. miRox-DR5:GUS and miRox-AtARR5:GUS constructs were obtained by cloning a DR5:GUS cassette (a kind gift from Dr. Tom Guilfoyle) or an AtARR5:GUS cassette (cloned by amplifying AtARR5 upstream sequences; D’Agostino et al., 2000) into the miRNA-overexpressing constructs above. Vectors were electroporated into Agrobacterium rhizogenes K599 cells, and transgenic composite plants for gene expression analyses and miRNA overexpression/misexpression were generated as described previously (Collier et al., 2005) using 2 week-old soybean (Glycine max ‘Williams82’) seedlings. Transgenic roots of interest were identified using GFP epifluorescence.
Hormone Response Assays
For physiological assays, GFP-positive transgenic composite plant roots were individually labeled with a Tough-Tags polyester label, and both root length and number of LR primordia + emerged roots were counted (0-d measurements). The use of GFP as a marker to identify transgenic roots enabled the identification of LR primordia with a fluorescence dissection microscope without clearing the roots. Composite plants were transferred to 4-inch pots containing a mixture of vermiculite:perlite (3:1) and watered with nutrient solution with or without 2,4-D. One week later, plants were uprooted, and root length and number of total LRs (primordia + emerged) were counted on tagged roots (7-d measurements). The difference in root length and number of total LRs (0-d versus 7-d measurements) was calculated for each root. Student’s t test was used to compare different treatments and genotypes using Microsoft Excel.
For DR5:GUS and marker gene expression assays (auxin response), transgenic plants with tagged roots (see above) were transferred to sterile deionized water with or without 0.2 or 1.0 μm 2,4-D and incubated for 12 h (DR5:GUS) or 6 h (quantitative PCR assays) at 25°C in the dark. For AtARR5:GUS and marker gene expression assays (cytokinin response), transgenic plants were treated with sterile deionized water with or without 5.0 μm BAP for 1 h at 25°C in the dark. Histochemical localization of GUS was performed as described before (Jefferson et al., 1987). For gene expression assays, whole roots were harvested after auxin/cytokinin treatment, blot dried, and immediately frozen in liquid nitrogen.
Bradyrhizobium japonicum Inoculation and Nodulation Assays
For nodulation assays, transgenic composite plants (3 weeks post transformation with the respective miRNA overexpression or misexpression construct) were transferred to 4-inch pots containing a mixture of vermiculite:perlite (1:3), allowed to grow for 1 week (16 h of light, 25°C, 50% relative humidity), and inoculated with a suspension of B. japonicum USDA110 (optical density at 600 nm = 0.08). Two weeks after inoculation, roots were harvested, and GFP-positive transgenic roots were separated and observed with a dissection microscope for nodulation. Nodules appearing as “bumps” were classified as emerging nodules, and those that were round and pink were classified as mature nodules. Mock-inoculated composite plants were used as an inoculation control in each experiment, and no nodules were observed on these plants. Nodule numbers between different overexpression and misexpression roots and the respective controls were examined for statistically significant differences, if any, using zero-inflated Poisson distribution analysis in the statistical analysis package R. To examine rhizobial colonization and infection thread development, composite plants were inoculated as above with B. japonicum transformed with an nptII:GUS construct (a kind gift from Dr. Gary Stacey). Tissue fixation and GUS staining to visualize rhizobial colonization and infection thread formation were performed as described previously (Loh et al., 2002). Nodule primordia were visualized by clearing roots in 10% bleach for 10 min. Since nodule primordium cell division occurs in the outer cortex in soybean, it is easy to distinguish nodule primordia from LR primordia. For gene expression assays, whole roots were harvested at appropriate time points, rinsed briefly in sterile deionized water to remove vermiculite/perlite particles, blot dried, and immediately frozen in liquid nitrogen.
Microscopy
To examine DR5:tdT expression, mock- or B. japonicum-inoculated transgenic composite plant roots were observed with a laser confocal microscope (Olympus FV300) or a fluorescence compound microscope (Olympus AX70) at 3, 7, 10, and 14 dpi. Images shown in Figure 1, A to D, were obtained using the confocal microscope with the following settings (channel 1, 488-nm excitation/515-nm emission for GFP; channel 2, 568-nm excitation/635-nm emission for tdT; 1.5% gain; Kalman acquisition). To examine DR5:GUS expression, mock- and B. japonicum-inoculated roots were stained for GUS activity (see above).
Gene Expression Assays
Total RNAs were isolated from transgenic roots using Trizol reagent and complementary DNAs prepared from 2 μg of total RNA using oligo(dT) and Moloney murine leukemia virus reverse transcriptase (New England Biolabs). Quantitative PCR assays for gene expression were performed using Stratagene MX3000P equipment and SYBR premix (Clontech). Gene expression levels were normalized to that of GmActin using the delta delta threshold cycle (ddCt) method (Livak and Schmittgen, 2001) and further confirmed using two additional housekeeping genes, GmCONS7 and GmCONS15 (data not shown; Libault et al., 2008). Statistical analyses for pairwise comparison of dCt = delta threshold cycle (dCt) values were done using Student’s t test on Microsoft Excel. Primers used for quantitative PCR assays are presented in Supplemental Tables S2 and S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. DR5 expression in hairy root composite soybean plants.
Supplemental Figure S2. Expression of miR393 and miR160 and their cognate targets in miRNA-overexpressing roots.
Supplemental Figure S3. ENOD40 expression in hairy root composite soybean plants.
Supplemental Figure S4. Rhizobial colonization tracked using B. japonicum-expressing GUS.
Supplemental Figure S5. Peptide sequence alignment of the “middle regions” of ARF10/16/17 family members.
Supplemental Figure S6. DR5:GUS expression in nodule initial cells.
Supplemental Figure S7. A hypothetical model of a feedback loop involving auxin and cytokinin during determinate nodule development.
Supplemental Table S1. Root growth and LR density in control, miR393ox, and miR160ox roots in response to auxin.
Supplemental Table S2. List of mRNA qPCR primers used in this study.
Supplemental Table S3. List of miRNA cDNA synthesis and qPCR primers used in this study.
Acknowledgments
We thank Dr. Gary Stacey (University of Missouri) for B. japonicum nptII:GUS cells and tdT vector, Dr. Tom Guilfoyle (University of Missouri) for the DR5 promoter, Dr. Claude Fauquet (Donald Danforth Plant Science Center) for the CsVMV promoter, Dr. Gemechis Djira (South Dakota State University) for suggestions on statistical analyses, Dr. Michael Hildreth (South Dakota State University) for guidance on microscopy, and Drs. Martin Crespi (CNRS), Ulrike Mathesius (Australian National University), and Takuya Suzaki (National Institute for Basic Biology) for critical comments and suggestions on the manuscript. Assistance from Ms. Natalie Krier and Ms. Holly Pueppke (South Dakota State University) for counting nodules is acknowledged.
Glossary
- Aux/IAA
auxin/indole-3-acetic acid
- ARF
auxin response factor
- miRNA
microRNA
- LR
lateral root
- 2,4-D
2,4-dichlorophenoxyacetic acid
- dpi
days post inoculation
- CsVMV
cassava vein mosaic virus
- BAP
6-benzylaminopurine
References
- Allen EK, Allen ON, Newman AS. (1953) Pseudonodulation of leguminous plants induced by 2-bromo-3,5-dichlorobenzoic acid. Am J Bot 40: 429–435 [Google Scholar]
- Bazin J, Bustos-Sanmamed P, Hartmann C, Lelandais-Brière C, Crespi M. (2012) Complexity of miRNA-dependent regulation in root symbiosis. Philos Trans R Soc Lond B Biol Sci 367: 1570–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot KJM, van Brussel AAN, Tak T, Spaink HP, Kijne JW. (1999) Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp nigra roots. Mol Plant Microbe Interact 12: 839–844 [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ. (2008) Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier R, Fuchs B, Walter N, Lutke WK, Taylor CG. (2005) Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J 43: 449–457 [DOI] [PubMed] [Google Scholar]
- Cui F, Wu S, Sun W, Coaker G, Kunkel B, He P, Shan L. (2013) The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol 162: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy F, Grosjean C, May S, Bennett M, Cullimore JV. (2001) Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact 14: 267–277 [DOI] [PubMed] [Google Scholar]
- Deinum EE, Geurts R, Bisseling T, Mulder BM. (2012) Modeling a cortical auxin maximum for nodulation: different signatures of potential strategies. Front Plant Sci 3: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Ding Y, Oldroyd GE. (2009) Positioning the nodule, the hormone dictum. Plant Signal Behav 4: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mathesius U. (2003) Signaling interactions during nodule development. J Plant Growth Regul 22: 47–72 [Google Scholar]
- Graham TL, Graham MY, Subramanian S, Yu O. (2007) RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol 144: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Grønlund M, Roussis A, Flemetakis E, Quaedvlieg NE, Schlaman HR, Umehara Y, Katinakis P, Stougaard J, Spaink HP. (2005) Analysis of promoter activity of the early nodulin Enod40 in Lotus japonicus. Mol Plant Microbe Interact 18: 414–427 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2012) Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci 190: 82–88 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Reid DE, Lorenc MT, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ. (2012) Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max [L.] Merr.) roots. Plant Biotechnol J 10: 995–1010 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. (2011) Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol Plant Microbe Interact 24: 1385–1395 [DOI] [PubMed] [Google Scholar]
- Hirsch AM. (1992) Developmental biology of legume nodulation. New Phytol 122: 211–237 [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Bhuvaneswari TV, Torrey JG, Bisseling T. (1989) Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc Natl Acad Sci USA 86: 1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Fang Y. (1994) Plant hormones and nodulation: what’s the connection? Plant Mol Biol 26: 5–9 [DOI] [PubMed] [Google Scholar]
- Huo X, Schnabel E, Hughes K, Frugoli J. (2006) RNAi phenotypes and the localization of a protein:GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J Plant Growth Regul 25: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilina EL, Logachov AA, Laplaze L, Demchenko NP, Pawlowski K, Demchenko KN. (2012) Composite Cucurbita pepo plants with transgenic roots as a tool to study root development. Ann Bot (Lond) 110: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns AN. (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282: 1114–1117 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kondorosi E, Schultze M, Savoure A, Hoffmann B, Dudits D, Pierre M, Allison L, Bauer P, Kiss GB, Kondorosi A (1993) Control of nodule induction and plant cell growth by Nod factors. In Nester EW, Verma DPS, eds, Advances in Molecular-Genetics of Plant-Microbe Interactions. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 143–150 [Google Scholar]
- Kuppusamy KT, Ivashuta S, Bucciarelli B, Vance CP, Gantt JS, Vandenbosch KA. (2009) Knockdown of CELL DIVISION CYCLE16 reveals an inverse relationship between lateral root and nodule numbers and a link to auxin in Medicago truncatula. Plant Physiol 151: 1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G. (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1: 44–54 [Google Scholar]
- Libault M, Zhang XC, Govindarajulu M, Qiu J, Ong YT, Brechenmacher L, Berg RH, Hurley-Sommer A, Taylor CG, Stacey G. (2010) A member of the highly conserved FWL (tomato FW2.2-like) gene family is essential for soybean nodule organogenesis. Plant J 62: 852–864 [DOI] [PubMed] [Google Scholar]
- Libbenga KR, Iren F, Bogers RJ, Schraag-Lamers MF. (1973) The role of hormones and gradients in the initiation of cortex proliferation and nodule formation in Pisum sativum L. Planta 114: 29–39 [DOI] [PubMed] [Google Scholar]
- Liu X, Huang J, Wang Y, Khanna K, Xie Z, Owen HA, Zhao D. (2010) The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in microRNA160a, in organogenesis and the mechanism regulating its expression. Plant J 62: 416–428 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loh J, Lohar DP, Andersen B, Stacey G. (2002) A two-component regulator mediates population-density-dependent expression of the Bradyrhizobium japonicum nodulation genes. J Bacteriol 184: 1759–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. (2004) Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J 38: 203–214 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA. (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Ni DA, Wang LJ, Ding CH, Xu ZH. (2001) Auxin distribution and transport during embryogenesis and seed germination of Arabidopsis. Cell Res 11: 273–278 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Op den Camp RH, De Mita S, Lillo A, Cao Q, Limpens E, Bisseling T, Geurts R. (2011) A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiol 157: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C, Schlaman HR, Boot K, Admiraal P, Langerak JM, Stougaard J, Spaink HP. (2003) Auxin distribution in Lotus japonicus during root nodule development. Plant Mol Biol 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup R, Jansen L, Devos G, Auguy F, Collin M, Santi C, Hocher V, Franche C, Bogusz D, et al (2007) Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol 144: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine-Walker F, Doumas P, Lucas M, Vaissayre V, Beauchemin NJ, Band LR, Chopard J, Crabos A, Conejero G, Péret B, et al. (2010) Auxin carriers localization drives auxin accumulation in plant cells infected by Frankia in Casuarina glauca actinorhizal nodules. Plant Physiol 154: 1372–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F, Jr, Vazquez F. (2011) miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol 157: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SA, Meyers BC, Sherrier DJ. (2009) MicroRNAs in the rhizobia legume symbiosis. Plant Physiol 151: 1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI. (2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol 174: 11–25 [DOI] [PubMed] [Google Scholar]
- Su YH, Liu YB, Zhang XS. (2011) Auxin-cytokinin interaction regulates meristem development. Mol Plant 4: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Graham MY, Yu O, Graham TL. (2005) RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol 137: 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Stacey G, Yu O. (2006) Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J 48: 261–273 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Stacey G, Yu O. (2007) Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci 12: 282–285 [DOI] [PubMed] [Google Scholar]
- Suzaki T, Ito M, Kawaguchi M. (2013) Genetic basis of cytokinin and auxin functions during root nodule development. Front Plant Sci 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. (2012) Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006 [DOI] [PubMed] [Google Scholar]
- Takanashi K, Sugiyama A, Yazaki K. (2011) Involvement of auxin distribution in root nodule development of Lotus japonicus. Planta 234: 73–81 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- van Noorden GE, Kerim T, Goffard N, Wiblin R, Pellerone FI, Rolfe BG, Mathesius U. (2007) Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol 144: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Yang WC, Katinakis P, Hendriks P, Smolders A, de Vries F, Spee J, van Kammen A, Bisseling T, Franssen H. (1993) Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J 3: 573–585 [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Stacey G, Yu O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J 57: 171–183 [DOI] [PubMed] [Google Scholar]