GA signaling controls seed germination in the sly1 mutant background, in which DELLA repressors cannot be destroyed by the ubiquitin-proteasome pathway.
Abstract
DELLA repression of Arabidopsis (Arabidopsis thaliana) seed germination can be lifted either through DELLA proteolysis by the ubiquitin-proteasome pathway or through proteolysis-independent gibberellin (GA) hormone signaling. GA binding to the GIBBERELLIN-INSENSITIVE DWARF1 (GID1) GA receptors stimulates GID1-GA-DELLA complex formation, which in turn triggers DELLA protein ubiquitination and proteolysis via the SCFSLY1 E3 ubiquitin ligase and 26S proteasome. Although DELLA cannot be destroyed in the sleepy1-2 (sly1-2) F-box mutant, long dry after-ripening and GID1 overexpression can relieve the strong sly1-2 seed dormancy phenotype. It appears that sly1-2 seed dormancy results from abscisic acid (ABA) signaling downstream of DELLA, since dormant sly1-2 seeds accumulate high levels of ABA hormone and loss of ABA sensitivity rescues sly1-2 seed germination. DELLA positively regulates the expression of XERICO, an inducer of ABA biosynthesis. GID1b overexpression rescues sly1-2 germination through proteolysis-independent DELLA down-regulation associated with increased expression of GA-inducible genes and decreased ABA accumulation, apparently as a result of decreased XERICO messenger RNA levels. Higher levels of GID1 overexpression are associated with more efficient sly1 germination and increased GID1-GA-DELLA complex formation, suggesting that GID1 down-regulates DELLA through protein binding. After-ripening results in increased GA accumulation and GID1a-dependent GA signaling, suggesting that after-ripening triggers GA-stimulated GID1-GA-DELLA protein complex formation, which in turn blocks DELLA transcriptional activation of the XERICO inhibitor of seed germination.
Gibberellins (GAs) are tetracyclic diterpenoid plant hormones that stimulate many developmental transitions in the plant life cycle, including the transition from seed dormancy to germination, the phase change from juvenile to adult plant development, stem elongation, and the transition to flowering and floral development (for review, see Sun and Gubler, 2004). The role of GA in these processes has been established through studies of GA biosynthesis mutants, like the mutation in copalyl synthetase in ga1-3. The failure to synthesize GA results in failure to germinate, extreme dwarfism, delayed transition to flowering, and poor fertility (Koornneef and van der Veen, 1980). All of these phentoypes are reversed by GA application, demonstrating the direct connection between GA signaling and physiological responses. This study examines the role of Arabidopsis (Arabidopsis thaliana) GIBBERELLIN-INSENSITIVE DWARF1 (GID1) GA receptors in sleepy1 (sly1)-independent GA signaling with a focus on seed after-ripening and germination.
Seeds are an important source of human nutrition and a vehicle for plant propagation and dispersal. Making the decision to germinate at the appropriate time is essential both to agriculture and to species survival. Seeds can be dormant when they are first released from the mother plant so that they fail to germinate under favorable conditions (for review, see Bewley and Black, 1994; Finkelstein et al., 2008). Seed dormancy prevents germination on the mother plant or out of season, reduces competition between siblings, and allows species to survive natural catastrophes as ungerminated seeds in the soil. Dormant seeds eventually acquire the ability to germinate through a period of dry storage called after-ripening and/or through a period of moist chilling called cold stratification.
Seed dormancy is associated with higher levels of the plant hormone abscisic acid (ABA), whereas the ability to synthesize GA is associated with dormancy-breaking treatments (Karssen et al., 1983; Jacobsen et al., 2002; Yamauchi et al., 2004; Lefebvre et al., 2006; Okamoto et al., 2006). ABA accumulation prevents premature germination during embryo maturation and establishes and later maintains seed dormancy (for review, see Finkelstein et al., 2008). Loss of dormancy through after-ripening is associated with ABA turnover (Okamoto et al., 2006). GA, on the other hand, accumulates during cold stratification and is required for Arabidopsis seed germination (Koornneef and van der Veen, 1980; Yamauchi et al., 2004). According to the hormone balance theory, the antagonism between these two hormones regulates seed dormancy, cold stratification, after-ripening, and germination (Karssen and Lacka, 1986; Finch-Savage et al., 2007). Seeds of the GA biosynthesis mutants ga1, ga2, and ga3 cannot germinate and do not after-ripen sufficiently to germinate without GA application (Koornneef and van der Veen, 1980). It is difficult to distinguish whether these seeds fail to germinate because they are highly dormant or because of a physical requirement for GA in seed germination. The GA-insensitive sly1 mutants also show increased seed dormancy, and the degree of seed dormancy is dependent on the severity of the sly1 allele. The seed dormancy phenotype of sly1-2 can be rescued by the ABA-insensitive mutation abi1-1 by removing the seed coat and by long after-ripening for 1 to 2 years (Steber et al., 1998; Ariizumi and Steber, 2007). The fact that the GA-insensitive sly1-2 mutant shows increased seed dormancy, but regains the ability to germinate with long after-ripening, provides a useful tool to examine the roles of GA and ABA in seed dormancy relief.
At the molecular level, GA hormone induces GA signal transduction by triggering proteasomal degradation of DELLA repressors of GA responses. DELLA (for Asp-Glu-Leu-Leu-Ala) family proteins are defined by the presence of the highly conserved D-E-L-L-A and V-H-Y-N-P amino acid sequences in the N-terminal DELLA regulatory domain and by the C-terminal functional domain with homology to the GRAS (for GAI, RGA and Scarecrow) family of putative transcription factors (for review, see Sun and Gubler, 2004). Mutations in the N-terminal DELLA regulatory domain result in varying degrees of gain-of-function GA-insensitive phenotypes (i.e. dwarfism), whereas mutations in the GRAS functional domain result in loss-of-function GA-hypersensitive phenotypes (i.e. increased height). The Arabidopsis genome contains five DELLA genes, RGA (for REPRESSOR OF GA1), GAI (for GA-INESENSITIVE), RGL1 (for RGA-LIKE1), RGL2, and RGL3 with partly overlapping functions. The roles of Arabidopsis DELLA genes were defined based on double mutant analysis with ga1-3 and depend largely on the location and timing of gene expression (Cheng et al., 2004; Tyler et al., 2004; Cao et al., 2005; Piskurewicz and Lopez-Molina, 2009; Gallego-Bartolomé et al., 2010). RGA, GAI, and RGL1 are the main repressors of stem elongation, whereas RGL2, RGA, and RGL1 repress the transition to flowering. RGL2 is the main repressor of seed germination, although RGA, GAI, and RGL3 also contribute. Mutations in RGL2 rescue the germination phenotypes of the ga1-3 biosynthesis mutant and of sly1 alleles (Lee et al., 2002; Tyler et al., 2004; Ariizumi and Steber, 2007).
GA lifts the DELLA repression of GA responses by triggering DELLA proteolysis through the ubiquitin proteasome pathway (Wang et al., 2009). The GA receptor GID1 undergoes a conformational change when it binds GA that stimulates DELLA protein binding to form the GID1-GA-DELLA complex (Murase et al., 2008; Shimada et al., 2008; Ueguchi-Tanaka et al., 2008; Hirano et al., 2010). GID1 binding to DELLA protein requires the N-terminal DELLA regulatory domain (Dill and Sun, 2001; Ueguchi-Tanaka et al., 2007). GID1-GA-DELLA complex formation stimulates DELLA recognition by the F-box protein SLY1, leading to DELLA polyubiquitinaiton, which in turn targets DELLA for destruction by the 26S proteasome (McGinnis et al., 2003; Dill et al., 2004; Fu et al., 2004). SLY1 is the F-box subunit of the SCF (for Skp, Cullin, F-box) E3 ubiquitin ligase that is the main SCF responsible for the recognition and proteasomal targeting of DELLA (Ariizumi et al., 2011). There are three GID1 homologs in Arabidopsis, GID1a, GID1b, and GID1c, all of which bind GA, form stable complexes with DELLAs, and show overlapping function in GA signaling (Griffiths et al., 2006; Willige et al., 2007). GID1b has a higher binding affinity for bioactive GA and shows limited ability to bind DELLA in the absence of GA (Nakajima et al., 2006; Suzuki et al., 2009; Yamamoto et al., 2010). Triple gid1a gid1b gid1c mutants and loss of sly1 function result in GA-insensitive phenotypes associated with an inability to degrade DELLA protein, indicating that these genes are positive regulators of GA signaling because they are negative regulators of DELLA repressors. Both sly1 and gid1a gid1b gid1c mutants result in either increased seed dormancy or a complete failure to germinate (Griffiths et al., 2006; Ariizumi and Steber, 2007; Iuchi et al., 2007; Willige et al., 2007). However, single and double gid1a, gid1b, and gid1c mutants cause no or little change in germination capacity (Willige et al., 2007).
The GA receptor GID1 can down-regulate DELLA repressors of GA signaling via both proteolysis-dependent and proteolysis-independent mechanisms (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). If GA signaling is controlled only by DELLA degradation, then we would expect the level of DELLA accumulation to positively correlate with the severity of GA signaling phenotypes. In contrast, the sly1 F-box mutants of Arabidopsis have weaker GA-insensitive seed dormancy, plant height, and infertility phenotypes than ga1-3 and gid1a gid1b gid1c mutants, but they accumulate higher levels of DELLA proteins (McGinnis et al., 2003; Griffiths et al., 2006; Willige et al., 2007; Ariizumi et al., 2008). This suggests that some GA signaling can occur in sly1 mutants even in the presence of DELLA repressors. This sly1 and proteolysis-independent GA signaling requires GA and the DELLA domain needed for the formation of the GID1-GA-DELLA complex. GID1 overexpression and GA application partly rescue sly1 dwarf and infertility phenotypes with no reduction in DELLA protein levels (Ariizumi et al., 2008). Thus, there appears to be a mechanism for GA signaling that does not require DELLA proteolysis but does require DELLA-GID1 protein-protein interaction.
This study examines the GA response mechanisms, especially those controlling germination, in the sly1 mutant background, where DELLA protein is not regulated by the ubiquitin-proteasome pathway. The sly1-2 mutant can be rescued by long after-ripening, abi1-1 mutations, and GID1 overexpression without any decrease in DELLA RGL2 protein levels. Rescue of sly1-2 by after-ripening and GID1 overexpression is associated with decreased ABA accumulation, suggesting that decreased ABA signaling is one mechanism allowing the rescue of sly1-2 by after-ripening and GID1 overexpression. After-ripening also leads to a marked increase in GA hormone accumulation. Both after-ripening and GID1 overexpression lead to an increase in GA-regulated gene expression, suggesting that proteolysis-independent GA signaling is also involved in sly1-2 rescue by after-ripening and GID1 overexpression. This notion is further supported by the fact that gid1 mutations interfere with the ability of sly1-2 to after-ripen as well as with other aspects of GA signaling in the sly1-2 mutant. Finally, this study finds a correlation between the capacity to germinate and GID1-GA-DELLA complex formation, providing, to our knowledge, the first in planta evidence that GID1 relieves seed dormancy through the down-regulation of DELLA via direct protein-protein interaction.
RESULTS
GA Receptor GID1 Overexpression and the abi1-1 Mutation Partially Rescue sly1 Mutant Seed Germination without Triggering DELLA Proteolysis
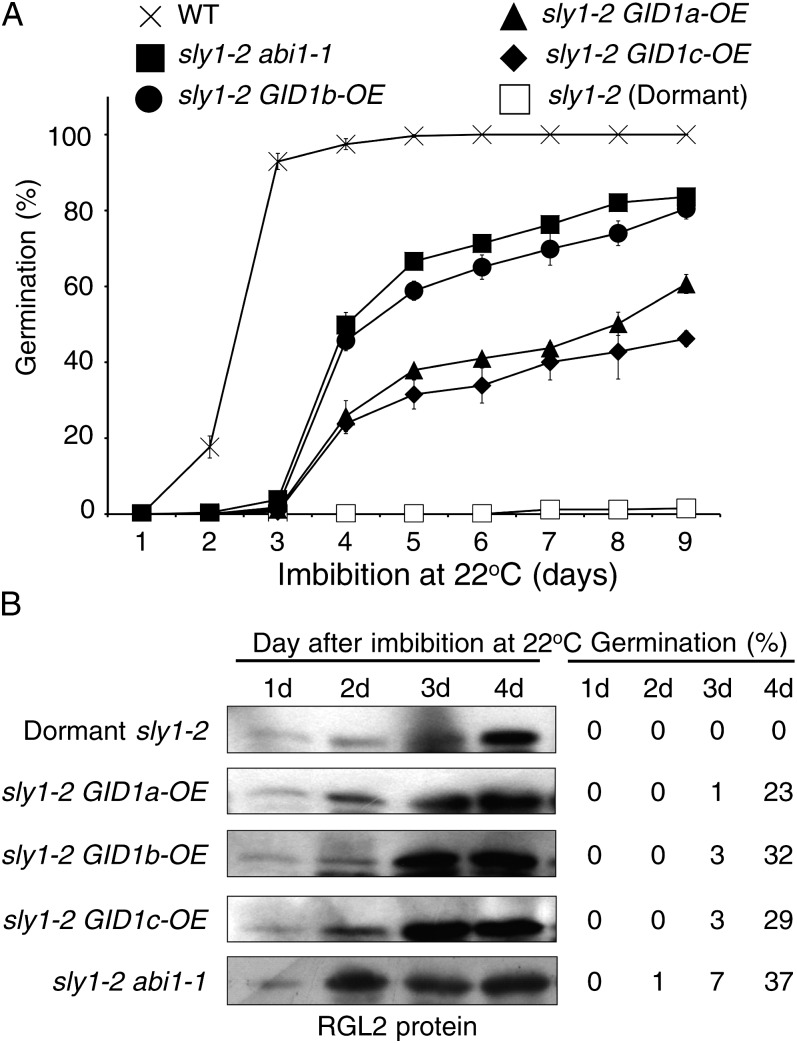
Previous research showed that GID1 overexpression can rescue plant height and flowering phenotypes of sly1 mutants (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). To explore the role of GID1 in controlling seed germination, we examined whether GID1 overexpression could rescue sly1 seed germination. The effect of HA:GID1a, HA:GID1b, and HA:GID1c overexpression on the constitutive cauliflower mosaic virus 35S promoter (lines called GID1a-OE, GID1b-OE, GID1c-OE) on seed germination was examined in sly1-2 seeds in the Landsberg erecta (Ler) background (Supplemental Fig. S1A). All seeds were after-ripened for 2 weeks. Dormant sly1-2 seeds showed very low germination over 9 d of imbibition, whereas GID1 overexpression resulted in partial restoration of seed germination (Fig. 1A). GID1b-OE in sly1-2 resulted in 81% seed germination, and GID1a-OE and GID1c-OE resulted in 61% and 46% germination, respectively. The rescue of sly1-2 seed germination by GID1b-OE was similar to rescue resulting from the abi1-1 ABA-insensitive mutation (84%; Fig. 1A; Steber et al., 1998). Previous research showed that abi1-1 causes increased germination of sly1-2 in the presence of ABA (Steber et al., 1998). GID1b-OE did not allow sly1-2 to germinate on 0.3 μm ABA, suggesting that abi1-1 and GID1b-OE rescue sly1-2 germination by different mechanisms (Supplemental Fig. S1B).
Figure 1.
Rescue of sly1 germination by GID1 overexpression is not associated with decreased RGL2 protein levels. A, Germination of 0.5-month-old wild-type (WT), sly1-2, sly1-2 abi1-1, sly1-2 GID1a-OE, sly1-2 GID1b-OE, and sly1-2 GID1c-OE seeds imbibed on 0.5× MS-moistened filter paper at 22°C. Percentage germination was scored daily. B, Accumulation of RGL2 protein during the time course of seed imbibition. Seeds were imbibed on filter paper at 22°C and harvested daily over 4 d for protein-blot analysis of RGL2 accumulation. Corresponding germination rates are shown.
Long after-ripening can rescue sly1-10 and sly1-2 seed germination without causing a decrease in DELLA RGL2 or RGA protein accumulation (Ariizumi and Steber, 2007). Protein-blot analysis was used to determine whether the rescue of sly1-2 seed germination by GID1 overexpression or by the abi1-1 mutation was associated with DELLA RGL2 protein disappearance during a time course of seed imbibition (Fig. 1B; Supplemental Fig. S1B). No decrease in DELLA RGL2 protein accumulation was observed in the sly1-2 abi1-1 line or in sly1-2 lines overexpressing HA:GID1a, HA:GIDb, and HA:GIDc over 4 d of seed imbibition. This result suggests that, like after-ripening, abi1-1 and GID1 overexpression suppression of sly1 dormancy is independent of DELLA proteolysis.
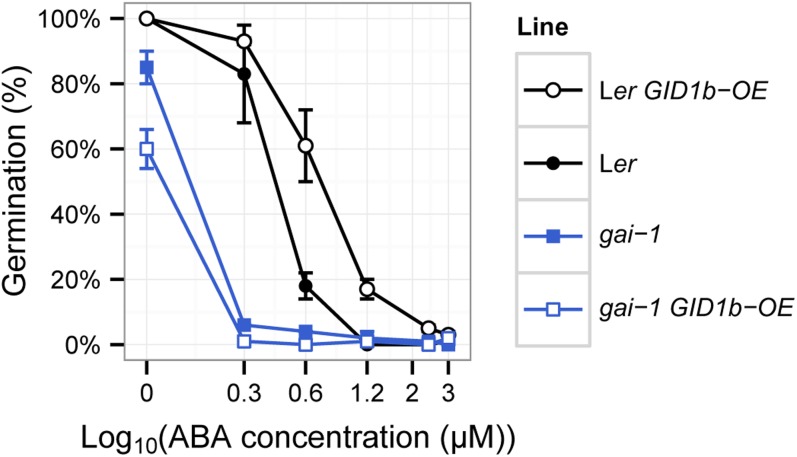
An ABA dose-response experiment was used to examine the effect of GID1b-OE on the germination potential of wild-type Ler and gai-1, the gain-of-function DELLA mutant that lacks the 17-amino acid DELLA domain required for interaction with GID1 proteins. While the gai-1 mutant shows 82% germination in the absence of ABA, it is hypersensitive to 0.3 and 0.6 μm ABA (Fig. 2). GID1b overexpression caused a significant increase in the ability to Ler to germinate in the presence of 0.6 μm ABA (20% versus 59% germination) and at 1.2 μm ABA. In contrast, GID1b-OE caused no increase in the germination of gai-1 in the presence of ABA. This result suggests that the increased germination potential resulting from GID1b-OE is not specific to the sly1-2 mutant background and that this increased germination potential depends, at least in part, on the DELLA domain of GAI.
Figure 2.
Overexpression of GID1b in the gai1-1 background does not reduce the ABA sensitivity of gai-1. An ABA dose-response curve was performed using wild-type Ler, Ler GID1b-OE, gai-1, and gai-1 GID1b-OE seeds. Seeds were incubated on MS-agar containing the indicated ABA concentrations at 4°C for 3 d followed by 96 h at 22°C. Error bars indicate se for three replicates (n = 30). [See online article for color version of this figure.]
Bioactive GA and ABA Hormone Measurements in Imbibing Seeds
If the increased seed dormancy of sly1-2 results mainly from elevated ABA levels, then dormant sly1-2 seeds (2 weeks after-ripened) should show higher ABA levels than sly1-2 after-ripened for 2 years or sly1-2 seeds (2 weeks after-ripened) whose germination capacity is restored by GID1 overexpression. Mass spectrometry was used to examine endogenous levels of ABA and bioactive GA (Fig. 3; Supplemental Fig. S2). Seeds of wild-type Ler, dormant and after-ripened sly1-2, and sly1-2 GID1b-OE were imbibed for 60 h at 4°C, followed by 12 h at 22°C, and then harvested for hormone measurement. Dormant sly1-2 seeds have higher endogenous ABA levels than the wild type (P < 0.001). ABA levels are lower in after-ripened sly1-2 and in sly1-2 GID1b-OE seeds than in dormant sly1-2 seeds (P < 0.05). This is a small, but statistically significant, decrease based on a two-sided Student’s t test, suggesting that the lower ABA levels contribute to the increased seed germination capacity (Supplemental Fig. S2). Dormant sly1-2 seeds accumulated significantly higher GA4 and GA1 levels than did wild-type Ler (P < 0.05). After-ripening for 2 years results in a highly significant increase in GA4 levels compared with dormant sly1-2 (P < 0.0001), suggesting that increased GA signaling contributes to the rescue of sly1-2 germination by after-ripening. GID1b-OE did not result in a significant change in GA hormone levels compared with dormant sly1-2. Thus, it appears that sly1-2 seed germination can be partly rescued through decreased ABA signaling, through increased GA signaling, or through both.
Figure 3.
Hormone measurement of imbibing seeds. Hormone measurements were conducted using seeds after-ripened for 2 weeks except for sly1-2(AR), which was after-ripened for 28 months. All seeds were imbibed on MS-agar at 4°C for 60 h followed by 22°C for 12 h. ABA and GA4 levels are averages of six to 10 samples. Error bars show se. DW, Dry weight.
After-ripening and Germination of sly1-2 Seeds Require GID1a
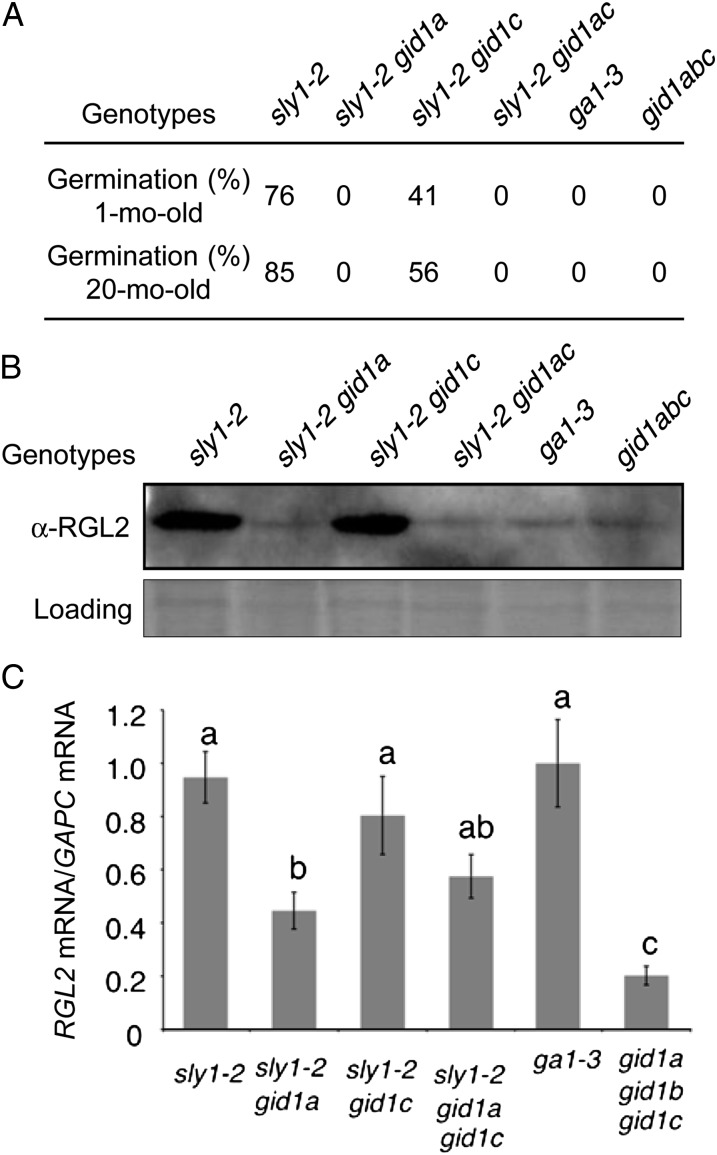
If after-ripening of sly1-2 results mainly from decreased ABA signaling, then sly1-2 after-ripening should be independent of the presence of the GA receptor GID1. Thus, we examined the effect of gid1 mutations on the seed dormancy phenotype of sly1-2 in the Columbia (Col-0) ecotype. Introduction of gid1a-1 and gid1c-2 mutations into sly1-2 resulted in decreased seed germination (Fig. 4A; Supplemental Fig. S3). While sly1-2 and sly1-2 gid1c-2 germination improved with 20 months of after-ripening, sly1-2 gid1a-1 and sly1-2 gid1a-1 gid1c-2 seed germination did not. The germination of these and other dormant seeds was rescued only by cutting the seed coat. The sly1-2 gid1b-1 double mutant also failed to germinate but was not analyzed carefully due to low fertility. Next, we examined whether the increased dormancy of the sly1-2 gid1a-1 double mutants was associated with increased accumulation of the DELLA RGL2 repressor. The increased seed dormancy of sly1-2 gid1a-1 and sly1-2 gid1a-1 gid1c-2 compared with sly1-2 was associated not with increased, but with decreased, levels of the DELLA RGL2 repressor (Fig. 4B). This situation, where a high degree of seed dormancy is accompanied by lower DELLA protein accumulation, resembles that in ga1-3 and the gid1a gid1b gid1c triple mutant (Fig. 4B; Willige et al., 2007). The reduced RGL2 protein levels in sly1-2 gid1a-1 and in sly1-2 gid1c-2 are associated with reduced RGL2 transcript accumulation, suggesting that reduced mRNA levels are at least partly responsible. It appears that while there is less DELLA RGL2 in sly1-2 gid1a-1 than in sly1-2, this DELLA protein is more active as a repressor of GA signaling and seed germination.
Figure 4.
GID1a is required for after-ripening and controls RGL2 protein accumulation in the sly1 mutant background. A, GID1a is needed for after-ripening of sly1-2. Germination is shown for seeds after-ripened for 1 and 20 months incubated on MS-agar plates for 3 d at 4°C followed by 7 d at 22°C. B and C, Seeds after-ripened for 20 months were incubated on MS-moistened filter paper for 3 d at 4°C followed by 48 h at 22°C and then used to examine RGL2 protein (B) and RGL2 mRNA (C) accumulation by protein-blot and RT-PCR analyses, respectively. Total protein (60 µg) was loaded, and Ponceau staining was used as a loading control.
GA Signaling in sly1 Shoots and Flowers Requires GID1
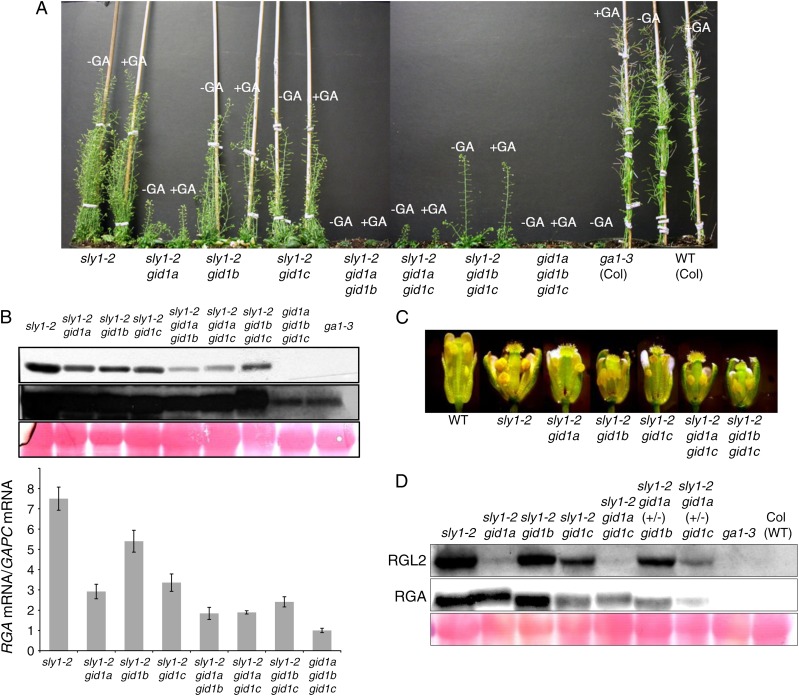
Although sly1 mutants have higher levels of DELLA repressor protein than either ga1-3 or the gid1a gid1b gid1c triple mutant, sly1 mutants show less extreme GA-insensitive phenotypes, suggesting that some GA signaling can occur in the absence of DELLA destruction (Ariizumi et al., 2008). If GA signaling in sly1 mutants results from a GID1-dependent mechanism, then we would expect gid1 mutations to increase the severity of sly1 plant height, flowering, and fertility phenotypes. To examine this, sly1-2 gid1a-1, sly1-2 gid1b-1, and sly1-2 gid1c-2 double and triple mutants lines were constructed in the Col-0 ecotype (Fig. 5). Both the sly1-2 gid1a-1 and sly1-2 gid1c-2 double mutants showed a significant decrease in final plant height and fertility compared with sly1-2, with sly1-2 gid1a-1 showing the strongest dwarfism of the double mutants (Fig. 5A; Supplemental Fig. S4). The gid1a-1 mutation significantly reduced the plant height and fertility of sly1-2, even in the heterozygous condition (sly1-2/sly1-2 GID1a+/gid1a-1), indicating that gid1a can be codominant (Supplemental Fig. S4). Even stronger plant height and infertility phenotypes were observed in triple mutant combinations. It appears that GID1b also contributes to stem elongation, since the sly1-2 gid1a-1 gid1b-1 triple mutant shows the most extreme dwarfism (Fig. 5A; Supplemental Fig. S5B).
Figure 5.
Additive effects of gid1 and sly1-2 mutations on plant height, fertility, and associated DELLA protein levels. A, Sixty-five-day-old wild-type (WT) and homozygous mutant plants with and without 100 µm GA4 treatment (every 3 d). B, Vegetative DELLA RGA protein accumulation appeared to depend on the gid1 genotype in protein-blot analysis of 40 µg of total protein extracted from 28-d-old whole plants. Two exposures of the same blot are shown with Ponceau staining as a loading control. Normalized RGA mRNA accumulation based on the mean values from three independent qRT-PCR experiments is plotted on the histogram. Error bars show se. C, Flower morphology at anthesis. D, DELLA RGA and RGL2 protein accumulation in these flowers depended on the presence of GID1 genes. [See online article for color version of this figure.]
Introduction of the gid1b-1 allele into sly1-2 resulted in the strongest infertility and floral development phenotypes. The sly1-2 gid1b-1 double mutant was almost completely infertile, and the sly1-2 gid1a gid1b and sly1-2 gid1b gid1c triple mutants were completely infertile (Supplemental Fig. S4). The increasing infertility of the sly1-2 gid1b-1 double and triple mutants was associated with increasingly defective floral development (Fig. 5C). Compared with sly1-2, the sly1-2 gid1b-1 and sly1-2 gid1b-1 gid1c-2 lines had smaller flowers with underdeveloped carpels, smaller petals, shorter filaments, and small anthers showing failed pollen dehiscence. Interestingly, the sly1-2 gid1b-1 and sly1-2 gid1b-1 gid1c-2 mutants were slightly taller than sly1-2 at maturity (Supplemental Fig. S5A). Possibly, these multiple mutants continued to grow taller and make flowers to compensate for extreme infertility. The sly1-2 gid1a-1 gid1b-1 triple mutant is similar to the gid1a-1 gid1b-1 gid1c-2 triple mutant in that it did not transition to flowering within 20 weeks of growth. Thus, it appears that GID1b makes a strong contribution to GA signaling in flowers.
Since DELLAs are negative regulators of GA responses, one would expect stronger GA-insensitive phenotypes to be associated with higher DELLA protein accumulation. Consistent with this, GA rescue of ga1-3 seed germination is associated with DELLA RGL2 protein disappearance (Supplemental Fig. S5C; Tyler et al., 2004; Ariizumi and Steber, 2007). Paradoxically, the ga1-3 and gid1a gid1b gid1c mutants accumulate less DELLA protein than sly1 mutants in spite of the fact that sly1 mutants are less infertile and less severely dwarfed (McGinnis et al., 2003; Griffiths et al., 2006; Willige et al., 2007; Ariizumi et al., 2008). Mutations in gid1 and sly1-2 had additive effects on DELLA RGA protein accumulation. The sly1 gid1 multiple mutants had higher levels of RGA protein accumulation than the gid1a gid1b gid1c and ga1-3 mutants but lower RGA levels than sly1-2 (Fig. 5B). This reduction was more apparent in sly1-2 gid1a-1 gid1b-1 and in sly1-2 gid1a-1 gid1c-2 than in sly1-2 gid1a-1, sly1-2 gid1b-1, and sly1-2 gid1c-2. Reduced RGA protein levels roughly correlated with decreased RGA mRNA accumulation, suggesting that the changes in protein level may result from changes in transcript level (Fig. 5B). Introduction of gid1 mutations into sly1-2 also resulted in decreased DELLA RGL2 and RGA protein accumulation in flowers (Fig. 5D). The RGL2 protein level was especially sensitive to the gid1a-1 mutation. Thus, gid1 mutations are partly epistatic to sly1 for the DELLA protein accumulation phenotype (Fig. 5, B and D).
The Level of GID1-DELLA Interaction Correlates with the Level of GID1 Suppression
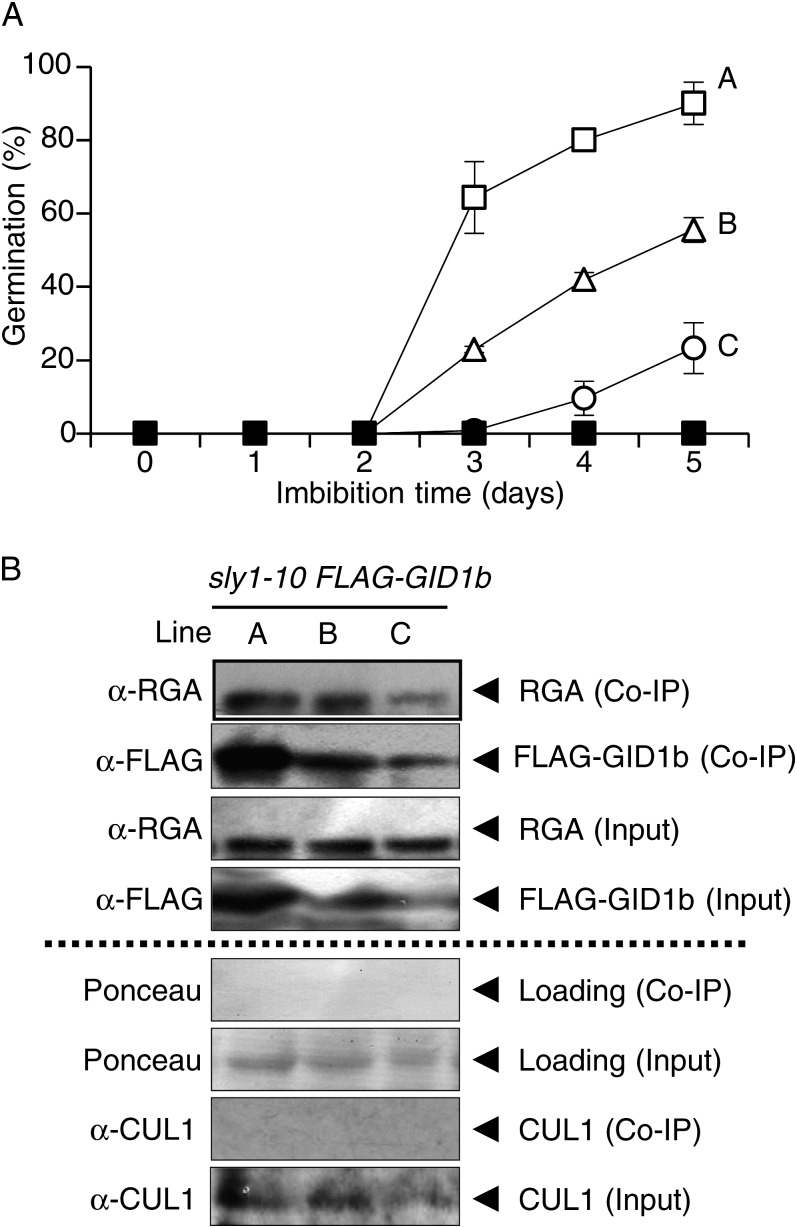
GID1 overexpression may rescue sly1 seed germination through two nonproteolytic mechanisms: by GA-stimulated inactivation of DELLA repressors through direct protein-protein interaction or by bypassing the need for DELLA destruction through direct down-regulation of ABA signaling. If GID1 blocks DELLA repression of sly1 seed germination through protein-protein interaction, then lines expressing higher levels of GID1b should show better rescue of seed germination associated with increased GID1-DELLA protein-protein interaction. To examine this, three sly1-10 35S:FLAG:GID1b overexpression lines were identified showing a range of seed germination potentials when incubated on Murashige and Skoog (MS)-moistened filter paper for 5 d at 22°C (Fig. 6A). Each line showed partial rescue of sly1-10 germination: line A showed the highest germination at 93% at 5 d, ranging down to line C at 21% germination after 5 d. Higher germination potential was correlated with higher FLAG:GID1b accumulation when FLAG:GID1b was immunoprecipitated from protein extracts of lines A through C (Fig. 6B). The lines showing higher germination potential and higher FLAG:GID1b accumulation also showed higher coimmunoprecipitation (co-IP) of DELLA RGA protein (Fig. 6B). This correlation suggests that GID1b overexpression rescues seed germination through increasing the fraction of DELLA RGA protein in complex with GID1 protein.
Figure 6.
Higher germination potential correlates with higher accumulation of FLAG-GID1b and formation of the GID1b-DELLA complex. A, Germination rates of sly1-10 (black squares) and three independent sly1-10 FLAG-GID1b seed lines (A, B, and C) sown on MS-moistened filter paper and incubated 5 d at 22°C. Under these conditions, sly1-10 does not germinate. B, A co-IP experiment performed using protein extracted from the indicated sly1-10 FLAG-GID1b seeds imbibed for 48 h at 22°C. Total protein was incubated with anti-FLAG matrix agarose. Immunoprecipitated FLAG-GID1b was detected by immunoblot with anti-FLAG, and DELLA RGA in complex with GID1b was detected with anti-RGA. Total protein (60 mg) was loaded (input). A Ponceau-stained membrane provided a loading control.
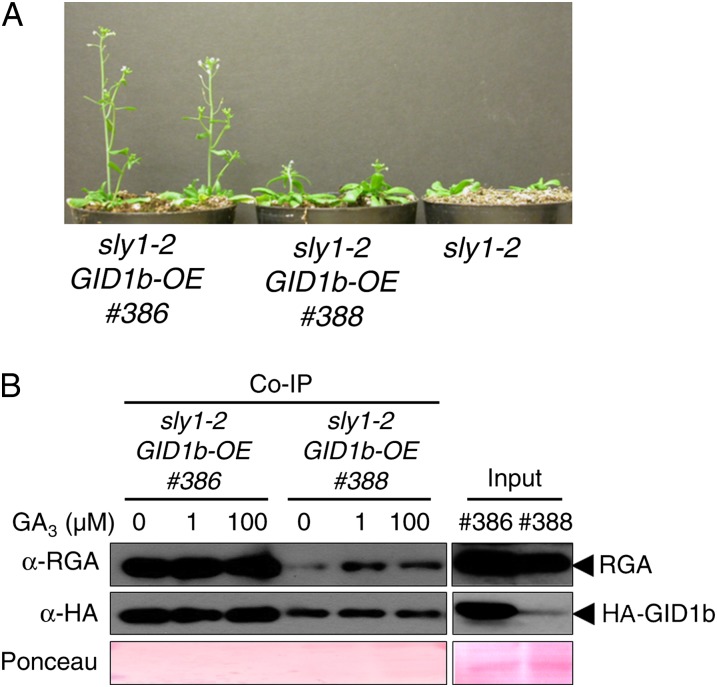
To examine whether this is also true for rescue of the sly1 plant height phenotype, we compared two independent sly1-2 HA-GID1b-OE lines that express HA-GID1b at different levels. Line 386 shows higher level HA-GID1b protein accumulation and faster vegetative plant growth than line 388 (Fig. 7; Supplemental Fig. S5D). The higher levels of HA-GID1b expression in line 386 also correlated with co-IP of more DELLA RGA protein from 12-d-old plants (Fig. 7B). The correlation of plant height and germination phenotypes with DELLA co-IP supports the model that GID1b overexpression suppresses sly1 phenotypes via direct GID1-DELLA protein-protein interaction. When the germination phenotype of the sly1-2 HA-GID1b lines was examined, higher level accumulation of HA:GID1b was associated with higher sly1-2 germination potential (Supplemental Fig. S6A). At day 7, the high line reached 66% germination, the low line reached 49%, and untransformed sly1-2 seeds reached 1% seed germination. After-ripening of sly1-2 seeds for 24 months resulted in 39% germination at day 7, indicating that germination rescue due to HA:GID1b overexpression was even more effective than after-ripening in rescuing sly1-2 seed germination. The higher level of HA:GID1b protein accumulation was also correlated with higher co-IP of DELLA RGL2 (Supplemental Fig. S6B). Thus, it appears that GID1b overexpression rescues sly1-2 and sly1-10 seed germination through increasing the fraction of DELLA RGL2 and RGA protein in complex with GID1 protein.
Figure 7.
HA-GID1b protein levels and HA-GID1b-RGA complex formation correlate with the suppression of sly1-2 dwarfism. A, sly1-2 plants expressing higher (line 386) or lower (line 388) levels of the HA-GID1b fusion protein during stem elongation. B, HA-GID1b was immunoprecipitated from protein extracted from 12-d-old sly1-2 plants expressing either high (line 386) or low (line 388) levels of HA-GID1b protein. Coimmunoprecipitated RGA was detected by protein-blot analysis. Protein extract was incubated with HA matrix agarose in the presence of 0.1% ethanol (mock), 1 µm GA3, and 100 µm GA3 and loaded on an SDS-PAGE gel. Protein-blot analysis was performed using anti-RGA and anti-HA. Protein extract (40 µg) was loaded (input). [See online article for color version of this figure.]
Suppression of the sly1-2 Germination Phenotype Is Associated with Increased GA-Responsive Gene Expression and Decreased GA20ox and XERICO Expression
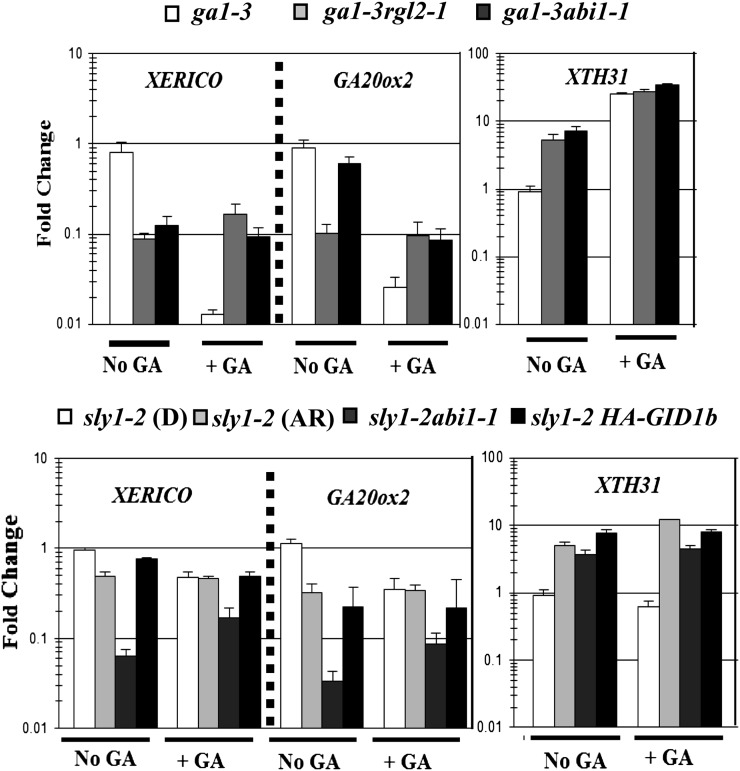
To examine whether GID1b overexpression rescues sly1-2 seed germination via increased GA signaling, the expression patterns of six GA-inducible genes were examined in seeds imbibed in the presence and absence of 10 μm GA3 by semiquantitative reverse transcription (RT)-PCR (Supplemental Fig. S7A), while the expression of the GA-induced gene XTH31 was examined by quantitative reverse transcription (qRT)-PCR (Fig. 8; Herzog et al., 1995; Ogawa et al., 2003; Yamauchi et al., 2004; Cao et al., 2006; Ariizumi and Steber, 2007). Seeds of ga1-3, ga1-3 rgl2-1, and ga1-3 abi1-1 were included as controls showing GA-responsive gene expression resulting from GA treatment, loss of DELLA RGL2 repression, and ABA insensitivity, respectively (Steber et al., 1998; Cao et al., 2005; Ariizumi and Steber, 2007). All of the seed stocks compared were after-ripened for 2 weeks, except for the after-ripened sly1-2 seed stock, which was after-ripened for 28 months. Dormant sly1-2 seeds fail to show GA-induced gene expression, and increased GA-inducible mRNA accumulation is associated with a loss of sly1-2 seed dormancy (Ariizumi and Steber, 2007; Supplemental Fig. S7A). After-ripening, GID1b overexpression, and the abi1-1 mutation all result in increased expression of GA-inducible genes in sly1-2 without exogenous GA3 application (Figs. 7A and 8). GA3 treatment results in increased XTH31 mRNA accumulation only in after-ripened sly1-2 seeds (Fig. 8). Thus, it appears that all three conditions rescue sly1-2 germination, at least in part, through increasing the expression of GA-inducible genes.
Figure 8.
Effect of GID1 overexpression on gene expression in seeds. The expression of the ABA biosynthesis gene, XERICO, GA20ox2, and XTH31 was examined by qRT-PCR. For each experiment, total was RNA extracted from seeds imbibed on MS-agar for 60 h at 4°C followed by 24 h at 22°C and then incubated for a further 12 h after the addition of a mock treatment or 10 mm GA3. All seeds were dry after-ripened for 2 weeks, including dormant sly1-2 (D), except for sly1-2 after-ripened (AR) for 28 months For each condition, there were three biological replicates, and each replicate was subsampled three times. Error bars represent sd. Statistical significance was determined with an ANOVA using Tukey’s adjustment. P values are given in Supplemental Figure S8.
The observation that the abi1-1 mutation results in increased accumulation of GA-inducible transcripts in ga1-3 and in sly1-2 raised the question of whether ABA could also repress the expression of these GA-inducible genes. To examine this, GA-inducible mRNA accumulation was compared in the same after-ripened sly1-2, sly1-2 abi1-1, and sly1-2 GID1b-OE seeds imbibed with and without 10 μm ABA (Supplemental Fig. S7B). ABA treatment repressed GA-inducible transcript accumulation in after-ripened sly1-2 and sly1-2 GID1b-OE but not in sly1-2 abi1-1. Thus, ABA and GA have opposing effects on the expression of these GA-responsive genes.
If changes in sly1 seed germination result from changes in DELLA repression, then we would expect this to result in changes in DELLA-regulated gene expression. The positive regulator of ABA biosynthesis, XERICO, and the GA biosynthesis gene GA20ox2 were identified as DELLA RGA-activated genes in vegetative tissue by chromatin immunoprecipitation (Zentella et al., 2007). Thus, the effect of after-ripening, GID1b-OE, and the abi1-1 mutation on the expression of these genes was examined by qRT-PCR in sly1-2 seeds (Fig. 8). Both GA treatment and the DELLA rgl2-1 mutation significantly decreased XERICO and GA20ox2 mRNA accumulation in ga1-3, indicating that DELLA RGL2 also activates the expression of these transcripts in seeds (Supplemental Fig. S8A). GA20ox2 and XERICO transcript levels are highest in dormant sly1-2 seeds and decrease significantly as a result of after-ripening (3.5- and 2-fold), GID1b overexpression (5- and 1.2-fold), and the abi1-1 mutation (15.5- and 37-fold; Fig. 8; for P values, see Supplemental Fig. S8). The abundance of GA20ox2 transcript in dormant and after-ripened sly1-2 seeds likely contributes to higher bioactive GA accumulation (Figs. 3 and 8). GA20ox2 and XERICO mRNA levels in dormant sly1-2 seeds are both reduced by GA treatment, suggesting that DELLA down-regulation does not require the SLY1 gene. XERICO and GA20ox2 mRNA levels in sly1-2 are lowered by conditions that rescue germination, including after-ripening, GID1b overexpression, and the abi1-1 mutation. Collectively, these data suggests that all three mechanisms that rescue sly1 seed germination interfere with the ability of DELLA to activate XERICO gene expression. Moreover, since this rescue does not require sly1, it appears that this involves down-regulation of the DELLA activator of XERICO and GA20ox2 transcription without DELLA proteolysis.
DISCUSSION
Nonproteolytic Regulation of DELLA Repressors of GA Signaling
This study examines the role of GID1-GA-DELLA protein complex formation in the nonproteolytic down-regulation of DELLA repressors of GA signaling. In the classic model of GA signaling, DELLA destruction by the ubiquitin-proteasome pathway lifts DELLA repression, thereby stimulating GA responses, including stem elongation, flowering, and seed germination. Based on this model, higher DELLA protein levels should always correlate with stronger GA-insensitive phenotypes. In contrast, the sly1 mutants accumulate more DELLA protein than ga1-3 or gid1a gid1b gid1c mutants but have weaker GA-insensitive phenotypes (Ariizumi et al., 2008). Previous research showed that some vegetative GA signaling occurs in sly1 mutants of Arabidopsis and in the corresponding gid2 mutants of rice (Oryza sativa) in the absence of DELLA protein destruction (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). GA signaling also occurs during the rescue of sly1 seed germination in the absence of DELLA destruction (Figs. 1 and 8). To our knowledge, this is the first study to demonstrate GID1-DELLA complex formation in an F-box mutant and the first study to show that the degree of sly1 rescue correlates with the degree of GID1b-DELLA complex formation (Figs. 6 and 7; Supplemental Fig. S6). Moreover, to our knowledge, this is the first study to show that after-ripening is associated not only with a decrease in ABA accumulation but with an increase in GA accumulation using the sly1-2 background (Fig. 3).
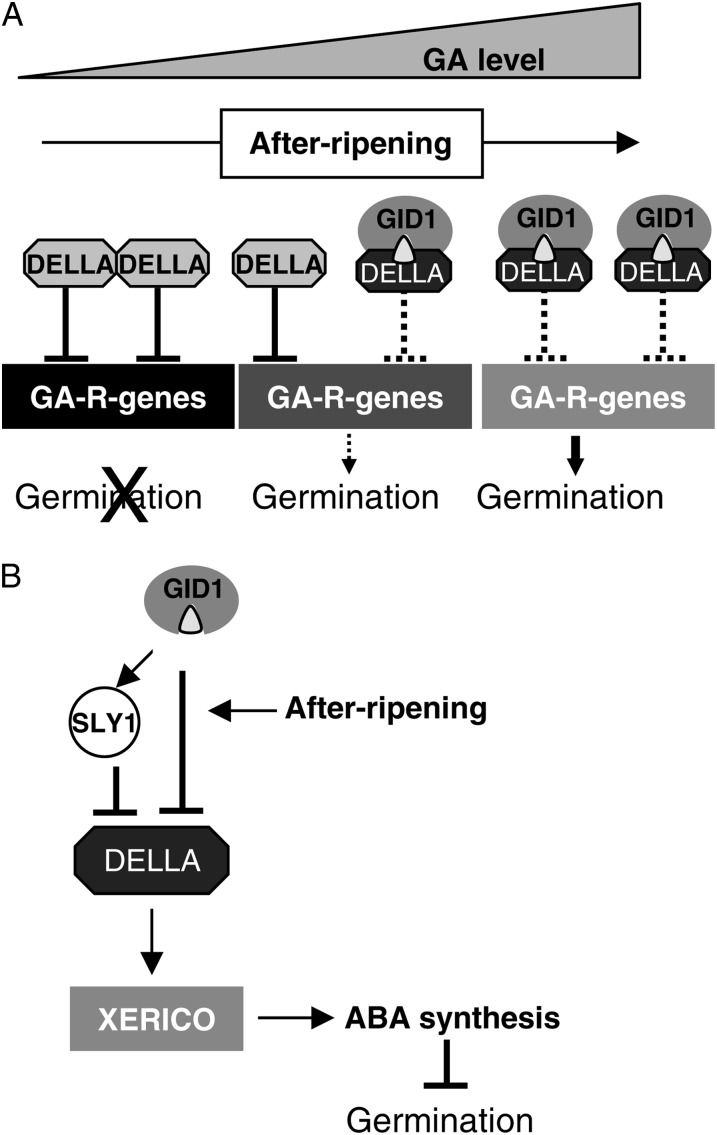
Seed germination was used as a system to examine nonproteolytic GA signaling, because the sly1-2 seed dormancy phenotypes are partly rescued not only by GID1 overexpression but by dry after-ripening and the abi1-1 mutation (Fig. 1). In all three cases, sly1 seed germination occurs in the presence of high levels of the DELLA RGL2 repressor of seed germination. It appears that the seed dormancy in sly1-2 seeds results from downstream ABA signaling, because dormant sly1-2 seeds have elevated ABA hormone levels, whereas nonproteolytic rescue of sly1-2 germination by GID1 overexpression and after-ripening are associated with reduced ABA levels (Fig. 3). The model that after-ripening relieves sly1-2 seed dormancy through increased GID1-GA-DELLA complex formation is based on the observations that GID1a is required for sly1 after-ripening and that GA levels rise with after-ripening (Fig. 9A). It appears that ABA signaling is at least partly responsible for sly1 dormancy, since germination rescue by after-ripening and GID1b-OE is associated with reduced ABA levels (Fig. 3). Since GID1a is required for sly1 after-ripening, and since GA levels rise with after-ripening, it appears that increased GID1-GA-DELLA complex formation may result in nonproteolytic rescue of sly1 germination through after-ripening (Fig. 9A). Increasing GA levels should stimulate GID1-GA-DELLA complex formation. The importance of GID1-GA-DELLA complex formation in lifting sly1 seed dormancy is demonstrated by the fact that the degree of GID1-GA-DELLA complex formation correlates with the degree of seed germination rescue (Fig. 6; Supplemental Fig. S6). Moreover, rescue of sly1-2 germination by after-ripening and GID1b-OE occurs via GA signaling, since rescue is associated with increased GA-inducible mRNA levels (Fig. 8; Supplemental Fig. S7). The high DELLA protein level in sly1-2 seeds is associated with high levels of the DELLA-induced transcripts XERICO and GA20ox2. Since XERICO is a positive regulator of ABA biosynthesis (Ko et al., 2006), this suggests that the strong dormancy in sly1 seeds results from DELLA-induced ABA biosynthesis as well as a lack of GA-inducible gene expression. Nonproteolytic rescue of sly1 germination by after-ripening and GID1b-OE is associated with decreased XERICO mRNA and ABA accumulation, suggesting that DELLA activation of XERICO can be prevented through GID1-GA-DELLA complex formation (Fig. 9B).
Figure 9.
Model for the rescue of sly1 seed germination by GID1-GA-DELLA complex formation. A, After-ripening of sly1-2 stimulates seed germination by increasing endogenous GA levels that promote GID1-GA-DELLA complex formation. GID1 overexpression also increases GID1-GA-DELLA formation, thereby inactivating DELLA repression of GA-responsive (GA-R) gene expression and responses. B, GID1 can prevent DELLA induction of XERICO transcription either via SLY1-directed DELLA degradation or directly through GID1-GA-DELLA complex formation. Reduced XERICO expression results in decreased ABA accumulation in after-ripened sly1 and sly1 GID1 overexpression lines.
GID1 and GA Signaling in sly1-2
Examination of sly1 gid1 multiple mutants provided information about the functional roles of the three Arabidopsis GID1 genes in proteolysis-independent GA signaling during multiple stages of development (Figs. 4 and 5). Proteolysis-independent GA signaling in sly1 can be augmented by GID1 overexpression, resulting in partial suppression of sly1 phenotypes (Fig. 1; Ariizumi et al., 2008). This suggests that DELLA repressors can be inactivated even when they cannot be degraded. Mutations in all of the GA receptors had an additive effect with sly1-2, resulting in increased seed dormancy, dwarfism, infertility, and underdeveloped flowers (Figs. 4 and 5). This is consistent with the analysis of plant height in the gid1 gid2 double mutants of rice (Ueguchi-Tanaka et al., 2008). Based on sly1 gid1 double mutant analysis, it appears that GID1b plays an important role in regulating floral development and fertility, while GID1a plays an important role in stimulating stem elongation in the absence of DELLA proteolysis (Fig. 5; Supplemental Fig. S4). Paradoxically, the increased severity of GA-insensitive phenotypes in sly1 gid1 double mutants was associated not with increased DELLA repressor accumulation but with decreased DELLA RGA or RGL2 protein accumulation in whole plants, flowers, and seeds (Figs. 4 and 5). The reduction in DELLA protein accumulation correlated with the number of gid1 mutant alleles (Fig. 5, B and D). The gid1a-1 gid1b-1 gid1c-2 triple and single mutants show far less DELLA protein accumulation than the sly1 mutants (Willige et al., 2007; Fig. 5D). The correlation between reduced DELLA protein and mRNA levels suggests that GID1 is needed for the high DELLA mRNA and protein levels seen in sly1 mutants (Fig. 5B). Down-regulation of DELLA protein activity by GID1 and GA may be followed by up-regulation of DELLA gene transcription as a feedback response.
The sly1-2 allele has increased seed dormancy and can require 2 years to after-ripen well (Ariizumi and Steber, 2007). Both the sly1-2 gid1a and sly1-2 gid1c double mutants show higher seed dormancy than the sly1-2 mutant (Fig. 4). Whereas sly1-2 typically after-ripens within 15 to 24 months, sly1-2 gid1a completely failed to germinate after 20 months of after-ripening. While the gid1a-1 gid1b-1 gid1c-2 triple mutant fails to germinate, all of the gid1 single and double mutant combinations are able to germinate (Griffiths et al., 2006; Willige et al., 2007). Thus, it is only in the context of a sly1 mutant that a single gid1a allele blocks after-ripening. Although the role of GID1b in sly1-2 after-ripening could not be well characterized due to sly1-2 gid1b-1 infertility, HA-GID1b overexpression stimulated sly1-2 seed germination more strongly than HA-GID1a or HA-GID1c (Fig. 1). It is known that GID1b protein has higher affinity for DELLA than GID1a and GID1c, even in the absence of GA (Suzuki et al., 2009; Yamamoto et al., 2010). The fact that sly1-2 after-ripening requires GID1a suggests that GID1 overexpression and after-ripening rescue sly1-2 seed germination by similar GID1-dependent mechanisms. Additional evidence supports a role for GID1 in seed germination: the gid1b single mutant shows decreased GA sensitivity during seed germination, while the gid1c, gid1a gid1b, and gid1a gid1c mutants show an apparent increase in ABA sensitivity and slower seed germination (Iuchi et al., 2007; Voegele et al., 2011).
After-ripening of sly1-2 Seeds Is Associated with Reduced ABA Levels Associated with Reduced XERICO Expression
Rescue of sly1-2 seed germination by both after-ripening and GID1b-OE is associated with decreased accumulation of the DELLA-activated mRNA XERICO and decreased ABA hormone levels (Figs. 8 and 9). It appears that the seed dormancy in GA-insensitive sly1-2 seeds partly results from increased ABA signaling, because imbibing dormant sly1-2 seeds accumulate significantly more ABA than wild-type Ler (Fig. 3). Rescue of sly1-2 seed germination by after-ripening and GID1b-OE is associated with decreased ABA hormone levels, suggesting that ABA acts downstream of SLY1, GID1, and DELLA. This is consistent with abi1-1 rescue of sly1 seed germination and with the fact that lack of ABA biosynthesis rescues the germination of GA biosynthesis mutants (Fig. 1; Koornneef et al., 1982). Previous research showed that DELLA RGA and GAI are activators of XERICO transcription in vegetative tissue (Zentella et al., 2007; Gallego-Bartolomé et al., 2011) and that mutations in DELLA RGL2 result in decreased ABA accumulation in seeds (Piskurewicz et al., 2008). This study shows that both GA and an rgl2 mutation decrease XERICO mRNA levels in the ga1-3 mutant, indicating that DELLA RGL2 is a positive regulator of XERICO in seeds. Thus, high ABA levels in dormant sly1-2 are likely a direct consequence of high DELLA protein levels, resulting in increased transcription of the inducer of ABA biosynthesis, XERICO (Figs. 8 and 9). After-ripening and GID1b-OE result in decreased XERICO mRNA and ABA levels, likely due to proteolysis-independent inactivation of DELLA as a positive regulator of XERICO (Figs. 8 and 9B).
Rescue of sly1-2 Seed Germination Is Associated with Increased GA Signaling
The GA signaling in sly1 mutants may result from increased accumulation of bioactive GA. The GA-insensitive sly1-2 mutant has higher bioactive GA levels than the wild type, consistent with the increased expression of GA biosynthesis genes such as GA3ox1 (McGinnis et al., 2003) and GA20ox2 (Fig. 8). A similar increase in bioactive GA levels is seen in the GA-insensitive DELLA gain-of-function mutant gai-1 and is likely a feedback response to decreased GA sensitivity (Talon et al., 1990). The elevated levels of GA20ox2 in dormant sly1-2 may result from the increased accumulation of active DELLA protein in sly1, since GA20ox2 was identified as a possible DELLA target by chromatin immunoprecipitation (Zentella et al., 2007). Our data lend support to this finding, as we see a significant decrease in GA20ox2 mRNA in ga1-3 rgl2-1, after-ripened sly1-2, s1y-2 abi1-1, and sly1-2 GID1b-OE (Fig. 8; Supplemental Fig. S8). After-ripening resulted in a further increase in GA4 and GA1 hormone levels in imbibing sly1-2 seeds (Fig. 3; Supplemental Fig. S2). This is, to our knowledge, the first evidence showing an increase in GA accumulation with after-ripening of Arabidopsis seeds. An increase in precursors of bioactive GA was observed with after-ripening of imbibed barley (Hordeum vulgare) grain, indicating that this phenomenon is likely not limited to Arabidopsis sly1 mutants (Jacobsen et al., 2002). Previous research showed that GA application mildly stimulates the germination of dormant sly1-2 seeds without DELLA degradation (Ariizumi and Steber, 2007). The increase in GA levels with after-ripening may lift the DELLA repression of germination by stimulating GID1-GA-DELLA complex formation (Fig. 9A; Griffiths et al., 2006; Ariizumi et al., 2008).
Rescue of sly1-2 seed germination by after-ripening and GID1b-OE is associated with increased expression of a diverse set of GA-inducible genes (Fig. 8; Supplemental Fig. S7A). Thus, both GID1 overexpression and after-ripening appear to act via increased GA signaling. It is interesting that with the exception of SCS10, the levels of all of the GA-inducible transcripts examined increased in sly1-2 abi1-1 compared with dormant sly1-2. Since ABA treatment of after-ripened sly1-2 and sly1-2 GID1b-OE seeds resulted in decreased expression of GA-inducible transcripts, it appears that many, but not all, GA-inducible genes of Arabidopsis are also ABA repressed (Supplemental Fig. S7B). This competition between GA and ABA at the level of gene expression is consistent with previous research showing that ABA and GA have opposing effects on α-amylase gene expression in the barley aleurone (for review, see Sun and Gubler, 2004).
Evidence That GID1 Interaction with DELLA Proteins Rescues sly1-2 Phenotypes
We propose a model in which after-ripening and GID1 overexpression both rescue sly1 seed germination by stimulating GID1-GA-DELLA complex formation, which in turn down-regulates DELLA protein activity (Fig. 9A). Rescue of sly1 dwarfism and infertility by GID1 overexpression depends upon GA biosynthesis and the presence of the DELLA protein domain, both required for GID1-DELLA protein-protein interaction (Ariizumi et al., 2008). GID1b-OE can increase the germination of wild-type Ler, but not of the DELLA deletion allele gai-1, on ABA (Fig. 2). This suggests that the ability of GID1b to stimulate seed germination is mediated by the DELLA domain required for GID1-DELLA protein interaction. Higher levels of HA:GID1b and FLAG:GID1b protein expression are associated with more efficient sly1 germination and increased FLAG:GID1b-GA-RGA and HA:GID1b-GA-RGL2 complex formation, suggesting that GID1 down-regulates DELLA RGA and DELLA RGL2 through direct protein binding (Fig. 6; Supplemental Fig. S6). To our knowledge, this is the first proof that GID1-DELLA complex formation can stimulate seed germination without DELLA proteolysis. We suggest that after-ripening also inactivates DELLA by stimulating GID1-GA-DELLA complex formation, thereby blocking DELLA activation of XERICO transcription, leading to lower ABA hormone accumulation (Fig. 9B). GID1 binding to DELLA may block DELLA binding to other transcription factors to regulate gene transcription. For example, DELLA has been shown to regulate dark-induced gene expression through interaction with the transcription factors PHYTOCHROME-INTERACTING FACTOR3 (PIF3) and PIF4 (de Lucas et al., 2008; Feng et al., 2008). Future work will need to examine DELLA interaction with transcription factors regulating seed dormancy and germination. DELLA can negatively regulate GA signaling by stimulating the expression of negative regulators of GA signaling like XERICO (Zentella et al., 2007). Consistent with this, both rice and Arabidopsis DELLA proteins activate transcription when fused to a DNA-binding domain in the yeast two-hybrid system, and GID1-GA-DELLA complex formation blocks DELLA SLR1 transcriptional activation in vegetative tissue (Fu et al., 2004; Hirano et al., 2012).
This study suggests that GA signaling functions to improve seed germination potential during after-ripening. Decreased ABA levels can be detected with after-ripening of dry Arabidopsis and barley seeds, but changes in bioactive GA accumulation are difficult to detect until after seeds are imbibed (Jacobsen et al., 2002; Okamoto et al., 2006; Yamauchi et al., 2007; Barrero et al., 2009). Nevertheless, after-ripening results in increased GA sensitivity, suggesting that GA signaling elements are involved (Karssen et al., 1989). That sly1-2 seeds undergo GID1a-dependent after-ripening suggests that this process does not require DELLA proteolysis but does require the GA receptor and GA signaling. Other lines of evidence suggest that GID1 and DELLA proteins regulate seed after-ripening and germination. DELLA appears to act in the covering tissues of the seed coat and endosperm to inhibit cell wall loosening and stimulate ABA production, since the DELLA rgl2 mutation results in reduced ABA hormone levels and an RGL2 promoter-GUS fusion is expressed in imbibed Arabidopsis seed coats (Piskurewicz et al., 2008). Analysis of GID1-GUS fusions showed that GID1a and GID1c are also expressed in the endosperm and in the embryo, where they may down-regulate DELLA RGL2 (Voegele et al., 2011).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions were cultivated in Conviron growth chambers according to McGinnis et al. (2003). The infertility and dwarfism of ga1-3 were rescued through the application of 100 µm GA4 every 3 d (Fig. 5). After harvest, seeds were stored in open tubes for 2 weeks at room temperature and then stored in closed tubes either at room temperature for after-ripening or at −20°C to preserve dormancy. The following lines were constructed in the Ler ecotype: sly1-2, sly1-10, gai-1, sly1-2 abi1-1, ga1-3 rgl2-1, ga1-3 abi1-1 sly1-2 GID1a-OE, sly1-2 GID1b-OE, sly1-2 GID1c-OE, gai-1 GID1b-OE, and wild-type Ler GID1b-OE as described previously (Steber et al., 1998; Steber and McCourt, 2001; Cao et al., 2005; Ariizumi et al., 2008). The gid1a-1 gid1b-1, gid1b-1 gic1c-2, and gid1a-1 gid1c-2 double mutants were in the Col-0 ecotype (gift of C. Schwechheimer; Willige et al., 2007). The ga1-3 allele in Col-0 was a gift of T. Sun (Tyler et al., 2004; Fig. 5). The sly1-2 mutant allele, originally isolated in Ler (Steber et al., 1998), was moved into the Col-0 background through three crosses to wild-type Col-0. The BC3F4 sly1-2 was crossed to the gid1a-1 gid1b-1, gid1b-1 gic1c-2, and gid1a-1 gid1c-2 double mutants in order to derive the sly1-2 gid1a-1, sly1-2 gid1b-1, and sly1-2 gid1c-1 double mutants and the sly1-2 gid1a-1 gid1b-1, sly1-2 gid1b-1 gid1c-2, and sly1-2 gid1a-1 gid1c-2 triple mutants, respectively. The genotypes containing single and double gid1 mutations in the sly1-2 background were selected in the F2 and F3 generations by PCR analysis (Supplemental Table S1; McGinnis et al., 2003; Willige et al., 2007). Triple mutants sly1-2 gid1a-1 gid1b-1 and sly1-2 gid1b-1 gid1c-2 were maintained as heterozygotes due to complete infertility. F3 plants were used in this study (Fig. 5). 35S:FLAG-GID1b and 35S:HA-GID1 plasmid and strain construction was described previously by Ariizumi et al. (2008).
Germination Experiments
For germination experiments (Figs. 1, 2, 4, and 6; Supplemental Figs. S1C and S6A), 30 to 100 seeds were sterilized with 10% (v/v) bleach and 0.01% (w/v) SDS for 15 to 20 min and plated on 0.5× MS salts (Sigma-Aldrich) and 5 mm MES, pH 5.5, and 0.8% (w/v) agar (referred to as MS-agar plates). Percentage germination was determined following 3 d of incubation at 4°C followed by 3 to 10 d of incubation under constant light at 22°C. The average germination rate was calculated using at least three independent replicates. For Figure 2, (+/−)-ABA (Phytotechnology) was added to 0.5× MS-agar medium. For protein preparation, seeds were sown on filter paper moistened with 0.5× MS salts buffered with 5 mm MES to pH 5.5. Based on the previous characterization of the sly1 germination phenotype, we know that conditions resulting in reduced availability of water, such as sowing on filter paper, PEG8000, or soil, reduce the germination efficiency of sly1 mutants (Ariizumi and Steber, 2007). Consistent with this, sly1 mutant seeds germinated on MS-moistened filter paper in this study show lower germination potential than those on MS-agar plates (Figs. 1 and 6; Supplemental Fig. S6A). For experiments in which the effect of GID1 overexpression on sly1 mutant seed germination was examined (Figs. 1 and 6; Supplemental Fig. S6A), seeds of similar age from plants grown side by side were used, including the T4 generation of sly1-2 GID1a-OE, sly1-2 GID1b-OE, and sly1-2 GID1c-OE and T3 sly1-10 FLAG-GID1b.
RT-PCR Analysis of mRNA Expression
RT-PCR analysis was used to analyze the mRNA levels of RGL2, RGA, and GA-regulated genes. For quantitative and semiquantitative analysis of GA-regulated gene expression during seed imbibition (Fig. 8; Supplemental Fig. S7), seeds were incubated for 60 h at 4°C to stimulate germination and then incubated at 22°C for 24 h, at which time seeds were treated with 10 µm GA3, 10 µm (+/−)-ABA, or a mock treatment and then incubated for a further 12 h. No seed germination was observed at this time point. GA-induced transcripts based on previous reports (Herzog et al., 1995; Ogawa et al., 2003; Yamauchi et al., 2004; Cao et al., 2006) were analyzed using published primer pairs (Supplemental Table S1; Ariizumi and Steber, 2007).
For semiquantitative RT-PCR (Supplemental Fig. S7), RNA was isolated from imbibing seeds using a modified SDS-phenol extraction according to Martin et al. (2005; H. Nonogaki, personal communication). Genomic DNA contamination was removed using the DNA-Free RNA kit (ZYMO Research). Complementary DNA (cDNA) was generated from 5 μg of total RNA using the First-Strand cDNA Synthesis Kit (GE Healthcare). cDNA was diluted 1:10 and then used as a template for PCR amplification with Mango-Taq DNA polymerase (Bioline) for 30 to 40 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 60°C, and extension for 1 min at 72°C, followed by final extension for 5 min at 72°C. Agarose gels were stained with SYBR gold.
For qRT-PCR analysis (Fig. 8), total RNA was isolated from imbibing seeds using the phenol-chloroform extraction method of Oñate-Sánchez and Vicente-Carbajosa (2008) with an additional chloroform extraction and the use of Phase Lock (5-PRIME) to prevent contamination of the aqueous phase. cDNA was synthesized using 1 µg of total RNA (ProtoScript M-MuLV First Strand cDNA Synthesis Kit; New England Biolabs), and qRT-PCR analysis was performed using the Roche LightCycler FastStart DNA Master SYBR Green I kit. PCR conditions were as follows: 10 min of denaturation at 95°C, followed by 45 cycles of 10 s of denaturation at 95°C, 5 s of annealing at 60°C, and a 10-s extension at 72°C. RGA and RGL2 mRNA levels were expressed relative to the constitutive GAPC mRNA (Griffiths et al., 2006). XERICO, GA20ox2, and XTH31 expression changes were calculated using the Delta-Delta Ct method relative to the constitutive IAP-LIKE PROTEIN1 mRNA control (Livak and Schmittgen, 2001; Graeber et al., 2011). An ANOVA with Tukey’s comparison adjustment was conducted using the SAS 9.3 statistical package.
Co-IP Experiment
Co-IP was performed to detect the interactions FLAG-GID1b and HA-GID1b with DELLA RGA as described previously (Figs. 6 and 7; Ariizumi et al., 2011). For Figure 6, T3 seeds of sly1-10 FLAG-GID1b were imbibed on 0.5× MS/MES-moistened filter paper for 48 h and then harvested and ground under liquid nitrogen with extraction buffer C (20 mm Tris, 150 mm NaCl, 0.5% (v/v) Triton, and 1× complete proteinase inhibitor [Roche]). T3 sly1-2 35S-HA-GID1b was used for detection of the HA-GID1b interaction with RGA protein using total protein extracted from 12-d-old seedlings (Ariizumi et al., 2008; Supplemental Fig. S6). Protein-blot analysis was used to detect immunoprecipitated FLAG-GID1b, HA-GID1b, and coimmunoprecipitated RGA.
The following method was devised for co-IP of RGL2 (Supplemental Fig. S6B). Imbibing T4 sly1-2 HA-GID1b seeds were ground under liquid nitrogen and resuspended in extraction buffer (50 mm potassium phosphate buffer, pH 7.0, 1× protease inhibitor cocktail [Sigma-Aldrich], and GA as indicated). Crude extracts were incubated on ice for 30 min. Total protein (500 µg) was incubated with 20 µL of anti-HA magnetic beads (MBIOL) for 24 h at 4°C. Beads were washed three times with extraction buffer, and the protein was resuspended in 30 µL of extraction buffer plus 30 µL of 2× loading buffer (Bio-Rad). Immunoprecipitated HA:GID1b and RGL2 after co-IP were detected by protein-blot analysis. Samples were also probed with anti-CUL1 to confirm the specificity of the HA:GID1b and RGL2 interaction (Supplemental Fig. S6, B and C).
Protein-Blot Analysis
Total protein was extracted from imbibed seeds by grinding in liquid nitrogen with extraction buffer A (50 mm potassium phosphate buffer, pH 6.8, with 1× protease inhibitor cocktail [Roche]). The protein concentration was determined using the Bradford protein assay (Bio-Rad). Total protein (60 µg) was separated on an 8% (w/v) SDS-PAGE gel and transferred onto a polyvinylidene difluoride membrane. Time points for protein extraction were taken at 24-h intervals during imbibition as indicated. Primary antibodies included rabbit polyclonal antibodies to RGL2 (1:10,000 [gift of J. Peng]; Hussain et al., 2005), RGA (1:10,000; Silverstone et al., 2001), anti-CUL1 (1:10,000 [gift of X.W Deng]; Wang et al., 2002), HA (1:5,000; Immuno Consultants Laboratory), and monoclonal antibody to FLAG (1:5,000; Sigma-Aldrich). The specificity of the RGA and RGL2 antibodies was confirmed using sly1 rga-24 and sly1 rgl2 double mutants (Ariizumi and Steber, 2007). Protein detection was performed using anti-rabbit IgG-horseradish peroxidase as a secondary antibody (1:200,000; GE Healthcare) and the ECL Advance chemiluminescence system (GE Healthcare).
Hormone Measurement
For hormone measurements, 200 mg of seeds was incubated at 4°C for 60 h followed by 12 h at 22°C and then ground in liquid N2 and lyophilized for 48 h (Fig. 3; Supplemental Fig. S2). ABA, GA4, and GA1 plant hormone levels were determined by liquid chromatography-tandem mass spectrometry using 100 to 150 mg dry weight for each biological replicate (n = 6–10) as described previously (Oh et al., 2007; Varbanova et al., 2007) A two-tailed Student’s t test was performed using the R statistics package (R Development Core Team, 2010).
Arabidopsis Genome Initiative locus identifiers for the genes in this study are as follows: GID1a (At3g05120), GID1b (At3g63010), GID1c (At5g27320), SLY1 (At4g24210), GAI (At1g14920), RGA (At2g01570), RGL2 (At3g03450), ABI1 (At4g26080), XTH5 (At5g13870), XTH31 (At3g44990), CP1 (At4g36880), SCS10 (At2g27920), GASA4 (At5g15230), EXP1 (At1g69530), GAPC (At3g04120), and IAP-LIKE PROTEIN1 (At1g17210).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects of the 35S:HA-GID1 overexpression construct.
Supplemental Figure S2. Bioactive GA1 levels and Student’s t test P values.
Supplemental Figure S3. Lack of GID1c slows down sly1-2 after-ripening.
Supplemental Figure S4. Effect of gid1 mutations on final plant height and fertility of sly1-2.
Supplemental Figure S5. Phenotypes of sly1-2 gid1 multiple mutants, DELLA RGL2 expression in ga1-3 and the gid1a gid1b gid1c triple mutant, and the specificity control for RGA co-IP with FLAG-GID1b.
Supplemental Figure S6. HA-GID1b protein levels and RGL2 co-IP correlate with seed germination efficiency.
Supplemental Figure S7. Effect of GID1 overexpression on gene expression in seeds.
Supplemental Figure S8. P values for qRT-PCR analysis in Figure 7.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank members of the Steber laboratory for helpful comments on the manuscript. We also thank Dr. T. Sun for providing access to the polyclonal RGA antibody, Dr. J. Peng for providing the RGL2 antibody, and Dr. C. Schwechheimer for providing gid1 mutant lines.
Glossary
- ABA
abscisic acid
- Ler
Landsberg erecta
- Col-0
Columbia
- MS
Murashige and Skoog
- co-IP
coimmunoprecipitation
- RT
reverse transcription
- qRT
quantitative reverse transcription
- cDNA
complementary DNA
References
- Ariizumi T, Lawrence PK, Steber CM. (2011) The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol 155: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Murase K, Sun T-P, Steber CM. (2008) Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Steber CM. (2007) Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Talbot MJ, White RG, Jacobsen JV, Gubler F. (2009) Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol 150: 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley DJ, Black M (1994). Seeds: Physiology of Development and Germination. Plenum Press, New York [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J. (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J. (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113 [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T-P. (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun T-P. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM. (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51: 60–78 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Reeves W, Ariizumi T, Steber CM. (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Alabadí D, Blázquez MA. (2011) DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. (2010) Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Wood ATA, Leubner-Metzger G. (2011) A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 23: 2045–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang Z-L, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-P, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog M, Dorne AM, Grellet F. (1995) GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol 27: 743–752 [DOI] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kouketu E, Katoh H, Aya K, Ueguchi-Tanaka M, Matsuoka M. (2012) The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J 71: 443–453 [DOI] [PubMed] [Google Scholar]
- Hussain A, Cao D, Cheng H, Wen Z, Peng J. (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44: 88–99 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim Y-C, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, et al. (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50: 958–966 [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN. (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Karssen CM, der Brinkhorst-van Swan DLC, Breekland AE, Koornneef M. (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies in abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Karssen CM, Laçka E (1986). A revision of the hormone balance theory of seed dormancy: studies on gibberellin and/or abscisic acid-deficient mutants of Arabidopsis thaliana. In M Bopp, ed, Plant Growth Substances 1985: Proceedings of the 12th International Conference on Plant Growth Substances. Springer-Verlag, New York, pp 315–323 [Google Scholar]
- Karssen CM, Zagorski S, Kepczynski J, Groot SPC. (1989) Key role for endogenous gibberellins in the control of seed germination. Ann Bot (Lond) 63: 71–80 [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, der Brinkhorst-van Swan DLC, Karssen CM. (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 61: 385–393 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45: 309–319 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Martin RC, Liu P-P, Nonogaki H. (2005) Simple purification of small RNAs from seeds and efficient detection of multiple microRNAs expressed in Arabidopsis thaliana and tomato (Lycopersicon esculentum) seeds. Seed Sci Res 15: 319–328 [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-P, Steber CM. (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T-P, Hakoshima T. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim Y-C, Park S-H, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee H-S, Sun T-P, Kamiya Y, Choi G. (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Lopez-Molina L. (2009) The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal Behav 4: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2010). R: A Language and Environment for Statistical Computing. http://www.R-project.org/ (February 3, 2012)
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P. (1998) Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, McCourt P. (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-P, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Park S-H, Okubo K, Kitamura J, Ueguchi-Tanaka M, Iuchi S, Katoh E, Kobayashi M, Yamaguchi I, Matsuoka M, et al. (2009) Differential expression and affinities of Arabidopsis gibberellin receptors can explain variation in phenotypes of multiple knock-out mutants. Plant J 60: 48–55 [DOI] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. (1990) Accumulation of C19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Heynh. Planta 182: 501–505 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun T-P. (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M. (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, et al. (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19: 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele A, Linkies A, Müller K, Leubner-Metzger G. (2011) Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J Exp Bot 62: 5131–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan L-M, Deng XW. (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kang D, Feng S, Serino G, Schwechheimer C, Wei N. (2002) CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: a structure-function study of CSN1 subunit of COP9 signalosome. Mol Biol Cell 13: 646–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Hirai T, Yamamoto E, Kawamura M, Sato T, Kitano H, Matsuoka M, Ueguchi-Tanaka M. (2010) A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell 22: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Takeda-Kamiya N, Hanada A, Ogawa M, Kuwahara A, Seo M, Kamiya Y, Yamaguchi S. (2007) Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol 48: 555–561 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang Z-L, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]