A metabolic/regulatory network of rice incorporates two important tissue types, germinating seeds and photorespiring leaves, is validated through experiments with rice suspension cultures, and applied to analyze metabolic capability under flooding and drought conditions.
Abstract
Rice (Oryza sativa) is one of the major food crops in world agriculture, especially in Asia. However, the possibility of subsequent occurrence of flood and drought is a major constraint to its production. Thus, the unique behavior of rice toward flooding and drought stresses has required special attention to understand its metabolic adaptations. However, despite several decades of research investigations, the cellular metabolism of rice remains largely unclear. In this study, in order to elucidate the physiological characteristics in response to such abiotic stresses, we reconstructed what is to our knowledge the first metabolic/regulatory network model of rice, representing two tissue types: germinating seeds and photorespiring leaves. The phenotypic behavior and metabolic states simulated by the model are highly consistent with our suspension culture experiments as well as previous reports. The in silico simulation results of seed-derived rice cells indicated (1) the characteristic metabolic utilization of glycolysis and ethanolic fermentation based on oxygen availability and (2) the efficient sucrose breakdown through sucrose synthase instead of invertase. Similarly, flux analysis on photorespiring leaf cells elucidated the crucial role of plastid-cytosol and mitochondrion-cytosol malate transporters in recycling the ammonia liberated during photorespiration and in exporting the excess redox cofactors, respectively. The model simulations also unraveled the essential role of mitochondrial respiration during drought stress. In the future, the combination of experimental and in silico analyses can serve as a promising approach to understand the complex metabolism of rice and potentially help in identifying engineering targets for improving its productivity as well as enabling stress tolerance.
Rice (Oryza sativa) is one of the prominent food crops in the world and is considered a major staple food in many Asian countries: each person consumes more than 100 kg of rice per year, on average (Nelson, 2011). Since the Green Revolution in 1960, there has been a progressive increase in the yield of rice. However, the growing population and adverse climatic changes pose huge challenges to sustaining the growing demand for rice. Moreover, of several abiotic stresses, flooding stress tremendously limits rice productivity (Xu et al., 2006), particularly in the rain-fed lowlands of Southeast Asia. In general, when plants are water-logged by flooding, they experience a lower oxygen availability (hypoxia) or a total absence of oxygen (anoxia), thus severely impairing energy generation through reduced/eliminated mitochondrial respiration. Nevertheless, rice is unique in its ability to survive up to 2 weeks in complete submergence conditions by prolonged flood (Jackson and Ram, 2003; Bailey-Serres and Voesenek, 2008). Rice seeds can germinate and grow up to the coleoptile even in anoxia through its distinctive metabolic adaptations. Under such conditions, predominant amounts of energy required for survival are produced by fermentative pathways, especially ethanolic fermentation (Atwell et al., 1982; Alpi and Beevers, 1983; Perata and Alpi, 1993; Guglielminetti et al., 1995; Gibbs et al., 2000; Magneschi and Perata, 2009). Besides flooding stress, the photosynthetic carbon-fixing capacity of rice and other C3 plants is limited by an energetically wasteful process, photorespiration. It is initiated by the partial replacement of oxygen instead of CO2 into the bifunctional enzyme Rubisco, which is the first reaction in the CO2-fixing pathway. This oxygenase side reaction of Rubisco produces one molecule of 3-phosphoglycerate (3-PGA) and one molecule of 2-phosphoglycolate (2-PG) for each molecule of oxygen fixed. Of these, 2-PG is a wasteful by-product, requiring sufficient amounts of energy for its salvation into 3-PGA through a series of enzymatic steps known as “the photorespiratory cycle” (Jordan and Ogren, 1984). In addition, the amount of CO2 available for photosynthesis is drastically reduced under drought conditions owing to the partial/full closure of leaf stomata, resulting in increased photorespiration (de Carvalho, 2008). Thus, photorespiration has always been considered as a major target for crop improvement in C3 plants such as rice to increase its productivity.
Over the last few decades, several efforts have been made to understand the cellular metabolism of rice by conventional experimental approaches. However, its metabolic adaptations under different stress conditions remain poorly characterized. Thus, with recent advancements in the modern genomic era, a systematic approach is required to improve our understanding of the metabolic changes of rice. In the postgenomic era, it can be achieved by utilizing the abundant high-throughput “omics” data sets in combination with in silico metabolic modeling. To this end, the development of a predictive in silico model based on the available biochemical, genomic, and regulatory information, and its subsequent examination by the well-established computational framework of constraints-based flux analysis, also known as flux balance analysis (FBA), is highly desired.
Interestingly, flux-based metabolic modeling of plant systems has recently gained more attention following its tremendous success in elucidating the metabolic capabilities of myriad microbial and mammalian species and rationally engineering them to achieve desirable phenotypes (Edwards et al., 2002; Lee et al., 2005; Lewis et al., 2012). In this regard, metabolic network models for several plants, such as Arabidopsis (Arabidopsis thaliana; Poolman et al., 2009; de Oliveira Dal’Molin et al., 2010a; Saha et al., 2011; Chung et al., 2012; Mintz-Oron et al., 2012), barley (Hordeum vulgare; Grafahrend-Belau et al., 2009), rapeseed (Brassica napus; Hay and Schwender, 2011; Pilalis et al., 2011), maize (Zea mays; Saha et al., 2011), and a general C4 plant model (de Oliveira Dal’Molin et al., 2010b) have already been developed, and some of them are even in genome scale. Nevertheless, to the best of our knowledge, the metabolic model of rice is not available to date. Therefore, in this study, we reconstructed a combined metabolic/regulatory network model representing the central metabolism of rice cells and subsequently utilized it for characterizing the cellular behavior in two different tissue types: germinating seeds and photorespiring leaves. In addition, we also simulated their metabolic states during abiotic stresses and investigated how they acclimatize to such conditions.
RESULTS AND DISCUSSION
Reconstruction of the Rice Central Metabolic Model
The central metabolic network of rice cells was reconstructed through a three-step process: (1) compilation of genes, reactions, and related information on rice metabolism from enzyme databases and literature; (2) verification of elemental balances in reactions and assignment of compartments; and (3) dead-end identification and network gap filling (see “Materials and Methods”). Steps 1 and 2 in the reconstruction process gave rise to a draft network model consisting of 298 reactions with several missing links, mainly in amino acid biosynthetic pathways. Such gaps could be filled by adding new reactions derived from other plants. For example, a metabolic gap existed in the His biosynthetic pathway of the draft model, since the gene coding for the enzyme, histidinol phosphatase (EC 3.1.3.15), was not found in the rice genome. Thus, we manually included it based on published references supporting its existence in Arabidopsis and plausibly in rice (Petersen et al., 2010). Similarly, we added nine more reactions in the amino acid biosynthetic pathways and seven reactions in folate metabolism to improve the network connectivity (Supplemental Data File S1). Furthermore, several intercompartmental transport reactions (48 between plastid and cytosol and 27 between mitochondrion and plastid) were also added along with their transport systems, such as free diffusion or proton symport.
The resulting metabolic network of rice accounts for 248 enzymes, catalyzing 326 reactions in the rice central metabolism, primarily from the following pathways: glycolysis/gluconeogenesis, tricarboxylic acid (TCA) cycle, pentose phosphate, Calvin cycle, photorespiratory, glyoxylate cycle, oxidative phosphorylation, starch and sucrose (Suc) metabolism, fermentation, cell wall metabolism, amino acid synthesis, and fatty acid synthesis. It should be noted that 52 direct and indirect regulatory interactions using 12 regulatory proteins for the discrimination between the photosynthetic and nonphotosynthetic cells under normal and stressed conditions were also incorporated into the model using Boolean logic formalism, on the basis of established procedures (Covert et al., 2001; see “Materials and Methods”). As a result, the activation of 40 light-specific metabolic reactions in the model can be controlled by a relevant logic statement. Cellular biomass composition is also an important prerequisite for subsequent in silico flux analysis, since the primary cellular objective is to maximize cell growth. Thus, we have developed biomass equations for describing the cellular growth of two tissue types. The biomass equation for the germinating cells of rice seeds was derived from the macromolecular composition of rice coleoptiles (Edwards et al., 2012), while the other biomass equation representing the photorespiring cells of rice leaves was developed using the rice straw composition (Juliano, 1985). Table I summarizes the general features of the reconstructed rice network; Supplemental Data File S1 provides complete details on the model, which is also available as a Systems Biology Markup Language file (level 2, version 1; http://sbml.org) in Supplemental Data File S2.
Table I. Properties of the reconstructed rice central metabolic network.
| Features | Cytosol | Plastid | Mitochondria | Total |
|---|---|---|---|---|
| Metabolic reactions | 139 | 153 | 34 | 326 |
| Transport and exchange reactions | 16a | 48b | 27b | 91 |
| Gene-enzyme reaction associations | 130 | 145 | 30 | 305 |
| Reactions with regulatory rules | 15 | 19 | 6 | 40 |
| Metabolites | 156 | 162 | 53 | 371 |
Exchange reaction. bIntercompartmental transport reactions.
In Silico Flux Analysis of Seed-Derived Rice Cells under Normal and Flooded Conditions
The reconstructed rice metabolic model was first utilized to simulate the phenotypic behavior of seed-derived cells in response to varying levels of oxygen. To do so, the coleoptile biomass equation was maximized while simultaneously constraining the uptake rate of Suc at 1 mmol g−1 dry cell weight d−1 and varying oxygen uptake rates gradually from 0 to 4 mmol g−1 dry cell weight d−1 (see “Materials and Methods”). It should be noted that the Boolean gene regulatory rules were utilized to consider active reactions in seed-derived cells (Supplemental Data File S3). The model simulations show a linear decrease in biomass growth and increase in ethanol fermentation with decreasing levels of oxygen supply, suggesting the predominance of ethanol fermentation in the absence of cellular respiration (data not shown). It should be highlighted that these observations are in good agreement with previous reports (Mohanty et al., 1993; Gibbs et al., 2000; Edwards et al., 2012). The model also predicted a sharp decline in cell growth without any production of ethanol beyond a certain value of oxygen influx (3.35 mmol g−1 dry cell weight d−1), demarking the stoichiometric optimal value of oxygen uptake to maximize cell growth for the imposed restriction in Suc uptake. Any excess oxygen above this value resulted in a futile energy cycle involving several redundant pathways to utilize the excess ATP produced, thus decreasing cellular growth.
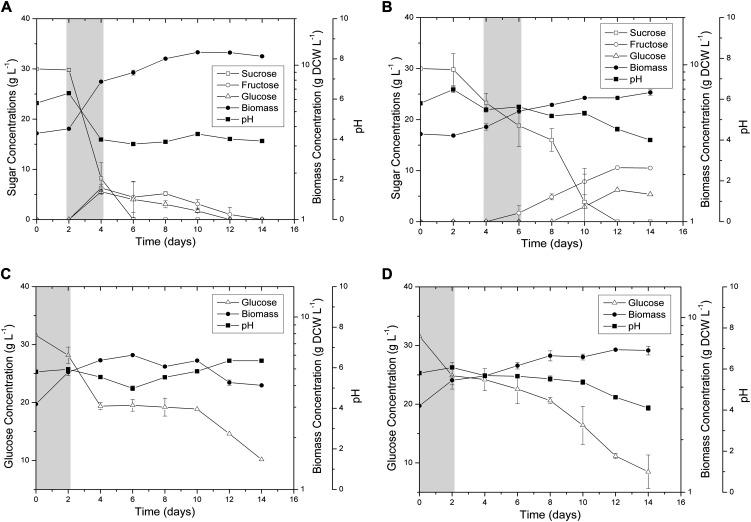
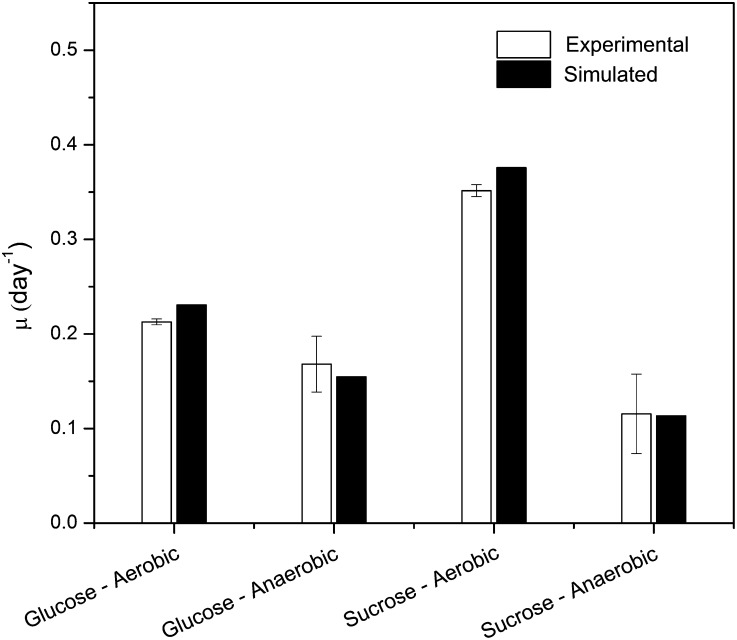
In order to further validate the model quantitatively, we also conducted batch cultures of rice cells growing on various carbon sources including Suc and Glc in the presence and absence of oxygen. The residual concentrations of supplemented sugars in the medium as well as the dry cell weight were monitored (Fig. 1). In case of Suc, a lag phase of 2 d was observed under both aerobic and anaerobic conditions (Fig. 1, A and B). Subsequently, the supplemented Suc was sharply consumed as well as hydrolyzed into Glc and Fru in the culture medium under aerobic conditions (Fig. 1A). This external accumulation of hexose sugars was also accompanied by a sharp decrease in medium pH, dropping from 6.3 to 4.0 at day 4, and was maintained for the remaining 10 d (Fig. 1A). Collectively, these results demonstrate the acidification of culture medium, thus promoting cell growth by increased cell wall-associated invertase activity (Amino and Tazawa, 1988; Shimon-Kerner et al., 2000). On the other hand, cells supplemented with Suc under anoxia showed a delayed increase in pH of the medium (Fig. 1B), possibly suggesting that the external invertase activity is dependent upon the presence of oxygen (Kwon et al., 2012). To explain this phenomenon clearly, further studies are required where the correlations among cell growth, invertase activity, and energy consumption must be investigated thoroughly. In the case of Glc, rice cells grew without a lag phase under both aerobic and anaerobic conditions, albeit with a short exponential growth in the former (Fig. 1, C and D). The supplemented Glc was steadily consumed in anoxia, whereas the concentration did not decline in aerobic conditions from day 4 to day 10 (Fig. 1C). Despite these differences in cell growth behavior, both Suc- and Glc-supplemented cells grew faster under aerobic conditions than those in anaerobic conditions (3.20- and 1.33-fold higher, respectively), confirming our earlier simulation results. Nevertheless, in order to examine the quantitative agreement between experimental and simulated growth rates, we again maximized the coleoptile biomass equation while constraining the uptake rate of the carbon source (i.e. Suc or Glc) at experimentally measured values in both aerobic and anaerobic conditions. Here, it should be noted that the carbon source uptake rates from the exponential phase of the cell cultures were used for simulations, since it is believed that cells typically evolve toward maximal growth in this phase (Schuster et al., 2008). Notably, the simulation results show good agreement between the in silico simulated growth rates and experimental observations within the acceptable error range for all four cases (Fig. 2).
Figure 1.
Profiles of cell biomass and residual concentrations of the carbon nutrient components in batch cultures of aerobic Suc (A), anaerobic Suc (B), aerobic Glc (C), and anaerobic Glc (D). Highlighted regions correspond to exponential growth phases of the cultures. DCW, Dry cell weight.
Figure 2.
Experimental and simulated growth rates during the exponential phase on different batch cultures.
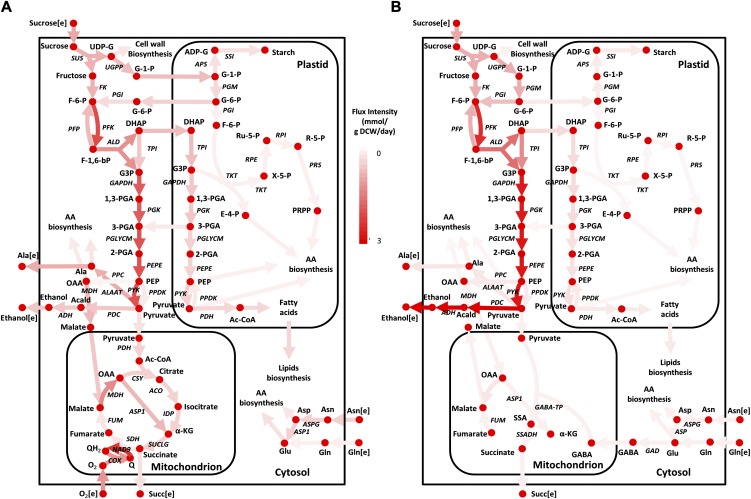
To further understand how oxygen influences cellular growth, we examined the internal flux distributions under aerobic and anaerobic conditions and observed significant differences among several metabolic pathways, including glycolysis, TCA cycle, fermentation, and glutaminolysis (Fig. 3). In order to confirm the confidence of the simulated metabolic states, flux variability analysis (FVA; Mahadevan and Schilling, 2003) was conducted for both conditions, thereby identifying commonly activated reactions, indicating the plausibility of such internal metabolic utilization (for complete results, see Supplemental Data File S4). The characteristic differences in flux distribution across various metabolic pathways between aerobic and anaerobic conditions are presented below.
Figure 3.
In silico flux maps of seed-derived suspension culture rice cells grown on Suc under aerobic (A) and anaerobic (B) conditions. The color intensity of the lines in the central carbon metabolic network corresponds to the normalized flux values with respect to the Suc uptake rates in each condition. DCW, Dry cell weight. Metabolite abbreviations not defined in the text are as follows: 1,3-PGA, 1,3-diphosphoglycerate; 2-PGA, 2-phosphoglycerate; AA, amino acids; Acald, acetaldehyde; Ac-CoA, acetyl-CoA; E-4-P, erythrose-4-phosphate; F-6-P, Fru-6-P; F-1,6-bP, Fru-1,6-bisP; G-1-P, Glc-1-P; G3P, glyceraldehyde-3-phosphate; G-6-P, Glc-6-P; PEP, phosphoenolpyruvate; PRPP, phosphoribosyl pyrophosphate; Q, ubiquinone; QH2, ubiquinol; R-5-P, Rib-5-P; Ru-5-P, ribulose-5-phosphate; SSA, succinic semialdehyde; UDP-G, UDP-Glc, X-5-P, d-xylulose-5-phosphate. Enzyme abbreviations not defined in the text are as follows: ACO, aconitase; ADH, alcohol dehydrogenase; ALAAT, Ala aminotransferase; ALD, aldolase; APS, Glc-1-P adenylyltransferase; ASP1, Asp aminotransferase; ASPG, asparaginase; COX, cytochrome c oxidase; CSY, citrate synthase; FK, fructokinase; FUM, fumarase; GABA-TP, γ-aminobutyrate aminotransferase; GAD, Glu decarboxylase; GAPDH, glyceraldehyde phosphate dehydrogenase; IDP, isocitrate dehydrogenase (NADP dependent); MDH, malate dehydrogenase; NAD9, NADH dehydrogenase; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; PEPE, phosphoenolpyruvate enolase; PFK, 6-phosphofructokinase; PFP, PPi-dependent phosphofructokinase; PGI, phosphoglucoisomerase; PGK, phosphoglycerate kinase; PGLYCM, phosphoglycerate mutase; PGM, phosphoglucomutase; PPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate orthophosphate dikinase; PRS, Rib phosphate diphosphokinase; PYK, pyruvate kinase; RPE, Rib-5-P epimerase; SDH, succinate dehydrogenase; SSADH, succinic semialdehyde dehydrogenase; SSI, starch synthase; SUCLG, succinyl-CoA ligase; TKT, transketolase; TPI, triose phosphate isomerase; UGPP, UDP-Glc pyrophosphorylase. [See online article for color version of this figure.]
Suc Metabolism
In both aerobic and anaerobic conditions, Suc synthase (SUS) was utilized for degrading Suc into Fru and UDP-Glc (Fig. 3), which is consistent with the previous experimental suggestions (Mohanty et al., 1993; Guglielminetti et al., 1995). The in silico analysis also confirmed the recycling of UTP to SUS, which occurs through the nucleoside diphosphate kinase (NDPK) and the UDP-Glc pyrophosphorylase (UGPP). Although the supply of UTP to SUS from UGPP was proposed by Guglielminetti et al. (1995), they did not clearly reveal the source of inorganic pyrophosphate (PPi); rather, they just hypothesized its origin from PPi-dependent phosphofructokinase (PFP). In this regard, our simulation results suggested that the PPi could be produced toward the gluconeogenesis direction via reactions catalyzed by either PFP or pyruvate orthophosphate dikinase (PPDK), forming a substrate cycle in glycolysis. Such combined utilization of SUS, NDPK, UGPP, and PFP/PPDK for the breakdown of Suc is energetically more efficient than invertase, since they consume 1 mol of ATP less for each mol of Suc degraded (Guglielminetti et al., 1995; Magneschi and Perata, 2009).
Glycolysis
Compared with aerobic conditions, anaerobic glycolytic fluxes in cytosol showed a significant increase (by more than 42%), where the majority (97.5%) was fermented to ethanol (Fig. 3B). Notably, such an increase in carbon flux through glycolysis and a sharp decline in coleoptile growth have already been reported (Mohanty et al., 1993; Gibbs et al., 2000; Magneschi and Perata, 2009), highlighting the importance of ethanolic fermentation in combination with glycolysis for energy production under anaerobiosis. On the other hand, aerobic glycolysis showed that significant amounts of pyruvate were utilized in the oxidative conversion to acetyl-CoA, thus enabling its entry into the TCA cycle for energy production (Fig. 3A). Furthermore, our simulations also indicated a certain amount of pyruvate (22%) being fermented into ethanol in aerobic conditions, in agreement with earlier reports (Mohanty et al., 1993; Edwards et al., 2012). In general, ethanol fermentation under aerobic conditions occurs mainly due to high glycolytic fluxes and/or limited oxygen availability, where the excess pyruvate that cannot be oxidized is fermented to regenerate the NAD+ lost in glycolysis. It should be noted that glycolytic fluxes are regulated by the hexokinase activity in plants, which is determined by the supernatant hexose concentrations (Lalonde et al., 1999). Therefore, it is possible that the excessive hexose concentrations resulting from the Suc breakdown via SUS/invertase may have led to higher activity of hexokinase enzyme, thereby resulting in an overflow metabolism via the glycolytic pathway. In plants, the plastidic glycolysis is more crucial than the cytosolic one, as most of the amino acids and fatty acids are synthesized in plastids. In this regard, flux analysis results indicated anaerobic glycolysis with highly reduced fluxes (by 46%); most of the carbon source is channeled via cytosolic glycolysis to feed fermentative pathways for energy generation, as mentioned earlier.
TCA Cycle
Under anoxic conditions, simulation results revealed a truncated TCA cycle operation between fumarate and oxaloacetate (OAA), mainly due to the limited regeneration of redox cofactors, since the mitochondrial respiration is impaired (Fig. 3A). These observations are in good agreement with earlier reports on plants, suggesting a partial TCA cycle activity under hypoxia and anoxia (Sweetlove et al., 2010). On the other hand, a fully operational TCA cycle was characterized under aerobic conditions, where the carbon flux enters the cycle at three different points: (1) acetyl-CoA from pyruvate; (2) malate from OAA; and (3) α-ketoglutarate (α-KG) from glutaminolysis (Fig. 3B). It should be noted that a part of this α-KG was also withdrawn from the cycle and utilized in amino acid biosynthesis to keep the cycle balanced, as any additional α-KG above the acetyl-CoA influx would cause an imbalance. Interestingly, the flux analysis also suggested the possibility of a γ-aminobutyric acid (GABA) shunt to be operational for the conversion of α-KG to succinate instead of α-KG dehydrogenase and succinate-CoA ligase (Fig. 3B). However, further analysis revealed that the operation of a GABA shunt is just an alternative solution, as confirmed by FVA (Supplemental Data File S4). Thus, the final determination of actual or plausible flux distributions in this case must await experimental verification through isotope-based internal flux measurements.
Glutaminolysis and Amino Acid Biosynthesis
During the model simulations, the rice cells were freely allowed to consume Gln and Asn as nitrogen sources for the amino acid biosynthesis (Bewley and Black, 1994). Under both aerobic and anaerobic conditions, the consumed Gln and Asn were completely converted into Glu and Asp, and subsequently into α-KG and OAA, for amino acid synthesis. Furthermore, the amounts of Asn consumed in aerobic conditions were reasonably higher, since an enhanced pyruvate pool facilitated the amino acid biosynthesis. Our simulation results also highlighted the functional ability of rice to synthesize all amino acids via biosynthetic pathways even under anoxia, rather than protein degradation, as often speculated.
Interestingly, the flux analysis also suggested the possibility for GABA, a nonprotein amino acid, to be synthesized under both aerobic and anaerobic conditions. As mentioned previously, although the synthesis of GABA and its subsequent utilization in the GABA shunt did not influence cellular growth under aerobic conditions, it enhanced the growth rate slightly under anaerobic conditions, owing to the crucial role in Gly biosynthesis. Under anoxia, GABA is first synthesized from Glu by Glu decarboxylase (GAD) and then converted into succinate via GABA aminotransferase and succinic semialdehyde dehydrogenase (SSADH). During these conversions, NADH is liberated in the SSADH step and recycled via a series of enzymes including Ser hydroxymethyltransferase (SHM1), producing net amounts of Gly. These observations are in very good agreement with earlier experiments (Shingaki-Wells et al., 2011) that reported the anaerobic accumulation of GABA along with an increase in the expression of SHM1.
In Silico Flux Analysis of Photorespiring Rice Leaf Cells under Normal and Stressed Conditions
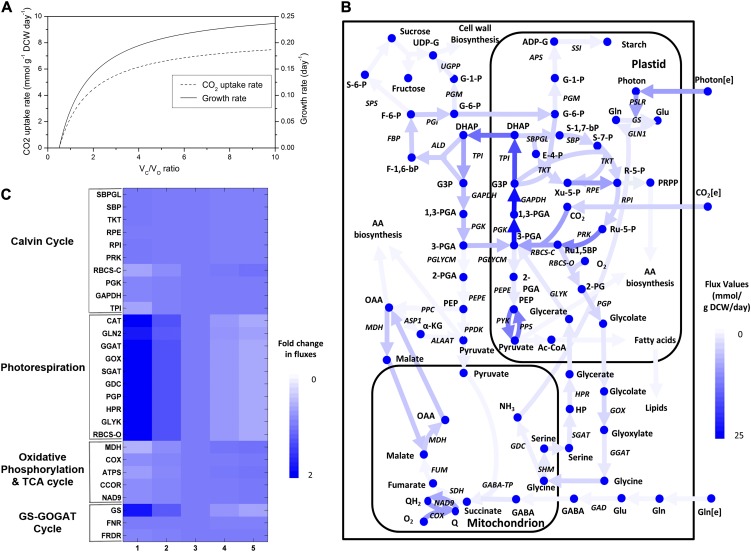
The reconstructed metabolic model was also used to understand metabolic behaviors of photorespiring rice leaf cells under normal and drought-stressed conditions. Photorespiration in rice leaf cells was simulated by maximizing the straw biomass equation while constraining the photon uptake rate at 100 mmol g−1 dry cell weight d−1 and fixing the carboxylation-to-oxygenation flux ratio (VC/VO) of Rubisco with a value between 1 and 10. Here, it should be noted that the simulations with VC/VO ratios greater than or equal to 3 represent photorespiration under normal conditions, while any lesser value corresponds to the drought conditions (Jordan and Ogren, 1984). Again, the Boolean gene regulatory rules were also utilized to consider active reactions in the leaf cells (Supplemental Data File S3). Model simulations indicated significant reductions in the CO2 uptake rates and leaf growth rates as VC/VO decreases, eventually reaching zero at the compensation point (VC/VO = 0.5; Fig. 4A). At severe photorespiration conditions, the carbon fixation rate is reduced significantly (decreased by 40% at VC/VO = 2 when compared with VC/VO = 10) while absorbing the same amounts of photon, since most of the energy is wasted in recycling the 2-PG. Overall, these results are highly consistent with the theoretical calculations by Heldt and Piechulla (2011), who suggested that “more than one third of the captured photons will be wasted because of photorespiration.” The resultant flux distributions in Figure 4B illustrate the classical photorespiratory metabolism linked to glycolysis, TCA cycle, Calvin cycle, photorespiratory cycle, and glutaminolysis reactions. To support this simulated metabolic behavior, FVA was conducted in photorespiring rice leaf cells (for details, see “Materials and Methods”), confirming the possible metabolic state (for complete results, see Supplemental Data File S4). Figure 4C depicts the flux changes through several important metabolic pathways under normal (VC/VO = 3) and drought-stressed (VC/VO = 1) conditions.
Figure 4.
A, Effect of VC/VO of Rubisco on leaf cellular growth and CO2 uptake while absorbing equal amounts of photon. B, Flux map of the central metabolism in photorespiring leaves under normal conditions (VC/VO = 3). C, Variation in fluxes through key enzymes upon varying levels of VC/VO. The color intensity of the lines in the central carbon metabolic network (B) corresponds to the flux values obtained from simulations. C was drawn by normalizing the flux values with respect to VC/VO = 3. DCW, Dry cell weight. Metabolite abbreviations not defined in the text are as follows: 1,3-PGA, 1,3-diphosphoglycerate; 2-PGA, 2-phosphoglycerate; Ac-CoA, acetyl-CoA; ADP-G, ADP-Glc; E-4-P, d-erythrose-4-phosphate; F-1,6-bP, Fru-1,6-bisP; F-6-P, Fru-6-P; G-1-P, Glc-1-P; G3P, glyceraldehyde-3-phosphate; G-6-P, Glc-6-P; HP, hydroxypyruvate; PEP, phosphoenolpyruvate; PRPP, phosphoribosyl pyrophosphate; Q, ubiquinone; QH2, ubiquinol; S-6-P, Suc-6-P; R-5-P, Rib-5-P; Ru-1,5-bP, ribulose-1,5-bisphosphate; Ru-5-P, ribulose-5-phosphate; S-1,7-bP, sedoheptulose-1,7-bisphosphate; S-7-P, sedoheptulose-7-phosphate; Xu-5-P, xylulose-5-phosphate; UDP-G, UDP-Glc. Enzyme abbreviations not defined in the text are as follows: ACO, aconitase; ALAAT, Ala aminotransferase; ALD, aldolase; APS, Glc-1-P adenylyltransferase; ASP1, Asp aminotransferase; COX, cytochrome c oxidase; CSY, citrate synthase; FBP, Fru-bisphosphatase; FK, fructokinase; FUM, fumarase; GABA-TK, γ-aminobutyrate aminotransferase; GAD, Glu decarboxylase; GAPDH, glyceraldehyde phosphate dehydrogenase; GDC, Gly decarboxylase; GGAT, Gly aminotransferase; GLYK, glycerate kinase; GLN1, Glu ammonia ligase; GOX, glycolate oxidase; GS, Glu synthase (ferredoxin dependent); HPR, hydroxypyruvate reductase; MDH, malate dehydrogenase; NAD9, NADH dehydrogenase; PEPE, phosphoenolpyruvate enolase; PFK, phosphofructokinase; PGI, Glc-6-P isomerase; PGK, phosphoglycerate kinase; PGLYCM, phosphoglycerate mutase; PGM, phosphoglucomutase; PGP, phosphoglycolate phosphatase; PPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate orthophosphate dikinase; PPS, pyruvate-water dikinase; PRK, phosphoribulokinase; PSLR, photosynthetic light reaction; PYK, pyruvate kinase; RBCS-C, ribulose-1,5-bisphosphate carboxylase; RBCS-O, ribulose-1,5-bisphosphate oxygenase; RPE, Rib-5-phosphate epimerase; SBP, sedoheptulose-bisphosphatase; SBPGL, sedoheptulose 1,7-bisphosphate d-glyceraldehyde-3-phosphate lyase; SDH, succinate dehydrogenase; SGAT, Ser glyoxylate aminotransferase; SHM, Ser hydroxymethyltransferase; SPP, Suc phosphatase; SPS, Suc phosphate synthase; SSADH, succinic semialdehyde dehydrogenase; SSI, starch synthase; TKT, transketolase; TPI, triose phosphate isomerase; UGPP, UDP-Glc pyrophosphorylase. [See online article for color version of this figure.]
Calvin Cycle
Despite the large differences in CO2 uptake between normal and drought-stressed photorespiration, the Calvin cycle did not exhibit any appreciable differences in terms of fluxes (Fig. 4C). In either case, it was driven by the supply of 3-PGA and CO2 from cytosol to plastid through the triose phosphate/phosphate translocator and subsequently was withdrawn in the form of dihydroxyacetone phosphate (DHAP) through a similar phosphate translocator, back to cytosol (Fig. 4B). The remaining DHAP in the cytosol was utilized in two routes: (1) carbon storage via Suc synthesis; and (2) energy production and biomass synthesis via cytosolic glycolysis and the TCA cycle, in agreement with earlier reports (Weber, 2007). The bifurcation of DHAP between carbon storage and energy production was regulated by Fru-1,6-bisP, which is recognized as a check point for the conversion of DHAP into Suc or starch under dark conditions by a marked increase in its concentration (Stitt, 1987).
Photorespiratory Pathway
The flux analysis also successfully simulated the utilization of the photorespiratory cycle from Rubisco to glycerate kinase, recycling the 2-PG back to 3-PGA (Fig. 4B). A sharp increase in fluxes along these pathways was observed with increasing levels of drought stress (Fig. 4C). Such enormous increases in the fluxes within photorespiratory pathways release large amounts of ammonia in the mitochondria during Gly oxidation via Gly decarboxylase and SHM1. Even though this excess ammonia can be recovered within mitochondria via Glu dehydrogenase, the Gln synthetase-Gln oxoglutarate aminotransferase cycle in plastids is utilized with the help of malate transporters to maintain the redox balance of plastids (Atkin and Macherel, 2009); the excessive amounts of NADPH generated from photosynthetic light reactions in plastids under drought conditions also need to be recycled.
TCA Cycle and Oxidative Phosphorylation
Unlike seed-derived rice cells, the model simulation observed a truncated TCA cycle operation between malate and OAA via malate dehydrogenase (MDH) during photorespiration (Fig. 4B), mainly to recycle the redox cofactors (Hanning and Heldt, 1993). Under such conditions, the redox cofactors generated from photosynthesis light reactions cannot be utilized in plastid due to the reduction in Calvin cycle fluxes. Thus, the cofactors are exported out of the cytosol in the form of DHAP and Glu, so that they can be further transmitted to mitochondrion via malate transporters for eventual utilization in oxidative phosphorylation. As photorespiration increased, the flux through MDH decreased accordingly, since the Gly oxidation predominantly supplies the required redox cofactors to mitochondrial respiration (Fig. 4C). On the other hand, interestingly, the flux through oxidative phosphorylation did not change much with increasing levels of photorespiration and indicated a sharp increase in the ratio of respiration to photosynthesis. This respiration-to-photosynthesis ratio is a common denominator for analyzing the magnitude of the negative impact caused by photorespiration (Atkin and Macherel, 2009); it is important for plants to ensure a balance between respiration and photosynthesis so that the carbon balance can be maintained. Furthermore, this result complements the experimental observations by Flexas et al. (2005) highlighting that respiration cannot be impaired during drought stress, in spite of the differences in the magnitude of fluxes. The simulation results also indicated that the ATP generated from respiration (i.e. oxidative phosphorylation) was exported to the plastids through the ATP/ADP translocator to feed the photorespiratory pathways, the Gln synthetase-Gln oxoglutarate aminotransferase cycle, and the Calvin cycle. Such behavior is essential to maintain cellular hemostasis, as the amounts of ATP generated from reduced photosynthesis under drought conditions are not sufficient (Keck and Boyer, 1974). In addition, the ATP transported from mitochondrion to plastid increases considerably in drought-stressed cells. Collectively, these observations on the TCA cycle and oxidative phosphorylation suggest that they are essential for producing energy rather than providing carbon skeletons for biomass synthesis under drought-stressed conditions.
Comparison of Our Model with Existing Plant Models, and Future Directions for Further Improvement
In this work, we have developed the central metabolic/regulatory network of rice cells. Compared with the existing plant models, our model is unique in incorporating the regulatory information into a FBA framework for investigating the effect of external signals on plant metabolic behaviors. This can be well exemplified while simulating the growth of germinating seed cells on starch as a carbon source under aerobic and anaerobic conditions (data not shown). Generally, when plant seeds germinate, they require the reserve carbohydrates such as starch to be metabolized to fuel the synthetic pathways of biomass precursors. In this regard, rice is a unique plant due to its ability to metabolize starch via α-amylase even under anaerobic conditions. It should be highlighted that our model clearly differentiated the induction of α-amylase by different regulatory routes via GA response elements and the calcineurin B-like protein-interacting protein kinases under aerobic and anaerobic conditions, respectively (Supplemental Data File S3). Furthermore, the incorporation of regulatory information also allowed us to simulate photosynthetic metabolism by eliminating nonactive enzymes under light conditions; our leaf simulations successfully eliminated the PFP and chose FBP for the synthesis of Fru-6-P during photorespiration/photosynthesis, as PFP is controlled by the allosteric regulator Fru-2,6-bisP, which is produced only under dark conditions. In addition, our newly reconstructed model is also distinct from other plant models in terms of representing the intercompartment metabolite transporters: it clearly presents the transport details of almost all the metabolites, such as free diffusion, proton symport/antiport, and redox shuttles based on extensive literature searches. Thus, the translocation of various metabolites across compartments can be properly described along with the relevant redox balance and energy requirements.
Unlike microbes or animals, the secondary metabolism of plants plays a significant role in orchestrating the cellular phenotype in response to abiotic stresses (Ramakrishna and Ravishankar, 2011). For example, ascorbic acid, glutathione, and α-tocopherol have been recognized for their role as antioxidants against the reactive oxygen species, which are produced during increased photorespiration (Jogaiah et al., 2012). Thus, it is highly required to expand the scope of our model into the genome-scale model of rice metabolism in order to better understand its metabolic characteristics. However, the reconstruction of such a large-scale plant metabolic network could be challenging, mainly attributable to the physiological differences in their tissues, the subcellular localization of reactions, and the annotation of genomic content whose functions remain unidentified (Sweetlove and Ratcliffe, 2011; Seaver et al., 2012). We believe these issues can be appropriately resolved by resorting to transcriptomic and/or proteomic data sets as surrogates for transcriptional regulation in developing tissue-specific models (Mintz-Oron et al., 2012), subcellular localization prediction software for compartmentalizing metabolic reactions (Mintz-Oron et al., 2012), and comparative genomics for annotating undiscovered genomic content (Seaver et al., 2012). Therefore, the comprehensive model developed in such a pipeline can be exploited to further enhance our understanding of the complex metabolic behavior of rice and potentially help in rationally designing the modified crops of the future.
CONCLUSION
In this study, what is to our knowledge the first metabolic/regulatory network of rice cells was reconstructed to elucidate the cellular phenotypes of two different rice tissues via intracellular metabolic flux profiles during abiotic stresses. Using the model, we showed how the regulatory information in plants can be used for simulating tissue-specific and/or condition-specific metabolic behavior in rice cells. In agreement with our experimental phenotypic results, the flux analysis of seed-derived rice cells revealed the importance of ethanolic fermentation together with glycolysis for ATP production under anaerobic growth conditions. The simulations also confirmed the crucial role of SUS ahead of invertase for breaking down Suc in an energetically efficient manner while growing under both aerobic and anaerobic conditions. Moreover, our model even suggested the possible role of GABA in Gly synthesis via SHM1 under anaerobic conditions. Similarly, flux analysis of photorespiring leaves highlighted the important role of malate transporters in balancing the redox cofactors across compartments and the active engagement of respiration in generating the required ATP for biosynthetic processes during the reduction in photosynthesis. Moving forward, we believe that in silico modeling of plant metabolism as highlighted in this work can be considered an effective tool to illustrate their intricate cellular traits and to aid plant scientists and engineers in postulating design strategies for improving the yield and/or stress tolerance of plants.
MATERIALS AND METHODS
Cell Line and Media Conditions
Rice (Oryza sativa) suspension cells were obtained from calli induced by rice seed on amino acid callus induction medium containing 2,4-dichlorophenoxyacetic acid (2 mg L−1), Suc (30 g L−1), and Gelrite (2 g L−1) for 2 months. The calli were then sieved for the establishment of suspension cells using a stainless screen mesh with a pore size of 1 mm. The suspension cells were cultured in 500-mL Erlenmeyer flasks. An Erlenmeyer flask contained 126 mL of amino acid (AA) medium consisting of 30 g L−1 Suc, 2 mg L−1 2,4-dichlorophenoxyacetic acid, and 0.02 mg L−1 kinetin. After sterilization, 14 mL of a 10-fold-concentrated AA mixture was sterilized using 0.22-μm syringe filters with 0.1 mg L−1 GA. Subculture was performed every 9 d. Both aerobic and anaerobic cell cultures were performed at 120 rpm and 28°C in a gyratory shaking incubator. For cultivation under anaerobic conditions, a disposable bag chamber was designed. To apply anaerobic conditions, nitrogen gas was supplied through an air filter (Sartorius), and dissolved oxygen (DO) levels were monitored using a dissolved oxygen electrode (Mettler-Toledo Process Analytical). In both conditions, 1 g of fresh cells was inoculated in 100-mL Erlenmeyer flasks with 30 mL of AA medium containing either 87.6 mmol of Suc or 175.3 mmol of Glc as carbon source.
Analytical Techniques
After sampling every 4 d, the suspension cells in cultured broth were filtered using Whatman No. 1 filter paper. The rice cells were washed three times using distilled water to remove remaining sugar on the cell surface, and the cells were then measured to determine fresh cell weight on a preweighed dish. To confirm dry cell weight, fresh cells were dried at 60°C for 2 d and then weighed. Extracellular Suc, Glc, and Fru concentrations were measured using an HPLC apparatus (Young Lin Instrument) with a Zorbax Carbohydrate Analysis column (Agilent Technologies). The mobile phase was an acetonitrile:water mixture (75:25), and the column temperature was maintained at 40°C. Samples were filtered with 0.45-μm syringe filters (Millipore). The sugars were detected with a refractive index detector (Waters). Sugar concentration was estimated from a standard curve with known concentrations of Glc, Fru, and Suc.
Metabolic Network Reconstruction
The central metabolic network of rice cells was reconstructed based on information collected from various biological and genomic databases, such as the Kyoto Encyclopedia of Genes and Genomes (Ogata et al., 1999), RiceCyc, and MetaCyc (Caspi et al., 2012). Initially, the biochemical reactions of primary metabolism, which are necessary to generate the biomass precursors, were identified and included in the preliminary draft model. These reactions were then corrected for any stoichiometric imbalances, mapped with appropriate genes to devise proper gene-reaction relationships, and assigned to respective cellular compartments based on extensive literature studies. Additionally, some spontaneous as well as non-gene-associated reactions, including that of metabolite transport, were also incorporated into the model based on the physiological relevance from literature and databases. Once the draft model was assembled with all necessary reactions, it was then checked for any connectivity issue (i.e. dead ends or blocked metabolites) by maximizing the production of individual metabolite precursors upon feeding it with various carbon sources such as Suc, Glc, and Fru. The identified missing links were then filled either by introducing sink reactions to allow for material exchange between the cell and its surrounding environment or by adding reactions from other similar plants to close in the knowledge gaps.
Following reconstruction of the metabolic network, we also performed an extensive literature search to collect information on the regulatory mechanisms of all the reactions in primary metabolic pathways. Finally, this information was represented as a set of regulatory rules using the Boolean formalism, which can guide whether relevant reactions are either active (ON) or not (OFF; Covert et al., 2001). Specifically, the activity/inactivity of a particular reaction in the presence of a certain regulatory protein(s) can be described using various logical operators such as IF, AND, OR, and NOT. For example, if a reaction, rxn1, is activated by either regulatory protein A or B, then the corresponding rule can be formulated as “IF (A OR B).” Similarly, if a reaction, rxn2, is inactivated in the presence of both proteins C and D, the regulatory rule can be written as “IF NOT (C AND D).”
Constraints-Based Regulatory Flux Analysis
In this study, we utilized constraints-based flux analysis to simulate rice metabolism under varying environmental conditions by manipulating the constraints. The biomass equation was maximized to obtain the optimal solution of the metabolic network as detailed elsewhere (Oberhardt et al., 2009; Orth et al., 2010). Mathematically, the problem specific to our study, the maximization of biomass subjected to stoichiometric and flux capacity constraints, can be represented as follows:
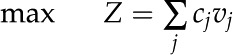 |
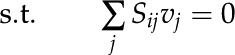 |
where Sij refers to the stoichiometric coefficient of metabolite i involved in reaction j, vj denotes the flux or specific rate of metabolic reaction j, 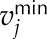 and
and 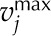 represent the lower and upper limits on the flux of reaction j, respectively, and Z corresponds to the cellular objective as a linear function of all the metabolic reactions, where the relative weights are determined by the coefficient cj.
represent the lower and upper limits on the flux of reaction j, respectively, and Z corresponds to the cellular objective as a linear function of all the metabolic reactions, where the relative weights are determined by the coefficient cj.
In order to simulate the cellular metabolism of the seed-derived rice cells growing on either Suc or Glc, we first applied the regulatory constraints to the network under steady-state conditions by evaluating whether metabolic enzymes were active or not for the given conditions using the Boolean rules and by constraining the fluxes of repressed enzymes to zero. If an enzyme is available, then its flux value was allowed to be determined by FBA. In the case of exchange reactions, we just constrained the carbon source uptake rates at the experimentally measured values. Additionally, for the aerobic simulations, the oxygen-exchange reaction was constrained at 3.312 mmol g−1 dry cell weight d−1 based on the literature (Wen and Zhong, 1995). For simulating the photorespiring metabolism of rice leaf cells, we again followed a similar procedure, by first constraining the fluxes of dark reactions to zero using Boolean regulatory rules as mentioned above. Subsequently, leaf cell growth was simulated by maximizing the leaf biomass while constraining the photon uptake at 100 mmol g−1 dry cell weight d−1. In addition, the ratio of flux through Rubisco was set with a value between 1 and 10 for simulating the photorespiratory behavior at different VC/VO values (Weber, 2007). In this study, all simulations were implemented by the General Algebraic Modeling System Integrated Development Environment, version 23.9. In order to compute the regulatory rules for each condition, we also used the Microsoft Excel spreadsheet package.
FVA
Since FBA is an optimization-based technique, it is often possible to have multiple solutions attaining the same objective value. Therefore, in order to confirm the phenotypic and metabolic states predicted by FBA, we performed FVA, which identifies all the active fluxes and their possible ranges given a particular physiological state. It can be represented using the same constraints of FBA as follows:
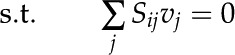 |
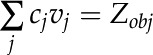 |
where Zobj denotes the value of the objective calculated from FBA and n is the number of fluxes. The upper range of fluxes is identified by maximizing the objective, whereas the lower range is obtained by minimizing the same. In this study, FVA was implemented in General Algebraic Modeling System Integrated Development Environment, version 23.9.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data File S1. The reconstructed rice metabolic network, detailed information on the biomass composition of rice seeds and straw, and list of references used to reconstruct the rice metabolic network.
Supplemental Data File S2. Systems Biology Markup Language file of the reconstructed rice metabolic model.
Supplemental Data File S3. List of reactions that are regulated in the seed-derived cells growing on various carbon sources and photorespiring leaf cells.
Supplemental Data File S4. Results of FVA and FBA.
Acknowledgments
We thank the anonymous reviewers for their useful comments, which helped us to improve the quality of this work significantly.
Glossary
- 3-PGA
3-phosphoglycerate
- 2-PG
2-phosphoglycolate
- FBA
flux balance analysis
- TCA
tricarboxylic acid
- PPi
inorganic pyrophosphate
- OAA
oxaloacetate
- α-KG
α-ketoglutarate
- GABA
γ-aminobutyric acid
- FVA
flux variability analysis
- VC/VO
carboxylation-to-oxygenation flux ratio
- DHAP
dihydroxyacetone phosphate
References
- Alpi A, Beevers H. (1983) Effects of O2 concentration on rice seedlings. Plant Physiol 71: 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amino SI, Tazawa M. (1988) Uptake and utilization of sugars in cultured rice cells. Plant Cell Physiol 29: 483–487 [Google Scholar]
- Atkin OK, Macherel D. (2009) The crucial role of plant mitochondria in orchestrating drought tolerance. Ann Bot (Lond) 103: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell BJ, Waters I, Greenway H. (1982) The effect of oxygen and turbulence on elongation of coleoptiles of submergence-tolerant and submergence-intolerant rice cultivars. J Exp Bot 33: 1030–1044 [Google Scholar]
- Bailey-Serres J, Voesenek LA. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York [Google Scholar]
- de Carvalho MHC (2008) Drought stress and reactive oxygen species. Plant Signal Behav 3: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. (2012) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 40: D742–D753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BK-S, Lakshmanan M, Klement M, Mohanty B, Lee DY. (2013) Genome-scale in silico modeling and analysis for designing synthetic terpenoid-producing microbial cell factories. Chem Eng Sci (in press) [Google Scholar]
- Covert MW, Schilling CH, Palsson B. (2001) Regulation of gene expression in flux balance models of metabolism. J Theor Biol 213: 73–88 [DOI] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. (2010a) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. (2010b) C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol 154: 1871–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JM, Roberts TH, Atwell BJ. (2012) Quantifying ATP turnover in anoxic coleoptiles of rice (Oryza sativa) demonstrates preferential allocation of energy to protein synthesis. J Exp Bot 63: 4389–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Covert M, Palsson B. (2002) Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol 4: 133–140 [DOI] [PubMed] [Google Scholar]
- Flexas J Galmes J, Ribas-Carbo M, Medrano H (2005) The effects of drought in plant respiration. In H Lambers, ed, Advances in Photosynthesis and Respiration, Vol 18. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 85–94 [Google Scholar]
- Gibbs J, Morrel S, Valdez A, Setter TL, Greenway H. (2000) Regulation of alcoholic fermentation in coleoptiles of two rice cultivars differing in tolerance to anoxia. J Exp Bot 51: 785–796 [PubMed] [Google Scholar]
- Grafahrend-Belau E, Schreiber F, Koschutzki D, Junker BH. (2009) Flux balance analysis of barley seeds: a computational approach to study systemic properties of central metabolism. Plant Physiol 149: 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A. (1995) Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol 108: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning I, Heldt HW. (1993) On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves: partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products. Plant Physiol 103: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J, Schwender J. (2011) Metabolic network reconstruction and flux variability analysis of storage synthesis in developing oilseed rape (Brassica napus L.) embryos. Plant J 67: 526–541 [DOI] [PubMed] [Google Scholar]
- Heldt H-W, Piechulla B (2011) In the photorespiratory pathway phosphoglycolate formed by the oxygenase activity of Rubisco is recycled. In Plant Biochemistry. Academic Press, London, pp 193–209 [Google Scholar]
- Jackson MB, Ram PC. (2003) Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann Bot (Lond) 91: 227–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogaiah S, Govind SR, Tran LS. (2012) Systems biology-based approaches toward understanding drought tolerance in food crops. Crit Rev Biotechnol 33: 23–39 [DOI] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. (1984) The CO2/O2 specificity of ribulose 1,5-bisphosphate carboxylase/oxygenase. Planta 161: 308–313 [DOI] [PubMed] [Google Scholar]
- Juliano BO (1985) Rice hull and rice straw. In BO Juliano, ed, Rice: Chemistry and Technology, Ed 2. American Association of Cereal Chemists, St. Paul, p 699 [Google Scholar]
- Keck RW, Boyer JS. (1974) Chloroplast response to low leaf water potentials. III. Differing inhibition of electron transport and photophosphorylation. Plant Physiol 53: 474–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, Lee KH, Chen SH, Ryu HN, Kim SJ, Kim DI. (2012) Adsorptive loss of secreted recombinant proteins in transgenic rice cell suspension cultures. Plant Cell Rep 31: 551–560 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM. (1999) The dual function of sugar carriers: transport and sugar sensing. Plant Cell 11: 707–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Lee DY, Kim TY. (2005) Systems biotechnology for strain improvement. Trends Biotechnol 23: 349–358 [DOI] [PubMed] [Google Scholar]
- Lewis NE, Nagarajan H, Palsson BO. (2012) Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol 10: 291–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magneschi L, Perata P. (2009) Rice germination and seedling growth in the absence of oxygen. Ann Bot (Lond) 103: 181–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH. (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic model. Metab Eng 5: 264–276 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Meir S, Malitsky S, Ruppin E, Aharoni A, Shlomi T. (2012) Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci USA 109: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B, Wilson PM, Aprees T. (1993) Effects of anoxia on growth and carbohydrate-metabolism in suspension-cultures of soybean and rice. Phytochemistry 34: 75–82 [Google Scholar]
- Nelson A (2011) Who eats the most rice? In Rice Today, Vol 10. International Rice Research Institute, Metro Manila, Philippines, pp 44–45 [Google Scholar]
- Oberhardt MA, Chavali AK, Papin JA. (2009) Flux balance analysis: interrogating genome-scale metabolic networks. Methods Mol Biol 500: 61–80 [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO. (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata P, Alpi A. (1993) Plant responses to anaerobiosis. Plant Sci 93: 1–17 [Google Scholar]
- Petersen LN, Marineo S, Mandala S, Davids F, Sewell BT, Ingle RA. (2010) The missing link in plant histidine biosynthesis: Arabidopsis myoinositol monophosphatase-like2 encodes a functional histidinol-phosphate phosphatase. Plant Physiol 152: 1186–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilalis E, Chatziioannou A, Thomasset B, Kolisis F. (2011) An in silico compartmentalized metabolic model of Brassica napus enables the systemic study of regulatory aspects of plant central metabolism. Biotechnol Bioeng 108: 1673–1682 [DOI] [PubMed] [Google Scholar]
- Poolman MG, Miguet L, Sweetlove LJ, Fell DA. (2009) A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol 151: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna A, Ravishankar GA. (2011) Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 6: 1720–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Suthers PF, Maranas CD. (2011) Zea mays iRS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 6: e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Pfeiffer T, Fell DA. (2008) Is maximization of molar yield in metabolic networks favoured by evolution? J Theor Biol 252: 497–504 [DOI] [PubMed] [Google Scholar]
- Seaver SM, Henry CS, Hanson AD. (2012) Frontiers in metabolic reconstruction and modeling of plant genomes. J Exp Bot 63: 2247–2258 [DOI] [PubMed] [Google Scholar]
- Shimon-Kerner N, Mills D, Merchuk JC. (2000) Sugar utilization and invertase activity in hairy-root and cell-suspension cultures of Symphytum officinale. Plant Cell Tiss Org 62: 89–94 [Google Scholar]
- Shingaki-Wells RN, Huang S, Taylor NL, Carroll AJ, Zhou W, Millar AH. (2011) Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol 156: 1706–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. (1987) Fructose 2,6-bisphosphate and plant carbohydrate metabolism. Plant Physiol 84: 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Ratcliffe RG. (2011) Flux-balance modeling of plant metabolism. Front Plant Sci 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM (2007) Synthesis, export and partitioning of the end products of photosynthesis. In RR Wise, JK Hoober, eds, The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 273–292 [Google Scholar]
- Wen Z-Y, Zhong J-J. (1995) A simple and modified manometric method for measuring oxygen uptake rate of plant cells in flask cultures. Biotechnol Tech 9: 521–526 [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]