Abstract
The ER is a continuous membrane system consisting of the nuclear envelope, flat sheets often studded with ribosomes, and a polygonal network of highly-curved tubules extending throughout the cell. Although protein and lipid biosynthesis, protein modification, vesicular transport, Ca2+dynamics, and protein quality control have been investigated in great detail, mechanisms that generate the distinctive architecture of the ER have been uncovered only recently. Several protein families including the reticulons and REEPs/DP1/Yop1p harbor hydrophobic hairpin domains that shape high-curvature ER tubules and mediate intramembrane protein interactions. Members of the atlastin/RHD3/Sey1p family of dynamin-related GTPases interact with the ER-shaping proteins and mediate the formation of three-way junctions responsible for the polygonal structure of the tubular ER network, with Lunapark proteins acting antagonistically. Additional classes of tubular ER proteins including some REEPs and the M1 spastin ATPase interact with the microtubule cytoskeleton. Flat ER sheets possess a different complement of proteins such as p180, CLIMP-63 and kinectin implicated in shaping, cisternal stacking and cytoskeletal interactions. The ER is also in constant motion, and numerous signaling pathways as well as interactions among cytoskeletal elements, the plasma membrane, and organelles cooperate to position and shape the ER dynamically. Finally, many proteins involved in shaping the ER network are mutated in the most common forms of hereditary spastic paraplegia, indicating a particular importance for proper ER morphology and distribution in large, highly-polarized cells such as neurons.
Keywords: Atlastin, Endoplasmic reticulum, Hereditary spastic paraplegia, Morphology, REEP, Reticulon, Shaping
1. Introduction
It is the pervading law of all things organic and inorganic,
Of all things physical and metaphysical,
Of all things human and all things super-human,
Of all true manifestations of the head,
Of the heart, of the soul,
That the life is recognizable in its expression,
That form ever follows function.This is the law.
■ Louis Sullivan, architect [1]
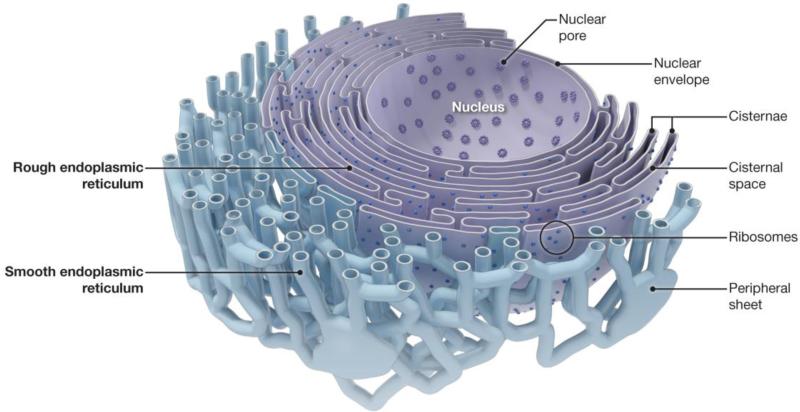
Cellular organelles have diverse but highly characteristic shapes that are typically conserved across species, suggesting that their structural features are intimately tied to their specific roles within the cell. The ER is particularly striking for its heterogeneity of form and function. It plays critical roles in the synthesis, modification, quality control, and trafficking of integral membrane proteins and soluble proteins destined for secretion, the mobilization and regulated release of Ca2+, sterol/lipid synthesis and distribution, signaling, carbohydrate metabolism, and detoxification of harmful substances [2]. Reflecting these diverse functions, the ER comprises a continuous membrane system that includes the inner and outer nuclear membranes, sheet-like cisternae, and a network of interconnected tubules extending promiscuously into the cell periphery (Figs. 1 and 2A) [3-8]. The ER is the largest continuous organelle, with its membranes comprising about half of the total membrane and its lumen enclosing about 10% of the volume of a typical eukaryotic cell [9].
Fig 1.
The ER network in cells. Schematic diagram showing the different structural features and morphologies of the continuous ER in animal cells.
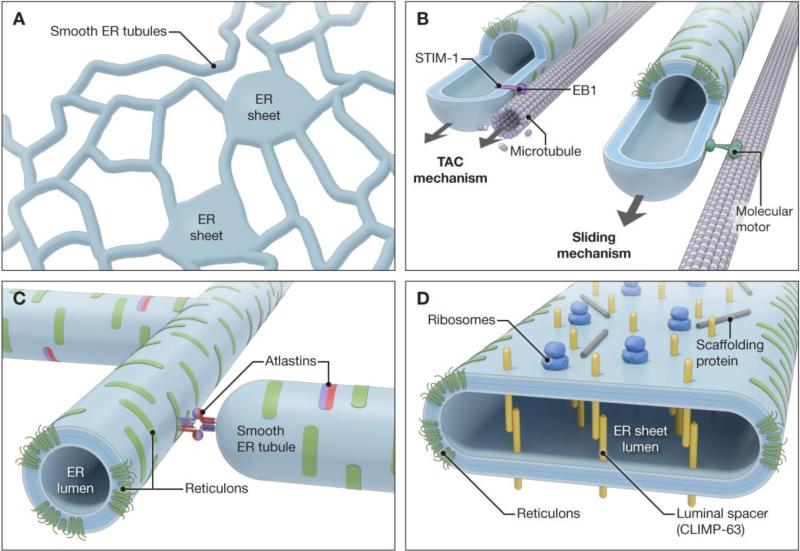
Fig 2.
Mechanisms involved in shaping the ER network. (A) Schematic diagram showing interconnected smooth ER tubules and peripheral sheets. (B) Sliding and TAC mechanisms involved in the formation and extension of ER tubules along stable, acetylated microtubule tracks and polymerizing microtubules, respectively. (C) ER shaping reticulons and atlastins on ER tubules, with a schematic depiction of atlastin-dependent tubule fusion. (D) ER proteins involved in formation and stabilization of ER sheets. Scaffolding proteins include p180 and kinectin. Adapted and modified from Lin et al. [8] and Pendin et al. [86].
Prominent functional specializations appear to correlate with the distinct morphologies of the ER. For instance, ER sheets studded with polyribosomes (so-called “rough” ER) are associated with the biosynthesis, modification, and quality control of secreted and integral membrane proteins. The initial membrane trafficking step in the biosynthetic secretory pathway, the export of proteins and lipids from the endoplasmic reticulum (ER), is mediated by COPII-coated vesicles arising from specialized ER exit sites for trafficking to the Golgi apparatus. By contrast with ER sheets, tubules are mostly smooth ER and associated with lipid synthesis and delivery, metabolism of carbohydrates, establishing contacts with other organelles, detoxification, and lipid droplet formation [3].
Befitting the distinct functional specializations of these domains, the ER can appear drastically different across cell types. For instance, professional secretory cells such as pancreatic acinar cells and plasma cells harbor abundant stacks of ribosome-studded ER sheets which are involved in the production and secretion of proteins, up to thousands of molecules per second. On the other hand, hepatocytes exploit an extensive smooth ER network mostly devoid of ribosomes but enriched in enzymes for the metabolism of carbohydrates and detoxification of biosynthetic products as well as exogenous substances such as drugs and poisons. Muscle cells possess a specialized form of smooth ER, the sarcoplasmic reticulum, that is important for the mobilization and regulated release of Ca2+ into the cytoplasm during muscle contractions. Retinal pigment epithelial cells exhibit stacked ER sheets mostly devoid of ribosomes, the organized smooth ER. As a last example, the nuclear envelope is pierced by several thousand large, multimeric nuclear pore complexes that regulate trafficking between the cytoplasm and the nucleoplasm and help to stabilize the internal structure of the nucleus [8, 10].
These morphologic variations still do not encompass fully the broad range of ER specializations, as the ER is highly dynamic. It undergoes prominent shape transitions during events such as cell division and differentiation, and fusion reactions of ER tubules to form new three-junctions have been observed in interphase cells across many cell types and species [11-15]. The ER interacts with the cytoskeleton and the plasma membrane as well as other dynamic cellular organelles such as endosomes and mitochondria [4, 16-18]. In highly-polarized and specialized neurons, ER shape changes in dendritic spines both influence and are regulated by synaptic signaling pathways [19-21]. Implicit in this dynamic variability is the general concept that many different types of cellular signals are able to trigger shape changes across different ER domains.
2. Mechanisms generating high membrane curvature in the ER
The different morphologies of the ER derive in part from variations in membrane curvature across different domains, enabling them to maintain distinct shapes while remaining physically continuous. In fact, such mechanisms are broadly conserved among eukaryotic cells. The luminal diameters of ER tubules are generally similar across a range of organisms and cell types, ranging from about 30-100 nm (for reference, the thickness of the phospholipid bilayer is ~4 nm). Likewise, ER sheets across most species are similar in luminal thickness (50-100 nm) to the diameter of tubules but can extend several micrometers and only curve prominently at the edges [22]. Finally, although the nuclear envelope appears spherical in cells, its large diameter of about 6-10 μm means that the inner and outer nuclear membranes take on a sheet-like appearance, with areas of high curvature mostly restricted to the edges of nuclear pores [3, 10, 23].
Although asymmetric distribution of lipids between the two leaflets of the lipid bilayer could introduce high membrane curvature, this mechanism seems not to be used in the ER. Instead, proteins are widely implicated in generating ER curvature [4, 6, 23, 24]. Generally speaking, proteins can generate and stabilize high-degree curvature in cellular membranes through a variety of means: membrane deformation by force-generating proteins (e.g., molecular motors); bending tubules through highly-curved protein scaffolds that interact with the phospholipid bilayer; protein-protein crowding; and hydrophobic insertion of proteins into the outer leaflet of the bilayer (hydrophobic wedging) [23, 25, 26]. ER tubules can be formed by membranes sliding along microtubules or attached to polymerizing microtubules. In this context, specialized tip attachment complexes (TACs) associated with polymerizing microtubules cooperate to pull the nascent tubules in the plus-end direction, while molecular motors mediate tubular extension along established, acetylated microtubules (Fig. 2B) [27, 28].
Under these and related scenarios, it is imperative to establish how ER tubules are stabilized. Several classes of proteins appear necessary and sufficient for the generation and maintenance of ER tubules in eukaryotic cells and lipid tubules in vitro [29-35]. These “ER-shaping,” curvature-stabilizing proteins comprise two main families, the DP1/REEPs/Yop1p proteins and the reticulons. Though these families show little overall sequence homology, they share an important structural signature—elongated hydrophobic segments that are predicted to form partially membrane-spanning hairpin (or wedge) domains (Fig. 2C) [29]. Within these extended hydrophobic domains are often charged amino acids or proline residues, and the composition and length may be important for both targeting to different ER domains and shaping them [29, 32, 33, 36]. The bulk of the hydrophobic portions of these ER-shaping proteins appear to occupy the outer leaflet of the phospholipid bilayer, possibly generating curvature via hydrophobic wedging. The reticulons form large, immobile oligomers [31, 33], so scaffolding, protein-protein crowding, or both might also play a role in curvature generation. Last, although the hydrophobic hairpins play key roles in mediating intramembrane interactions required for the formation of these large structures [29, 31], specificity may be also provided by interactions among cytoplasmic domains [3].
Multiple types of evidence indicate that these ER-shaping proteins are able to generate highly-curved ER morphologies. Overexpression of reticulon-4a (Rtn4a) in mammalian cells leads to more prominent tubules and the depletion of ER sheets [23, 29]. Similarly, in plant cells overexpression of reticulons dramatically reduces the luminal diameter of ER tubules, most likely by increasing curvature [34]. Conversely, depleting ER-shaping proteins of the DP1/REEP/Yop1p and reticulon families from mammalian or yeast cells drives an increase in sheets at the expense of tubules [29, 37]. A seminal demonstration of the tubule-shaping properties of these proteins was provided by Hu et al. [30], who showed that recombinant yeast Yop1p and Rtn1p are sufficient to deform proteoliposomes in vitro into tubules with a diameter of 15-17 nm. On the other hand, simultaneous depletion of reticulons and Yop1p in yeast does not prevent ER tubules from being pulled from a cisterna into the bud, emphasizing a primary in vivo role in curvature stabilization and maintenance of peripheral ER tubules [35].
3. Formation and stabilization of ER sheets
Flat ER sheets do not require stabilization and can form spontaneously, but they have a degree of curvature along their edges similar to that of ER tubules, possibly due to the localized enrichment of curvature-stabilizing reticulons [23, 38]. The relatively constant luminal thickness of extensive ER sheets appears to be maintained through a number of means, with proteins including CLIMP-63, p180, and kinectin playing important roles. Each of these proteins is enriched in ER sheets, and their overexpression induces sheet proliferation. Simultaneous depletion of all of these proteins does not eliminate ER sheets, though those that remain exhibit a decrease in luminal width [38]. Additional players such as polyribosome complexes may also play a role in sheet stabilization [8, 38].
The mechanisms underlying sheet stabilization are becoming increasingly clear. CLIMP-63, kinectin, and p180 each contain coiled-coil domains, and the large domains of CLIMP-63 within the ER lumen form intraluminal bridges that can stabilize a constant sheet width [39]. By contrast, the coiled-coil domains of p180 and kinectin are cytoplasmic, and may form long rods which stabilize the flatness of the sheets (Fig. 2D) [8]. Stacks of sheets may be further stabilized through protein interactions at the cytoplasmic face (Fig. 1D) [40].
4. Enveloping the nucleus
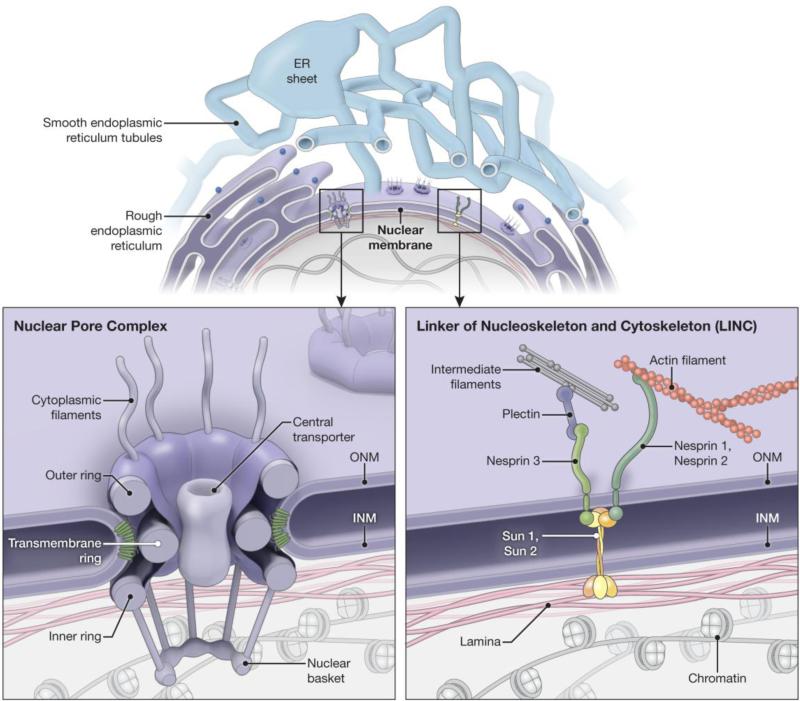
The nuclear envelope consists of the outer and inner nuclear membranes that connect at the sites of nuclear pores but are otherwise separated from one another in sheet-like structures [10, 41]. This separation is about 40-50 nm, thinner than the distance separating the two membranes of a typical peripheral ER sheet. Proteins of the reticulon and DP1/REEP/Yop1p families are required for nuclear pore formation [42], likely because of their membrane-curving and stabilizing functions, and the nucleoporin core complex is also likely involved (Fig. 3) [23]. Flatter areas of the nuclear envelope are stabilized by interactions of nuclear membrane proteins with chromatin and the nuclear lamina [10]. In particular, the linker of nucleoskeleton and cytoskeleton (LINC) complex plays a key role in holding the distance between the outer and inner nuclear membranes constant. This complex consists of Sad1 and UNC-84 (SUN) in the inner nuclear membrane and KASH (klarsicht, ANC-1, and syne/nesprin homology) domain proteins that are inserted into the outer nuclear membrane. SUN proteins (e.g., Sun 1 and Sun 2) interact with the KASH domain proteins (e.g., Nesprins), generating a protein bridge that connects the outer and inner nuclear membranes and may link the nuclear lamina to the cytoplasmic cytoskeleton (Fig. 3) [8, 43].
Fig 3.
Specialized nuclear envelope protein complexes. Top, Schematic diagram showing the continuous lumen between the nuclear membrane and the ER network. Bottom, Enlargements of the boxed areas in the top panel, showing the organization of the nuclear pore complex (left) and LINC complex (right). ER-shaping proteins are shown in green in the left panel. INM, inner nuclear membrane; ONM, outer nuclear membrane.
5. Fusing the network
Previous sections of this review have emphasized how the formation and maintenance of ER morphology depends on the proper segregation of various types of shaping proteins. But producing highly-curved ER tubules is only one part of shaping the peripheral ER network, and the formation of three-way junctions to generate the characteristic polygonal array requires the fusion of tubules. Abundant evidence suggests that the atlastin family of dynamin-related GTPases plays a critical role in this process. The atlastins are large, multimeric, integral membrane GTPases that localize predominantly to highly-curved ER membranes, including tubular ER and edges of ER sheets [44-48]. In mammals, there are three closely related atlastins, each of which harbors an N-terminal GTP-binding domain, a middle assembly domain, two very closely spaced hydrophobic segments near the C-terminus, and a C-terminal tail [44, 46]. Atlastin-related GTPases appear to be ubiquitous and include the functional orthologs Sey1p in S. cerevisiae and RHD3 in Arabidopsis [47]. These large GTPases interact with ER-shaping proteins of both the DP1/REEP/Yop1p and reticulon families and mediate the formation of three-way junctions in the ER (Fig. 2C) [46, 47, 49].
Concordant with their role in ER network formation, the atlastins and Sey1p are distributed in puncta along ER tubules including at three-way junctions [3, 46, 47], where they mediate homotypic fusion of ER membranes [48, 50]. Recent structural and biochemical studies support a model whereby atlastin oligomers in one membrane interact in trans with atlastin oligomers in an apposing membrane through GTP binding, forming a tethered complex. Upon GTP hydrolysis, conformational changes pull the membranes into closer proximity, with curvature generated by the atlastin membrane domains and the C-terminal domain, destabilizing the membrane and allowing fusion to occur. In the post-fusion state, GDP is released from atlastin, rebooting the fusion machinery [8, 51-53].
The means for dynamically regulating atlastin-mediated fusion have remained less clear, as has the functional role of atlastin interactions with the ER shaping proteins. However, a recent study in S. cerevisiae by Chen et al. [54] has indicated that Lnp1p, a member of the conserved Lunapark family defined by several hydrophobic domains and a Zn2+ finger motif, also plays a role in ER network formation. Lnp1p binds to the reticulons and Yop1p and resides at ER tubule three-way junctions in both yeast and mammalian cells. In yeast, the interaction of Lnp1p with the reticulon Rtn1p as well as the localization of Lnp1p to ER junctions are regulated by Sey1p. The authors proposed that Lnp1p might counterbalance Sey1p-directed polygon formation by promoting polygon loss through ring closure [54]. Last, it remains unclear whether atlastins account for all fusion of ER tubules, or whether other protein classes are involved. In fact, sey1Δ yeast exhibit some residual ER-ER fusion that requires the ER SNARE [50], indicating that other mechanisms may be uncovered in the future.
6. Cytoskeletal and organellar interactions
Another means by which ER morphological domains and their spatial distribution in the cell might be stabilized is through interactions with the cytoskeleton, plasma membrane, or other organelles (such as mitochondria). Organelle interactions with the ER are a focus of another review in this volume as well as several other recent reviews, and won't be discussed further here [4, 18]. Instead, we will focus our discussion on the roles of plasma membrane and cytoskeletal interactions in shaping the ER network.
Though the ER appears intimately associated with the cytoskeleton in all eukaryotic cells, there are important differences among cell types and across species. In animal cells, there is a particularly close association of the ER with the microtubule cytoskeleton, and the ER is formed along microtubules through a number of distinct mechanisms [27, 28, 55]. In yeast, movements of nuclear ER during the cell cycle are dependent on microtubules, while cortical ER movement and morphology depend on actin fibers [56]. Actin-based mechanisms also predominate in plants [6, 12, 57]. Notably, the cytoskeleton is not absolutely required for ER tubule formation, and an interconnected tubular network can be generated from a Xenopus microsomal membrane fraction in a GTP-dependent manner in the absence of microtubules [58]. Even so, cytoskeletal interactions are important for the characteristic appearance of ER in cells, since disruption of the microtubule cytoskeleton with nocodazole causes collapse of the ER by retraction from the cell periphery and conversion of peripheral ER tubules to extended sheet-like structures [14, 59].
Links between cytoskeletal elements and ER proteins provide important insights into how the ER is dynamically regulated. In addition to interacting with the ER shaping proteins, atlastins also bind the microtubule-severing, AAA (ATPases associated with a variety of cellular activities) protein spastin [60, 61], physically linking membrane and cytoskeletal remodeling proteins. This interaction occurs between the paired hydrophobic membrane domain of the atlastins and a hydrophobic hairpin domain present near the N-terminus only in the larger M1 spastin isoform; the M87 spastin isoform lacks this domain. Atlastins and M1 spastin also interact with DP1/REEPs within the tubular ER [62]. These latter proteins comprise 6 members in humans: REEP1-6—REEP5 is also known as DP1. Both structurally and phylogenetically there are clear distinctions between two main clades, comprising REEP1-4 and REEP5-6. The yeast ER-shaping protein Yop1p is most similar to REEP5-6, while REEP1-4 proteins also interact with microtubules through an extended C-terminal cytoplasmic domain enriched in basic amino acids [62]. Though coordinated interactions among REEPs, atlastins, and M1 spastin via hydrophobic segments provide a compelling mechanism for coupling ER membrane remodeling to cytoskeletal dynamics, it remains unclear whether these proteins are components of structures such as TACs. In a complementary manner, CLIMP-63 may mediate the attachment of ER sheets to microtubules [63]. In yeast, Estrada et al. [64] demonstrated that cortical ER relies on actin, the myosin V motor Myo4p, and the adaptor protein She3p, a component of the exocyst tethering complex. During bud growth, ER tubules are anchored to the bud tip, and they extend from there in a manner regulated by the cell wall integrity MAP kinase pathway [56].
Stabilization and proper distribution of the ER network also depends on interactions with the plasma membrane, and in fact ER-plasma membrane junctions are highly-conserved structures across species. A recent study by Emr and colleagues [17] found that wholesale depletion of three classes of ER-plasma membrane tethering proteins in yeast -- Ist2 (related to mammalian TMEM16 channels), the tricalbins (orthologs of the extended synaptotagmins), and the vesicle-associated membrane protein-associated proteins Scs2 and Scs22 [65-67] -- caused untethering of the ER from the plasma membrane and the aberrant accumulation of ER in the cytoplasm. Furthermore, phosphoinositide signaling was misregulated at the plasma membrane, and the ER unfolded protein response was constitutively activated [17].
7. Dynamic alterations in ER shape
Thus far, we have emphasized how proteins establish and maintain the different morphologies of ER domains. However, the ER is highly dynamic and undergoes continuous fusion reaction, interactions with the cytoskeleton and other organelles, and transitions between tubules and sheets. Numerous regulatory schemes have been described that move ER, often in conjunction with molecular motors (e.g., kinesin-1 and myosin V) and actin-and microtubule-based cytoskeletal elements.
One regulatory protein is the cytoplasmic protein p22, an EF-hand Ca2+-binding protein that binds microtubules in a manner dependent on N-myristoylation and forms Ca2+-dependent interactions with microsomes [68]. Another example of dynamic ER regulation is provided by the stromal interaction molecule (STIM) proteins, which act as Ca2+sensors in the ER, oligomerizing in response to Ca2+depletion and translocating to the plasma membrane via a polybasic C-terminal domain, then activating highly Ca2+-selective Orai channels to trigger Ca2+dependent signaling events [69]. In addition, STIM1 also functions in ER morphogenesis through interactions with the microtubule +TIP protein end binding 1 (EB1). Protein phosphorylation of STIM1 triggers its dissociation from EB1 during mitosis and represents a major regulatory mechanism that results in the exclusion of concentric ER sheets from the mitotic spindle [70].
Even more dramatic morphological changes occur during cellular events such as fertilization and cell division. Within minutes after fertilization, the ER in starfish eggs fragments, accompanied by a release of Ca2+ from internal stores [71]. In fact, Ca2+-induced reversible ER fragmentation has also been reported in cell lines and neurons [72, 73], prefiguring another role for signaling pathways in the modification of ER morphology. A prominent change in ER morphology also occurs during cell division, though the character of these changes remains a matter of debate. Puhka et al. [13] have reported that in mitotic cells the ER undergoes a spatial reorganization and transformation of sheets toward more fenestrated and tubular forms, while Lu et al. [14] have countered that from prometaphase to telophase in various mammalian cell types, most ER is organized as extended cisternae.
Though ER dynamics in animal cells rely heavily on microtubules, short-range movements utilize the myosin V motor. For instance, myosin-Va acts as an organelle transporter to pull ER as cargo into the dendritic spines of cerebellar Purkinje neurons. The myosin-Va accumulates at ER tips as the ER moves into spines, and ATP hydrolysis by myosin-Va is required for this spine ER targeting. Thus, an actin-based motor moves ER within animal cells to regulate synaptic plasticity [20].
Another example of prominent ER dynamics in dendrites was recently reported by Cui-Wang et al. [21]. These authors demonstrated that membrane proteins including AMPA-type glutamate receptors rapidly diffuse within the continuous network of dendritic ER, but are confined by zones of increased ER complexity at dendritic branch points and near spines. The spatial range of receptor mobility was restricted by metabotropic glutamate receptors in a protein kinase C- and CLIMP-63-dependent manner. Moreover, these local zones of ER complexity compartmentalized ER export and were associated with sites of new dendritic branch formation, thus spatially scaling secretory trafficking within complex dendritic arbors [21].
Since ER tubules can be observed to fuse during interphase in a variety of cell types, mechanisms must exist to regulate this process. Very recently, English and Voeltz [15] identified Rab10 as a modulator of ER morphology at dynamic ER-associated structures that track along microtubules and marks areas of nascent ER tubule growth. This was unexpected, since Rab10 has been studied mainly for its roles in endocytic recycling, especially the basolateral recycling pathways in polarized epithelial cells, as well as for GLUT4 storage vesicle transport to the plasma membrane and dense core vesicle secretion [74]. However, Rab10 depletion or expression of a Rab10 GDP-locked mutant clearly altered ER morphology [15], resulting in fewer ER tubules due to a reduced ability of dynamic ER tubules to grow out and successfully fuse with adjacent ER. This Rab10 domain is enriched in at least two ER enzymes that regulate phospholipid synthesis, phosphatidylinositol synthase (PIS) and choline/ethanolamine phosphotransferase 1 (CEPT1). Since the formation and function of this Rab10/PIS/CEPT1 dynamic domain could be inhibited by expression of a GDP-locked Rab10 mutant or by depletion of Rab10, these dynamic morphology changes may be coupled to phospholipid synthesis at these leading edge domains. However, a luminal ER marker did not distribute to these domains, raising the possibility that this constitutes a distinct membrane compartment. In fact, photoactivation experiments performed by Kim et al. [75] had previously indicated that a dynamic domain similarly enriched in PIS is ER-derived, but distinct and highly mobile. They described this structure as a PIPERosome (PI-producing ER-derived organelle) which makes contact with a variety of organelles. Thus, the mechanism by which Rab10 influences ER structure remains unclear.
Interestingly, the mostly early-endosomal-associated Rab5 has also been implicated in ER shaping [76], and an intriguing link between Rab5 and Rab10 in another context has been described in C. elegans, where RAB-5 and RAB-10 recruit one another's Tre2/Bub2/Cdc16 domain-containing GAP molecules in a reciprocal exclusion cascade at a Golgi apapratus-endosomal interface to regulate neuropeptide release [77]. Also, a C. elegans rab-10 mutant exhibited increased endosomal phosphatidylinositol-4,5-bisphosphate levels, with subsequent alterations in the distribution of the membrane-bending proteins RME-1/Ehd and SDPN-1/Syndapin/Pacsin. This likely occurs via RAB-10 interactions with the Arf6GAP CNT-1 because Arf6 activates type I phophatidylinositol-4-phosphate 5 kinase, which is involved in the synthesis of phosphatidylinositol-4,5-bisphosphate [78]. Together, these endosomal studies prefigure a general involvement of Rab10 within specialized domains linked to phospholipid synthesis and protein recruitment.
The involvement of Rabs in modulating ER morphology is an emerging area of research, and some effects may not be direct or conserved. At a fundamental level, the involvement of Rab GTPases may not be surprising, since they provide directionality and specificity to tubule formation and budding in a variety of cellular contexts, and Balch and colleagues implicated Rabs in ER membrane fusion years ago [79]. Even so, there are over 60 human Rabs known, yet only 11 in S. cerevisiae, and most of the dozen or so human Rabs associated with various ER functions lack clear yeast orthologs, including Rab10 [80]. Arabidopsis also does not have a clear Rab10 ortholog, though the tubular ER network in plant cells is also highly dynamic. Thus, it is not clear how phylogenetically conserved the Rab10-dependent process is for regulating ER dynamics. This stands in contrast to the atlastin/RHD3/Sey1p GTPases and most ER-shaping reticulons/REEPs, which have orthologs in all eukaryotes. One possible explanation is that although ER movement and remodeling in animal cells is mostly microtubule-based, in yeast and plants actinomyosin dependence predominates [6]; thus different organisms may have evolved different means for the regulation of ER distribution and dynamics.
8. Insights from neurological disease
Highly-polarized cells such as neurons present special challenges for shaping and distributing the ER network because of their large size and complexity [81]. Acompelling example of the importance of ER morphology and distribution in neurons is provided by a class of human diseases known as the hereditary spastic paraplegias, whose cardinal feature is a length-dependent axonopathy of the corticospinal motor neurons, the axons of which can reach up to one meter in length [82]. Though these disorders comprise over 50 distinct genetic loci (SPG1-55), over half of all affected individuals, including those with the three most common forms, harbor autosomal dominant mutations in one of four proteins already discussed for their roles in ER network formation: spastin (SPG4), atlastin-1 (SPG3A), and REEP1 (SPG31) and reticulon-2 (SPG12) [68]. These proteins interact with one another and mediate ER shaping and interactions of the tubular ER network with the microtubule cytoskeleton [3, 82].
Additional studies have suggested a role for ER shaping mechanisms in the pathogenesis of related neurologic disorders such as amyotrophic lateral sclerosis (ALS). In the superoxide dismutase 1 (SOD1) G93A transgenic mouse model for ALS, which involves both corticospinal and lower motor neurons, reticulon-4A selectively redistributed the ER chaperone protein disulfide isomerase, and reticulon-4A overexpression protected against neurodegeneration. Conversely, knock out of reticulon-4A,B on this SOD1 G93A background worsened disease symptoms in mice [83]. Further supporting a role for ER morphogenesis in ALS disease pathogenesis, the VAP-B mutant P56S that underlies ALS8 is associated with the production of a novel form of organized smooth ER [84] as well as a nuclear envelope defect associated with impaired transport of nucleoporins and emerin to the nuclear envelope [85]. Taken together, these studies identify tantalizing links that may support a general neurologic disease mechanism centered on aberrant alterations in ER shape.
9. Conclusion
A unifying model for ER organization and dynamics posits that the ER network can be formed and maintained by ubiquitious proteins such as atlastin GTPases and reticulons/REEPs, with interactions between the ER and the cytoskeleton, other organelles, and the plasma membrane also playing a role. Highly-selective Rab GTPases, Lunaparks, SNAREs, and other protein classes may also be involved. Seemingly, the distinct morphologies of the ER network mask an even more complex heterogeneity of lipid- and protein-defined functional domains that will continue to illuminate how ER form follows function.
With an expanding number of proteins now identified that mediate the generation and shaping of the ER network, the stage is now set for investigations into the mechanisms regulating ER morphology and distribution within the cell. Signaling pathways linked to modifications of these proteins seem likely to play key roles in modulating the structure of the ER within cells and dynamically positioning functional ER domains. Finally, since ER morphology defects appear to be the prominent mechanism underlying the hereditary spastic paraplegias and related disorders, the identification of additional disease genes may provide new insights to untangle the form and elucidate the functions of the ER.
Highlights.
The ER is a continuous organelle comprising sheets, tubules and the nuclear envelope.
Protein families including reticulons and REEPs/DP1/Yop1p shape high-curvature ER tubules.
Atlastin GTPases mediate the fusion of ER tubules to form a polygonal network.
Multiple ER network proteins are mutated in hereditary spastic paraplegias.
Acknowledgments
Ethan Tyler (NIH Division of Medical Arts) prepared the figures. The authors were supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health (C.B.) and the Howard Hughes Medical Institute-National Institutes of Health Research Scholars Program (U.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Sullivan LH. The tall office building artistically considered. Lippincott's Magazine #57. 1896:403–409. [Google Scholar]
- 2.Lynes EM, Simmen T. Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim. Biophys.Acta. 2011;1813:1893–1905. doi: 10.1016/j.bbamcr.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SH, Blackstone C. Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 2010;11:515–521. doi: 10.1038/embor.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 2013;5:a013227. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011;147:1226–1231. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparkes I, Hawes C, Frigerio L. FrontiERs: movers and shapers of the higher plant cortical endoplasmic reticulum. Curr. Opin. Plant Biol. 2011:658–665. doi: 10.1016/j.pbi.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Doyle C, Qi X, Zheng H. The endoplasmic reticulum: a social network in plant cells. J. Integr. Plant Biol. 2012;54:840–850. doi: 10.1111/j.1744-7909.2012.01176.x. [DOI] [PubMed] [Google Scholar]
- 8.Lin S, Sun S, Hu J. Molecular basis for sculpting the endoplasmic reticulum membrane. Int. J. Biochem. Cell Biol. 2012;44:1436–1443. doi: 10.1016/j.biocel.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. fourth ed. Garland Science; New York: 2002. [Google Scholar]
- 10.Walters AD, Bommakanti A, Cohen-Fix O. Shaping the nucleus: forces and factors. J. Cell. Biochem. 2012;113:2813–2821. doi: 10.1002/jcb.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 12.Prinz WA, Grzyb L, Veenhuis M, Kahan JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell. 2012;23:2424–2432. doi: 10.1091/mbc.E10-12-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 2013;(15):169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 2010;29:2715–2723. doi: 10.1038/emboj.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondrial contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J.Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner W, Brenowitz SD, Hammer JA., 3rd Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat. Cell Biol. 2011;13:40–48. doi: 10.1038/ncb2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voeltz GK, Prinz WA. Sheets, ribbons and tubules - how organelles get their shape. Nat. Rev. Mol. Cell Biol. 2007;8:258–264. doi: 10.1038/nrm2119. [DOI] [PubMed] [Google Scholar]
- 23.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;25:329–354. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 25.Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat. Cell Biol. 2012;14:634–639. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- 26.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 27.Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 28.Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JWCC, Jr., Hoogenraad A. Akhmanova, STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 30.Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 31.Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparkes I, Tolley N, Aller I, Svozil J, Osterrieder A, Botchway S, Mueller C, Frigerio L, Hawes C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolley N, Sparkes I, Craddock CP, Eastmond PJ, Runions J, Hawes C, Frigerio L. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 2010;64:411–418. doi: 10.1111/j.1365-313X.2010.04337.x. [DOI] [PubMed] [Google Scholar]
- 34.Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic. 2008;9:94–102. doi: 10.1111/j.1600-0854.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- 35.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronchi P, Colomo S, Francolini M, Borgese N. Transmembrane domain-dependent partitioning of membrane proteins within the endoplasmic reticulum. J. Cell Biol. 2008;181:105–118. doi: 10.1083/jcb.200710093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri H-P. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal α-helical segment. J. Cell Biol. 2001;153:1287–1300. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. Formation of stacked cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 42.Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu P-P, Patterson A, Lavoie B, Stadler J, Shoeb M, Patel R, Blackstone C. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 2003;278:49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
- 45.Zhu P-P, Soderblom C, Tao-Cheng J-H, Stadler J, Blackstone C. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet. 2006;15:1343–1353. doi: 10.1093/hmg/ddl054. [DOI] [PubMed] [Google Scholar]
- 46.Rismanchi N, Soderblom C, Stadler J, Zhu P-P, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 2008;17:1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J, Shibata Y, Zhu P-P, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 49.Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, Frigerio L. An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol. 2013;197:481–489. doi: 10.1111/nph.12038. [DOI] [PubMed] [Google Scholar]
- 50.Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 2012;197:209–217. doi: 10.1083/jcb.201111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moss TJ, Andreazza C, Verma A, Daga A, McNew JA. Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11133–11138. doi: 10.1073/pnas.1105056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2146–E2154. doi: 10.1073/pnas.1208385109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Byrnes LJ, Singh A, Szeto K, Benvin NM, O'Donnell JP, Zipfel WR, Sondermann H. Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J. 2013;32:369–384. doi: 10.1038/emboj.2012.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S, Novick P, Ferro-Novick S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 2012;14:707–716. doi: 10.1038/ncb2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terasaki M, Chen LB, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J. Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr. Opin. Cell Biol. 2013;25 doi: 10.1016/j.ceb.2013.02.006. doi:pii: S0955-0674(13)00029-X. 10.1016/j.ceb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bola B, Allan V. How and why does the endoplasmic reticulum move? Biochem. Soc. Trans. 2009;37:961–965. doi: 10.1042/BST0370961. [DOI] [PubMed] [Google Scholar]
- 58.Dreier L, Rapoport TA. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J. Cell Biol. 2000;148:883–898. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terasaki M, Reese TS. Interactions among endoplasmic reticulum, microtubules, and retrograde movements of the cell surface. Cell Motil. Cytoskeleton. 1994;29:291–300. doi: 10.1002/cm.970290402. [DOI] [PubMed] [Google Scholar]
- 60.Evans K, Keller C, Pavur K, Glasgow K, Conn B, Lauring B. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanderson CM, Connell JW, Edwards TL, Bright NA, Duley S, Thompson A, Luzio JP, Reid E. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 2006;15:307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park SH, Zhu P-P, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klopfenstein DR, Kappeler F, Hauri H-P. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Estrada P, Kim J, Coleman J, Walker L, Dunn B, Takizawa P, Novick P, Ferro-Novick S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schulz TA, Creutz CE. The tricalbin C2 domains: lipid-binding properties of a novel, synaptotagmin-like yeast protein family. Biochemistry. 2004;43:3987–3995. doi: 10.1021/bi036082w. [DOI] [PubMed] [Google Scholar]
- 66.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 67.Wolf W, Kilic A, Schrul B, Lorenz H, Schwappach B, Seedorf M. Yeast Ist2 recruits the endoplasmic reticulum to the plasma membrane and creates a ribosome-free membrane microcompartment. PLoS One. 2012;7:e39703. doi: 10.1371/journal.pone.0039703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrade J, Zhao H, Titus B, Timm Pearce S, Barroso M. The EF-hand Ca2+-binding protein p22 plays a role in microtubule and endoplasmic reticulum organization and dynamics with distinct Ca2+-binding requirements. Mol. Biol. Cell. 2004;15:481–496. doi: 10.1091/mbc.E03-07-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smyth JT, Beg AM, Wu S, Putney JWNM., Jr. Rusan, Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr. Biol. 2012;22:1487–1493. doi: 10.1016/j.cub.2012.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terasaki M, Jaffe LA, Hunnicutt GR, Hammer JA., 3rd Structural change of the endoplasmic reticulum during fertilization: evidence for loss of membrane continuity using the green fluorescent protein. Dev. Biol. 1996;179:320–328. doi: 10.1006/dbio.1996.0263. [DOI] [PubMed] [Google Scholar]
- 72.Subramanian K, Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89:963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- 73.Kucharz K, Krogh M, Ng AN, Toresson H. NMDA receptor stimulation induces reversible fission of the neuronal endoplasmic reticulum. PLoS One. 2009;4:e5250. doi: 10.1371/journal.pone.0005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang J, Blackstone C. Rab10 joins the ER social network. Nat. Cell Biol. 2013;(15):135–136. doi: 10.1038/ncb2682. [DOI] [PubMed] [Google Scholar]
- 75.Kim YJ, Guzman-Hernandez ML, Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Audhya A, Desai A, Oemega K. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 2007;178:43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasidharan N, Sumakovic M, Hannemann M, Hegermann J, Liewald JF, Olendrowitz C, Koenig S, Grant BD, Rizzoli SO, Gottschalk A, Eimer S. RAB-5 and RAB-10 cooperate to regulate neuropeptide release in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18944–18949. doi: 10.1073/pnas.1203306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi A, Liu O, Koenig S, Banerjee R, Chen CC, Eimer S, Grant BD. RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2306–E2315. doi: 10.1073/pnas.1205278109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turner MD, Plutner H, Balch WE. A Rab GTPase is required for homotypic assembly of the endoplasmic reticulum. J. Biol. Chem. 1997;272:13479–13483. doi: 10.1074/jbc.272.21.13479. [DOI] [PubMed] [Google Scholar]
- 80.Sandoval CA, Simmen T. Rab proteins of the endoplasmic reticulum: functions and interactors. Biochem. Soc. Trans. 2012;40:1426–1432. doi: 10.1042/BST20120158. [DOI] [PubMed] [Google Scholar]
- 81.Renvoisé B, Blackstone C. Emerging themes of ER organization in the development and maintenance of axons. Curr. Opin. Neurobiol. 2010;20:531–537. doi: 10.1016/j.conb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang YS, Harel NY, Strittmatter SM. Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J. Neurosci. 2009;29:13850–13859. doi: 10.1523/JNEUROSCI.2312-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fasana E, Fossati M, Ruggiano A, Brambillasca S, Hoogenraad CC, Navone F, Francolini M, Borgese N. A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J. 2010;24:1419–1430. doi: 10.1096/fj.09-147850. [DOI] [PubMed] [Google Scholar]
- 85.Tran D, Chalhoub A, Schooley A, Zhang W, Ngsee JK. A mutation in VAPB that causes amyotrophic lateral sclerosis also causes a nuclear envelope defect. J. Cell Sci. 2012;125:2831–2836. doi: 10.1242/jcs.102111. [DOI] [PubMed] [Google Scholar]
- 86.Pendin D, McNew JA, Daga A. Balancing ER dynamics: shaping, bending, severing, and mending membranes. Curr. Opin. Cell Biol. 2011;23:435–442. doi: 10.1016/j.ceb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]