Abstract
We conducted a translational genomics pilot study to evaluate the impact of genomic information related to colorectal cancer (CRC) risk on psychosocial, behavioral and communication outcomes. In 47 primary care participants, 96% opted for testing of three single nucleotide polymorphisms (SNPs) related to CRC risk. Participants averaged 2.5 of 6 possible SNP risk alleles (10% lifetime risk). At 3-months, participants did not report significant increases in cancer worry/distress; over half reported physical activity and dietary changes. SNP risk scores were unrelated to behavior change at 3-months. Many participants (64%) shared their SNP results, including 28% who shared results with a physician. In this pilot, genomic risk education, including discussion of other risk factors, appeared to impact patients' health behaviors, regardless of the level of SNP risk. Future work can compare risk education with and without SNP results to evaluate if SNP information adds value to existing approaches.
Keywords: translational genomic research, SNP testing, colorectal cancer risk, genomics education, behavior change
1. Introduction
Effective clinical translation of genomic information from low-penetrance genes into meaningful health improvements remains elusive. Research to date indicates that genomic information from single nucleotide polymorphisms (SNPs) may have a limited impact on long-term behavior change. For example, within the context of smoking cessation, genomic feedback influenced smoking quit attempts [1,2] but these quit attempts do not appear to translate into sustained smoking cessation [3-5]. Likewise, the evidence for other types of health behavior change following provision of genomic risk information is equivocal [6-8]. Large scale studies such as those conducted by The Coriell Personalized Medicine Collaborative (CPMC) [9] and the NIH Multiplex Initiative [10] suggest that although participants understood the genomic information and did not report negative emotional responses [6,11], behavior change was minimal. Similarly, among participants who obtained genomic testing through a Direct-to-Consumer (DTC) commercial company, intentions to improve physical activity, diet and cancer screening were high, but few actual behavior changes were noted [12]. In contrast, in a different DTC study sample, participants changes in physical activity, diet and communication of genomic results to healthcare providers [7], particularly among people who had personal or family history of disease. Likewise, research conducted with individuals selected for family history reported changes in vitamin intake after disclosure of APOEe4 status, a SNP related to Alzheimer's disease [13].
Behavioral research has hastened the translation of genetic discoveries into clinical practice [14]. Currently, the “vision” for genomics includes use of genomic information to facilitate health behavior change efforts [15]. The continuum of translational genomic research described by Khoury and colleagues [16] includes the translation of genomic discoveries to improve public health. However, to date, more than 95% of funded cancer genomic research has focused on the early stages of genomic discovery. The potential for genomic advances to impact population health will not be realized unless and until we engage in translational research on implementation and outcomes of genomic testing [16].
Few studies to date have examined multiple health behavior change outcomes within the context of pre- and post-test cancer genomics education and testing [9,10,13]. We conducted a pilot study with primary care patients to evaluate people's responses to SNP testing for colorectal cancer (CRC) risk. We elected to examine risk for CRC given that it occurs in both men and women, has effective screening/prevention guidelines, has modifiable lifestyle risk factors (e.g., physical activity, diet), and has a growing evidence base of identified SNPs related to CRC risk [17].
2. Materials and Methods
2.1. Participants
We invited male and female patients from Georgetown University Hospital's Division of General Internal Medicine to participate in a study offering free genomic testing for a research panel of three SNPs related to CRC risk. We recruited participants in-person in the clinic waiting room and through mailed study invitation letters. Eligibility criteria included age ≥40 years, ability to read and understand English and ability to provide informed consent. We did not exclude anyone based on prior personal or family cancer history. All study procedures were approved by the Institutional Review Board at Georgetown University/MedStar Health. The recruitment materials clearly explained that SNP testing was optional. The consent document emphasized the uncertain clinical utility of SNP testing and described the risks and benefits of study participation and SNP testing [18].
2.2. Procedures
2.2.1. Overview
After providing written informed consent, participants attended pre- and post-test education sessions with a certified genetic counselor. Sessions were held in person or by telephone and were audio-recorded if participants agreed. We conducted four brief assessments. Participants completed two assessments at the time of the first education session, one as baseline (pre-education and pre-testing) and the other immediately post-education, but before testing. Immediately after the second education session in which participants learned their SNP risk scores, they completed a post-test assessment. Finally, participants completed follow-up assessments 3-months after receipt of their SNP results. Participants received gift cards valued at $30 for completion of the initial education session and baseline survey, $25 for completion of the results education session and immediate post-test survey and $10 for completion of the 3-month follow-up survey.
2.2.2. Genomic Education Sessions
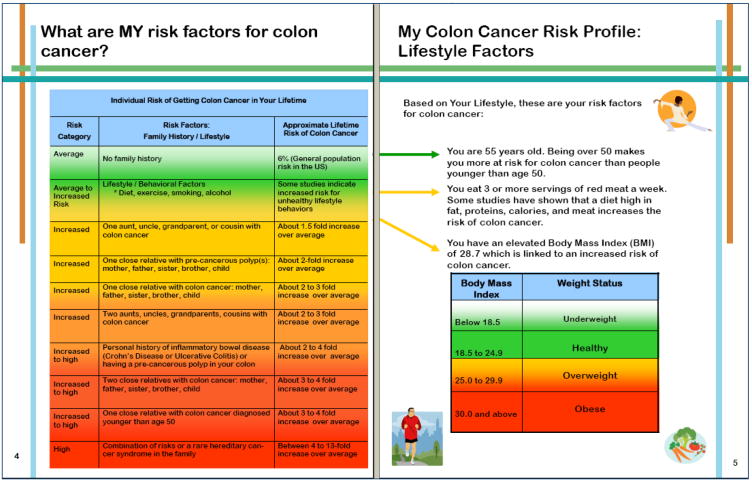
Our transdisciplinary research team developed the content of the printed materials utilized by the genetic counselor during the education sessions. Details of the material content have been published elsewhere [18]. Briefly, the pre-test materials included descriptions of risk factors for CRC; definitions of SNPs and how SNPs might be related to CRC risk; the benefits, limitations and risks of SNP testing and steps to reduce risk of CRC. We emphasized the uncertain clinical utility of SNP testing for CRC as well as the uncertainties surrounding how to best combine risk estimates from different SNPs. At the end of the pre-test education session, participants were given the option of SNP testing for three research SNPs related to CRC Risk (Table 1). Interested participants provided a DNA sample using a mouthwash oral rinse solution by standard collection procedures. CLIA-approved genotyping was performed in the Genomics and Epigenomics Shared Resource at the Lombardi Comprehensive Cancer Center at Georgetown University. We disclosed results to participants 8 to 10 weeks later via an in-person or telephone session with the genetic counselor using individually-tailored, printed booklets. Participants who completed the education sessions by telephone received the pre- and post-test printed booklets by mail prior to the scheduled sessions. The post-test materials provided individual lifetime risk estimates based on SNP results and reviewed participants' other risk factors for CRC (e.g., family history, lifestyle risk factors; see Figure 1).
Table 1. Selected Panel of Three CRC SNPs.
| SNP/ Closest Gene | Alleles | Approximate Risk Odds Ratio; 95% Confidence Interval | SNP Frequencies [36] | % of Individuals with Each Genotype by CRC Family History | ||||
|---|---|---|---|---|---|---|---|---|
| GUH AA | GUH Cauc. | dbSNP AA | dbSNP Cauc. | CRC Family History | No CRC Family History | |||
| 8q24.21 rs6983267 [19] POU5F1P1, MYC [48] |
GG | OR= 1.68; 1.21-2.33 | 0.79 | 0.44 | 0.79 | 0.20 | 0.15 | 0.03 |
| GT | OR = 1.39; 1.03-1.88 | 0.21 | 0.52 | 0.21 | 0.50 | 0.31 | 0.31 | |
| TT | OR = 1.0 | 0 | 0.04 | 0 | 0.30 | 0.54 | 0.66 | |
| 15q13.3 rs4779584 [20] CRAC1 |
TT | OR= 1.70; 1.41-2.04 | 0.36 | 0 | 0.17 | 0.07 | 0 | 0.19 |
| CT | OR= 1.23; 1.13-1.33 | 0.36 | 0.30 | 0.58 | 0.19 | 0.46 | 0.28 | |
| CC | OR=1.0 | 0.28 | 0.70 | 0.25 | 0.74 | 0.54 | 0.53 | |
| 11q23.1 rs3802842 [21] LOC120376 [48] |
CC | OR=1.35; 1.22-1.49 | 0 | 0.11 | 0.13 | 0.17 | 0.69 | 0.47 |
| AC | OR=1.18; 1.11-1.25 | 0.21 | 0.33 | 0.57 | 0.31 | 0.23 | 0.47 | |
| AA | OR=1.0 | 0.79 | 0.55 | 0.30 | 0.52 | 0.08 | 0.06 | |
Figure 1. Genomic Education Materials: Lifestyle and SNP Risk Information.
2.2.3. SNP Panel and Genotyping
We selected three SNPs for inclusion in the research panel based on a review of the literature at the time of study initiation and our a priori selection criteria: rs6983267 (8q24.21) [19], rs4779584 (CRAC1) [20], and rs3802842 (11q23.1) [21] (Table 1). Selection criteria included: 1) number of published studies examining the association (≥ 3 studies), 2) sample size of the studies (≥ 5,000 cases and 5,000 controls), 3) sample demographics that were similar to the demographics of our study population, and 4) statistical strength of the results. The three SNPs that met our criteria were part of commercially available DTC CRC panels at the time of study initiation [22-24].
We processed and stored mouthwash samples as pellets at −80°C until analysis. We used allelic discrimination techniques based on real time PCR methods with Taqman® probes for SNP analysis. We performed PCR reactions on the ABI 7900HT sequence detection system (Applied Biosystems, Foster City, CA). To ensure consistency, we randomly selected 20% of selected samples for repeat analysis.
2.3. Study Assessments
In the baseline assessment conducted prior to the first genomics education session and testing, we assessed demographics, family and personal history of cancer, prior CRC screening and polyp and bowel disease history. We also assessed psychosocial and health behavior variables at baseline, post-education, immediately post-test and at the 3-month follow-up. We assessed communication of SNP results variables at the 3-month follow-up.
2.3.1. Psychosocial variables
Included perceived risk for CRC (3-items: absolute risk, comparative risk [25], numeric risk [26]) and cancer worry [27]). At the 3-month follow-up, we measured psychological responses to SNP testing using an adapted version of the Multidimensional Instrument of Cancer Risk Assessment [28]. Behavioral variables at baseline, post-test and 3-months included face-valid items adapted from behavioral screeners to assess self-reported behaviors related to CRC risk: alcohol consumption, diet, physical activity [29,30] and smoking. We also assessed whether participants had seen their primary care physician, scheduled or completed appointments for CRC screening, and intentions for screening [31]. At 3-months, we assessed communication variables of participants' communication of SNP results to family members or physicians.
3. Calculation
3.1. SNP Risk Estimation
We generated lifetime risk estimates using a multiplicative model [23,32,33], first multiplying the odds ratios (OR) of each genotype and then multiplying the product by 6%, the average CRC population risk [18-21]. With no consensus standard for combining SNP risk estimates [34], we used the multiplicative model due to 1) the strong correlation between results from alternative and multiplicative models [33], 2) GWAS evidence that increasing numbers of risk alleles are associated with greater risk [35], 3) use of the multiplicative model for estimating increased risk of common genetic variants in other cancers [32] and 4) use of this model by DTC testing companies [23].
3.2. Statistical Analyses
We computed descriptive statistics to characterize the demographics of the sample and generated means, standard deviations and frequencies of study variables. We used Pearson and Spearman correlations, t-tests and χ2-tests to examine relationships between the SNP risk scores and psychosocial, behavioral and communication variables in bivariate analyses. We used simultaneous multiple linear regression models to evaluate the independent impact of SNP test results, demographic and clinical characteristics on the psychosocial, behavioral and communication outcomes.
4. Results
Of 157 primary care patients we approached, 47 (30%) chose to participate. Primary reasons for non-participation were lack of interest or time. Study decliners did not differ from participants on age, gender or race. Participants had a mean age of 58.3 years (SD = 10.4 years; Range 40 – 84 years) and 21% had a personal history of cancer (n = 10; cancers included breast, prostate, skin, sarcoma, thyroid, endometrial, bladder and leukemia). Slightly more than one-quarter (27%) had a family history of CRC (See Table 2). All participants reported having some form of health insurance. Table 2 includes information about participant characteristics.
Table 2. Participant Characteristics and Bivariate Predictors of Selected Psychosocial, Communication and Behavioral Outcomes at 3-months.
| Variable | CRC Worry M (SD) | Genetic Testing Distress M (SD) | Intended to have CRC Screening % Yes | Positive Dietary Changes % Yes | Positive Exercise Changes % Yes | Shared Results: Family % Yes | Shared Results: Physician % Yes |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Age (M=58.3, SD=10.4yrs) | t=2.67* | t= -0.23 | X2=3.20± | X2=0.02 | X2=0.02 | X2=0.20 | X2=1.95 |
| < 50 years (n=9, 20%) | 0.11 (0.33) | 5.11 (3.10) | 44.4% | 44.4% | 37.5% | 55.6% | 20.0% |
| ≥50 years (n=3 8,80%) | 0.57 (0.78) | 5.43 (3.77) | 13.8% | 47.1% | 40.0% | 47.4% | 54.6% |
|
| |||||||
| Sex | t = -0.60 | t = -0.62 | X2=0.63 | X2=0.64 | X2=0.32 | X2=0.56 | X2=0.90 |
| Female (n=26, 56%) | 0.42 (0.65) | 5.04 (3.72) | 63.6% | 52.2% | 43.5% | 53.9% | 40.0% |
| Male (n=21, 44%) | 0.55 (0.83) | 5.75 (3.80) | 75.0% | 40.0% | 35.0% | 42.9% | 58.3% |
|
| |||||||
| Race | t = 0.17 | t = -0.04 | X2=0.03 | X2=1.71 | X2=1.21 | X2=2.24 | X2=4.49* |
| Caucassian (n=27,57%) | 0.46 (0.65) | 5.39 (3.71) | 68.0% | 38.5% | 46.2% | 59.3% | 31.3%#x00025; |
| Non-Caucasian(n=20,43%) | 0.50 (0.86) | 5.33 (3.87) | 70.6% | 58.8% | 29.4% | 36.8% | 72.7% |
|
| |||||||
| Education | t = -0.19 | t = 1.63 | X2=0.02 | X2=0.38 | X2=0.04 | X2=5.12* | X2=3.63± |
| < College (n=8, 17%) | 0.43 (0.79) | 7.43 (3.91) | 71.4% | 57.1% | 42.9% | 12.5% | 100% |
| ≥ College (n=39, 83%) | 0.49 (0.73) | 4.97 (3.62) | 68.6% | 44.4% | 38.9% | 56.4% | 41.7% |
|
| |||||||
| Annual Income (n=42)a | t = -0.57 | t = 1.87± | X2=1.59 | X2=0.53 | X2=1.10 | X2=0.94 | X2=4.80* |
| < $50,000 (n=9, 21%) | 0.38 (0.74) | 7.63 (5.01) | 50.0% | 57.1% | 25.0% | 33.3% | 100% |
| ≥ $50,000 (n=33, 79%) | 0.55 (0.77) | 4.83 (3.40) | 73.3% | 41.9% | 45.2% | 51.5% | 40.0% |
|
| |||||||
| Personal History: Cancer | t = -1.40 | t = -0.54 | X2=0.16 | X2=0.37 | X2=1.22 | X2=0.01 | X2=0.34 |
| No (n=37; 79%) | 0.40 (0.74) | 5.23 (4.10) | 67.7% | 44.1% | 35.3% | 48.7% | 45.5% |
| Yes (n=10; 21%) | 0.78 (0.67) | 5.78 (1.97) | 75.0% | 55.6% | 55.6% | 50.0% | 60.0% |
|
| |||||||
| Personal History: Polyp | t = 0.47 | t = 0.43 | X2=4.11* | X2=4.74* | X2=5.63* | X2=0.51 | X2=0.02 |
| No (n=29, 62%) | 0.52 (0.70) | 5.52 (3.96) | 57.7% | 59.3% | 53.9% | 44.8% | 47.1% |
| Yes (n=18, 38%) | 0.41 (0.80) | 5.12 (3.43) | 87.5% | 25.0% | 17.7% | 55.6% | 50.0% |
|
| |||||||
| Family History: CRC | t = -2.60* | t = -2.03* | X2=1.14 | X2=1.16 | X2=0.03 | X2=0.17 | X2=0.11 |
| No (n=34, 72%) | 0.31 (0.59) | 4.69 (3.51) | 64.5% | 51.6% | 38.7% | 47.1% | 50.0% |
| Yes (n=13, 28%) | 0.92 (0.90) | 7.17 (3.83) | 81.8% | 33.3% | 41.7% | 53.9% | 42.9% |
|
| |||||||
| BMI(M=27.2,SD=5.3) | t= -2.60** | t= -1.68* | X2=0.50 | X2=0.97 | X2=0.60 | X2=1.27 | X2=0.11 |
| < 25 (n=16, 34%) | 0.14 (0.36) | 6.71 (4.41) | 61.5% | 35.7% | 30.8% | 37.5% | 42.9% |
| > 25 (n=31, 66%) | 0.63 (0.81) | 4.73 (3.26) | 72.4% | 51.7% | 43.3% | 54.8% | 50.0% |
|
| |||||||
| SNP Score (n = 45 tested) | t = -0.87 | t = 1.24 | X2=1.70 | X2=4.48* | X2=0.89 | X2=0.20 | X2=0.01 |
| Low (> 12; n=36; 80%) | 0.43 (0.70) | 5.71 (3.97) | 73.5% | 38.2% | 42.9% | 47.4% | 47.6% |
| High (< 12; n=9; 20%) | 0.67 (0.87) | 4.00 (2.23) | 50.0% | 77.8% | 25.0% | 55.6% | 50.0% |
Note: CRC = Colorectal Cancer.
5 participants chose not to answer this question. Results for 3-month outcomes presented as means and standard deviations. BMI = Body Mass Index.
p < .10,
p < .05.
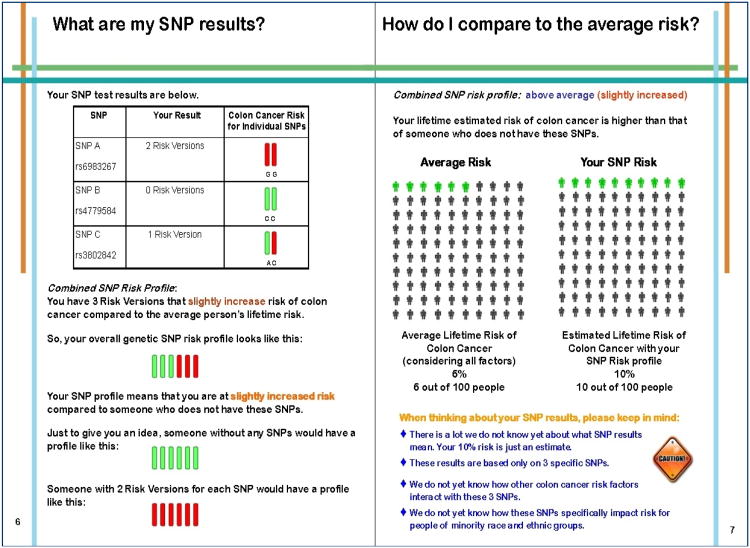
Forty-five of the 47 participants (96%) opted for SNP testing after a genomics education session with a certified genetic counselor. Participants averaged 2.5 of 6 possible SNP risk alleles with an estimated 10% lifetime risk (SD=2.3%, Sample Range=6.0% to 15.0%; Possible Range = 6% to 23%). Twenty percent of the sample had a risk at or above 12% (twice average risk). Table 1 presents the allele frequencies for each of the three SNPs in both our sample and population estimates from dbSNP [36].
4.1. Psychosocial Outcomes
Immediately post-test, SNP risk scores were unrelated to perceived CRC risk or CRC worry. At 3-months post-test, bivariate analyses identified relationships between numeric perceived CRC risk and SNP risk scores (r = .22, p = .06), family history of CRC [Satterthwaite t (df = 13.5) = -2.68, p = .02], personal history of cancer [t (40) = -2.60, p = .01] and baseline numeric perceived risk (r = .76, p < .0001). Perceived risk scores by personal cancer history and SNP risk score categories are shown in Table 3. Age, gender and race were not related to perceived CRC risk 3-months post-test. Bivariate associations between SNP risk score categories (high, meaning twice average risk, vs. low) and study outcomes are presented in Table 2. In multivariate analyses, higher SNP risk scores no longer remained an independent significant predictor of higher numeric perceived CRC risk at 3-months (β = .15, p = .08). Baseline pre-education numeric perceived CRC risk and both personal history of cancer and family history of CRC contributed significantly to the final model of perceived numeric risk at 3-months post-results, F (4,36) = 23.8, p < .001, R2 = .70. SNP risk scores were not related to absolute or comparative perceived risk, CRC worry or genetic-testing distress at the post-test or 3-month assessments. Overall, using our adapted measure, participants reported very low levels of distress related to genomic testing (M = 5.4, SD = 3.8; Possible Score Range 0 – 16) [28].
Table 3. Perceived Numeric Risk of Colorectal Cancer Before and After Genomic Risk Education and Testing.
| Baseline | Post-Education | Post-Test | 3-Month Follow-up | |
|---|---|---|---|---|
| M (SD) Possible Range = 0 – 100 | ||||
|
| ||||
| Overall Sample (n = 45) | 32.45 (25.0) | 23.24 (21.9) | 23.7 (24.7) | 24.1 (25.1) |
| Personal Cancer History | ||||
| Yes (n = 8)a | 43.1 (25.2) | 39.9 (23.0) | 43.3 (21.9) | 43.0 (32.2) |
| Ca Hx and High SNP Score (n=2) | 50.0 (0) | 50.0 (0) | 35.0 (21.2) | 50.0 (14.1) |
| Ca Hx and Low SNP Score (n=6) | 40.8 (29.4) | 36.5 (26.2) | 46.6 (23.6) | 40.7 (37.2) |
| No (n = 35) | 27.8 (24.2) | 18.3 (18.4) | 19.8 (22.9) | 19.1 (21.0) |
| No Ca Hx and High SNP Score (n=7) | 28.0 (23.9) | 23.0 (25.3) | 27.1 (24.0) | 25.2 (21.5) |
| No Ca Hx and Low SNP Score (n=28) | 27.8 (24.8) | 16.7 (16.2) | 17.7 (22.6) | 17.8 (21.1) |
| Family History of CRC | ||||
| Yes (n = 13) | ||||
| Family Hx and High SNP Score (n = 1) | 10.0 | 8.0 | 20.0 | 40.0 |
| Family Hx and Low SNP Score (n = 12) | 41.3 (27.9) | 32.8 (26.1) | 38.3 (28.9) | 42.6 (33.8) |
| No (n =31)b | ||||
| No Family Hx and High SNP Score (n=8) | 38.3 (21.4) | 32.8 (25.0) | 30.0 (23.7) | 30.1 (23.6) |
| No Family Hx and Low SNP Score (n=23) | 23.4 (22.5) | 17.9 (17.6) | 15.8 (20.1) | 11.9 (11.8) |
Note: Possible Score Range on Perceived Numeric Risk is 0 (Definitely won't get CRC) to100 (Definitely will get CRC). Ca Hx = Cancer History. High SNP Score = ≥ 12% Lifetime CRC risk estimate. Low SNP Score = >12% Lifetime CRC risk estimate.
Missing data for one participant with a personal history of cancer and another participant with cancer opted not to puruse genomic testingve.
Missing numeric perceived risk data from one person without a family history of disease.
4.2. Behavioral Outcomes
Most participants (89%) were currently adherent to CRC screening guidelines. Of the remaining 11% of participants (n = 5) who were non-adherent at baseline, four reported intentions to screen immediately post-test. None of these individuals had obtained screening by the 3-month follow-up, although three participants maintained intentions to screen within the next couple of years.
Immediately post-test, about half of the sample reported plans to improve physical activity (64%) and nutrition (48%). At the 3-month follow-up, 56% and 55% reported actual changes to exercise and eating behaviors, respectively, since the second genomic education session and receipt of SNP results. Reported changes in eating behavior included reduced red meat and increased vegetable and fish consumption. Reported changes in exercise behavior included increased walking or increased frequency of physical activity. In bivariate analyses, SNP risk score was related to participants' reported engagement in moderate exercise at 3-months (r = .48, p < .001). In multivariate logistic regression analyses, SNP risk score was not an independent predictor of changes in exercise at 3-months (OR = 1.43; 95% CI = 0.97 – 2.10) in a regression model that also included baseline levels of exercise behavior, personal history of having a polyp, personal history of cancer, family history of CRC, and body mass index. Only not having a personal history of a colon polyp was statistically significantly associated with reported changes in exercise at 3-months (OR = 10.4; 95% CI = 1.5 – 74.1). SNP risk scores were not related to reported changes in eating behavior.
Only 2 participants reported smoking at baseline; immediately post-test, both participants reported intentions of quitting smoking within the next 6 months. We did not assess intentions to quit smoking at the 3-month follow-up. No consistent changes were noted for alcohol consumption.
4.3. Communication Outcomes
By the 3-month follow-up, more than half of participants (64%) reported talking about their SNP results with other people. Among participants who shared their SNP results, 68% talked with a spouse/significant other, 42% with children, 32% with a sister, 20% with a brother, and 13% with parents. Among participants who reported at the 3-month follow up that they had an appointment with their primary care physician since receipt of the SNP test results (n=26), 50% shared results with their physician, representing 28% of all study participants. More African American participants reported sharing SNP results with their physicians than Caucasian participants, χ 2 = 4.5, p = .009. Sharing of SNP results was not related to SNP risk score, cancer worry or CRC perceived risk. In bivariate analyses, only family history of CRC was related to sharing of results with brothers (r = .56, p = .007), although this relationship was not evident for the sharing of results to any other family members or physicians and was not significant in multivariate analyses.
5. Discussion
For genomic discoveries to improve public health, clinical and translational research must begin to apply genomic risk information in ways that evaluate a range of outcomes [15]. A major goal of translational research is to establish whether genomic risk information has utility in the area of disease prevention [16]. We conducted an innovative pilot study to prospectively evaluate psychosocial, behavioral and communication outcomes following genomic risk education and testing of a research panel of three SNPs related to CRC risk. Study participants had a high rate of SNP-test uptake, reported increases in physical activity and healthy eating following risk education, and had moderate rates of disclosure of results to family and physicians. We found no evidence that the receipt of genomic risk information increased cancer worry or distress related to testing. To our knowledge, this is the first study to report a range of outcomes following genomics education and testing for CRC risk among a diverse sample of primary care patients.
The small but growing literature on outcomes following genomic testing for risk related to common, complex diseases such as cancer suggests that people are interested in obtaining this type of information, even when the risks, benefits and uncertainty of the information are carefully explained [13,18]. Approximately one-third of the individuals approached to participate in the study agreed and among study participants, almost all of them opted for genomic testing. The high rate of test uptake among study participants is consistent with recent research related to interest in genomic testing [37,38]. These high rates are likely because individuals who agree to participate in genetics and genomics research are likely interested in learning more about genomic risk. We were unable to examine predictors of test uptake as only two participants declined testing.
We found that participants in our study had an average lifetime CRC risk of 10%. Moreover, over one-quarter of our sample had a family history of CRC and one-fifth of participants were themselves cancer survivors (although no participants had a history of CRC). These factors indicate that our sample includes individuals at increased risk of CRC. Personal and family history of disease may lead to greater salience of genomic risk information [7]. Our study findings that SNP risk estimates were unrelated to reports of behavior change provide information about how people interpret and use different types of risk information. In the current study, the mechanism of change for improvements in self-reported health behaviors is likely the risk education provided by certified genetic counselors rather than the SNP results themselves. For example, genetic counselors emphasized how individual risk factors, such as high body mass index, can impact future risk. The present results that participants with a personal history of colon polyps reported positive changes in exercise behavior at 3-months controlling for other factors appear to support the impact of the risk education on later health behaviors.
Results from this pilot study suggest that participants who agree to a study about genomic risk for CRC do not experience increased distress or cancer worry. These findings are consistent with other studies indicating that genomic testing for susceptibility alleles does not lead to significant emotional concerns [6]. Likewise, these findings are not surprising given that the receipt of genetic test results for much higher penetrance genes such as BRCA1/BRCA2 does not appear to cause substantially increased distress over time [39].
Our findings that SNP risk scores were not independently associated with numeric perceived risk contrasts with earlier literature on genomic feedback and smoking behavior which indicates that genomic information impacts perceived risk of lung cancer [1]. Recent results from vignette-based research indicate that higher hypothetical SNP-based risk translated into higher perceptions of disease risk [40]. No changes in perceived risk related to SNP risk scores may be a reflection of the genetic counselors' emphasis on the lack of clinical utility of SNP risk information. We used several visual, text-based and graphical representations of genomic risk [41]. An important area of future study will be to investigate ways to combine other risk factors (e.g., lifestyle, family or personal history) and genomic risk information into a single metric to appropriately communicate more complete risk information. Relevant to perceived risk, we elected to include only SNPs that increased risk as part of our research SNP-panel and thus no participants received risk scores below average risk.
Limited evidence to date supports behavior change following genomic risk testing. Our approach combining education about lifestyle, family history and genomic risk factors yielded modest self-reported behavior change at 3-months post-test. Reduced red meat consumption, increased fish and vegetable intake and increased walking and other exercise are behavior changes that relate to CRC risk reduction [42]. Behavior change was evident despite the lack of clinical utility of the SNP results and the lack of impact of SNP risk itself on behavior change. These findings suggest that 1) risk education sessions with certified genetic counselors that discussed personal, family history and lifestyle risk feedback increased the salience of improving certain behaviors for CRC risk reduction and/or 2) more definitive genomic risk information, including higher levels of increased risk or less uncertainty surrounding the meaning of SNP-risk information, might be needed for a stronger catalyst of behavior change. As the genomic risk information was presented together with individual and family history risk feedback, the present results do not resolve whether genomic risk information alone has personal utility for people. These data suggest that it may be crucial to identify intervention approaches for leveraging the apparent motivational increase of risk feedback and to test the impact of such feedback when it is given with and without genomic risk information. A test of risk feedback with and without genomic risk information would provide evidence of whether genomic risk information has clinical utility in terms of sustained behavior change [38]. Future research also will need to confirm if participants' self-reported health behavior change reflects actual sustained changes using objective behavioral and health outcomes measures (e.g., weight loss, improved fitness). Our initial report provides preliminary evidence suggesting that CRC risk education may lead to intentions and actual efforts to modify behavior, separate from the level of genomic risk. This is consistent with research on risk feedback related to tobacco, in which there is evidence for increased quit attempts but more limited evidence for long-term behavior change.
Beyond the reported changes in physical activity and dietary behavior, our results contribute to the literature as among the first studies to report on communication of results following genomic testing for a common complex disease. Prior research in this area has explored whether early adopters of DTC genomic testing shared results with healthcare providers [7]. Our finding that the sharing of SNP results was more common among African American participants compared to Caucasian participants is interesting. Perhaps this increased sharing of results is due to greater salience of CRC given the higher rate of CRC incidence and mortality in African Americans [43]. Future work can explore the possible reasons for this differential rate of disclosure by race. Our finding that almost two-thirds of the sample discussed their SNP results with family or a health care provider suggests additional avenues for research. First, future studies can explore the specific topics related to genomic risk information that people share, including whether the information impacts the perceived risk of family members. Second, investigators can examine how genomic risk information is received by physicians and whether this information influences clinical care [44]. Participants appeared to discuss their SNP results regardless of their SNP risk score, suggesting that level of genomic risk may not be related to result communication.
A number of caveats should be considered when interpreting study findings. First, this was a pilot study with a small sample size of which 20% had been previously affected with cancer. Our sample may have been more open to the receipt of information about cancer risks and may also have been more familiar with the concepts of genetic or genomic risk for cancer. These biases could limit the generalizability of results to other samples. Likewise, with 45 individuals who tested, we were underpowered to detect small to moderate relationships (e.g., effect sizes below 0.36) between SNP risk scores and our study outcomes. Despite the small size, our sample was diverse in terms of participants' self-reported race. Although we captured general reasons for non-participation among study decliners, we did not specifically ask if patients declined because they were not interested in genomic testing. Our recruitment from a primary care clinic extends prior work that has largely focused on early-adopters of DTC genomic testing or individuals from a managed care organization [10,45]. Clinic-based recruitment also likely influenced the high baseline rate of adherence to CRC screening guidelines (89%) in our sample, a finding which could reflect a high level of engagement with health care and interest in health-related information such as genomic risk for CRC. Second, we offered free testing and education sessions with a certified genetic counselor within the context of the study, likely influencing the rate of test uptake. Our inclusion of a genetics professional for pre- and post-test education makes the present findings different from most DTC genomic-testing models [44] and may have contributed to the lower rates of worry and perhaps greater understanding of the limited utility of SNP-based genomic information. Third, behavior change and communication of results were assessed through participant self-report and thus subject to the potential biases associated with self-reports of certain health behavior [46,47]. Future research can include more objective measures of these outcomes. Finally, we did not adjust the lifetime CRC risks by participants' age when providing the SNP risk information. For example, an individual who is 65 years old with an identical SNP profile to a 45 year old would have a lower lifetime risk of CRC. In our study, the genetic counselors discussed this issue, but we did not provide an age-adjusted quantitative risk estimate. Adjusting genomic risk estimates by age or providing risk estimates for a defined interval is straightforward and should be included in future work.
6. Conclusions
Our study is among the first to explore the impact of genomic risk information on a range of outcomes in a sample of primary care patients. Exploring ways to appropriately translate genomic discoveries into clinical applications and preventive health interventions will accelerate our ability to improve the clinical and personal utility of this information.
Highlights.
We conduct a genomic testing pilot study about colorectal cancer risk.
We examine psychosocial, behavioral and communication outcomes.
Participants report modest behavior change without increased distress.
Many participants communicated genomic results with their family and physicians.
Results highlight potential use of genomic risk education and testing.
Acknowledgments
Thank you to Clinton Finch for his time and assistance with creating the study database system. We also thank the participants who provided their time and shared their experiences with us. Finally, our appreciation goes to Ms. Susan Marx for her assistance with manuscript preparation.
Grant Support: This project was supported by funding through NCI K07CA131172-S2 (KG), the Fisher Center for Familial Cancer Research and NCI P30CA051008, which partially supports The Genomics and Epigenomics Shared Resource at Lombardi Comprehensive Cancer Center. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lerman C, Gold K, Audrain J, Lin TH, Boyd NR, Orleans CT, Wilfond B, Louben G, Caporaso N. Incorporating biomarkers of exposure and genetic susceptibility into smoking cessation treatment: effects on smoking-related cognitions, emotions, and behavior change. Health Psychol. 1997;16:87–99. doi: 10.1037//0278-6133.16.1.87. [DOI] [PubMed] [Google Scholar]
- 2.McBride CM, Bepler G, Lipkus IM, Lyna P, Samsa G, Albright J, Datta S, Rimer BK. Incorporating genetic susceptibility feedback into a smoking cessation program for African-American smokers with low income. Cancer Epidemiol Biomarkers Prev. 2002;11:521–528. [PubMed] [Google Scholar]
- 3.Audrain J, Boyd NR, Roth J, Main D, Caporaso NF, Lerman C. Genetic susceptibility testing in smoking-cessation treatment: one-year outcomes of a randomized trial. Addict Behav. 1997;22:741–751. doi: 10.1016/s0306-4603(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter MJ, Strange C, Jones Y, Dickson MR, Carter C, Moseley MA, Gilbert GE. Does genetic testing result in behavioral health change? Changes in smoking behavior following testing for alpha-1 antitrypsin deficiency. Ann Behav Med. 2007;33:22–28. doi: 10.1207/s15324796abm3301_3. [DOI] [PubMed] [Google Scholar]
- 5.Ito H, Matsuo K, Wakai K, Saito T, Kumimoto H, Okuma K, Tajima K, Hamajima N. An intervention study of smoking cessation with feedback on genetic cancer susceptibility in Japan. Prev Med. 2006;42:102–108. doi: 10.1016/j.ypmed.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Gordon ES, Griffin G, Wawak L, Pang H, Gollust SE, Bernhardt BA. “It's not like judgment day”: public understanding of and reactions to personalized genomic risk information. J Genet Couns. 2012;21:423–432. doi: 10.1007/s10897-011-9476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufman DJ, Bollinger JM, Dvoskin RL, Scott JA. Risky business: risk perception and the use of medical services among customers of DTC personal genetic testing. J Genet Couns. 2012;21:413–422. doi: 10.1007/s10897-012-9483-0. [DOI] [PubMed] [Google Scholar]
- 8.Marteau TM, French DP, Griffin SJ, Prevost AT, Sutton S, Watkinson C, Attwood S, Hollands GJ. Effects of communicating DNA-based disease risk estimates on risk-reducing behaviours. Cochrane Database Syst Rev. 2010:CD007275. doi: 10.1002/14651858.CD007275.pub2. [DOI] [PubMed] [Google Scholar]
- 9.Coriell Personalized Medicine Collaborative. [Accessed January 26, 2012]; Available at http://cpmc.coriell.org/
- 10.National Human Genome Research Institute and National Institutes of Health. The Multiplex Initiative. [Accessed November 15, 2012]; Available at http://www.genome.gov/27539449.
- 11.Kaphingst KA, McBride CM, Wade C, Alford SH, Reid R, Larson E, Baxevanis AD, Brody LC. Patients' understanding of and responses to multiplex genetic susceptibility test results. Genet Med. 2012;14:681–687. doi: 10.1038/gim.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts JS, Christensen KD, Green RC. Using Alzheimer's disease as a model for genetic risk disclosure: implications for personal genomics. Clin Genet. 2011;80:407–414. doi: 10.1111/j.1399-0004.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride CM, Guttmacher AE. Commentary: trailblazing a research agenda at the interface of pediatrics and genomic discovery--a commentary on the psychological aspects of genomics and child health. J Pediatr Psychol. 2009;34:662–664. doi: 10.1093/jpepsy/jsn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green ED, Guyer MS. National Human Genome Research Institute, Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 16.Schully SD, Benedicto CB, Gillanders EM, Wang SS, Khoury MJ. Translational research in cancer genetics: the road less traveled, Public Health Genomics. 2011;14:1–8. doi: 10.1159/000272897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Cancer Institute. Variant GPS. SNP500Cancer. [Accessed November 30, 2012]; Available at http://variantgps.nci.nih.gov/cgfseq/pages/snp500.do.
- 18.Nusbaum R, Leventhal KG, Hooker GW, Peshkin BN, Butrick M, Salehizadeh Y, Tuong W, Eggly S, Mathew J, Goerlitz D, Shields PG, Schwartz MD, Graves KD. Translational genomic research: protocol development and initial outcomes following SNP testing for colon cancer risk. Transl Behav Med. 2013;3:17–29. doi: 10.1007/s13142-012-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, CORGI Consortium. Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 20.Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, Walther A, Spain S, Pittman A, Kemp Z, Sullivan K, Heinimann K, Lubbe S, Domingo E, Barclay E, Martin L, Gorman M, Chandler I, Vijayakrishnan J, Wood W, Papaemmanuil E, Penegar S, Qureshi M, CORGI Consortium. Farrington S, Tenesa A, Cazier JB, Kerr R, Gray J, Peto J, Dunlop M, Campbell H, Thomas H, Houlston R, Tomlinson I. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 21.Pittman AM, Webb E, Carvajal-Carmona L, Howarth K, Di Bernardo MC, Broderick P, Spain S, Walther A, Price A, Sullivan K, Twiss P, Fielding S, Rowan A, Jaeger E, Vijayakrishnan J, Chandler I, Penegar S, Qureshi M, Lubbe S, Domingo E, Kemp Z, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop T, Gray R, Maher ER, Lucassen A, Kerr D, Evans GR, CORGI Consortium. van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, EPICOLON Consortium. Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la HM, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen LA, Cazier JB, Tomlinson IP, Houlston RS. Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum Mol Genet. 2008;17:3720–3727. doi: 10.1093/hmg/ddn267. [DOI] [PubMed] [Google Scholar]
- 22.23andMe. Colorectal Cancer. [Accessed December 1, 2011]; Available at https://www.23andme.com/health/Colorectal-Cancer/
- 23. [Accessed December 1, 2011];deCODE genetics, deCODEme risk calculations. Available at https://www.decodeme.com/health-watch-information/risk-calculation.
- 24.Navigenics. [Accessed March 7, 2013]; Available at http://www.navigenics.com/
- 25.Lipkus IM, Kuchibhatla M, McBride CM, Bosworth HB, Pollak KI, Siegler IC, Rimer BK. Relationships among breast cancer perceived absolute risk, comparative risk, and worries. Cancer Epidemiol Biomarkers Prev. 2000;9:973–975. [PubMed] [Google Scholar]
- 26.Kelly KM, Graves KD, Harper FW, Schmidt JE, Dickinson SL, Andrykowski MA. Assessing perceptions of cancer risk: does mode of assessment or numeracy matter? Cancer Detect Prev. 2007;31:465–473. doi: 10.1016/j.cdp.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Lerman C, Trock B, Rimer BK, Boyce A, Jepson C, Engstrom PF. Psychological and behavioral implications of abnormal mammograms. Ann Intern Med. 1991;114:657–661. doi: 10.7326/0003-4819-114-8-657. [DOI] [PubMed] [Google Scholar]
- 28.Cella D, Hughes C, Peterman A, Chang CH, Peshkin BN, Schwartz MD, Wenzel L, Lemke A, Marcus AC, Lerman C. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21:564–572. [PubMed] [Google Scholar]
- 29.Blair SN, Haskell WL, Ho P, Paffenbarger RS, Jr, Vranizan KM, Farquhar JW, Wood PD. Assessment of habitual physical activity by a seven-day recall in a community survey and controlled experiments. Am J Epidemiol. 1985;122:794–804. doi: 10.1093/oxfordjournals.aje.a114163. [DOI] [PubMed] [Google Scholar]
- 30.Block G, Gillespie C, Rosenbaum EH, Jenson C. A rapid food screener to assess fat and fruit and vegetable intake. Am J Prev Med. 2000;18:284–288. doi: 10.1016/s0749-3797(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 31.McQueen A, Vernon SW, Myers RE, Watts BG, Lee ES, Tilley BC. Correlates and predictors of colorectal cancer screening among male automotive workers. Cancer Epidemiol Biomarkers Prev. 2007;16:500–509. doi: 10.1158/1055-9965.EPI-06-0757. [DOI] [PubMed] [Google Scholar]
- 32.Sawyer S, Mitchell G, McKinley J, Chenevix-Trench G, Beesley J, Chen XQ, Bowtell D, Trainer AH, Harris M, Lindeman GJ, James PA. A role for common genomic variants in the assessment of familial breast cancer. J Clin Oncol. 2012;30:4330–4336. doi: 10.1200/JCO.2012.41.7469. [DOI] [PubMed] [Google Scholar]
- 33.Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, Zeggini E, Rayner NW, Shields B, Owen KR, Hattersley AT, Frayling TM. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med. 2006;3:e374. doi: 10.1371/journal.pmed.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson ME, Storm CD, Weitzel J, Wollins DS, Offit K. American Society of Clinical Oncology, American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2010;28:893–901. doi: 10.1200/JCO.2009.27.0660. [DOI] [PubMed] [Google Scholar]
- 35.Kupfer SS, Anderson JR, Hooker S, Skol A, Kittles RA, Keku TO, Sandler RS, Ellis NA. Genetic heterogeneity in colorectal cancer associations between African and European Americans. Gastroenterology. 2010;139:1677–85. 1685. doi: 10.1053/j.gastro.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Center for Biotechnology Information. dbSNP short genetic variations. [Accessed November 30, 2012]; Available at http://www.ncbi.nlm.nih.gov/SNP/
- 37.Gollust SE, Gordon ES, Zayac C, Griffin G, Christman MF, Pyeritz RE, Wawak L, Bernhardt BA. Motivations and perceptions of early adopters of personalized genomics: perspectives from research participants. Public Health Genomics. 2012;15:22–30. doi: 10.1159/000327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant RW, Meigs JB, Florez JC, Park ER, Green RC, Waxler JL, Delahanty LM, O'Brien KE. Design of a randomized trial of diabetes genetic risk testing to motivate behavior change: the Genetic Counseling/lifestyle Change (GC/LC) Study for Diabetes Prevention. Clin Trials. 2011;8:609–615. doi: 10.1177/1740774511414159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graves KD, Vegella P, Poggi EA, Peshkin BN, Tong A, Isaacs C, Finch C, Kelly S, Taylor KL, Luta G, Schwartz MD. Long-term psychosocial outcomes of BRCA1/BRCA2 testing: differences across affected status and risk-reducing surgery choice. Cancer Epidemiol Biomarkers Prev. 2012;21:445–455. doi: 10.1158/1055-9965.EPI-11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graves KD, Peshkin BN, Luta G, Tuong W, Schwartz MD. Interest in genetic testing for modest changes in breast cancer risk: implications for SNP testing. Public Health Genomics. 2011;14:178–189. doi: 10.1159/000324703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han PK, Klein WM, Killam B, Lehman T, Massett H, Freedman AN. Representing randomness in the communication of individualized cancer risk estimates: effects on cancer risk perceptions, worry, and subjective uncertainty about risk. Patient Educ Couns. 2012;86:106–113. doi: 10.1016/j.pec.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harriss DJ, Atkinson G, George K, Cable NT, Reilly T, Haboubi N, Zwahlen M, Egger M, Renehan AG CLEAR group. Lifestyle factors and colorectal cancer risk (1): systematic review and meta-analysis of associations with body mass index. Colorectal Dis. 2009;11:547–563. doi: 10.1111/j.1463-1318.2009.01766.x. [DOI] [PubMed] [Google Scholar]
- 43.American Cancer Society. Colorectal Cancer Facts & Figures 2011-2013. Atlanta: American Cancer Society; 2011. [Accessed November 27, 2012]. Available at http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-028323.pdf. [Google Scholar]
- 44.Bellcross CA, Page PZ, Meaney-Delman D. Direct-to-consumer personal genome testing and cancer risk prediction. Cancer J. 2012;18:293–302. doi: 10.1097/PPO.0b013e3182610e38. [DOI] [PubMed] [Google Scholar]
- 45.Bloss CS, Darst BF, Topol EJ, Schork NJ. Direct-to-consumer personalized genomic testing. Hum Mol Genet. 2011;20:R132–R141. doi: 10.1093/hmg/ddr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natarajan L, Pu M, Fan J, Levine RA, Patterson RE, Thomson CA, Rock CL, Pierce JP. Measurement error of dietary self-report in intervention trials. Am J Epidemiol. 2010;172:819–827. doi: 10.1093/aje/kwq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warner ET, Wolin KY, Duncan DT, Heil DP, Askew S, Bennett GG. Differential accuracy of physical activity self-report by body mass index. Am J Health Behav. 2012;36:168–178. doi: 10.5993/AJHB.36.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, Edlund CK, Haile RW, Gallinger S, Zanke BW, Lemire M, Rangrej J, Vijayaraghavan R, Chan AT, Hazra A, Hunter DJ, Ma J, Fuchs CS, Giovannucci EL, Kraft P, Liu Y, Chen L, Jiao S, Makar KW, Taverna D, Gruber SB, Rennert G, Moreno V, Ulrich CM, Woods MO, Green RC, Parfrey PS, Prentice RL, Kooperberg C, Jackson RD, Lacroix AZ, Caan BJ, Hayes RB, Berndt SI, Chanock SJ, Schoen RE, Chang-Claude J, Hoffmeister M, Brenner H, Frank B, Bezieau S, Kury S, Slattery ML, Hopper JL, Jenkins MA, Le ML, Lindor NM, Newcomb PA, Seminara D, Hudson TJ, Duggan DJ, Potter JD, Casey G. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131:217–234. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]