Abstract
A fundamental question in cell biology is how cells determine membrane compartment identity and the directionality with which cargoes pass through the secretory and endocytic pathways. The discovery of so-called “Rab cascades” provides a satisfying molecular mechanism that helps to resolve this paradox. One Rab GTPase has the ability to template the localization of the subsequent acting Rab GTPase along a given transport pathway. Thus, in addition to determining compartment identity and functionality, Rab GTPases are likely able to order the events of membrane trafficking. This review will highlight recent advances in our understanding of Rabs and Rab cascades.
Since the discovery of the first yeast, Ras-related GTPase (YPT1) gene [1] and the landmark paper reporting the discovery of the SEC4 gene sequence [2] that first suggested a key role for these Ras-like GTPases in the control of membrane traffic, we now know that there are about 66 human Rab proteins and 11 yeast Rab-related Ypt proteins [3,4] that are master regulators of the secretory and endocytic pathways. Zerial and coworkers [5] were the first to provide evidence that each membrane compartment in the cytoplasm is likely to be decorated with distinct Rab proteins. This was an incredibly important finding because Rabs became the first true molecular markers for different membrane compartments of the endocytic and secretory pathways. Today we know that Rabs recruit discrete sets of effector proteins to the surfaces of different membranes. These effectors can drive the formation of transport vesicles, link to motor proteins for vesicle motility and/or recognize docking factors for delivery to target membranes (see [6–8] for excellent reviews).
Rabs are comprised of a compact, globular, GTP binding and hydrolysis domain, linked to an unstructured, hypervariable C-terminal domain [9]. The hypervariable domain is the most divergent region between Rab GTPase sequences. Active Rabs carry GTP; inactive Rabs carry GDP. Rabs are activated by guanine nucleotide exchange factors (GEFs) that enhance release of GDP; they are inactivated by GTPase activating proteins (GAPs) [9,10]. By definition, effector proteins bind Rabs with preference for their GTP-bound conformations, and carry out the downstream functions of individual Rab proteins.
In a few rare cases, it has been reported that Rabs bind to putative effectors in their GDP-forms or may not show preference for one nucleotide over another. An important cautionary note: some GDP-preferring mutant Rab proteins are sticky in vitro because they become nucleotide-free, so these reports must be evaluated with great care. The most reliable experiment is to utilize a wild type Rab protein and compare directly, the binding of GDP with that of GTP. Also, a lack of preference for GTP over GDP can be an artifact of incomplete nucleotide exchange in vitro. As would be expected for an enzyme:substrate interaction, Rab GEFs bind Rabs with preference for the GDP-bound forms, as do cytosolic GDIs (GDP dissociation inhibitors; see below) [9,10].
Rabs associate with membranes via one or two stable prenyl groups that are covalently attached to C-terminal cysteine residues. GDIs recognize GDP-bearing Rabs and can extract them from membranes and redeliver those Rabs to the appropriate target membrane [6–8]. The structure of GDI includes a pocket for the hydrophobic prenyl groups that normally anchor Rab proteins to membranes [9]. This extraction process can correct mistakes in Rab delivery and also retrieve Rab proteins from target membranes after a vesicular transport event.
Because effector proteins show preference for GTP-Rab proteins, they by definition interact with the so-called “switch” regions of Rab proteins that are the only parts of the Rab that change conformation between GTP- and GDP-bound states [9,11]. Comparative analysis of the three dimensional electrostatic and hydrophobic molecular interaction fields of 62 human Ran proteins adds new clues that may help explain the logic of Rab effector binding selectivity [12]. One might have imagined that Rab hypervariable domains would be very important for effector binding, as their variability would provide specificity in Rab binding interaction. Surprisingly, the importance of hypervariable domains in effector binding has not been widely investigated; the crystal structures of Rab proteins bound to their effectors often has been determined using truncated Rab proteins that are missing the hypervariable domains [9].
In the case of two Rab9A effectors, p40 and TIP47, the Rab9A hypervariable domain is an important determinant of effector binding and for TIP47, and is sufficient to confer binding capacity to Rab5 and Rab1 protein chimeras [13]. In contrast, the Rab5 effectors, Rabaptin 5 and EEA1 are much less dependent upon the hypervariable domain for Rab binding; similarly, GM130 binds Rab1 in the absence of the Rab1 hypervariable domain, however p115 is highly dependent upon the presence of the Rab1 hypervariable domain for binding [13]. Because available structures of Rab:effector complexes have been solved successfully using truncated Rab proteins, hypervariable domains may be less important for a number of Rab:effector interactions. Surprises are also possible: Rab6 binding to the GCC185 tethering protein C-terminus requires the presence of the Rab6 hypervariable domain, even though the structure model interface of Rab6 bound to a GCC185 coiled coil encompasses a relatively large surface and does not reveal information regarding the orientation of the essential hypervariable domain present in the crystals [14].
Rab microdomains
The discovery that Rabs occupy distinct domains of a given compartment represents an important landmark in our understanding of how the endocytic pathway is defined. Zerial and colleagues first reported that Rab4, Rab5 and Rab11 appear segregated on a single endosome when visualized in live cells [15]. Later it was reported that Rab7 and Rab9 define distinct microdomains on late endosomes [16]. Presumably, each Rab recruits a set of effector proteins onto the surface of membranes and assembles the appropriate molecular machinery to mediate membrane docking and fusion or linkage to specific motor proteins. A given Rab has multiple effector proteins and may template the assembly of multiple membrane microdomains. That has been shown for Rab5, which can interact with APPL or EEA1 in separate locations [17].
The identification of effector proteins that bind two different Rab GTPases revealed the potential of such effectors to link two, subsequently acting Rab microdomains to one another [18]. There are now multiple examples of proteins that contain multiple Rab binding sites. In the case of the multi-subunit HOPS tethering factor, the beautiful structure determined by electron microscopy revealed two independent Rab7 binding sites at opposite ends, contributed by Vps41 and Vps39 subunits [19]. In this case, the Rab binding sites bring together two Rab7-bearing membrane compartments. An additional HOPS subunit is responsible for SNARE binding—organizing all the components needed to drive fusion after successful docking. Thus, Rab GTPases organize this complex docking and fusion process: tethers often bind to SNAREs and often use Rabs to mediate their membrane localization.
Rab GTPase localization
Rabs are first delivered to membranes in their GDP bound forms by GDI [20,21] and then quickly activated by cognate guanine nucleotide exchange factors (Figure 1). In at least some cases, the release of a Rab from GDI can be facilitated by a membrane bound, GDI displacement factor or GDF [22,23]. Two recent papers show that the process of accurate Rab GTPase delivery can be hijacked if exogenous GEFs are mislocalized (and anchored) to an inappropriate membrane compartment [24,25]. Using a drug-inducible dimerization motif to mislocalize the Rab5 GEF, Rabex-5, Itzen and colleagues were able to relocalize Citrine-tagged Rab5 from early endosomes to mitochondria [24]. Rabex-5 alone is not able to act on Rab5 that is part of a GDI complex [26]. Thus, these findings suggest that Citrine-Rab5 is first mis-delivered to mitochondria by GDI and then stabilized there after activation by mitochondrially-localized Rabex-5. It is important to note that only exogenously expressed Rab5 was monitored; it is possible that extra Rab5 copies are targeted to membranes with somewhat lower fidelity than endogenous Rab5, as key effector proteins and Rab5 modulators become limiting. Presumably, once bound there, Rab5-GTP on mitochondria should have the capacity to recruit Rab5 effectors to that site. Anchoring of a Rab32/Rab38 GEF to mitochondria similarly led to Rab32 relocalization [25].
Figure 1.
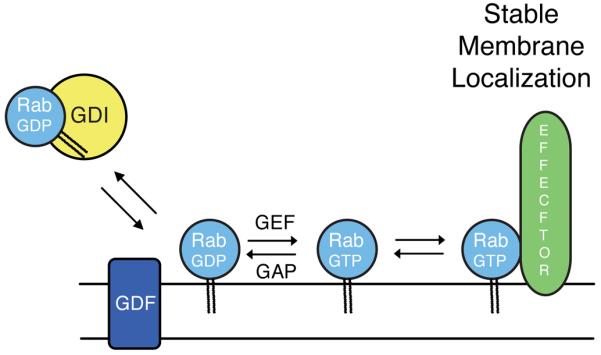
Prenylated Rab GTPases are delivered to membranes by GDI in their GDP-bound forms in a process that may be facilitated by a GDI-displacement factor (GDF). Guanine nucleotide exchange factors (GEFs) catalyze the release of bound GDP to permit GTP binding. Rab-GTP can be stabilized on membranes by binding to cognate effectors. GDI in cytosol can retrieve Rabs delivered to incorrect membranes unless they encounter their cognate GEF at that location.
GEFs are generally not anchored to membranes and thus cannot serve as classical targeting receptors. These experiments also reveal some unanticipated promiscuity in Rab GTPase membrane delivery and suggest that cytosolic GDI retrieves mis-localized Rabs from mitochondrial membranes to some extent at steady state. In addition, these findings confirm previous work demonstrating the importance of Rab effector interactions in the stabilization of Rab GTPases on specific membrane compartments [13]. By activating a Rab at a particular membrane location, that Rab becomes resistant to removal by GDI and is also capable of effector binding that will further stabilize it at that location. Indeed, Itzen and coworkers [24] found that a GTP-locked version of Rab5 was mis-localized with lower frequency than wild type Rab5 protein, consistent with an important role for GDI in the Rab delivery process. In summary, GDI, in some cases GDFs, GEFs and effectors are all important in Rab delivery. An important puzzle remains how GEFs and GAP proteins are able to modulate Rab activity at the correct place and time [10].
Ordered GEF recruitment creates Rab cascades
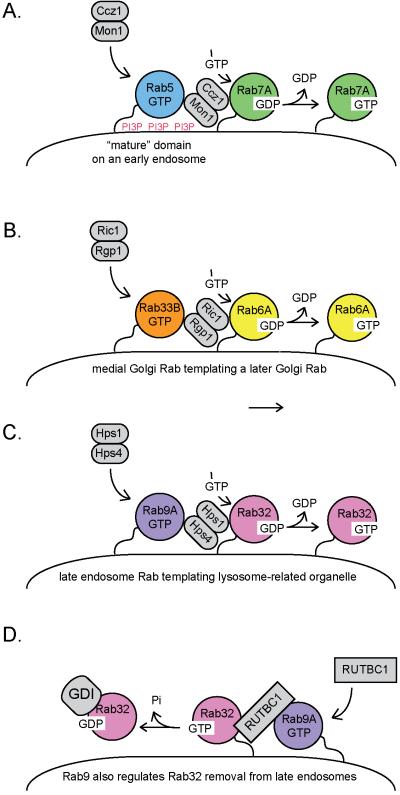
In 2002, Ortiz and Novick [27] showed that the Golgi Rab, Ypt32 recruits the Sec2 guanine nucleotide exchange factor to activate the secretory granule Rab protein, Sec4. This provided a molecular mechanism by which a Golgi-localized machinery could initiate the creation of a nascent secretory vesicle. Since that discovery, a number of proteins have been identified that use one Rab GTPase to target a modifier of another, adjacently-acting Rab GTPase. For example, the dimeric Ric1/Rgp1 GEF for the late Golgi Rab6 GTPase has an independent binding site for the medial Golgi Rab33B protein [28], and this interaction may template a late Golgi microdomain in relation to the Rab33B compartment (Figure 2). Similarly, the dimeric Mon1/Ccz1 GEF for Rab7 (Ypt7; [29]) has a binding site (on Mon1) for the previous acting Rab5 [30] (Figure 2).
Figure 2.
Heterodimeric Rab GEFs recruit subsequent acting Rabs as part of Rab cascades. A. On early endosomes, Rab5 recruits the kinase that generates phosphatidylinositol 3-phosphate (PI3P); Ccz1/Mon1 is then recruited to membranes containing Rab5-GTP [30] and PI3P [31]. Ccz1/Mon1 catalyzes the release of GDP from Rab7A [29; 25], thereby permitting the entry of GTP. Presumably a Rab5 GAP will be identified that is a Rab7A effector to help clear Rab5 from a newly formed, Rab7A membrane microdomain. Ccz1/Mon1 also displaces the Rab5 GEF, Rabex-5, from membranes [31]. B. On the Golgi, medial Golgi Rab33B recruits the Ric1/Rgp1 GEF for Rab6A [28]. On late endosomes, the Hps1/Hps4 GEF for Rab32 [25] is recruited to late endosomes by Rab9A [32]. D. Rab9A can also recruit RUTBC1, a GTPase activating protein that inactivates Rab32 [33]. Rab32 GDP would be a target for membrane extraction by cytoplasmic GDI protein. This would counteract the reaction in C and may be restricted to specific tissues and/or cell types.
Mon1 does more than help activate Rab7: it also displaces the Rab5 GEF, Rabex-5, from early endosomes [31]. Rab5 recruits the kinase that generates phosphatidylinositol 3-phosphate (PIP3) on early endosomes. Because the Mon1 protein also recognizes PI3P, Spang and colleagues [31] propose that Mon1 might only be recruited onto more “mature” endosomes that contain higher levels of this lipid. In this model, the Mon1/Ccz1 GEF would seek membranes rich in both Rab5 and PI3P (Figure 2). Coincidence detection in membrane recruitment of key protein modulators provides selectivity to the process, and is a common theme throughout membrane trafficking: it likely explains how cells displace Rab5 to permit Rab7 recruitment, only at the right time and place, using factors that are in equilibrium between the cytosol and specific membranes.
A somewhat more confusing circuit was revealed by the discovery that the Hps1-Hps4 “BLOC-3” complex that comprises a GEF for Rab32/Rab38 [25] is an effector of the late endosomal protein, Rab9 [32]. As described below, Rab9 also recruits the Rab32 GAP, RUTBC1 [33]. Thus, Rab9 appears to template both the activation and inactivation of Rab32 proteins (Figure 2). In addition, the VARP protein, a GEF for Rab21, also binds what were thought to be later acting Rab32 and Rab38 proteins [34]. Obviously more work is needed to explain this conundrum, which may include tissue-specific, differential expression of these Rab modulatory factors.
Rab domains may grow with GEF action and subsequent effector binding until some key constituent becomes limiting; at that point Rab-GAP action could predominate, triggering local catastrophy and Rab microdomain disassembly. As more details become available regarding the identity of the requisite players and determinations of their abundance, interaction affinities, and catalytic rates, it should become possible to model the process of Rab conversion with physiological relevance [35,36]. Beautiful recent structural and kinetic analyses continue to reveal the detailed mechanisms of an increasing number of Rab GEF and GAP proteins (cf. [37–39]).
Ordered GAP recruitment enhances fidelity of Rab segregation
In the yeast secretory pathway, the Ypt32 Rab acts after Ypt1 Rab protein. Rivera-Molina and Novick have shown that the Gyp1 GAP that acts on Ypt1 protein relies on the later acting, Ypt32 protein for its localization and stability [40]. This activity will clear Ypt1 protein from a region that is rich in Ypt32 protein. Similarly, the late endosomal Rab9A GTPase binds to RUTBC1 and RUTBC2 proteins but is not a substrate for these GAP proteins: in cells, these proteins display GAP activity for Rab32 and Rab36, respectively [33,41]. In both cases, the Rab9 binding site is entirely separate from the GAP catalytic (“TBC”) domain [33, 41–43]. Localization of RUTBC1 and RUTBC2 by Rab9 protein will serve to remove Rab32 and Rab36 from Rab9A-enriched membrane areas, and indicates that they function in membrane microdomains adjacent to Rab9 protein.
Large scale screening for novel Rab GTPase binding partners has identified a number of Rab GAPs that contain binding sites for non-substrate Rab GTPases [43]. How these binding partners contribute specifically to possible Rab cascades awaits further investigation.
More Fruit from large screens
Broad-reaching interaction and modifier screens continue to provide critical information to our understanding of the functions of the large family of Rab proteins and all their functions in secretion and endocytosis. Francis Barr and his colleagues have been very successful using such screens and recently identified FAM116A as a GEF for Rab14 GTPase [44]. This unbiased screen for Rab GTPases and Rab GEFs involved in cell migration provided a function for an under-studied Rab and identified Rab14 as a GTPase that acts after Rab4 and Rab5 but before Rab11 in a recycling compartment that has a function distinct from the other Rabs and is key for cell migration [44].
Rab conversion need not reflect compartment maturation
Recently two models for transport from early to late endosomes have been discussed [45–49]. In one, called Rab conversion, a Rab5 compartment matures into a Rab7 compartment; Rab5 falls off as Rab7 comes on [45]. In another model, a Rab7 compartment is seen to bud off of a Rab5 compartment [46,49]. Both of these models can be driven by a common, Rab-based mechanism. The important principle is that a Rab5 compartment assembles, nearby, a Rab7 compartment. The added component of the second model is some type of fission reaction: Rab5 and Rab7 link to different motor proteins, and if one imagines that endosomes undergo Rab-linked motor protein and cytoskeleton-dependent fission and fusion events, Rab7 domains can be segregated from Rab5 domains. Important to this discussion is the molecular mechanism by which a Rab5 compartment is converted into a Rab7 compartment.
A similar discussion is underway in our thinking about transport through the Golgi complex: if one overlays a classic Rab cascade on top of a set of membranes that can undergo homotypic fusion and cytoskeleton-dependent fission, forward transport of proteins through a stable Golgi complex can be achieved [50]. For both the Golgi and endosome systems, elucidation of the key Rabs and their modifiers will help shed light on the mechanisms by which proteins move through these compartments. Whichever models turn out to be correct, Rabs are surely templating the directionality of transport through the secretory and endocytic pathways.
Highlights
Rab GTPases recruit distinct sets of effector proteins to membrane microdomains
Rab-bound effectors confer unique functionalities to membrane microdomains
Activity-modulators are localized by non-substrate Rabs and/or Rab modulators
Rab GTPases template the sequence of membrane traffic events
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Schmitt HD, Wagner P, Pfaff E, Gallwitz D. The ras-related YPT1 gene product in yeast: a GTP-binding protein that might be involved in microtubule organization. Cell. 1986;47:401–412. doi: 10.1016/0092-8674(86)90597-0. [DOI] [PubMed] [Google Scholar]
- [2].Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–38. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- [3].Diekmann Y, Seixas E, Gouw M, Tavares-Cadete F, Seabra MC, Pereira-Leal JB. Thousands of rab GTPases for the cell biologist. PLoS Comput Biol. 2011;7(10):e1002217. doi: 10.1371/journal.pcbi.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Elias M, Brighouse A, Gabernet-Castello C, Field MC, Dacks JB. Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J Cell Sci. 2012;125:2500–2508. doi: 10.1242/jcs.101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–29. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- [6].Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–59. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- [9].Itzen A, Goody RS. GTPases involved in vesicular trafficking: structures and mechanisms. Semin Cell Dev Biol. 2011;22:48–56. doi: 10.1016/j.semcdb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- [10].Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–70. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stroupe C, Brunger AT. Crystal structures of a Rab protein in its inactive and active conformations. J Mol Biol. 2000;304:585–598. doi: 10.1006/jmbi.2000.4236. [DOI] [PubMed] [Google Scholar]
- [12].Stein M, Pilli M, Bernauer S, Habermann BH, Zerial M, Wade RC. The interaction properties of the human Rab GTPase family--comparative analysis reveals determinants of molecular binding selectivity. PLoS One. 2012;7:e34870. doi: 10.1371/journal.pone.0034870. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper presents a comparative analysis of Rab protein electrostatic and hydrophobic molecular interaction fields that provide clues to molecular distinctions between Rab family members.
- [13].Aivazian D, Serrano RL, Pfeffer S. TIP47 is a key effector for Rab9 localization. J Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper includes a rare analysis of the importance of Rab hypervariable domains in effector binding.
- [14].Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–98. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–14. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Barbero P, Bittova L, Pfeffer SR. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J Cell Biol. 2002;156:511–8. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–56. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- [18].de Renzis S, Sönnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat Cell Biol. 2002;4:124–33. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- [19].Bröcker C, Kuhlee A, Gatsogiannis C, Balderhaar HJ, Hönscher C, Engelbrecht-Vandré S, Ungermann C, Raunser S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci USA. 2012;109:1991–6. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Electron microscopy is used to determine the structure of the multi-subunit HOPS complex that mediates late endosome fusion. Using antibodies and immunogold labeling, the authors are able to identify the location of subunits that bind Ypt7 and requisite SNARE proteins to drive endosome docking and fusion.
- [20].Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–160. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- [21].Soldati T, Shapiro AD, Svejstrup AB, Pfeffer SR. Membrane targeting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature. 1994;369:76–78. doi: 10.1038/369076a0. [DOI] [PubMed] [Google Scholar]
- [22].Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425:856–859. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- [23].Ohya T, Miaczynska M, Coskun U, Lommer B, Runge A, Drechsel D, Kalaidzidis Y, Zerial M. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- [24].Blümer J, Rey J, Dehmelt L, Mazel T, Wu Y-W, Bastiens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013 doi: 10.1083/jcb.201209113. 2013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This is an interesting paper that demonstrates the contribution of Rab GEFs to Rab localization. The work does not prove that GEFs are sufficient for this process; rather, it reveals some level of mis-localization of prenylated Rabs that can be trapped by anchoring of a GEF at an inappropriate cellular location. Presumaby, Rabs are stabilized there after GEF activation and effector binding.
- [25].Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–9. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–59. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- [27].Ortiz D, Medkova M, Walch-Solimena C, Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol. 2002;157:1005–15. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper presents the first indication of the existence of Rab cascades to template the order of events in the secretory pathway.
- [28].Pusapati GV, Luchetti G, Pfeffer SR. Ric1-Rgp1 Complex Is a Guanine Nucleotide Exchange Factor for the Late Golgi Rab6A GTPase and an Effector of the Medial Golgi Rab33B GTPase. J Biol Chem. 2012;287:42129–42137. doi: 10.1074/jbc.M112.414565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]; •• This paper presents an elegant biochemical and genetic proof of the identity of the Ypt7 GEF and explains why others mistakenly thought that the activity was a constituent of the HOPS complex.
- [30].Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper presents a thorough analysis of the Rab5-to-Rab7 switch in phagosome maturation and shows that the Mon1/Ccz1 GEF for Rab7 is recruited onto membranes by Rab5.
- [31].Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]; •• This paper reports the discovery that SAND1/Mon1 displaces Rabex5 from membranes as part of the conversion from a Rab5 structure to a Rab7 structure. The authors propose further that “mature” early endosomes are distinguished by their content of PI3P.
- [32].Kloer DP, Rojas R, Ivan V, Moriyama K, van Vlijmen T, Murthy N, Ghirlando R, van der Sluijs P, Hurley JH, Bonifacino JS. Assembly of the biogenesis of lysosome-related organelles complex-3 (BLOC-3) and its interaction with Rab9. J Biol Chem. 2010;285:7794–804. doi: 10.1074/jbc.M109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nottingham RM, Ganley IG, Barr FA, Lambright DG, Pfeffer SR. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J Biol Chem. 2011;286:33213–22. doi: 10.1074/jbc.M111.261115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ohbayashi N, Yatsu A, Tamura K, Fukuda M. The Rab21-GEF activity of VARP, but not its Rab32/38 effector function, is required for dendrite formation in melanocytes. Mol Biol Cell. 2012;23:669–78. doi: 10.1091/mbc.E11-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Del Conte-Zerial P, Brusch L, Rink JC, Collinet C, Kalaidzidis Y, Zerial M, Deutsch A. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol Syst Biol. 2008;4:206. doi: 10.1038/msb.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Foret L, Dawson JE, Villaseñor R, Collinet C, Deutsch A, Brusch L, Zerial M, Kalaidzidis Y, Jülicher F. A general theoretical framework to infer endosomal network dynamics from quantitative image analysis. Curr Biol. 2012;22:1381–1390. doi: 10.1016/j.cub.2012.06.021. [DOI] [PubMed] [Google Scholar]
- [37].Wu X, Bradley MJ, Cai Y, Kümmel D, De La Cruz EM, Barr FA, Reinisch KM. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci U S A. 2011;108:18672–7. doi: 10.1073/pnas.1110415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chin HF, Cai Y, Menon S, Ferro-Novick S, Reinisch KM, De La Cruz EM. Kinetic analysis of the guanine nucleotide exchange activity of TRAPP, a multimeric Ypt1p exchange factor. J Mol Biol. 2009;389:275–288. doi: 10.1016/j.jmb.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gavriljuk K, Gazdag EM, Itzen A, Kötting C, Goody RS, Gerwert K. Catalytic mechanism of a mammalian Rab-RabGAP complex in atomic detail. Proc Natl Acad Sci USA. 2012;109:21348–53. doi: 10.1073/pnas.1214431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci USA. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper visualizes a Rab GAP cascade on the Golgi in living yeast cells and provides a molecular mechanism to explain membrane traffic through the Golgi complex.
- [41].Nottingham RM, Pusapati GV, Ganley IG, Barr FA, Lambright DG, Pfeffer SR. RUTBC2 protein, a Rab9A effector and GTPase-activating protein for Rab36. J Biol Chem. 2012;287:22740–8. doi: 10.1074/jbc.M112.362558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- [43].Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–68. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- [44].Linford A, Yoshimura S, Nunes Bastos R, Langemeyer L, Gerondopoulos A, Rigden DJ, Barr FA. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012;22:952–66. doi: 10.1016/j.devcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper reports a comprehensive screen of Rab GTPases and GEFs that participate in cell migration and reports a novel role of Rab14 in the cycling of ADAM10, a protease important for cell surface cadherin shedding.
- [45].Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- [46].Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki Forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cabrera M, Ungermann C. Guiding endosomal maturation. Cell. 2010;141:404–406. doi: 10.1016/j.cell.2010.04.013. [DOI] [PubMed] [Google Scholar]
- [49].Mesaki K, Tanabe K, Obayashi M, Oe N, Takei K. Fission of tubular endosomes triggers endosomal acidification and movement. PLoS One. 2011;6:e19764. doi: 10.1371/journal.pone.0019764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pfeffer SR. How the Golgi works: a cisternal progenitor model. Proc Natl Acad Sci USA. 2010;107:19614–19618. doi: 10.1073/pnas.1011016107. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper presents a model that invokes a Rab cascade on the Golgi, overlaid by a process of membrane fusion and fission, to explain protein transport through the Golgi complex. This type of model is analogous to that proposed by Helenius and colleagues [44,45] for transport through the endocytic pathway.