Abstract
Ethanol induces hypoxia and elevates HIF-1α in the liver. CYP2E1 plays a role in the mechanisms by which ethanol generates oxidative stress, fatty liver and liver injury. The current study evaluated whether CYP2E1 contributes to ethanol-induced hypoxia and activation of HIF-1α in vivo and whether HIF-1α protects against or promotes CYP2E1-dependent toxicity in vitro. Wild type (WT), CYP2E1-knockin (KI) and CYP2E1 knockout (KO) mice were fed ethanol chronically; pair fed controls received isocaloric dextrose. Ethanol produced liver injury in the KI mice to a much greater extent than in the WT and KO mice. Protein levels of HIF-1α and downstream targets of HIF-1α activation were elevated in the ethanol-fed KI mice compared to the WT and KO mice. Levels of HIF prolylhydroxlase 2 which promotes HIF-1α degradation were decreased in the ethanol-fed KI mice in association with the increases in HIF-1α. Hypoxia occurred in the ethanol-fed CYP2E1 KI mice as shown by an increased area of staining using the hypoxia-specific marker pimonidazole. Hypoxia was lower in the ethanol-fed WT mice and lowest in the ethanol fed KO mice and all the dextrose-fed mice. In situ double staining showed that pimonidazole and CYP2E1 were co-localized to the same area of injury in the hepatic centrilobule. Increased protein levels of HIF-1α were also found after acute ethanol treatment of KI mice. Treatment of HepG2 E47 cells which express CYP2E1 with ethanol plus arachidonic (AA) acid or ethanol plus buthionine sulfoximine (BSO) which depletes GSH caused loss of cell viability to greater extent than in HepG2 C34 cells which do not express CYP2E1. These treatments elevated protein levels of HIF-1α to a greater extent in E47 cells than C34 cells. 2-Methoxyestradiol, an inhibitor of HIF-1α, blunted the toxic effects of ethanol plus AA and ethanol plus BSO in the E47 cells in association with inhibition of HIF-1α. The HIF-1α inhibitor also blocked the elevated oxidative stress produced by ethanol/AA or ethanol/BSO in the E47 cells. These results suggest that CYP2E1 plays a role in ethanol-induced hypoxia, oxidative stress and activation of HIF-1α and that HIF-1α contributes to CYP2E1-dependent ethanol-induced toxicity. Blocking HIF-1α activation and actions may have therapeutic implications for protection against ethanol/CYP2E1-induced oxidative stress, steatosis and liver injury.
Keywords: Ethanol, CYP2E1, Hepatotoxicity, Oxidative Stress, HIF-1α
Introduction
Hypoxia inducible factor (HIF) is activated by hypoxia and is master regulator of oxygen homeostasis as it regulates the expression of many genes involved in glycolysis, glucose transport, synthesis of nitric oxide and cytokines such as TNFα, blood flow, inflammation, and cell death (1–3). Expression and activity of the HIF-1α subunit is regulated by cellular oxygen levels, primarily at the level of protein stability (4,5). The HIF-1α protein is rapidly degraded under normoxic conditions as oxygen-dependent proline hydroxylation of HIF-1α by proline hydroxylases promotes ubiquitination of HIF-1α followed by rapid proteosome–mediated degradation. Hypoxia enhances HIF-1α levels by inhibiting proline hydroxylation and therefore the degradation of HIF-1α (1–5). The accumulated HIF-1α can dimerize with the aryl hydrocarbon receptor nuclear translocator, move to the nucleus, and bind to the hypoxia-responsive element in the promoter of its target genes (6–8). HIF-1α has been implicated in the toxicity found in many models of liver injury, including alcohol-induced liver injury (9–11).
Chronic ethanol consumption by rats was shown to cause hypoxia due to increasing oxygen consumption; the latter reflected the requirement for oxygen to reoxidize reducing equivalents, NADH, produced by the oxidation of ethanol by alcohol dehydrogenase and the oxidation of acetaldehyde by the low Km mitochondrial aldehyde dehydrogenase (12–14). Acute ethanol administration also produced hypoxia(14–16). Chronic intake of ethanol by intragastric infusion caused hypoxia and oxidative stress in rat liver and pancreas (17–19). The production of hypoxia by ethanol administration has been reviewed (20). Since ethanol-induced hypoxia was most pronounced in the pericentral zone of the liver acinus reflecting the gradient of oxygen across the liver, the ethanol-induced hypoxia was hypothesized to play a role in the ethanol-induced liver injury (12,13,20). Li et al (21) showed that HIF-1α mRNA and protein were elevated after intragastric administration of ethanol to rats for 24 weeks. Recent studies have assessed the possible role of HIF-1α in ethanol-induced steatosis. Nath et al (22) reported that feeding mice for 4 weeks with the Lieber-DeCarli diet increased HIF-1α mRNA, protein and DNA binding activity in wild type mice and produced the typical fatty liver associated with ethanol consumption. Ethanol-induced fatty liver was intensified in mice engineered to express high levels of hepatic HIF-1α but was decreased in mice deficient in hepatic HIF-1α (22). Nath et al concluded that HIF-1α plays an important role in ethanol-induced fatty liver and liver injury (22). Conversely, Nishiyama et al (23) found that activation of HIF-1α suppresses ethanol-induced fatty liver. Using the same Lieber-DeCarli model of ethanol feeding, they found that hepatocyte-specific HIF-1α knockout mice developed a more pronounced fatty liver and elevated triglycerides than did wild type mice and concluded that HIF-1α is protective against ethanol induction of fatty liver. Possible reasons for these different conclusions were discussed (24) but remain unclear and additional studies on this are needed.
CYP2E1 metabolizes and activates many toxicologically important substrates to more toxic products (25–28). Levels of CYP2E1 are elevated by ethanol (29) and during its catalytic reaction with molecular oxygen, reactive oxygen species such as superoxide and hydrogen peroxide are produced (30). In some, but not all studies the ethanol-induced liver pathology correlated with CYP2E1 levels and the generation of ROS and oxidative stress (31–36). We have shown that ethanol-induced steatosis and oxidative stress was lower in CYP2E1 knockout mice fed ethanol chronically as compared to wild type mice (37). The fatty liver produced in wild type mice was blunted by inhibitors of CYP2E1 (37). Ethanol-induced fatty liver and oxidative stress was restored in CYP2E1 knockin mice (38) in which the human CYP2E1 was expressed in the CYP2E1 knockout mice. Ethanol and other prooxidants were more toxic to HepG2 cells expressing CYP2E1 than control HepG2 cells not expressing CYP2E1 (31,39,40). CYP2E1 appears to play an important role in the mechanisms by which ethanol generates oxidative stress and is hepatotoxic (31,39,40). Given the different results on the role of HIF-1α in ethanol-induced fatty liver, the goal of the current study was to evaluate whether CYP2E1 contributes to ethanol-induced hypoxia and activation of HIF-1α and whether HIF-1α potentiates or prevents CYP2E1 –dependent toxicity.
Materials and Methods
In Vivo Mouse Models
SV129 background CYP2E1 knockout (KO) mice (41) and humanized transgenic CYP2E1 knockin (KI) mice (42,43) were kindly provided by Dr. Frank J. Gonzalez (Laboratory of Metabolism, National Cancer Institute, Bethesda, MD) and breeding colonies of these mice were established at Mount Sinai. SV129 wild type (WT) mice were purchased from Charles River Laboratory. All mice were housed in temperature-controlled animal facilities with 12-hour light/12-hour dark cycles. The mice received humane care, and experiments were carried out according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals and with approval of the Mount Sinai Animal Care and Use Committee.
WT, KO and KI mice, total of 72 mice (male 48, female 24), 8–10 weeks of age, weighing 22–28 g were initially fed the control liquid dextrose diet (Bio-Serv, Frenchtown, NJ) for 3 days to acclimate them to the Lieber and DeCarli liquid diet (44). Afterward, the mice were fed either the ethanol diet or the dextrose diet as follows.. The content of ethanol was gradually increased every 7 days from 10% (1.77% [vol/vol]) of total calories to 20% (3.54% [vol/vol]), 30% (5.31% [vol/vol]), and finally 35% of total calories (6.2% [vol/vol]) for four weeks. The control mice were pair-fed the dextrose diet on an isoenergetic basis. The amount of diet consumed by the knockout, the knockin and the wild type mice was approximately the same. Mice were sacrificed between 3 and 5 PM on the 28th day of ethanol (6.2% vol/vol) feeding. For acute ethanol treatment, 24 male WT, KO and KI mice, 6–8 weeks old, body weight of 20–25g, were gavaged with 30% ethanol at a dose of 3g/kg body weight, twice a day for four days or were gavaged with saline. Mice were fasted for 18 hr prior to sacrifice after the fourth day of acute ethanol or saline administration.
At the end of treatment, the mice were sacrificed and serum and liver were collected. The liver was rapidly excised into small fragments and washed with cold saline. One aliquot of tissue was placed in 10% formalin solution for paraffin processing and one aliquot of tissue was placed in RNAlater solution for RNA isolation (Ambion, Grand Island, NY). The remaining aliquots were stored at −80°C for further assays. Liver homogenates were prepared in ice-cold 0.15 M KCl. Nuclear extract was freshly prepared according to a nuclear extract kit protocol (Active Motive, Carlsbad, CA). All samples were stored at −80°C in aliquots.
Serum ALT and Ethanol Assay
Serum alanine aminotransferase (ALT) levels were measured using a diagnostic kit (Pointe Scientific Inc., Brussels, Belgium) and kinetically following changes in absorbance at 340 nm using a UV-160 U Recording Spectrophotometer (Shimadzu, Kyoto, Japan). Serum ethanol was measured using an Abcam’s Ethanol Assay Kit based on oxidizing ethanol by alcohol oxidase to generate H2O2 which then reacts with the provided probe to generate a specific chromophore color (λmax= 570 nm).
Organelle preparations and assays
Mitochondria were isolated after centrifugation of homogenates at 8,500×g for 30 min and suspended in 0.25 M sucrose-10 mM Tris pH 7.4 buffer and washed once. The postmitochondrial supernatant was centrifuged at 100,000×g at 4°C for 60 min to obtain the microsomal pellets and the cytosolic supernatant fraction, respectively. The protein concentration of the different fractions was determined using a protein assay kit based on the Lowry Assay (BioRad, Hercules, CA). CYP2E1 activity was measured in liver microsomes by the spectrophotometric analysis at 546 nm of the oxidation of p-nitrophenol to p-nitrocatechol in the presence of NADPH and oxygen (45). The production of thiobarbituric acid reactive substances (TBARS), expressed as malondialdehyde (MDA) equivalents, was assayed in liver mitochrondrial fractions by the spectrophotometric analysis at 535 nm of the formation of thiobarbituric acid-reactive components. The concentration of malondialdehyde was calculated using an extinction coefficient of 156 × mmol/L/cm and expressed as picomoles per milligram of protein (46). Reduced glutathione (GSH) was analyzed in liver mitochondria by a glutathione reductase assay (47).
HE Staining and Inflammatory Cell Detection
The fixed liver tissue was processed as paraffin blocks and sections were used for hematoxylin-eosin (HE) staining and inflammatory cell detection. The morphological changes of liver tissues were observed by two pathologists who were blinded from the experimental information. All changes of fatty degeneration, inflammatory infiltration and ischemic necrosis were graded as none (0), mild (<25%), moderate (25%–50%), and severe (>75%).
Immunohistochemistry (IHC)
Immunohistochemical staining was performed on paraffin slides with the Histostain Plus Broad Spectrum LAB-SA Kit (Invitrogen, Camarillo, CA) using either polyclonal rabbit-anti-4-hydroxy-2-nonenal michael adducts (4-HNE) antibody (1:200) (Calbiochem, La Jolla, CA) or polyclonal rabbit–anti–3-nitrotyrosine (3-NT) antibody (1:100) (Chemicon, Temecula, CA) or polyclonal rabbit-anti-HIF-1α antibody (1:100) (Millipore, Temecula, CA). Slides were visualized with 3,3′-diaminobenzidine (DAB), and positive staining was reflected by brownish yellow color. In each case, a negative control (nonimmune serum) was used. The immunostaining results were semi-quantified and evaluation of a specific positive reaction was marked as negative (−), weakly positive (+), moderately positive (++), and strongly positive (+++).
Western Blot Analysis
Levels of CYP2E1, HIF-1α, HPH-2, Bcl-2, P21 and LDHA protein in 20–100 μg of protein samples from freshly prepared liver microsomal or nuclear extract, or homogenate fractions were determined using western blot analysis with anti-CYP2E1 antibody (1:10,000) (a gift provided by Dr. Jerome Lasker), anti-HIF-1α antibody (1:1000), anti-HPH-2 antibody (1:500), anti-LDHA antibody (1:1000) (Thermo Scientific, Waltham, MA), anti-Bcl-2 antibody (1:500) and anti-P21 antibody (1:500) (Santa Cruz Biotechnology, Santa Cruz, CA), respectively, followed by incubation with anti-rabbit or anti-mouse Odyssey secondary antibody (1:10,000). Blots were scanned using an Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE). All specific bands were quantified with the Automated Digitizing System (ImageJ gel programs, version 1.34S, National Institute of Health). β-actin was assayed by Western blot and results were expressed as the sample/actin ratio, which are shown below the specific blots.
In Situ Detection of Liver Tissue Hypoxia
To detect liver hypoxia in situ, the Hypoxyprobe-1 Plus kit (Hypoxyprobe, Burlington, MA) was used after injecting pimonidazole hydrochloride in vivo followed by immunochemical staining. Pimonidazole is reductively activated in hypoxic cells and forms stable adducts with thiol goups in proteins (48). After four-weeks on the ethanol or dextrose diets, WT or KO or KI mice were injected IP with pimonidazole at a dose of 60 mg/kg body weight. After one hr, mice were sacrificed and liver tissue was collected for processing paraffin slides. Slides were immunostained for hypoxyprobe-1 adducts using a specific FITC-MAb1 primary antibody (1:50) and a peroxidase conjugated anti-FITC secondary antibody (1:50) according to the suggested kit protocol. Slides were visualized with 3-amino-9-ethylcarbazole (AEC), and positive staining was reflected by red color.
RNA Isolation and Real-Time PCR
Total RNA was extracted from liver tissues previously treated with RNAlater solution using TRIzol Reagent according to the protocol provided by the manufacturer (Invitrogen, Grand Island, NY). Reverse transcription was carried out using a RT system (Applied Systems, Carlsbad, Ca) as follows: 25°C for 10 min, 37°C for 120 min, 85°C for 5 min, and the product (the first strand of cDNA) was stored at −20°C. Real-time PCR was performed in 25 μl of reaction solution containing 5 μl of 2X Maxima SYBR Green/Fluorescein qPCR Master Mix (Fermentas, Waltham, MA), 300 nM primers, and cDNAs. The cycles for PCR were as follows: 95°C for 7 min, 40 cycles of 95°C for 20 s, 54°C for 30 s, and 72°C for 30 s (Mount Sinai Core Facility). The primers were as follows: Mouse HIF-1a-Forward, GAAGACAACGCGGGCACCGA, Mouse HIF-1a-Reverse, TGCTTCGCCGAGATCTTGCTGC. GAPDH was used as an internal control. mRNA levels were expressed as -fold changes after normalization with GAPDH.
In Vitro Cell Culture Model
E47 cells, a human hepatoma cell line that constitutively expresses CYP2E1 (HepG2 cells transfected with plasmid pCI-neo containing CYP2E1 cDNA in the sense orientation), and C34 cells (HepG2 cells transfected with pCI-neo), which do not express CYP2E1 (49) were used as an vitro model to evaluate the role of HIF-1α in CYP2E1-dependent toxicity. Cells were grown in MEM containing 10% fetal bovine serum and 0.4 mg/ml of G418 supplemented with 100 units/ml of penicillin and 100 mg/ml of streptomycin and 0.01% fungizone antibiotics in a humidified atmosphere in 5% CO2 at 37°C. Cells were subcultured at a 1:5 ratio once a week. The content of CYP2E1 was assayed by western blot analysis, and catalytic activity was determined by measuring the oxidation of p-nitrophenol (45). E47 cells and C34 cells were treated with ethanol (100 mM) or ethanol plus AA (30 μM), or ethanol plus BSO (300 μM) in the absence or presence of 2-ME (45 μM), an inhibitor of HIF-1α, for 2 days in a 5% CO2 incubator at 37°C. The water tray in the incubator contained added ethanol at a final concentration of 100 mM. The culture medium was replaced every 24 hr with fresh medium and reagents. The water tray was also replaced every 24 hr with fresh filtered water plus ethanol. The Cell Titer Assay was used to assess cell viability (50). Mitochondrial production of reactive oxygen species, mainly superoxide radical was assayed by incubating with MitoSox (5 μM, Invitrogen) for 30 min. Staining was observed under the fluorescence microscope (x100). Lipid peroxidation was determined by assay of formation of TBARS (46).
Statistical Analysis
Values reflect means±SD. One-way ANOVA with subsequent post-hoc comparisons by Tukey HSD and corrected for multiple group comparisons were performed by SPSS analysis software (version 10.0). P values of less than 0.05 were considered statistically significant and results are from experiments using 8–12 mice of each genotype in the chronic model and 4–6 mice of each genotype in the acute model. Results with the HepG2 cells are from 3 dishes.
Results
Chronic Ethanol- Induced-Hepatotoxicity
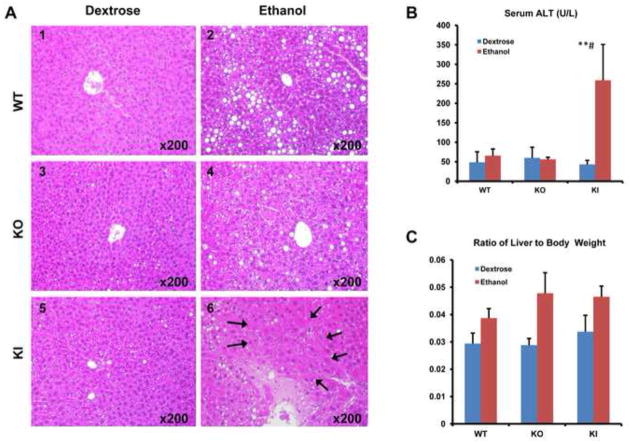
Wild type SV129 mice (WT), CYP2E1 knockout mice (KO) and CYP2E1 knockin mice (KI) in which the human CYP2E1 was added to replace the knocked out mouse CYP2E1 were fed ethanol chronically as described. Pair-fed controls received isocaloric dextrose. Serum ALT levels were elevated in the CYP2E1 KI mice fed with ethanol; no increase was found in the WT or KO mice fed with ethanol (Fig. 1B). Pathological observation revealed distinct steatosis but no necrosis in the ethanol-fed WT mice (Fig. 1A2). Steatosis was lower and necrosis was not observed in the ethanol-fed KO mice (Fig 1A4). In the ethanol-fed KI mice, cell degeneration, inflammatory infiltration and ischemic necrosis were observed (Fig. 1A6 arrows). Steatosis in the ethanol-fed KI mice was lower than in the ethanol-fed WT mice as found previously (38). This may reflect that the ethanol-fed KI mice already passed the maximal stage of steatosis to reach an inflammatory stage with liver necrosis. Time course kinetic experiments will be needed to evaluate this possibility. The ratio of liver to body weight was increased in all the ethanol-fed mice compared to the dextrose-fed mice (Fig. 1C). No pathological changes were found in any of the dextrose-fed mice (Fig 1A1,1A3,1A5).. Serum ethanol levels ranged between 6.4 and 7 mM at the time of sacrifice for all three genotypes fed ethanol and were below 1.2 mM for all three genotypes fed dextrose. After feeding mice for 4 weeks, the body weight was slightly decreased in the ethanol-fed WT and KI mice but not in the ethanol-fed KO mice. There was no significant change of body weight in any of the dextrose-fed mice.
Fig. 1. Metabolic Changes After Chronic Ethanol Feeding.
WT, CYP2E1 KO and CYP2E1 KI mice were fed ethanol or dextrose for 4 weeks. (A) Histopathology. Panel A6 show severe pathological changes including hepatocyte ischemic necrosis and infiltration of inflammatory cells in central zone of the hepatic lobule (arrows, HE×200). Panels A2 and A4 show mild pathological changes including simple liver steatosis. Panels A1, A3 and A5 show minimal steatosis and no obvious pathological changes (HE×200). (B) serum ALT. (C) Ratio of liver to body weight. * p<0.05, ** p<0.01 compared to the CYP2E1 KI group fed dextrose. # p<0.05 compared to CYP2E1 KO or WT group fed ethanol or dextrose.
Ethanol-Induced CYP2E1 Expression and Oxidative Stress
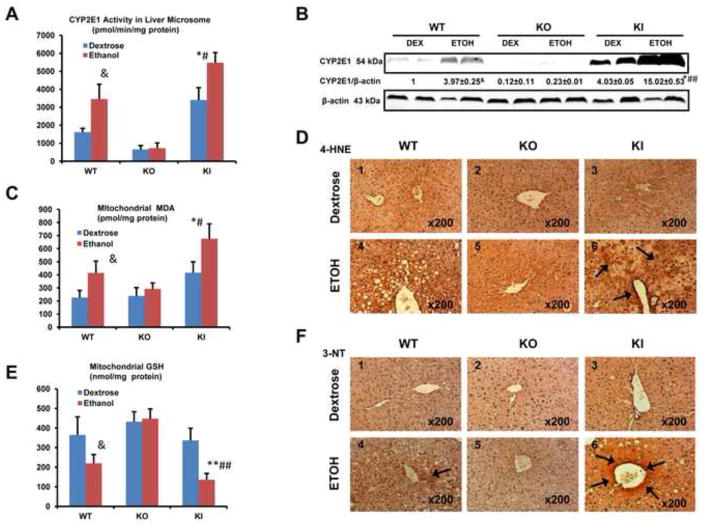
Chronic ethanol feeding elevates oxidative stress and induction of CYP2E1 plays an important role in these increases (28,29,31,51). Ethanol increased mouse hepatic CYP2E1 levels and catalytic activity in the WT mice (Fig. 2A,2B). CYP2E1 levels and PNP activity were very low or not detectable in the dextrose or ethanol-fed KO mice (Fig. 2A,2B). Human CYP2E1 and PNP activities were elevated in the ethanol-fed KI mice to an even greater extent than levels and activity of mouse CYP2E1 found in the ethanol-fed WT mice (Fig. 2A,2B), as previously described (38). MDA levels, a breakdown product of lipid peroxidation, were increased in hepatic mitochondria of the WT mice and further increased in mitochondria from the KI mice fed with ethanol compared to the mice fed with dextrose (Fig. 2C). No increase of MDA in liver mitochondria was found in the KO mice fed with ethanol. GSH levels were lowered about 40% in liver mitochondria from WT mice fed with ethanol and about 60% in liver mitochondria from the KI mice fed with ethanol (Fig. 2E). Ethanol had no effect on GSH levels in liver mitochondria from KO mice. Immunohistochemical staining for 4-HNE and 3-NT protein adducts was strongly positive(+++) in liver of the KI mice (arrows, Fig 2D6,2F6), and weakly positive in WT mice (+) fed with ethanol. (Fig 2F4). There were no obvious HNE or 3NT adducts observed in the mice (−) fed with dextrose (Fig. 2D1,2D2, 2D3,2F1,2F2,2F3) or the KO mice fed with ethanol (Fig 2D5,2F5). Thus, levels of ethanol induced oxidative stress correlated to the induction of CYP2E1 in mice fed with ethanol, being highest in the KI mice, intermediate in the WT mice and lowest in the KO mice, respectively
Fig. 2. CYP2E1 Levels and Oxidative Stress After Chronic Ethanol Feeding.
(A) CYP2E1 catalytic activity assayed with para-nitrophenol. (B) CYP2E1 protein levels. (C) MDA level in mitochondria. (D) Immunohistochemical staining for 4-HNE in liver. Panel D6 shows strong 4-HNE positive staining (arrows, IHC×200). Other panels show little or no obvious positive staining. (E) GSH level in mitochondria. (F) Immunohistochemical staining for 3-NT in liver. Panel D6 show strong 3-NT positive staining (arrows, IHC×200). Panel D4 show weak 3-NT positive staining (arrow, IHC×200). Other panels show no obvious positive staining * p<0.05, ** p<0.01 compared to the CYP2E1 KI groups fed dextrose. # p<0.05, ## p<0.01 compared to CYP2E1 KO or WT groups fed ethanol or dextrose. & p<0.05 compared to WT group fed dextrose.
Expression of HIF-1α
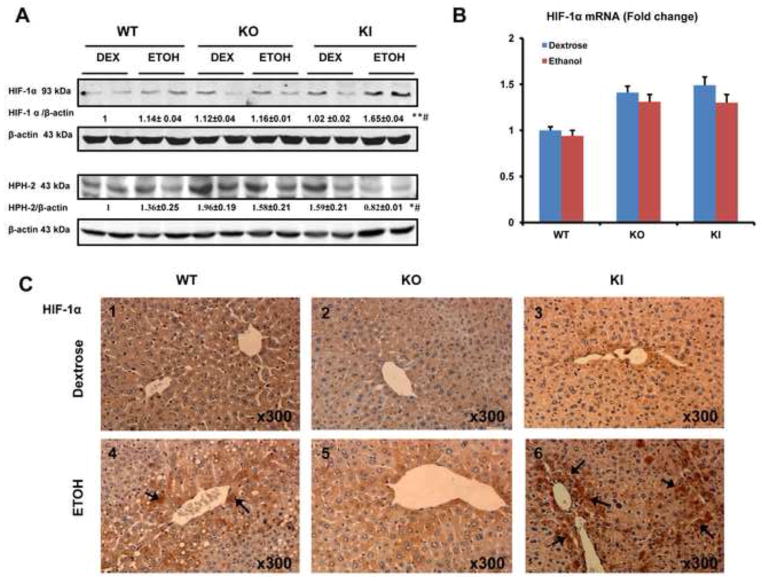
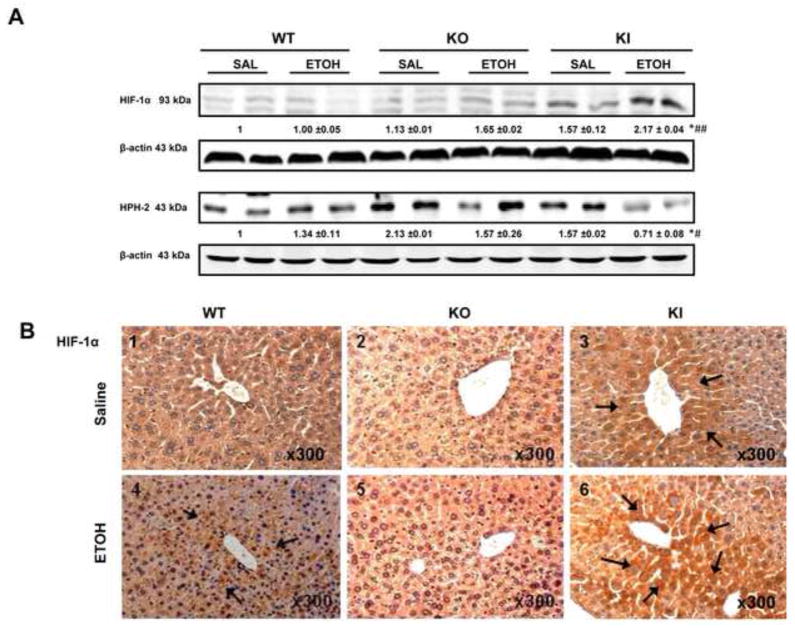
The levels of HIF-1α in nuclear extracts were significantly increased in the ethanol-fed KI mice compared to the ethanol-fed WT and KO mice or to the dextrose-fed mice (Fig. 3A). The levels of HPH-2, a proline hydroxylase of HIF-1α which targets HIF-1α for proteosome –mediated degradation, were significantly decreased in the ethanol-fed KI mice compared to the ethanol-fed WT and KO mice or to the dextrose-fed mice (Fig. 3A). The decrease in levels of HPH-2 may correlate with the increase in levels of HIF-1α in the ethanol-fed KI mice. Ethanol treatment lowers the catalytic activity of the proteasome complex (52) and this may also play a role in the ethanol-induced elevation of HIF-1α in addition to the decrease in HPH-2. Immunohistochemical detection of HIF-1α showed strongest staining in the livers of the KI mice (+++) fed ethanol (arrows, Fig. 3C6), moderately positive staining in WT mice (++)(Fig. 3C4) and no or weakly positive staining in KO mice fed with ethanol (Fig. 3C5). No obvious staining was observed in the dextrose-fed mice (−) (Fig. 3C1,3C2,3C3)). There were no significant changes in HIF-1α mRNA levels between all groups of mice (Fig. 3B), suggesting that the activation of HIF-1α may be at the level of posttranscription.
Fig. 3. Levels of HIF-1α and HPH-2 protein After Chronic Ethanol Feeding.
(A) Nuclear protein levels of HIF-1α and HPH-2. (B) HIF-1α mRNA level. Note no significant difference between all groups. (C) Immunohistochemical staining for HIF-1α in the liver. Panel C6 shows strong HIF-1a staining in the KI mice (+++) fed with ethanol (arrows, Fig. 3C), moderate HIF-1a staining in the WT mice (++)(Fig. 3C4, arrows, IHC×300) and weak or no positive staining in KO mice (Fig. 3C5, IHC×300) fed with ethanol. No obvious positive staining was observed in the dextrose-fed mice (−) (Fig. 3C1,3C2,3C3). * p<0.05, ** p< 0.01 compared to the CYP2E1 KI groups fed dextrose. # p< 0.05 compared to CYP2E1 KO or WT groups fed ethanol or dextrose.
HIF-1α Downstream Targets and Hepatic Hypoxia
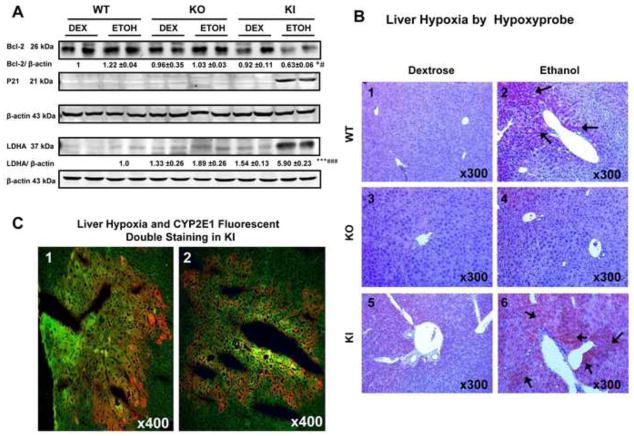
Hypoxia can induce expression or repression of many genes and HIF-1α-dependent targets include Bcl-2, P21 and LDHA (2,3,9,53). Protein levels of Bcl-2 were decreased about 40% in the ethanol-fed KI mice compared to the ethanol-fed WT and KO mice or to any of the dextrose-fed mice (Fig. 4A). Protein levels of P21 were very low or not reproducibly detectable in the ethanol-fed WT and KO mice or in the dextrose-fed mice but were highly expressed in the ethanol-fed KI mice (Fig. 4, lanes 11 and 12). LDHA was highly expressed in the ethanol-fed KI mice compared to the other groups (Fig. 4 lanes 11 and 12). These data suggest that Bcl-2 is negatively correlated and P21 and LDHA are positively correlated to the activation of HIF-1α in the ethanol fed KI mice. To evaluate whether hypoxia occurred in the ethanol-fed mice, liver slices were stained with the hypoxia-specific marker pimonidazole followed by immunohistochemical detection. Immunohistochemical staining was strongly positive in liver of the KI mice (arrows, +++) (Fig 4B6) and weakly positive in WT mice (+) fed with ethanol (Fig. 4B2). There were no staining in the KO mice (−) fed with ethanol (Fig. 4B4) or in any of the dextrose-fed mice (−) (Fig. 4B1,4B3, 4B5). In situ double staining of livers from the ethanol-fed KI mice showed that pimonidazole and CYP2E1 were co-localized to the same centrilobular area of the liver (Fig. 4C), suggesting that CYP2E1 may play an important role in ethanol-induced hypoxic liver injury.
Fig. 4. Levels of HIF-1α Downstream Targets and Pimonidazole Staining After Chronic Ethanol Feeding.
(A) The levels of Bcl-2, P21 and LDHA protein. Levels of p21 were very low and could not be reproducibly detected except for the ethanol-fed KI mice. Levels of LDHA could not be reproducibly detected in the dextrose-fed WT mice. The ratio of LDHA/β-actin for the ethanol-fed WT mice was taken as 1.0, to be compared to the other groups. (B) Immunohistochemical staining of the hypoxia-marker pimonidazole in liver. Panel 4B6 shows strongly hypoxia positive staining in liver of the KI mice (+++) (arrows, IHCx300). Panel 4B2 shows hypoxia positive staining in WT mice (+) (arrows, IHCx300) fed with ethanol. Panel 4B4 or 4B1,4B3,4B5 show no positive staining in the KO mice (−) fed with ethanol or in any of the dextrose-fed mice. (C) Fluorescent double staining of hypoxia-specific pimondazole and CYP2E1 in hepatic centrilobule. Panels 1 and 2 show co-location (yellow) of hypoxia (yellowish green, IHCx400) and CYP2E1 (red, IHCx400) in the same centrilobular area of the liver in the KI mice fed with ethanol * p< 0.05, *** P <0.001 compared to the CYP2E1 KI groups fed dextrose. # p<0.05, ### p< 0.001 compared to CYP2E1 KO or WT groups fed ethanol or dextrose.
Protein Expression of HIF-1α in an Acute Ethanol Model
The effect of acute ethanol-treatment on HIF-1α levels was evaluated in the three mouse genotypes. Levels of HIF-1α protein were increased in the KI mice gavaged with ethanol compared to the WT and KO mice gavaged with ethanol or to the mice gavaged with saline (Fig. 5A). The levels of HPH-2 protein were lowest in the KI mice gavaged with ethanol compared to the WT and KO mice gavaged with ethanol or to any of the mice gavaged with saline (Fig. 5A). Immunohistochemistry showed strong staining (+++) for HIF-1α in pericentral areas of livers of the ethanol-gavaged KI mice (arrows, Fig 5B6); although weaker, there was also staining (+) for HIF-1α in the ethanol-gavaged WT mice (Fig. 5B4) and the saline gavaged KI mice (Fig. 5B3). There was weak or no staining in the ethanol or saline treated WT or KO mice. Most of the staining appeared to be in the cytosol in the acute ethanol model. There were no increases in serum ALT or AST levels in any of the three genotypes treated acutely with ethanol (data not shown) despite the increases in CYP2E1, oxidant stress and HIF-1α. Most likely a more prolonged period of exposure to ethanol, than four days e.g as in the chronic ethanol-fed KI mice, may be needed for significant liver injury to develop.
Fig. 5. HIF-1α and HPH-2 Protein Levels After Acute Ethanol Treatment.
(A) Nuclear protein levels of HIF-1α and HPH-2. (B) Immunohistochemical staining for HIF-1α in the liver of WT, CYP2E1 KO and CYP2E1 KI mice treated with acute ethanol or saline. Panel B6 shows strong staining for HIF-1α in pericentral areas of livers of the ethanol-gavaged KI mice (+++) (arrows, IHCx300). Panel B4 show moderate staining for HIF-1α in the ethanol gavaged WT mice (arrows, IHCx300). Panel B3 shows weaker staining in the saline gavaged KI mice (arrows, IHCx300). Panels B1, B2 or B5 show no obvious HIF-1α positive staining in ethanol or saline treated KO mice or saline treated WT mice. * p<0.05 compared to the CYP2E1 KI groups treated with saline. # p<0.05, ## p<0.01 compared to CYP2E1 KO or WT groups treated with ethanol or saline.
Effects of Inhibition of HIF-1α on CYP2E1-dependent Toxicity in E47/C34 HepG2 Cells
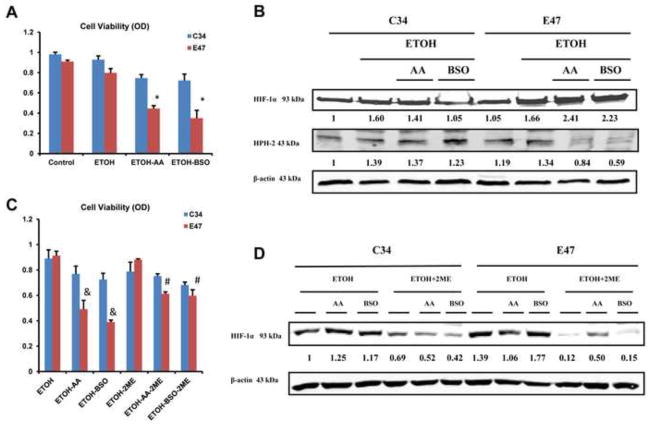
Experiments were carried out to evaluate the effect of HIF-1α on the toxicity of AA plus ethanol or BSO plus ethanol treatment in CYP2E1-expressing E47 cells and control C34 cells which do not express CYP2E1. Previous studies with these cells showed that polyunsaturated fatty acids such as AA (54,55) or depletion of GSH with BSO (56,57) produced greater toxicity to the E47 cells as compared to the C34 cells. This elevated toxicity was associated with more pronounced increases in oxidative stress in the E47 cells than the C34 cells (54–57). Fig 6A confirms that the combination of ethanol plus AA or ethanol plus BSO produced greater toxicity in E47 cells compared to C34 cells. Protein levels of HIF-1α were increased in E47 cells compared to C34 cells after treatment with ethanol plus AA or BSO plus ethanol (Fig. 6B). Protein levels of HPH-2 were decreased in the E47 cells compared to the C34 cells after treatment with ethanol plus AA or ethanol plus BSO (Fig. 6B). To evaluate a possible role for HIF-1α in the toxicity found in the E47 cells treated with AA or BSO, 2-methoxy-estradiol (2-ME), an inhibitor of HIF-1α (58,59) was added to the cells. After treatment with 2-ME, AA plus ethanol or BSO plus ethanol produced less toxicity in the E47 cells compared to the toxicity found in the absence of 2-ME (Fig. 6C). Little or no toxic effects were found with the C34 cells incubated with ethanol plus AA or ethanol plus BSO in the absence or presence of 2ME (Fig. 6C). The levels of HIF-1α protein were decreased both in E47 cells and C34 cells upon 2-ME treatment (Fig. 6D) validating the effectiveness of 2ME in inhibiting HIF-1α. These results suggest that HIF-1α contributes to the toxicity of AA and BSO in CYP2E1-expressin HepG2 cells.
Fig. 6. Toxicity of Ethanol, Ethanol + AA or Ethanol + BSO in E47/C34 HepG2 Cells in the Absence and Presence of the HIF-1α Inhibitor 2-ME.
E47 cells which express CYP2E1 and C34 cells which do not were treated for 48 hours with 100 mM ethanol alone or ethanol plus 30 μM AA or ethanol plus 300 μM BSO in the absence or presence of the HIF-1α inhibitor 45 μM 2-ME. (A,C): Cell viability was assayed by a MTT assay. (B, D): Protein levels of HIF-1α and HPH-2 from nuclear extracts of E47/C34 cells. * p<0.05 compared to ethanol treatment alone in E47 cells. # p<0.05 compared to E47 cells treated with ethanol plus AA or ethanol plus BSO in the absence of 2-ME. & p<0.05 compared to E47 cells treated with ethanol alone or to C34 cells treated with ethanol plus AA or ethanol plus BSO in the absence of 2-ME.
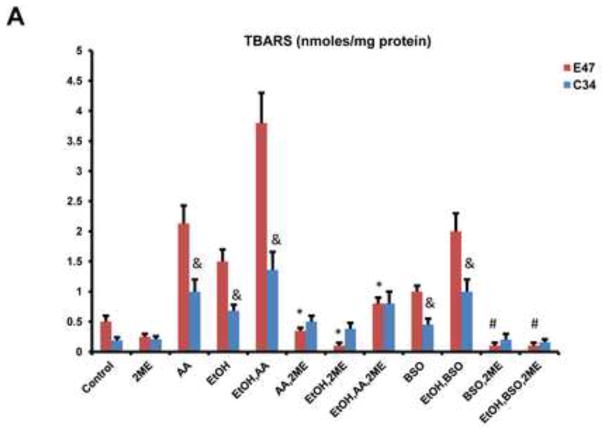
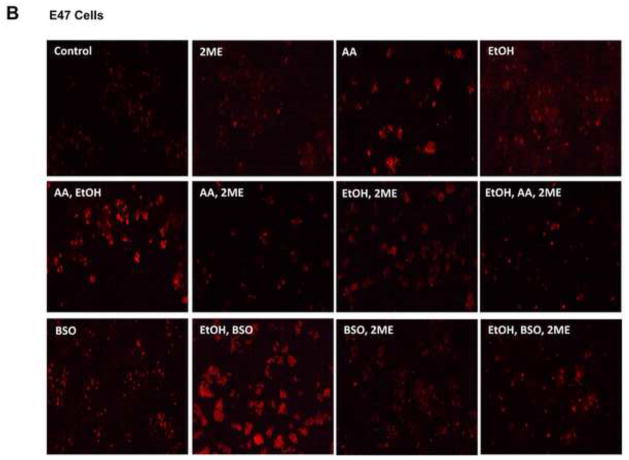
We next evaluated whether HIF-1α modulated AA- or BSO- CYP2E1-dependent oxidant stress since such oxidant stress is central to the toxicity produced by these compounds (54–57). Lipid peroxidation was elevated by treatment with ethanol alone or AA alone and especially by ethanol plus AA combined treatment to a greater extent in the E47 cells than the C34 cells (Fig. 7A). Similarly, lipid peroxidation was elevated to a greater extent in the E47 cells after treatment with BSO and especially ethanol plus BSO as compared to the C34 cells (Fig. 7A). Treatment with 2-ME lowered the levels of TBARS in E47 cells treated with AA or BSO in the absence or presence of ethanol as compared to the levels in the E47 cells without 2-ME treatment (Fig. 7A). Mitochondrial superoxide production was assayed via fluorescence of the mitochondrial localized probe, Mitosox. As shown in Fig. 7B, Mitosox fluorescence in the E47 cells was elevated by the addition of ethanol or AA or BSO and further increased by the combination of ethanol plus AA or ethanol plus BSO. Smaller increases were found with the C34 cells (Fig. 7B). Treatment of the E47 cells with 2ME blunted the increases in Mitosox fluoresecence produced by all these additions (Fig. 7B).
Fig. 7. Oxidative Changes in E47/C34 HepG2 Cells in the Absence and Presence of the HIF-1α inhibitor, 2-ME.
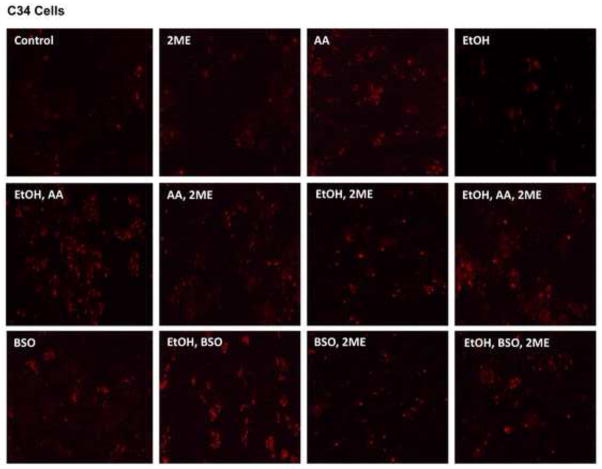
(A) The level of TBARS in E47 or C34 cells treated with 100 mM ethanol alone or 30 μM AA alone or 300 μM BSO alone or AA plus ethanol or BSO plus ethanol in the absence or presence of the HIF-1α inhibitor 45 μM 2-ME. * p<0.05 compared to E47 cells treated with AA alone or ethanol alone or AA plus ethanol in the absence of 2-ME. # p<0.05 compared to E47 cells treated with BSO alone or BSO plus ethanol in the absence of 2-ME. & p<0.05 compared to C34 cells control without treatment. (B) The positive staining of 5 μM Mitosox in E47/C34 cells treated with AA or ethanol or BSO or AA plus ethanol or BSO plus ethanol in the absence or presence of 2ME was observed under the fluorescence microscope. (Red color, ×100)
Discussion
The pioneering studies of Israel and co-workers showed that chronic ethanol administration increased oxygen consumption by the liver, consistent with the requirement for oxygen for the overall metabolism of ethanol (60,61). Ethanol-induced hypoxia occurs primarily in the pericentral zone of the liver, where oxygen levels are lowest. One important consequence of hypoxia is the induction and activation of the transcription factor HIF-1α (1,2). A major mode of regulation of HIF-1α is at the level of protein stabilization. Under normoxic conditions, the prolyl hydroylases use molecular oxygen to hydroxylate specific proline residues of HIF-1α. This results in ubiquitination of HIF-1α followed by proteosome-mediated degradation. Under hypoxia, the limitation in concentrations of oxygen lowers prolyl hydroxylase activity and prevents hydroxylation of HIF-1α which thereby stabilizes HIF-1α protein, followed by activation of HIF-1 target genes (1,2,4,5). Chronic ethanol consumption increases hepatic HIF-1α levels (13,14,17–19). What is not clear is what are the functional consequences of the increased levels of HIF-1α for the actions of ethanol. While hypoxia is thought to promote cellular toxicity, especially in the pericentral zone of the liver with low concentrations of oxygen, induction of HIF-1α may serve as a metabolic adaptation to hypoxia as HIF-1α upregulates many genes which can serve to protect against hypoxia. Two recent studies evaluated whether induction of HIF-1α protected against or played a critical role in promoting ethanol-induced liver steatosis and injury and for unexplained reasons came to opposite conclusions (22,23). As discussed by Mehal (24), in some other models of liver injury, HIF-1α appears to be a key step in driving the pathology rather than producing adaptive changes to protect against the liver injury. With respect to ethanol-induced liver injury, further studies are required to assess the role of HIF-1α in protecting against or contributing to the liver injury process.
CYP2E1 potentiates ethanol-induced oxidative stress and fatty liver and at high levels, plays a role in ethanol liver damage as shown by experiments with CYP2E1 inhibitors (33,34,37), CYP2E1 knockout and knockin mice (37,38) and transgenic mice overexpressing CYP2E1 (62). HIF-1α expression and activation is regulated by oxidative stress, redox state and inflammatory cytokines (63–65). ROS have been suggested to decrease hydroxylation of HIF-1α by prolyl hydroxylases by oxidizing the ferrous iron necessary for catalytic activity of these enzymes (65). This would promote stabilization of HIF-1α. For example, ROS produced by hypoxia from complex III of the respiratory chain stabilized HIF-1α by decreasing prolyl hydroxylase activity (66) and a decrease in manganese superoxide dismutase which increased superoxide levels potentiated hypoxic induction of HIF-1α (67). Exogenous addition of hydrogen peroxide to mouse embryonic cells stabilized HIF-1α (68). In view of these considerations, we evaluated the possibility that increased oxygen uptake, especially in the pericentral zone of the liver, the area where ethanol induction of CYP2E1 primarily occurs (29), may contribute to ethanol-induced hypoxia; and that increased production of ROS as a result of ethanol induction of CYP2E1 would elevate hepatic HIF-1α levels.
As shown by others, chronic ethanol consumption caused liver hypoxia as detected by pimonidazole staining. Intensity of staining was greatest in ethanol-fed CYP2E1 KI mice, intermediate in the ethanol-fed WT mice and weakest in the ethanol-fed KO mice. In the liver of the KI mice, the pimonidazole staining co-localized with CYP2E1. HIF-1α levels were elevated by the chronic ethanol feeding with highest amounts in the KI mice. Interestingly, in the KI mice, levels of HIF-1α mRNA were not increased after ethanol feeding but protein levels of prolyl hydroxlase 2 were decreased by the ethanol feeding suggesting that the increase in HIF-1α protein is posttranscriptional and likely due to prevention of HIF-1α rapid degradation. Although ethanol-induced hypoxia was more prominent in the WT than the KO mice, we could not detect significant differences in HIF-1α protein levels between these 2 groups. These results suggest only when CYP2E1 levels are very high, as in the ethanol-fed KI mice, do levels of HIF-1α correspondingly increase. Of note, although ethanol-induced fatty liver and oxidative stress was higher in the WT mice compared to the KO mice, liver injury or necrosis was minimal in both genotypes. However, liver injury did occur in the ethanol-fed KI mice with very high levels of CYP2E1 and HIF-1α. Additional experiments with direct inhibitors of HIF-1α or activators of prolyl hydroxylases to decrease HIF-1α or generating double CYP2E1KI/HIF-1α knockouts are needed to evaluate if the elevated HIF-1α levels play a role in the increased liver injury in vivo in the ethanol-fed KI mice. Future experiments with antioxidants are needed to assess the role of ROS in the elevated levels of HIF-1α in the KI mice.
The possible role of HIF-1α in CYP2E1-dependent toxicity was studied in vitro in HepG2 cells. As reviewed elsewhere (31,39,40), many hepatotoxins are more toxic in the E47 cells which express CYP2E1 than in the control C34 cells which do not express significant CYP2E1. Combined treatment with ethanol plus AA or with BSO plus ethanol caused the E47 cells to lose viability to a greater extent than did the C34 cells. This enhanced toxicity was associated with increases in ROS production in the E47 cells. Treatment with ethanol plus AA or ethanol plus BSO increased HIF-1α protein levels in the E47 cells but not the C34 cells. Levels of HPH-2 were decreased by the combined additions in the E47 cells but not the C34 cells. We speculate that increased ROS production by the combined treatments in the CYP2E1-expressing cells may have inactivated HPH-2, resulting in the stabilization of HIF-1α. Does the increased HIF-1α contribute to the enhanced toxicity produced by the combined treatments in the E47 cells ? HIF-1α levels in both the E47 and C34 cells were decreased by 2-methoxyestradiol, an inhibitor of HIF-1α (58,59), in the presence of ethanol alone or ethanol plus AA or ethanol plus BSO. This inhibition of HIF-1α resulted in protection of the E47 cells against loss of viability caused by the combined treatments. Inhibition of HIF-1α also resulted in blunting the increase in oxidative stress in the E47 cells produced by the combined treatments, a likely factor in the protection against loss of cell viability. These results suggest that HIF-1α plays a role in potentiating CYP2E1-generated oxidative stress and cellular toxicity in these HepG2 cells. Further studies such as assays of antioxidant defense and antioxidant transcription factors, mitochondrial dysfunction, autophagy, cell signaling pathway such as MAP kinase pathways, are required to understand mechanisms by which HIF-1α and CYP2E1 synergize to increase oxidative stress and cause loss of cell viability.
As described above, hypoxia and HIF-1α were elevated by chronic ethanol consumption to the greatest extent when CYP2E1 levels were highest. Under these conditions, ethanol-induced liver injury occurs. HIF-1α potentiated CYP2E1 toxicity and oxidative stress in vitro. Further studies to define and characterize potential interactions between CYP2E1 and HIF-1α would be important. For example, inducible nitric oxide synthase is elevated by HIF-1α (69). Increased nitric oxide production coupled to increased superoxide production from elevated CYP2E1 can produce peroxynitrite, (70) which has been implicated in the toxic actions of ethanol (71). Indeed 3-nitrotyrosine protein adducts (an indirect assay for peroxynitrite) were detected in livers of the ethanol-fed KI mice with elevated CYP2E1 and HIF-1α (Fig 2F). Since CYP2E1 is elevated by many chemicals besides ethanol, and is increased in diabetes, obesity and in non alcoholic fatty liver, whether HIF-1α and hypoxia potentiate CYP2E1 oxidative stress and toxicity would be of broad clinical significance to determine and to provide a rationale to develop hepatoprotective therapies which target HIF-1α or HIF-1α downstream mediators.
HIGHLIGHTS.
Ethanol induces hypoxia and elevates HIF-1α in the liver.
Ethanol increased liver injury and oxidative stress in CYP2E1 overexpresing KI mice compared to wild type and CYP2E1 knockout KO mice.
HIF-1 and hypoxia were elevated in the ethanol-fed KI mice compared to WT and KO mice. Inhibition of HIF-1α, blunted the toxic effects of ethanol in CYP2E1-expressing HepG2 cells
CYP2E1 plays a role in ethanol-induced hypoxia, oxidative stress and activation of HIF-1α. HIF-1α contributes to CYP2E1-dependent ethanol-induced toxicity.
Acknowledgments
We thank Dr. Stephen C. Ward, Department of Pathology, Mount Sinai School of Medicine, for his valuable observation and analysis of tissue slides and Mr. Leon An for technical assistance on some of the immunoblots. These studies were supported by USPHS grants RO1 AA018790 and R21 AA021362 from the National Institute on Alcohol Abuse and Alcoholism, NIH and by a grant from the National Natural Science Foundation of China (81270482, L. Gan).
Abbreviations
- AA
arachidonic acid
- AEC
3-amino-9-ethylcarbazole
- ALT
alanine aminotransferase
- BSO
l-buthionine-sulfoximine
- CYP2E1
cytochrome P450 2E1
- DAB
3,3′-Diaminobenzidine
- GSH
reduced glutathione
- HE
hematoxylin-eosin
- 4-HNE
4-hydroxynonenal
- HIF-1α
hypoxia inducible factor-1α
- HPH-2
HIF prolylhydroxylase-2
- IHC
immunohistochemistry
- KI
knock in
- KO
knock out
- LDHA
lactate dehydrogenase A
- MDA
malondialdehyde
- 2-ME
2-methoxyestradiol
- 3-NT
3-nitrotyrosine
- PNP
p-nitrophenol
- ROS
reactive oxygen species
- WT
wild type
- TBARs
thiobarbituric acid reactive substrates
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Semenza GL. Regulation of mammalian oxygen homeostasis by hypoxia- inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Formenti F, Constantin-Tedusoiv D, Emmanuel Y, Cheeseman J, Durrington KL, Edwards LM, Humphreys SM, Lappin TR, McMullin MF, McNamara CJ, Mills W, Murphy JA, O’Connor DF, Percy MJ, Ratcliffe PJ, Smith TG, Treacy M, Frayn KN, Greenhaff PL, Karpe F, Clarke K, Robbins PA. Regulation of human metabolism by hypoxia-inducible factor. Proc Natl Acad Sci USA. 2010;107:12722–12727. doi: 10.1073/pnas.1002339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juan M, Kondo K, Yang H, Kim W, Valiando J, Ohn M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for oxygen sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Majmudar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxia stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinag BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding and transcription properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 7.Chan DA, Giaccia AJ. Hypoxia, gene expression and metastasis. Cancer Metastasis Rev. 2007;26:333–339. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- 8.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1α. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 9.Copple BL, Bai S, Burgoon LD, Moon JO. Hypoxia – inducible factor 1 alpha regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int. 2010;31:230–244. doi: 10.1111/j.1478-3231.2010.02347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon J, Welch T, Gonzalez F, Copple B. Reduced liver fibrosis in hypoxia-inducible factor-1 alpha-deficient mice. Am J Physiol GI Liver Physiol. 2009;296:G582–592. doi: 10.1152/ajpgi.90368.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nath B, Szabo G. Hypoxia and hypoxia inducible factors: diverse roles in liver diseases. Hepatology. 2012;55:622–633. doi: 10.1002/hep.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thurman RG, Ji S, Matsumura T, Lemasters JJ. Is hypoxia involved in the mechanisms of alcohol-induced liver injury? Fund Appl Toxicol. 1984;4:125–133. doi: 10.1016/0272-0590(84)90112-x. [DOI] [PubMed] [Google Scholar]
- 13.French SW, Benson NC, Sun PS. Centrilobular liver necrosis induced by hypoxia in chronic ethanol-fed rats. Hepatology. 1984;4:912–917. doi: 10.1002/hep.1840040521. [DOI] [PubMed] [Google Scholar]
- 14.Sato N, Kanada T, Kawario S, Hayash N. Effect of acute and chronic ethanol consumption on hepatic oxygen tension in rats. Pharmacol Biochem Behav. 1983;18:443–447. doi: 10.1016/0091-3057(83)90215-0. [DOI] [PubMed] [Google Scholar]
- 15.Yuki T, Thurman RG. The swift increase in alcohol metabolism. Biochem J. 1980;186:119–126. doi: 10.1042/bj1860119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arteel GE, Raleigh JA, Bradford BU, Thurman RG. Acute alcohol produces hypoxia directly in rat liver tissue in vivo: role of Kupffer cells. Am J Physiol. 1996;271:G494–G500. doi: 10.1152/ajpgi.1996.271.3.G494. [DOI] [PubMed] [Google Scholar]
- 17.Arteel GE, Iimuro Y, Yin M, Raleigh JA, Thurman RG. Chronic enteral ethanol treatment causes hypoxia in rat liver tissue in vivo. Hepatology. 1997;25:920–926. doi: 10.1002/hep.510250422. [DOI] [PubMed] [Google Scholar]
- 18.McKim SE, Uesugi T, Raleigh JA, McClain CJ, Arteel GE. Chronic intragastric alcohol exposure causes hypoxia and oxidative stress in the rat pancreas. Arch Biochem Biophys. 2003;417:34–43. doi: 10.1016/s0003-9861(03)00349-7. [DOI] [PubMed] [Google Scholar]
- 19.Li J, French B, Wu Y, Vankatesh R, Montgomery R, Bardag-Gorce F, Kitto J, French SW. Liver hypoxia and lack of recovery after reperfusion at high blood alcohol levels in the intragastric feeding model of alcohol liver disease. Exp Mol Pathol. 2004;77:184–192. doi: 10.1016/j.yexmp.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 20.French SW. The role of hypoxia in the pathogenesis of alcoholic liver disease. Hepatol Res. 2004;29:69–74. doi: 10.1016/j.hepres.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Chen SH, Zhang Y, Yu CH, Li SD, Li YM. Is the hypoxia-inducible factor 1 alpha mRNA expression activated by ethanol-induced injury, the mechanism underlying alcoholic liver disease? Hepatabol Pancr Diseases Internat. 2006;5:560–563. [PubMed] [Google Scholar]
- 22.Nath B, Levin I, Csak T, Petrasek J, Mueller C, Kodys K, Catalano D, Mandrekar P, Szabo G. Hepatocyte-specific hypoxia-inducible factor-1α is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology. 2011;53:1526–1537. doi: 10.1002/hep.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishiyama Y, Goda N, Kanai M, Niwa D, Osani K, Yamamoto Y, Senov-Matsuda N, Johnson RS, Miura S, Kabe Y, Suematsu M. HIF-1α induction suppresses excessive lipid accumulation in alcoholic fatty liver in mice. J Hepatol. 2012;56:441–447. doi: 10.1016/j.jhep.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 24.Mehal WZ. HIF-1α is a major and complex player in alcohol-induced liver diseases. J Hepatol. 2012;56:311–312. doi: 10.1016/j.jhep.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Bolt M, Koos PH, Their R. The cytochrome P450 isoenzyme CYP2E1 in the biological processing of industrial chemicals. Int Arch Occup Environ Health. 2003;76:174–185. doi: 10.1007/s00420-002-0407-4. [DOI] [PubMed] [Google Scholar]
- 26.Koop DR. Oxidative and reductive metabolism by cytochrome P4502E1. FASEB J. 1992;6:724–730. doi: 10.1096/fasebj.6.2.1537462. [DOI] [PubMed] [Google Scholar]
- 27.Song BJ, Cederbaum AI, Koop DR, Ingelman-Sundberg M, Nanji A. Ethanol-inducible cytochrome P450 (CYP2E1): Biochemistry, molecular biology and clinical relevance. Alcoholism: Clin Exp Res. 1996;20 (Suppl):138A–146A. doi: 10.1111/j.1530-0277.1996.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 28.Gonzelez FJ. Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat Res. 2005;569:101–110. doi: 10.1016/j.mrfmmm.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Lieber CS. Cytochrome P4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- 30.Gorsky LD, Koop DR, Coon MJ. On the stoichiometry of the oxidase and monoxygenase reactions catalyzed by liver microsomal cytochrome P450. J Biol Chem. 1984;259:6812–6817. [PubMed] [Google Scholar]
- 31.Caro AA, Cederbaum AI. Oxidative stress, Toxicology and Pharmacology of Cyp2E1. Annual Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- 32.Nanji A, Zhao S, Sadrzadeh SMH, Dannenberg AJ, Tahan SR, Waxman DJ. Markedly enhanced cytochrome P4502E1 induction and lipid peroxidation is associated with severe liver injury in fish oil-ethanol-fed rats. Alcoholism: Clin Exp Res. 1994;18:1280–1285. doi: 10.1111/j.1530-0277.1994.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto M, Hagbjork AL, Wan YJ, Fu PC, Clot P, Albano E, Ingleman-Sundberg M, French SW. Modulation of experimental alcohol-induced liver disease by cytochrome P450 2E1 inhibitors. Hepatology. 1995;21:1610–1617. [PubMed] [Google Scholar]
- 34.Gouillon Z, Lucas D, Li J, Hagbjork AL, French BA, Fu P, Fang C, Ingelman-Sundberg M, Donohue TM, Jr, French SW. Inhibition of ethanol-induced liver disease in the intragstric feeding rat model by chlormethiazole. Proc Soc Biol Med. 2000;224:302–308. doi: 10.1046/j.1525-1373.2000.22435.x. [DOI] [PubMed] [Google Scholar]
- 35.Koop DR, Klopfenstein B, Iimuro Y, Thurman RG. Gadolinium chloride blocks alcohol-dependent liver toxicity in rats treated chronically with intragastric alcohol despite the induction of CYP2E1. Mol Pharmacol. 1997;51:944–950. doi: 10.1124/mol.51.6.944. [DOI] [PubMed] [Google Scholar]
- 36.Kono H, Bradford BU, Yin M, Sulik KK, Koop DR, Peters JM, Gonzalez FJ, McDonald T, Dikalova A, Kadiiska MB, Mason RP, Thurman RG. CYP2E1 is not involved in early alcohol-induced liver injury. Am J Physiol. 1999;277:G1259–G1267. doi: 10.1152/ajpgi.1999.277.6.G1259. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Zhuge J, Wang X, Bai J, Cederbaum AI. Cytochrome P4502E1 contributes to enthanol-induced fatty liver in mice. Hepatology. 2008;47:1483–1494. doi: 10.1002/hep.22222. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y, Wu D, Wang X, Ward SC, Cederbaum AI. Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in cytochrome P4502E1 knockin mice. Free Radical Biol Med. 2010;49:1406–1416. doi: 10.1016/j.freeradbiomed.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessova I, Cederbaum AI. CYP2E1: Biochemistry, Toxicology and Regulation. Current Molec Med. 2003;3:509–518. doi: 10.2174/1566524033479609. [DOI] [PubMed] [Google Scholar]
- 40.Jimenez-Lopez JM, Cederbaum AI. CYP2E1-dependent oxidative stress and toxicity: role in ethanol-induced liver injury. Expert Opinion Drug Metab Toxicol. 2001;1:671–685. doi: 10.1517/17425255.1.4.671. [DOI] [PubMed] [Google Scholar]
- 41.Lee SS, Buter JT, Pineau T, Fernandez-Salguero P, Gonzalez FJ. Role of CYP2E1 in the hepatotoxicity of acetaminophen. J Biol Chem. 1996;271:12063–12067. doi: 10.1074/jbc.271.20.12063. [DOI] [PubMed] [Google Scholar]
- 42.Cheun C, Yu AM, Ward JM, Krausz JW, Akiyama TE, Feigenbaum L, Gonzalez FJ. The CYP2E1-humanized transgenic mouse: role of CYP2E1 in acetaminophen hepatoxicity. Drug Metab Disp. 2005;33:449–457. doi: 10.1124/dmd.104.002402. [DOI] [PubMed] [Google Scholar]
- 43.Cheung C, Gonzalez FJ. Humanized mouse lines and their application for prediction of human drug metabolism and toxicological risk assessment. J Pharmacol Exp Ther. 2008;327:288–299. doi: 10.1124/jpet.108.141242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lieber CS, DeCarli LM. Animal models of chronic ethanol toxicity. Methods Enzymol. 1994;23:585–594. doi: 10.1016/s0076-6879(94)33061-1. [DOI] [PubMed] [Google Scholar]
- 45.Reinke LA, Moyer MJ. P-Nitrophenol hydroxylase: A microsomal oxidation which is highly inducible by ethanol. Drug Metab Disp. 1985;13:548–552. [PubMed] [Google Scholar]
- 46.Esterbauer H, Cheeseman KH. Determination of aldehydic lipid peroxidation products: malondiadehyde and 4-hydroxynonenal. Methods Enzymol. 1990;186:407–421. doi: 10.1016/0076-6879(90)86134-h. [DOI] [PubMed] [Google Scholar]
- 47.Mari M, Cederbaum AI. CYP2E1 overexpression in HepG2 cells induces glutathione synthesis by transcriptional activation of gamma-glutamylcysteine synthase. J Biol Chem. 2000;275:15563–15571. doi: 10.1074/jbc.M907022199. [DOI] [PubMed] [Google Scholar]
- 48.Terado N, Ohno N, Saitoh S, Ohno S. Immunohistochemical detection of hypoxia in mouse liver tissues treated with pimonidazole using “in vivo cryotechnique”. Histochem Cell Biol. 2007;128:253–261. doi: 10.1007/s00418-007-0324-4. [DOI] [PubMed] [Google Scholar]
- 49.Wu D, Cederbaum AI. Development and properties of HepG2 cells that constitutively express CYP2E1. Methods Mol Biol. 2008;447:137–150. doi: 10.1007/978-1-59745-242-7_11. [DOI] [PubMed] [Google Scholar]
- 50.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 51.Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Rad Biol Med. 2008;44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaija HM, Sarkioja T, Kortelainen ML, Vuoristo JT, Huikuri HV, Porvari KS. Stress-specific responses of p21 expression: implication of transcript variant p21 alta in long-term hypoxia. J Cell Biochem. 2012;113:544–552. doi: 10.1002/jcb.23377. [DOI] [PubMed] [Google Scholar]
- 53.Donohue TM, Cederbaum AI, French SW, Barve S, Gao B, Osna NA. Role of the proteasome in ethanol-induced liver injury. Alcoholism: Clin Exp Research. 2007;31:1446–1459. doi: 10.1111/j.1530-0277.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen Q, Galleano M, Cederbaum AI. Cytoxicity and apoptosis produced by arachidonic acid in HepG2 cells over-expressing human cytochrome P4502E1. J Biol Chem. 1997;272:14532–14541. doi: 10.1074/jbc.272.23.14532. [DOI] [PubMed] [Google Scholar]
- 55.Gong P, Cederbaum AI. Transcription factor Nrf2 protein protects HepG2 cells against CYP2E1 plus arachidonic acid-dependent toxicity. J Biol Chem. 2006;281:14573–14579. doi: 10.1074/jbc.M600613200. [DOI] [PubMed] [Google Scholar]
- 56.Wu D, Cederbaum AI. Removal of glutathione produces apoptosis and necrosis in HepG2 cells overexpressing CYP2E1. Alcoholism: Clin Exp Res. 2001;25:619–628. [PubMed] [Google Scholar]
- 57.Bai J, Cederbaum AI. Overexpression of CYP2E1 in mitochondria sensitizes HepG2 cells to the toxicity caused by depletion of glutathione. J Biol Chem. 2006;281:5128–5136. doi: 10.1074/jbc.M510484200. [DOI] [PubMed] [Google Scholar]
- 58.Semenza GL. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discovery Today. 2007;12:853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Xia Y, Choi HK, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur J Med Chem. 2012;49:24–40. doi: 10.1016/j.ejmech.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein JL, Videla L, Israel Y. Metabolic alterations produced in the liver by chronic ethanol administration. Biochem J. 1973;134:515–521. doi: 10.1042/bj1340515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Israel Y, Kalant H, Orrego H, Khanna JM, Videla LA, Phillips JM. Experimental alcohol-induced hepatic necrosis: suppression by propylthiouracil. Proc Natl Acad Sci USA. 1975;72:1137–1141. doi: 10.1073/pnas.72.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butura A, Nilsson K, Morgan K, Morgan TR, French S, Johansson I, Schuppe-Koistinen I, Ingleman-Sundberg M. The impact of CYP2E1 on the development of alcoholic liver disease as studied in a transgenic mouse model. J Hepatol. 2008;50:572–583. doi: 10.1016/j.jhep.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 63.Klimova T, Chandel NS. Mitochondrial Complex III regulates hypoxia activation of HIF. Cell Death Differ. 2008;15:660–666. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- 64.Walmsley SR, Cadwallader KA, Chilvers ER. The role of HIF-13 in myeloid cell inflammation. Trends Immunol. 2005;26:434–439. doi: 10.1016/j.it.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 65.Wheaton WW, Chandel NS. Hypoxia. 2 Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol. 2011;300:C385–393. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chua YL, Dufour E, Dassa EP, Rustin P, Jacobs HT, Taylor CT, Hagen T. Stabilization of hypoxia-inducible factor-13 protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J Biol Chem. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaewpila S, Venkataraman S, Buettner CR, Oberlely LW. Manganese superoxide dismutase modulates hypoxia-inducible factor-13 induction via superoxide. Cancer Res. 2008;68:2781–2788. doi: 10.1158/0008-5472.CAN-07-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon MC. Mitochondrial reactive oxygen species are required for hypoxic HIF alpha stabilization. Adv Exp Med Biol. 2006;588:165–170. doi: 10.1007/978-0-387-34817-9_15. [DOI] [PubMed] [Google Scholar]
- 69.Zhang XL, Yan ZW, Sheng WW, Xiao J, Zhang ZX, Ye ZB. Activation of hypoxia-inducible factor-1 ameliorates postischemic renal injury via inducible nitric oxide synthase. Mol Cell Biochem. 2011;358:287–295. doi: 10.1007/s11010-011-0979-y. [DOI] [PubMed] [Google Scholar]
- 70.Ischiropoulos H, al-Mehdi AB, Fisher AB. Reactive species in ischemic rat lung injury: contribution of peroxynitrite. Am J Physiol. 1995;269:L158–L164. doi: 10.1152/ajplung.1995.269.2.L158. [DOI] [PubMed] [Google Scholar]
- 71.McKim SE, Gabele E, Isayama F, Lambert JC, Tucker LM, Wheeler MD, Connor HD, Mason RP, Doll MA, Hein DW, Arteel GW. Inducible nitric oxide synthase is required in alcohol-induced liver injury: studies with knockout mice. Gastroent. 2003;125:1834–1844. doi: 10.1053/j.gastro.2003.08.030. [DOI] [PubMed] [Google Scholar]