Abstract
The neural crest is a population of mesenchymal cells that after migrating from the neural tube give rise to a structures and cell-types: jaw, part of the peripheral ganglia and melanocytes. Although much is known about neural crest development in jawed vertebrates, a clear picture of trunk neural crest development for elasmobranchs is yet to be developed. Here we present a detailed study of trunk neural crest development in the bamboo shark, Chiloscyllium punctatum. Vital labeling with DiI and in situ hybridization using cloned Sox8 and Sox9 probes demonstrated that trunk neural crest cells follow a pattern similar to the migratory paths already described in zebrafish and amphibians. We found shark trunk neural crest along the rostral side of the somites, the ventromedial pathway, branchial arches, gut, sensory ganglia and nerves. Interestingly, Chiloscyllium punctatum Sox8 and Sox9 sequences aligned with vertebrate SoxE genes, but appeared to be more ancient than the corresponding vertebrate paralogs. The expression of these two SoxE genes in trunk neural crest cells, especially Sox9, matched the Sox10 migratory patterns observed in teleosts. Interestingly, we observed DiI cells and Sox9 labeling along the lateral line, suggesting that in C. punctatum, glial cells in the lateral line are likely of neural crest origin. Though this has been observed in other vertebrates, we are the first to show that the pattern is present in cartilaginous fishes. These findings demonstrate that trunk neural crest cell development in Chiloscyllium punctatum follows the same highly conserved migratory pattern observed in jawed vertebrates
Keywords: shark embryo, sox8, sox9, neural crest, Chiloscyllium punctatum
Introduction
Neural crest cells in vertebrates are a population of cells which originate in the dorsal neural tube, but delaminate and migrate throughout the embryo to give rise to a significant portion of the peripheral nervous system, consisting of neurons and glia, craniofacial structures and even endocrine organs (Northcutt and Gans, 1983). Neural crest cells give rise to a significant portion of the peripheral nervous system, consisting of neurons and glia, craniofacial structures and even endocrine organs (Baker, 2005; Le Douarin et al., 2007). Because they are such a versatile group of cells, neural crest cells are involved in critical embryonic “breakthroughs” in vertebrate embryogenesis such as jaw and peripheral sensory ganglia formation (Gans and Northcutt, 1983; Northcutt, 2005). Understanding their diversity is critical for a better understanding of the key components in vertebrate evolution (Baker and Schlosser, 2005; Holland et al., 2008; Sauka-Spengler and Bronner-Fraser, 2008a).
The cartilaginous fishes (chondrichthyans) belong to one of two extant lineages of gnathostomes (jawed vertebrates). The bamboo shark, Chiloscyllium punctatum, stems from the basal lineage of gnathostomes and offers insight into the evolution of the ancestral condition for gnathostomes. The comparison between their developmental genetic mechanisms and those in the sister lineage, osteichthyans (bony fishes) can also provide essential insight into the developmental events that progressed into the common ancestor of jawed vertebrates (Gans and Northcutt, 1983; Northcutt, 2005). The common ancestor of cartilaginous and bony fishes lived some 400 million years ago (Cole and Currie, 2007).
In the 1800s, chondrichthyans held a pre-eminent position in the field of comparative embryology. However, in the 20th century chondrichthyans were replaced by model organisms; frogs, mice and zebrafish. These model organisms sustained life better in laboratory environments and responded better to genetic manipulations (Balfour, 1876; Cole and Currie, 2007). Although scientists have concluded that chondrichthyans have trunk neural crest cells due to the appearance of dorsal root ganglia and melanocytes, most of the past research focused on the development of the head and placodes, where little is known about trunk neural crest (Adachi et al., 2012; Freitas et al., 2006; Gillis et al., 2012; Horigome et al., 1999; Kuratani, 2005; Kuratani and Horigome, 2000; Ota et al., 2007). From these studies, we learned the migratory pathways of cranial neural crest cells and lateral line placode are highly conserved among amniotes but none have been studied in the development of trunk neural crest cells.
Trunk neural crest cells have been studied extensively in more recent amniotes, especially in avian and mammals (Kulesa and Gammill, 2010; Le Douarin, 2004; Minoux and Rijli, 2010; Sauka-Spengler and Bronner-Fraser, 2008a). Their migratory behavior has been divided into two main pathways: 1) ventromedial along the rostral portion of the somites where these cells will give rise to sensory ganglia and other tissues, 2) dorsolateral between the somites and ectoderm for the cells that will make future melanocytes.
In the last 20 years, the field of evolution and development exploded with new techniques attributed to orthologous gene identification and in situ hybridization. However, there have been few molecular studies on the early development of the nervous system in sharks, specifically research focused on the expression of some key, orthologous, transcription factors such as Otx (Sauka-Spengler et al., 2001), Pax, NeuroD and Phox2B (Derobert et al., 2002; O'Neill et al., 2007) and FoxD (Wotton et al., 2008). These studies demonstrated a majority of sharks show similar patterns in the formation of their nervous systems and are highly conserved among agnathans and other gnathostomes. Furthermore, the roles of HMG domain transcription factors in brain regionalization are highly conserved across vertebrate evolution (Derobert et al., 2002). SoxE are among the best described group of transcription factors because their critical role in glia development but also in neural crest development. The SoxE family members (Sox8, Sox9 and Sox10) expression patterns have been well studied across teleosts (Cheng et al., 2000; Dutton et al., 2001; Kim et al., 2003). However, although Sox8 has been cloned in spotted catshark Scyliorhinus canicula (Freitas et al., 2006), we still do not know if chondrichthyes neural crest cells express SoxE genes as is known for teleosts (Cheng et al., 2000; Freitas et al., 2006; Kuhlbrodt et al., 1998; Lakiza et al., 2011).
In order to characterize in detail the development of elasmobranch trunk neural crest, in particular in shark, we took advantage of vital labeling techniques of neural crest progenitor cells as well as molecular biology tools to clone neural crest SoxE transcription factors. Vital labeling with fluorescent dyes has been a preferred approach for years when studying the migration of neural crest cells during their development (Kulesa and Fraser, 1998; Serbedzija et al., 1992). This method has been used successfully in fish, amphibians, lampreys (Collazo et al., 1993; Epperlein et al., 2007; McCauley and Bronner-Fraser, 2003; Raible and Eisen, 1994 2) and more recently in snakes (Reyes et al., 2010). However, although it has been recently successfully used to follow lateral line development in the little skate, Leucoraja erinacea (Gillis et al., 2012), it has never been used in shark embryos to look at neural crest. Here we show for the first time the migration pattern of trunk neural crest cells in the bamboo shark Chiloscyllium punctatum by observing DiI cells and Sox8 and Sox9 expression patterns.
Materials and methods
Collection and Staging of Embryos
The Bamboo shark, Chiloscyllium punctatum,(Müller and Henle, 1838) egg cases, or mermaid's purses were harvested from the Long Beach Aquarium kindly provided by Chris Plante, reared at 25°C in sea water, and collected at different developmental stages. Embryos were removed from egg cases and staged according to Ballard's developmental table (Ballard, 1993) when feasible, or by their length in cm. The youngest embryos collected were stage 23, 3cm long, with most of their forebrain in full development. The oldest embryos were 10cm long, a stage at which they showed much physical activity and looked similar to their adult counterparts. Embryos were either fixed in Carnoy's solution (70% ethanol, 20% formaldehyde and 10% glacial acetic acid) or in 4% paraformaldehyde overnight at 4°C and kept in 70% ethanol at -20°C until histological preparation. For sufficient paraffin penetration, embryos needed extensive dehydration steps (about 1 day per alcohol grade) and two full days in histosol for clearing. The tissues were then immersed in hot paraffin (McCormick Scientific Paraplast Plus) and placed in a vacuum oven for two days before preparing the blocks and sectioning. Embryos were sectioned (7-12μm) with a microtome, collected on Super-Frost slides and dried overnight at 37°C on a slide warmer. DiI injected embryos were cryo-protected in 15% sucrose, 30% sucrose overnight, then embedded in gelatin for 3hrs at 38°C slowly freezing in liquid nitrogen and sectioned at 12μm. These experiments with Chiloscyllium punctatum sharks were approved by the Institutional Animal Care and Use Committee at CSUN.
DiI vital labeling
For live labeling, stage 23-29 shark embryos were partially immobilized with tricaine and injected with DiI (cell tracker CM-DiI, C-7001, Invitrogen/Molecular Probes) and diluted in ethanol (1/10) and 10% sucrose. Vital labeling was performed by injecting the DiI from the hindbrain region until the DiI reached the tail end. Embryos were placed in a Petri dish after a thorough rinsing in sterile seawater and incubated with 5ml of DMEM, 10% FBS, penicillin and streptomycin at 37°C for 12hrs or by placing the egg cases with the labeled embryos in a humidified chamber overnight at 25°C. At total of 10 embryos survived and were fixed first by immersing the case in PFA for 1hr after which embryos were removed and fixed further in 4% PFA overnight fixing at 4°C.
Scanning electron microscopy
Embryos were treated with Dispase for 30min in order to loosen the ectoderm, rinsed in PBS and fixed in 4% PFA overnight. The ectoderm was removed from embryo pieces with fine needles and post-fixed in Karnovsky (5mL 8% paraformaldehyde, 2mL 25% glutaraldehyde, 1mL, 0.2M/2N PBS and 3mL distilled water). After this step, embryos were post-fixed again in 4% osmium-tetroxide fixative for one hour then washed in PBS. Dehydrated embryos were coated in propylene oxide and resin mixtures by gradually increasing the concentration of resin and cured at 60°C for one day before scanning.
Production of cDNA
RNA from a pre-hatching embryo (~7cm) was used to make cDNA under RNAse-free conditions following Ambion Poly(A)Purist™ mRNA Purification Kit protocol. The reverse transcriptase reaction was performed using Invitrogen's directions. Briefly, 10ul of cDNA library was mixed with 0.5ug random hexamers and 1ul of SuperScript II RT; incubated at 42°C for 50 min then 70°C for 15 min. RNAse H was added and incubated at 37°C for 20 min to remove all remaining RNA.
Primer design, PCR and sequencing
Degenerate primers were designed manually from sequence alignments (5’ GAY AAR AGR CCN TTY ATH and 3’ CC DAT RTC NAC RTT NCC). The amplified fragments were purified by running the entire PCR product on a 1.5% agarose gel, excising the bands using a clean scalpel and purifying the DNA using a Qiagen MinElute Gel Extraction kit. Then, 4ul of purified cDNA fragments were ligated into the TOPO vector (Invitrogen), according to manufacturer's instructions. The ligations were then used to transform TOPO into electrocompetent E. coli. Plasmids were purified using a Qiagen Miniprep kit. Sequence results were analyzed using BLAST to determine cloned sequence identities.
Isolation of neural crest markers Sox8 and Sox9
From our Chiloscyllium punctatum cDNA library, we specifically targeted and amplified two neural crest markers, Sox8 and Sox9, by using degenerate PCR primers. The fragments were sequenced then their identities were verified by a BLAST search for the presence of the HMG protein motif of KRPMNAFMVWAQAARRK. To determine the open reading frame (ORF) from the Chiloscyllium punctatum sequenced clones, we used the software from Sequence Manipulation Suite (Stothard, 2000) and the dataset was built using complete protein sequences selected from bilaterian vertebrates from GenBank (Table 1).
Table 1. Protein Sequences used in Phylogenetic Analysis for C. punctatum Sox8 and Sox9 clones.
Protein sequences used for phylogenetic analysis of Sox8, Sox9 and Sox10 HMG domains were compared to the three C. punctatum Sox8 and Sox9 clones. The outgroup used were from the Sox4 subfamily.
| Organism of Interest | Common Name | Clone ID # | HMG Domain |
|---|---|---|---|
| Chiloscyllium punctatum | Bamboo Shark | A2.7 | Sox 8a |
| Chiloscyllium punctatum | Bamboo Shark | A2.16 | Sox 8b |
| Chiloscyllium punctatum | Bamboo Shark | A2.5 | Sox 9 |
| Species Name | Common Name | Protein Accession # | HMG Domain |
|---|---|---|---|
| Scyliorhinus canicula | Small-spotted catshark | ABA10785 | Sox 8 |
| Trachemys scripta | Red-eared slider turtle | AAP59791 | Sox 8 |
| Dicentrarchus labrax | European seabass | CBN81184 | Sox 8 |
| Sparus aurata | Gilt-headed sea bream | AEV53629 | Sox 8 |
| Tetraodon nigroviridis | Green spotted pufferfish | AAT42231 | Sox 8 |
| Oryzias latipes | Japanese medaka | NP_001158342 | Sox 8 |
| Epinephelus coioides | Orange spotted grouper | AFF57873 | Sox 8 |
| Misgurnus anguillicaudatus | Oriental weatherfish | ACZ65966 | Sox 8 |
| Anolis carolinensis | Green anole lizard | XP_003224806 | Sox 8 |
| Salmo salar | Salmon | NP_001117071 | Sox 8 |
| Gallus gallus | Chicken | NP_990062 | Sox 8 |
| Mus musculus | Mouse | NP_035577 | Sox 8 |
| Xenopus laevis | African clawed frog | NP_001083964 | Sox 8 |
| Danio rerio | Zebrafish | NP_001020636 | Sox 8 |
| Takifugu rubripes | Pufferfish | NP_001072112 | Sox 8 |
| Paralichthys olivaceus | Olive flounder | ACO40490 | Sox 9 |
| Xenopus laevis | African clawed frog | AFK08429 | Sox 9 |
| Mus musculus | Mouse | NP_035578 | Sox 9 |
| Gadus morhua | Atlantic cod | ADV03670 | Sox 9 |
| Trachemys scripta | Red-eared slider turtle | ACG70782 | Sox 9 |
| Aspidoscelis inornata | Little striped whiptail | ABQ44208 | Sox 9 |
| Epinephelus coioides | Orange spotted grouper | ACZ51153 | Sox 9 |
| Alligator mississippiensis | American alligator | AAD17974 | Sox 9 |
| Lepidochelys olivacea | Olive ridley sea turtle | ACT82009 | Sox 9 |
| Crocodylus palustris | Mugger crocodile | ACU12296 | Sox 9 |
| Oreochromis aureus | Blue tilapia | ABY66377 | Sox 9 |
| Scyliorhinus canicula | Small-spotted catshark | ABY71239 | Sox 9 |
| Oryzias latipes | Japanese medaka | NP_001098555 | Sox 9 |
| Gallus gallus | Chicken | NP_989612 | Sox 9 |
| Danio rerio | Zebrafish | NP_571718 | Sox 9 |
| Takifugu rubripes | Pufferfish | AF329945_3 | Sox 9 |
| Oryctolagus cuniculus | European rabbit | XP_002723578 | Sox 10 |
| Mus musculus | Mouse | NP_035567 | Sox 10 |
| Danio rerio | Zebrafish | NP_571950 | Sox 10 |
| Gallus gallus | Chicken | NP_990123 | Sox 10 |
| Xenopus laevis | African clawed frog | NP_001082358 | Sox 10 |
| Paramisgurnus dabryanus | Carp | AFD97051 | Sox 10 |
| Misgurnus anguillicaudatus | Oriental weatherfish | AFD97052 | Sox 10 |
| Oryzias latipes | Japanese medaka | NP_001158343 | Sox 10 |
| Epinephelus coioides | Orange spotted grouper | AFF57872 | Sox 10 |
| Ambystoma mexicanum | Salamander | ABI97016 | Sox 10 |
| Xenopus tropicalis | Western clawed frog | AAI36048 | Sox 10 |
| Cynoglossus semilaevis | Tongue sole | ABW87298 | Sox 10 |
| Outgroups | Common Name | Protein Accession # | HMG Domain |
|---|---|---|---|
| Mus musculus | Mouse | CAA49779 | Sox 4 |
| Xenopus laevis | African clawed frog | NP_001165672 | Sox 4 |
| Takifugu rubripes | Japanese medaka | AAQ18501 | Sox 4 |
| Danio rerio | Zebrafish | CAE18168 | Sox 4 |
Whole-mount in situ hybridization
Antisense RNA probes were transcribed using T7 or SP6 RNA polymerases (Roche) in conjunction with digoxigenin- or biotin-conjugated dUTPs (Roche or Fermentas) using the corresponding commercial protocols. For in situ hybridization, Chiloscyllium p. embryos were removed from the eggs, stripped of their membranes and fixed in 4% paraformaldehyde overnight before being stored in 100% methanol at -20°C. Embryos were prepared for hybridization by slow re-hydration in PBS-Tween (0.5%) series and pre-treatment with Proteinase K (10ug/ml) for 15 min. After post-fixing the embryos to prevent damage during hybridization for 20 min in PFA/0.5% glutaraldehyde, they were pre-hybridized at 65°C. After a minimum of 2 hrs, of pre-hybridization, embryos were hybridized with Sox8 or Sox9 probe overnight at 65°C. Following day after extensive hot then cold long washes in hybridization buffer and MABT embryos were blocked with special blocking from BoehringerManheim for 2hrs minimum, then anti-digoxygenin antibodies (Roche) conjugated with alkaline phosphatase (1:2000) was added overnight at 4°C. Next day embryos were extensively washed in MABT (Maleic acid, NaCl and tween-20) then Tris buffer and visualized by adding BCIP/NBT developers (Roche). A detailed protocol is publicly available in: http://neuro.bcm.edu/groveslab/ (Henrique et al., 1995).
In situ hybridization on slides
In situ hybridization of RNA probes to Chiloscyllium punctatum sections were performed as described above, after adapting the method to sections. This time embryos were fixed in modified Carnoy's solution and then dehydrated in an ethanol series, followed by two changes of histosol, then paraffin and sectioning. Prior to in situ, slides were dewaxed in histosol and then rehydrated by passing through a series of ethanol rinses, rinsed in water, PBS and 2xSSC (Saline-Sodium citrate). Hybridization was performed by using 1.5ng/ul of ShSox8 and ShSox9 probes from two different clones.
Immunohistochemistry on tissue sections
Chiloscyllium punctatum tissue sections were de-waxed in histosol and re-hydrated in a graded series of ethanol washes, then equilibrated in PBS before blocking in PBS containing 10% fetal bovine serum (FBS) and 1% Triton X-100. Primary antibodies were added at 1:500 dilution in PBS overnight at 4°C followed by extensive PBS washes. Secondary antibodies Alexa fluoroprobes, Invitrogen, were added for 30 min with DAPI (to label the nuclei) then washed in PBS 3 times for 5 min and coverslipped with Permount. Pictures of sections were taken using Axiovision LE software (Zeiss™) with an AxioCam camera attached to a Zeiss AxioimagerA1 upright fluorescent microscope and assembled into figures using Adobe Photoshop 7 by adjusting each color channel level (increasing contrast and reducing background to make images clearer) and reducing image size to a 300dpi and 3×3” size.
Antibody characterization
Antibodies details are described in Table 2.
- Monoclonal Tuj1 (catalog #MMS-435P Covance), this antibody was raised against microtubules derived from rat brain and recognizes a 50 kDa protein in Western blot. It is well characterized and highly reactive to neuron specific Class III ß-tubulin. TUJ1 does not identify ß-tubulin found in glial cells.
- Monoclonal HNK1 (catalog #3H5, DSHB, Developmental Studies Hybridoma Bank, UI), is derived from the VC1.1 hybridoma which recognizes the HNK-1 epitope, an N-linked carbohydrate. It is a well-known avian neural crest marker (Bronner-Fraser, 1986) and lateral line neuromasts (Ghysen and Dambly-Chaudiere, 2004).
- Monoclonal 7B3/transitin (was a kind gift from Jim Weston, Univ. of Oregon), recognizes a 300 kDa nestin-like intermediate filament in stem cells (Wakamatsu et al., 2007); it can now be purchased from DSHB #A2B11).
- FoxD3 polyclonal was courtesy of David Raible, raised against purified zebrafish FoxD3 and its specificity characterized it recognized on Western blots after immonoprecipitation of translated protein against control (Lister et al., 2006).
Table 2.
of Primary Antibodies used
| Antigen | Immunogen (what the antibody was raised against; full sequence and species) | Manufacturer, species antibody was raised in, mono- vs. polyclonal, catalog or lot number | Dilution used |
| TuJ1 | This antibody was raised against microtubules derived from rat brain. It is well characterized and highly reactive to neuron specific Class III β-tubulin (βIII) | Covance Research Products Inc (San Diego), Purified mouse IgG2a monoclonal Antibody, Unconjugated, Clone TUJ1, #MMS-435P-100 | 1:500 |
| HNK1 | E10 chick optic nerve | Developmental Studies Hybridoma Bank, (Univ. Iowa). Mouse monoclonal VC1.1 hybridoma. | 1:500 |
| 7B3/transitin | E7 chick retinal cells. Genbank accession no. X80877 | Developmental Studies Hybridoma Bank, (Univ. Iowa). Rat monoclonal. #A2B11 | 1:500 |
| FoxD3 | purified in vitro translated FoxD3. | Laboratory of David Raible (Univ. of Washington, Seattle). Antibody was raised in rabbit. | 1:500 |
Sox8, Sox9 and Sox10 HMG Domain Multiple Sequence Alignment and Construction of Phylogenetic Trees
We performed a multiple sequence alignment (MSA) to confirm the three unknown Chiloscyllium punctatum clones belonged to the Sox 8 and Sox9 sub-families rather than Sox10 sub-family. We used Sox4 as the MSA outgroup; Mouse Sox4, Xenopus laevis Sox4, Takifugu rubripes Sox4 and Danio rerio Sox4. The MSA (Supp. Figure 1) was performed using MUSCLE with its default parameters (Edgar, 2004). Only full-length HMG box domains were included in the alignment of the three Chiloscyllium punctatum clones. For the rooted phylogenetic tree, the MUSCLE “profile-profile alignment” algorithm was used between our three Chiloscyllium punctatum clones and the 43 BLAST verified protein sequences against the Sox4 outgroup protein sequences.
The distance matrix, FastTree was carried out on the MSA, which uses the Maximum-Likelihood (ML) method and Nearest-Neighborhood Interchanges (NNIs) (Price et al., 2010). FastTree creates trees using 1,000 replicates and utilizes the Jones-Taylor-Thorton (JTT) and/or the Whelan Goldman (WAG) models to determine amino acid evolution as well as uses the unbiased Shimodaira-Hasegawa test (SH) (Shimodaira, 2002). Both phylogenetic trees were viewed using FigTree (http://tree.bio.ed.ac.uk/software/figtree). The rooted tree was aligned and analyzed against the established Sox4 outgroup; Mouse Sox4, Xenopus laevis Sox4, Takifugu rubripes Sox4 and Danio rerio Sox4.
Results
Vital labeling of trunk neural crest
In our experiments, we used the classic chicken vital labeling method in Chiloscyllium punctatum by injecting DiI into the neural tubes of live Chiloscyllium punctatum shark embryos then incubated them for 24hrs in their own egg white. Due to the scarcity of shark embryos and the low survival rate with this method (~50%), we had positive results from relatively few embryos (N=10).
We performed these experiments in embryos ranging between st.23-29. From the best-labeled DiI injected embryos (now at stages 25-26, N=7/10) we observed two major trends. First, one set of labeled cells migrated outside the neural tube following the classic segmental pattern of trunk neural crest cells in other species (Gammill and Roffers-Agarwal, 2010) (Fig.1). In some instances we observed DiI cells in the developing heart (Fig. 1D) and branchial arches (Fig.1F). Sections through these embryos confirmed the presence of DiI cells migrating along the classic ventro-medial pathway of trunk neural crest cells, reaching the lateral sides of the aorta (Fig.2).
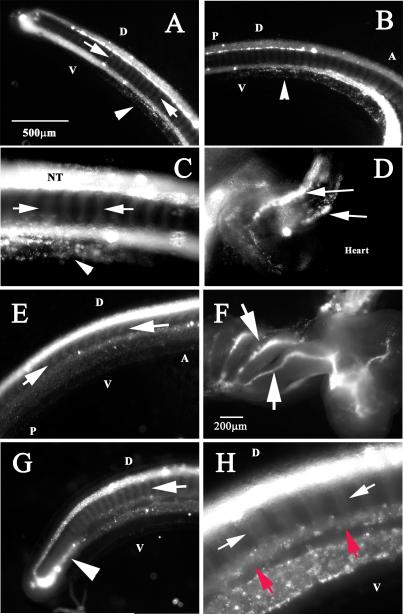
Figure 1. DiI labels migrating neural crest in Stage 25-27 C. punctatum embryos.
Shark embryos neural tubes were vitally labeled with DiI for 24hrs. Stage 25 embryo: Tail end (A) and caudal trunk (B, C) show segmental migration of neural tube derived cells (arrows) as well as a ventral line of DiI cells (arrowheads). (D) Shows 2 sets of migrating DiI cells in the out-tract of the heart (arrows). Stage 27 embryo: DiI this older embryo shows labeled cells in neural tube and segmentally migrating cells (arrows in E, G, H), ventral locations (red arrows in H), branchial arches (arrows in F) and turning rostrally (arrowhead in G).
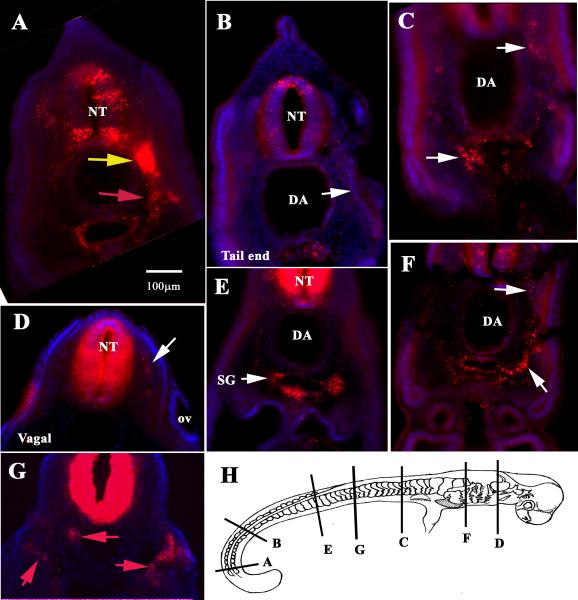
Figure 2. Sections through stage 25 C. punctatum vitally labeled with DiI.
Sections from different levels of a st.25 DiI labeled embryo showed: (A) Neural crest migrating as a sheet of cells at the tail (yellow arrow) and the “returning” stream (red arrow). (B-G) Show sections at different rostral to caudal levels in the embryo. (H) Cartoon of stage 25 embryo showing level of sections.
In addition, another set of DiI cells were observed along what would correspond to the lateral line (a group of sensory neurons of placodal origin used for detecting water movements) at the tail end (Fig.1A-C, H, at higher magnification). This observation suggested that some neural crest cells were able to migrate within 24hrs into the developing lateral line. Interestingly, it seemed that once this line of trunk DiI-positive cells reached the tailmost end, it continued migrating rostrally along the lateral line (arrowhead in Fig.1G and 3A-C). We observed this pattern in 6/10 DiI labeled embryos.
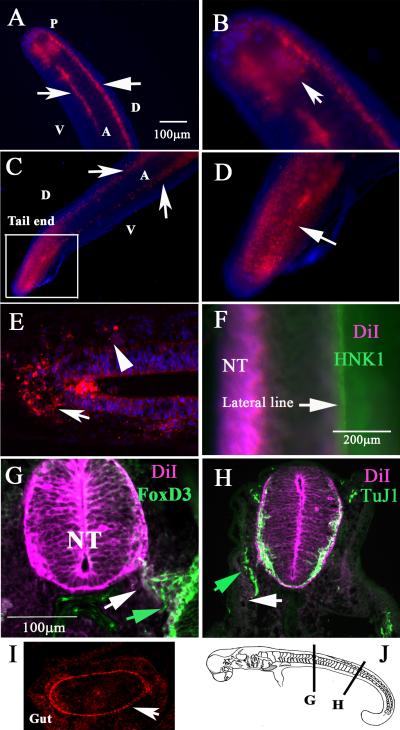
Figure 3. DiI labels migrating neural crest in Stage 26 & 29 C. punctatum embryos.
Shark embryo neural tubes were vitally labeled with DiI for 24hrs. Stage 26 shark embryo shows DiI cells as two bright lines (arrows in A) and as a sheet at tail end (arrow in B). Stage 29 shark embryo shows DiI cells as two bright lines (arrows in C) and as a sheet at the tail end (arrow in magnified area in D). Pseudo-longitudinal section showed delaminated DiI cells on top of the neural tube (arrow in E) and on one portion (rostral) of the somite (arrowhead in E). (F) shows whole-mount image of the embryo after HNK1 immunostaining of lateral line placodal neurons (arrow). (G) FoxD3 antibody stain (green arrow) co-localized with DiI cells (white arrow). (H) Anti-beta III tubulin labeling (TuJ1) neurons (green arrow) along the lateral line did not overlap with DiI cells (white arrow). (I) The developing gut had a line of DiI cells (arrow). Abbreviations: NT: neural tube; D: dorsal, V: ventral, A: anterior; P: posterior.
Although most of the embryos were already too old for complete labeling of cranial neural crest, we observed DiI-positive cells in cranial structures. We found that at the level of the forebrain and hindbrain, there were few DiI-positive cells that migrated under the ectoderm as well as into cranial ganglia (data not shown). Interestingly, we observed also DiI-positive cells migrating along the branchial arches (6/10), around the eye (2/10) and in the heart (2/10). More importantly, we observed in some embryos (N=4/10) a large number of DiI-positive cells in the developing gut (red arrow in Fig.3I).
After counter staining with HNK1, which labels lateral line sensory neurons, then sectioning these embryos, we were able to corroborate that those medial DiI-positive cells were along the lateral line both at hindbrain and trunk levels (arrows in Fig.2A-C and Fig.3F, G). However, the total amount of DiI-positive cells migrating into the lateral line was trivial as shown in the whole-mount double labeling with HNK1 in stage 26 embryos (Fig.3F). Furthermore, while we observed few DiI cells along the lateral line in conjunction with FoxD3 which labels glial cells (Fig.3G), we never saw overlap of DiI with TuJ1 that labels placodal sensory cells along the lateral line.
DiI labeling of an older embryo (stage 27) corroborated the presence of migrating trunk neural crest cells as a broad wave (N=6/10), indicating a classic ventral pathway (Fig. 3D). More importantly, we observed DiI-positive cells coming out of the dorsal part of the neural tube and migrating along the rostral side of the somites (arrowhead in Fig.3E).
Scanning electron microscopy of neural crest
Because neural crest cells migrate under the ectoderm, it is easy to observe this group of cells after removing the tissue. We did scanning electron microscopy (SEM) of Chiloscyllium punctatum embryos at stages 25, 27 and 29. The results showed in early embryos, stages 25 and 27 there is a population of cells that migrate beneath the ectoderm and over the rostral neural tube, similar to cranial neural crest cells migrating in amphibians, fish, birds and mammals. At stage 25 embryos, we observed migrating cells over the hindbrain rostrally with other cells migrating ventrally (Fig.4A). SEM of a stage 27 embryo showed cells delaminating from the neural tube and along the ventral pathway at the junction of hindbrain and vagal region (Fig.4B). Finally, SEM in the caudal trunk of an older, stage 29 embryo showed a significant number of cells migrating outside the neural tube ventrally (Fig. 4C and pseudo colored one in D).
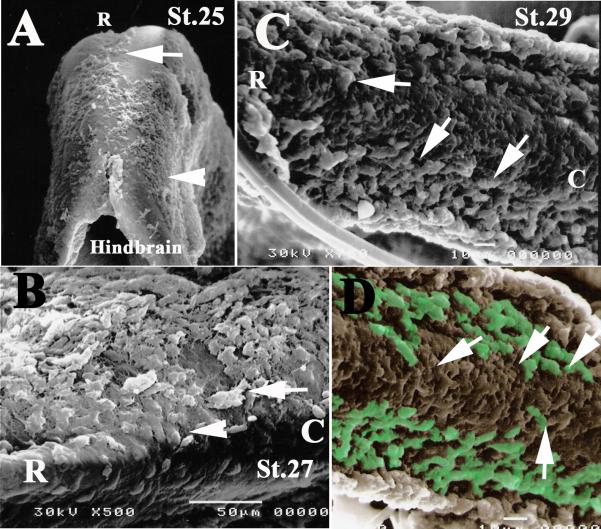
Figure 4. Scanning electron microscopy of C. punctatum embryo neural crest.
SEM of Chiloscyllium punctatum embryos at stages 25, 27 and 29 showed that in early embryos there is a population of cells that migrated beneath the ectoderm and over the rostral neural tube, such as cranial and trunk neural crest cells do in amphibians, fish, birds and mammals. (A) In stage 25 embryos, we observed migrating cells over the hindbrain, migrating rostrally (arrow in A) and others migrating ventrally (arrowhead in A). (B) SEM of a stage 27 embryo showed cells delaminating from the neural tube (arrowhead) along the ventral pathway and at the junction of hindbrain and trunk (arrows). (C, D) SEM in the caudal trunk of older, stage 29 embryo, showed a good number of cells migrating outside the neural tube ventrally (arrows in C and pseudo colored one in D).
Isolation, Alignment and Phylogenetic Analysis of Chiloscyllium punctatum neural crest markers Sox8 and Sox9
The protein sequence motif RPMNAFMVWAQ is conserved in all Sox8, Sox9 and Sox10 sequences as well as some sequence similarity seen in the Sox4 outgroups (Yusuf et al., 2012). From a cDNA library of Chiloscyllium punctatum, we isolated fragments of two neural crest markers, Sox8 and Sox9, by using degenerate PCR primers. The sequencing of three Chiloscyllium punctatum clones and posterior BLAST, showed 43 complete coding HMG domain sequences from the Sox8, Sox9 and Sox10 subfamilies, as illustrated in the colored alignment (Supp. Fig.1). The HMG domain is where the transcription factors for Sox8, Sox9 and Sox10 bind onto the DNA minor groove (Bowles, et. al., 2000).
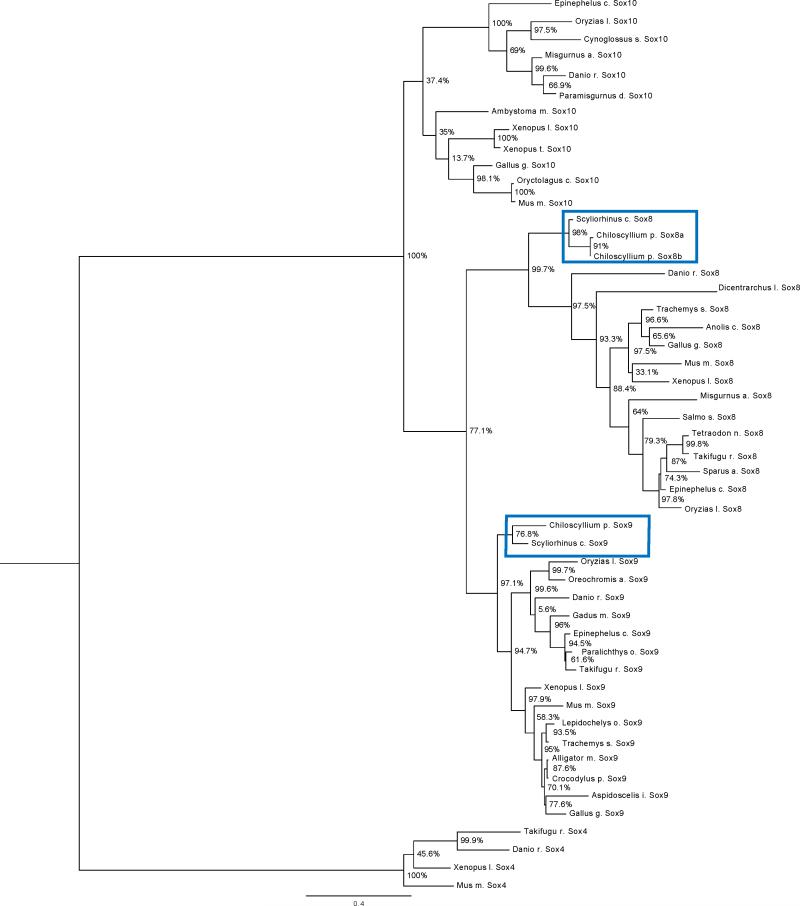
C. punctatum Sox clones named here as Sox8a, Sox8b, and Sox9, were compared to known Sox8 and Sox9 sequences. Sequences were aligned using MUSCLE and phylogenies were inferred with FastTree. The tree was rooted using Sox4 as an outgroup, which is considered reliable because Sox4 is part of the SoxC family and contains the HMG protein domain. C. punctatum sequences clustered with metazoan Sox8, Sox9 and Sox10 sub-families, demonstrating that they are true members of the metazoan Sox8 and Sox9 sub-families (Fig.5). Clustering C. punctatum Sox8a, Sox8b and Sox9 genes with their respective groups in the rooted tree showed that Chiloscyllium punctatum Sox8 and Sox9 are true orthologs in their sub-families (see also Supp. Fig.2). The Shimodaira-Hasegawa (SH) test of alternative topologies showed strong support of these groupings (Shimodaira, 2002; Shimodaira and Hasegawa, 2001).
Figure 5. Rooted phylogenetic tree for Sox8, Sox9 and Sox10 HMG domains.
The rooted phylogenetic tree was obtained from the Nearest-Neighborhood Interchanges program performed on FastTree. The Sox8, Sox9 and Sox10 HMG domain protein sequences were found in various vertebrate species and used in construction of the tree. The bar range located on the bottom of the rooted phylogenetic tree represents the branch length of 0.4 units. The branch length is proportionally related to the evolutionary distances and reflects the divergence between the Sox8, Sox9 and Sox10 genes. The unbiased SH test values are indicated on the branch nodes as percent similarities between the clades. The outgroup used was Sox4, which is part of the SoxC sub-family. C. punctatum Sox8 and Sox9 clones were positioned at the beginning of the phylogenetic tree indicating that our C. punctatum clones are more ancient than the rest of vertebrates with Sox8, Sox9 and Sox10 sub-families.
Sox8 and Sox9 expression in Chiloscyllium punctatum embryos
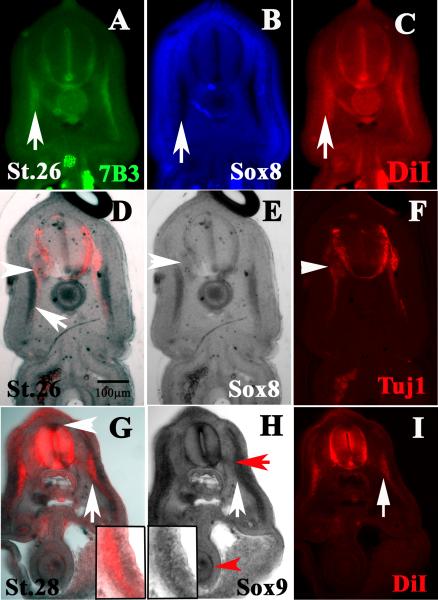
At stage 25 and stage 27, Chiloscyllium punctatum embryos were injected with DiI and then undergone in situ hybridization with Sox8 and Sox9 probes. We observed that Sox8 co-localized robustly with DiI along the lateral line (Fig.6A-C). Importantly it co-localized with Transitin/7B3, which labels neural crest and glial precursors along the lateral line (Rotenstein et al., 2009) (Fig.6A). During double labeling with Tuj1, which marked neurons, we did not observe co-localization of Tuj1 with Sox8 along the lateral line and did not observe Sox8 cells in the developing gut. On the other hand, we did observe Tuj1 labeled ventral motor axons where both Tuj1 and Sox8 co-localized and robustly labeled sensory ganglia (Fig.6D-F). Sox8 labeled a different set of cells located more laterally than the Tuj1-positive axons (white arrow in Fig.6D). Sox9 partly co-localized with DiI labeled neural crest cells along the lateral line (white arrow in Fig.6G and inserts) and weakly with peripheral nerves (red arrow in Fig.6H). Importantly, Sox9 was observed in the dorsal neural tube, gut and developing kidneys (Fig.6H).
Figure 6. C. punctatum Sox8 and Sox9 follows neural crest path and locations.
Mid-Trunk sections were observed through a DiI injected embryo after Sox8 (A-F) and Sox9 (G-I) whole-mount in situ hybridization. These mid-trunk sections, Sox8 and Sox9, co-localized with DiI cells. (A-C) Sections were immunostained with 7B3, which labels neural crest cells. Arrows point to cells in the ventromedial path and co-stained for all three markers: 7B3, Sox8 and DiI. (D-F) This section show both Sox8 and TuJ1 (red) staining. (E, F) Shows the single channel of same section. Sox8 labeled myotome (arrow) and notochord. (F) Arrowheads point to Tuj1. (G) Section shows both Sox9 and DiI staining. (G-I) Shows the single channel of same section. White arrows point to cells positive for both Sox9 and DiI along ventromedial path (higher magnification in inserts). (G) White arrowhead points to Sox9 on dorsal neural tube where pre-migratory neural crest cells reside. (H) Red arrow points to Sox9 positive cells along ventral spinal nerve. Red arrowhead marks Sox9 cells in developing gut.
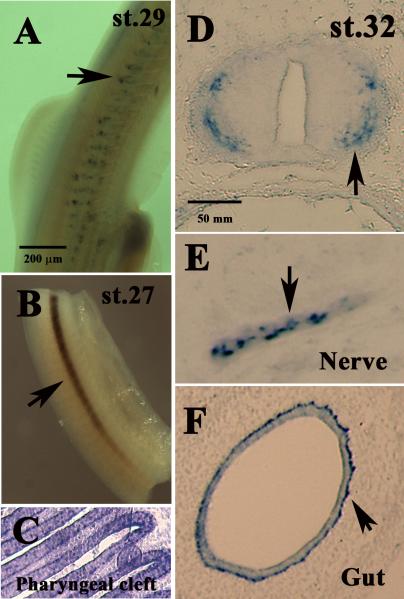
Whole-mount in situ for Sox8 of Chiloscyllium punctatum stage 27 embryos only labeled the lateral line and notochord (Fig.7A). However, in older Chiloscyllium punctatum stage 29 embryos, some cells migrated beneath the ectoderm on the rostral side of the somites were and disclosed the appearance of being true neural crest cells (Fig.7B). At stage 32, when the Chiloscyllium punctatum embryos displayed the least morphological and physical resemblance to other vertebrate embryos, Sox8 was found in tissues usually associated in other vertebrate species. We observed Sox8 in pharyngeal cleft cells (Fig.7C), differentiating neurons in the young spinal cord (Fig.7D), spinal nerves (Fig.7E), cells surrounding the lumen, enteric neurons (Fig.7F) and in developing cartilage (data not shown).
Figure 7. C. punctatum Sox8 expression at different stages of development.
(A) Whole-mount of stage 29 embryos show a stream of Sox8 cells along the trunk (arrows). (B) Whole-mount of a stage 27 embryo shows a strong line of Sox8 cells corresponding to notochord and ventromedial cells (arrow). (C) Sox8 labels cells in the pharyngeal cleft in a stage 32 embryo. (D) Cross-section through a stage 32 embryo shows Sox8 cells in the periphery of the neural tube (arrow). Sox8 labels Schwann cells in spinal nerve (arrow in E) and enteric neurons in st.32 embryo (arrow in F).
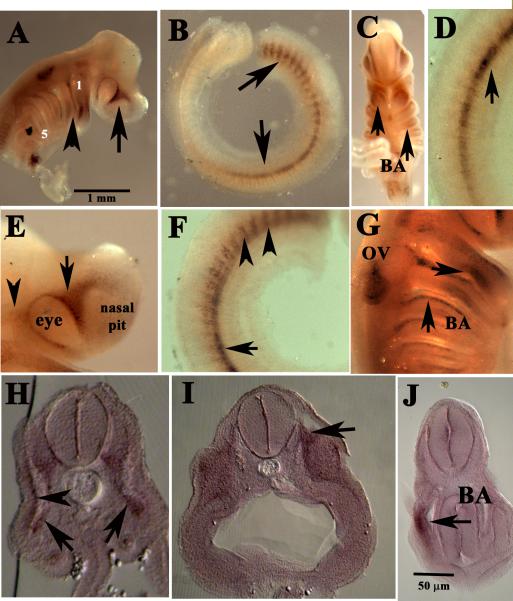
Whole-mount in situ hybridization of Sox9 at stage 26 embryos, showed a more definite neural crest pattern (Fig.8). We observed cells migrating into branchial arches, around the eye and nasal pit (Fig.8A, C, E, G). Along the trunk, we observed two groups of cells with Sox8 at stage 29 Chiloscyllium punctatum embryos. The first group was the most caudal ones along the lateral line and 20 somites up. The second group was a set of cells migrating on the rostral side of the somites beneath the ectoderm (Fig.8 B,D,F). Sections through an older embryo, stage 28, tested positive with Sox9 cells in sensory ganglia especially in the spinal nerves as well as a few positive cells for Sox9 along the lateral line (Fig.8H, I) and in the branchial arches (Fig.8J).
Figure 8. C. punctatum Sox9 expression at different stages of development.
(A-G) Whole-mount of stage 26 embryo shows Sox9 along classic neural crest cells pathways along branchial arches (arrowheads in B, G; arrows in C), along the somites (arrows in B, D F), around the eye (arrows in A, E) and otic vesicle (G). Sections through a stage 26 embryo highlights Sox9 cells along the ventromedial path (arrowhead in H), spinal nerves (arrows in H), cranial ganglia (arrow in I) and branchial arch (arrow in J).
Discussion
Identification of neural crest cells that have delaminated and migrated away from the neural tube requires both the use of vital labeling and classical markers within the developing embryo. Using a combination of these techniques, we are the first to describe the migration pattern of trunk neural crest cells in a shark embryo.
Classic, vital labeling of trunk neural tube cells in developing Chiloscyllium punctatum embryos was accomplished by injecting DiI in the neural tubes between stages 25-29. During this period, trunk neural crest cells actively developed and expanded. Though survivability was low, some DiI-labeled embryos at stages 25-29 (N=10) persisted. The remaining embryos showed migrating neural crest cells along the ventromedial pathway and into the lateral line. These migratory patterns of neural crest cells are unique in teleosts and elasmobranchs (Gillis et al., 2012).
The striking similarity of migration patterns detected using vital labeling and results obtained from Sox8 and Sox9 in situ hybridization underscores an emerging paradigm. Specifically, that neural crest cell development is highly conserved across vertebrate lineages (Holland and Holland, 2001; Meulemans and Bronner-Fraser, 2004). Past studies have shown that Sox8 and Sox9 sequences are highly conserved among fish, amphibians, reptiles and mammals. Even more striking than sequence conservation, however, is the conserved patterns of migration and expression in vertebrate neural crest cells and their derivatives (Bell et al., 2000; Guth et al., 2010; Horigome et al., 1999; Reyes et al., 2010; Sauka-Spengler and Bronner-Fraser, 2008b). This was especially remarkable in the trunk regions of C. punctatum embryos, since segmental migration through the rostral portion of the somites is practically undistinguishable from other vertebrates. We also observed a reduced number of migrating trunk neural crest cells in comparison with the large number of amniotes. This finding is consistent with observations in other aquatic vertebrates (fishes and amphibians) (Carmona-Fontaine et al., 2008; Dutton et al., 2001).
Our present study demonstrates that, in C. punctatum, glial cells in the lateral line are likely of neural crest origins. Though this has been observed in other vertebrates, we are the first to show that the pattern is present in cartilaginous fishes. Previous studies using DiI labeling in Xenopus (Collazo et al., 1993) and zebrafish neural tubes (Collazo et al., 1994) showed that neural crest cells contributed both to lateral line organs and other structures. Nusslein-Volhard and Eisen labs demonstrated that neuromasts are of placodal origin, while the Schwann cells along the lateral line are of neural crest origin (Gilmour et al., 2002; Kelsh et al., 2000). Here we demonstrate conservation of these patterns in C. punctatum (Gillis et al., 2012; O'Neill et al., 2007). Though we only double-labeled lateral line DiI cells with the glia-specific markers. Sox8 Sox9 and FoxD3, we hypothesize them to be glia under the light of our past research showing the presence of glial marker along the lateral line in Chiloscyllium punctatum shark (Rotenstein et al., 2009). Though Baker and co-workers extensively studied the shark's placodal development in the lateral line, they did not focus on the glia component in the ganglia (Baker et al., 2008; Gillis et al., 2012; Gillis et al., 2011; O'Neill et al., 2007). Our current findings strongly supports a neural crest contribution to the lateral line in C. punctatum.
In addition to observing DiI cells in segmented streams along the lateral line in the trunk, we also observed a group of DiI cells at the tail end migrating as a sheet and a smaller number as a stream migrating rostrally on the ventral portion of the tail. Though we do not know the ultimate destiny of these “returning” populations, the same neural crest migratory pattern has been observed in fish (Collazo et al., 1994) and amphibians (Collazo et al., 1993) but not in reptiles (Reyes et al., 2010) or mammals (Kuhlbrodt et al., 1998). This suggests that the migratory path became lost in non-aquatic gnathostomes.
It is well known that, in vertebrates, the neural crest gives rise to the enteric nervous system (Elworthy et al., 2005; Kuhlman and Eisen, 2007; Le Douarin and Teillet, 1973; Newgreen and Young, 2002; Young and Newgreen, 2001). Our observation of neural crest cells in the developing gut strongly supports what has been, until now, suspected based on the development of other vertebrates: that neural crest cells are responsible for generating the elasmobranch enteric nervous system. Thus, this neural crest pathway is very ancient and developmentally shares SoxE genes among gnathostomes (Elworthy et al., 2005; Hoff et al., 2008).
Neural crest cells are known to migrate on the rostral side of somites while avoiding caudal portions that have a set of repellants (e.g., ephrins, Slits, Sema) (Gammill and Roffers-Agarwal, 2010; Kulesa and Gammill, 2010). We observed that trunk neural crest cells avoided the caudal portion of the somites, suggesting that this migratory route/behavior is very old and likely due to a conserved structure in the somites. This observation has also been made in chordates such as lampreys, hagfish (Gostling and Shimeld, 2003; McCauley, 2008; Meulemans and Bronner-Fraser, 2007; Ota et al., 2007).
Evolution of the Sox genes is of keen interest given their role in stem cell fate and cancer progression (Scott et al., 2010). The presence of Sox8 and Sox9 in trunk neural crest in C. punctatum, stands in contrast to current findings of SoxE phylogeny for lampreys (Lakiza et al., 2011) and amphioxus (Yu et al., 2008). C punctatum Sox8, Sox9, Sox10 Sox4 and MSA sequences, did not cluster with teleost SoxE genes. Neither did they group with SoxE genes from lamprey (data not shown). The clade containing C. punctatum Sox8a, Sox8b and Sox9 sequences nested squarely between Scyliorhinus canicula Sox8 and Sox9 genes rather than grouping with the rest of the vertebrate Sox8, Sox9 and Sox10 sequences. This arrangement provides strong phylogenetic support to the hypothesis that shark Sox8 and Sox9 genes can be considered paralogs of amniotes Sox8 and Sox9 rather than orthologs (Cui et al., 2011). In other words, phylogenetic analysis of these sequences suggest that Sox8 and Sox9 genes appeared first in common ancestor of chondrichthyans and teleosts rather than in the common ancestor of teleosts.
More importantly, we showed the expression of these two SoxE genes in trunk neural crest cells was highly conserved. C. punctatum's Sox9 expression matched the Sox10 migratory patterns observed in teleosts. Expression profiles were particularly similar in delaminated migrating neural crest cells along the trunk, around the developing eye, branchial arches, gut, and peripheral nerves (Bell et al., 2000; Guth et al., 2010; Horigome et al., 1999; Reyes et al., 2010; Sauka-Spengler and Bronner-Fraser, 2008b). Expression patterns seen in this study differ from observations in Xenopus, zebrafish, chicken and mammals. In these systems, Sox9 expression has not been observed in the peripheral nervous system (Hong and Saint-Jeannet, 2005; Li et al., 2002). However, hagfish trunk neural crest cells appear to express Sox9, although these cells do not enter the somites, as was observed in C punctatum and osteichthyans (Ota et al., 2007). Though Sox8 and Sox9 expression has been fully characterized in the lamprey cranial neural crest, little is known about expression in the trunk (Lakiza et al., 2011; Uy et al., 2012). Preliminary results indicate that lamprey trunk neural crest migration is segmental, similar to our observations in shark (Dr. Marianne Bronner personal communication).
Sox8 expression in the C punctatum neural crest has been previously characterized. (O'Donnell et al., 2006). Our observations matched expectations based on these studies. However, we also observed Sox8 expression in developing chondrocytes (Schmidt and Patel, 2005) which was similar to Sox9 patterns observed in the mouse (Mori-Akiyama et al., 2003) chicken dermomyotome (Bell et al., 2000; Hall and Gillis, 2013). Although we were not able to clone Sox10 from C. punctatum cDNA, there is strong evidence that it exists (Dr. Clare V. Baker personal communication. Therefore, we conclude that C. punctatum has a full set of osteichthyan Sox8, 9 and 10 genes. This stands in contrast to earlier vertebrates, such as lampreys, which contain SoxE1-3 (Meulemans and Bronner-Fraser, 2004; Uy et al., 2012).
In summary, we describe the migration of trunk neural crest cells in C punctatum through vital labeling and in situ hybridization . To our knowledge, this is the first time that this approach has been utilized to study the trunk neural crest, in elasmobranchs. Sox8 and Sox9 migration though C. punctatum's neural crest and derivatives suggests that, once these genes were co-opted by neural crest cells, they proved to be highly conserved across vertebrates (Martinez-Morales et al., 2007). In addition, our observations with DiI labeling indicate that the migration of trunk neural crest cells has not changed much during evolution.
Supplementary Material
Supplementary Figure 1. Colored MSA for Sox 8, Sox9 and Sox10 HMG domains.
The sequences are aligned accordingly to nucleotide base similarity with complete HMG domains. The protein sequences were generated through Gblocks (Castresana, 2000). The symbol “#” represents highly conserved amino acids found in the Multiple Sequence Alignment (MSA). Chiloscyllium punctatum; Cp (name colored red), Scyliorhinus canicula; Sc (name colored blue). The outgroup used was Sox4, which is part of the SoxC sub-family (name colored green): Mus musculus Sox4; MmSox4, Xenopus laevis Sox4; XlSox4, Danio rerio Sox4; DrSox4, Takifugu rubripes; TrSox4.
Since the HMG domain is highly conserved in the Sox8, Sox9 and Sox10 subfamilies, the alignment within the MSA had very few gaps and insertions upstream (Bowles et al., 2000). The number symbol “#” in represents homology with the Sox8, Sox9, Sox10 and Sox4 consensus sequences (Castresana, 2000).
Acknowledgements
We would like to give a special thanks to Chris Plante and Michelle Malme from the Long Beach Aquarium for providing us the shark embryos. A special thanks goes to John Reiss for his helpful comments. Also, a special thank you to Kent Coleman for his valuable revisions to this manuscript. Partial support for M. Juarez on this project was provided by NIH GM 2 T34 GM008959 to ME Zavala. This work was partly supported by an NIH/NINDS AREA grant 2R15NS060099-02A1 to MEdB.
Role of Authors:
Marilyn Juarez: undergraduate, performed in situ hybridizations
Michelle Reyes: undergraduate, performed DiI injections
Tiffany Coleman: graduate, performed DNA/protein analyses, designed phylogenetic figures, bioinformatics analysis and revised the manuscript
Lisa Rotenstein: undergraduate, performed SEM
Sothy Sao: undergraduate, performed sectioning
Darwin Martinez and Matthew Jones: Sox8 and Sox9 cloning
Rachel Mackelprang: oversaw bioinformatics analysis
Maria Elena de Bellard: PI, wrote the manuscript, performed Sox8 and Sox9 cloning and DiI injections
This work supported by an NIH/NINDS AREA grant 2R15NS060099-02A1 and 5SC3GM096904-02 to MEdB.
Footnotes
Competing Interest:
Authors do not have any competing interests or conflicts.
References
- Adachi N, Takechi M, Hirai T, Kuratani S. Development of the head and trunk mesoderm in the dogfish, Scyliorhinus torazame: II. Comparison of gene expression between the head mesoderm and somites with reference to the origin of the vertebrate head. Evolution & development. 2012;14(3):257–276. doi: 10.1111/j.1525-142X.2012.00543.x. [DOI] [PubMed] [Google Scholar]
- Baker CV. In: Neural Crest and Cranial Ectodermal Placodes. Jacobson M, Rao MS, editors. Springer, New York; New York: 2005. pp. 67–127. [Google Scholar]
- Baker CV, O'Neill P, McCole RB. Lateral line, otic and epibranchial placodes: developmental and evolutionary links? Journal of experimental zoology Part B. 2008;310(4):370–383. doi: 10.1002/jez.b.21188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CV, Schlosser G. The evolutionary origin of neural crest and placodes. Journal of experimental zoology Part B. 2005;304(4):269–273. doi: 10.1002/jez.b.21060. [DOI] [PubMed] [Google Scholar]
- Balfour FM. The Development of Elasmobranch Fishes. Journal of anatomy and physiology. 1876;11(Pt 1):128–172. 121. [PMC free article] [PubMed] [Google Scholar]
- Ballard WW, Mellinger Jean, Lechenault Henri. A Series of Normal Stages for Development of Scyliorhinus canicula, the Lesser Spotted Dogfish (Chondrichthyes: Scyliorhinidae). J Exp Zool. 1993;267:318–336. [Google Scholar]
- Bell KM, Western PS, Sinclair AH. SOX8 expression during chick embryogenesis. Mechanisms of development. 2000;94(1-2):257–260. doi: 10.1016/s0925-4773(00)00296-3. [DOI] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227(2):239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. Analysis of the early stages of trunk neural crest migration in avian embryos using monoclonal antibody HNK-1. Dev Biol. 1986;115(1):44–55. doi: 10.1016/0012-1606(86)90226-5. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456(7224):957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular biology and evolution. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res Dev Brain Res. 2000;121(2):233–241. doi: 10.1016/s0165-3806(00)00049-3. [DOI] [PubMed] [Google Scholar]
- Cole NJ, Currie PD. Insights from sharks: evolutionary and developmental models of fin development. Dev Dyn. 2007;236(9):2421–2431. doi: 10.1002/dvdy.21268. [DOI] [PubMed] [Google Scholar]
- Collazo A, Bronner-Fraser M, Fraser SE. Vital dye labelling of Xenopus laevis trunk neural crest reveals multipotency and novel pathways of migration. Development (Cambridge, England) 1993;118(2):363–376. doi: 10.1242/dev.118.2.363. [DOI] [PubMed] [Google Scholar]
- Collazo A, Fraser SE, Mabee PM. A dual embryonic origin for vertebrate mechanoreceptors. Science (New York, NY. 1994;264(5157):426–430. doi: 10.1126/science.8153631. [DOI] [PubMed] [Google Scholar]
- Cui J, Shen X, Zhao H, Nagahama Y. Genome-wide analysis of Sox genes in Medaka (Oryzias latipes) and their expression pattern in embryonic development. Cytogenetic and genome research. 2011;134(4):283–294. doi: 10.1159/000329480. [DOI] [PubMed] [Google Scholar]
- Derobert Y, Baratte B, Lepage M, Mazan S. Pax6 expression patterns in Lampetra fluviatilis and Scyliorhinus canicula embryos suggest highly conserved roles in the early regionalization of the vertebrate brain. Brain Res Bull. 2002;57(3-4):277–280. doi: 10.1016/s0361-9230(01)00695-5. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development (Cambridge, England) 2001;128(21):4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S, Pinto JP, Pettifer A, Cancela ML, Kelsh RN. Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mechanisms of development. 2005;122(5):659–669. doi: 10.1016/j.mod.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Epperlein HH, Selleck MA, Meulemans D, McHedlishvili L, Cerny R, Sobkow L, Bronner-Fraser M. Migratory patterns and developmental potential of trunk neural crest cells in the axolotl embryo. Dev Dyn. 2007;236(2):389–403. doi: 10.1002/dvdy.21039. [DOI] [PubMed] [Google Scholar]
- Freitas R, Zhang G, Albert JS, Evans DH, Cohn MJ. Developmental origin of shark electrosensory organs. Evolution & development. 2006;8(1):74–80. doi: 10.1111/j.1525-142X.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Roffers-Agarwal J. Division of labor during trunk neural crest development. Dev Biol. 2010;344(2):555–565. doi: 10.1016/j.ydbio.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural Crest and the Origin of Vertebrates: A New Head. Science (New York, NY. 1983;220(4594):268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C. Development of the zebrafish lateral line. Current opinion in neurobiology. 2004;14(1):67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Gillis JA, Modrell MS, Northcutt RG, Catania KC, Luer CA, Baker CV. Electrosensory ampullary organs are derived from lateral line placodes in cartilaginous fishes. Development (Cambridge, England) 2012;139(17):3142–3146. doi: 10.1242/dev.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis JA, Rawlinson KA, Bell J, Lyon WS, Baker CV, Shubin NH. Holocephalan embryos provide evidence for gill arch appendage reduction and opercular evolution in cartilaginous fishes. Proc Natl Acad Sci U S A. 2011;108(4):1507–1512. doi: 10.1073/pnas.1012968108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nusslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34(4):577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Gostling NJ, Shimeld SM. Protochordate Zic genes define primitive somite compartments and highlight molecular changes underlying neural crest evolution. Evolution & development. 2003;5(2):136–144. doi: 10.1046/j.1525-142x.2003.03020.x. [DOI] [PubMed] [Google Scholar]
- Guth SI, Bosl MR, Sock E, Wegner M. Evolutionary conserved sequence elements with embryonic enhancer activity in the vicinity of the mammalian Sox8 gene. The international journal of biochemistry & cell biology. 2010;42(3):465–471. doi: 10.1016/j.biocel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Hall BK, Gillis JA. Incremental evolution of the neural crest, neural crest cells and neural crest-derived skeletal tissues. Journal of anatomy. 2013;222(1):19–31. doi: 10.1111/j.1469-7580.2012.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375(6534):787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Hoff S, Zeller F, von Weyhern CW, Wegner M, Schemann M, Michel K, Ruhl A. Quantitative assessment of glial cells in the human and guinea pig enteric nervous system with an anti-Sox8/9/10 antibody. J Comp Neurol. 2008;509(4):356–371. doi: 10.1002/cne.21769. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutierrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, Ferrier DE, Garcia-Fernandez J, Gibson-Brown JJ, Gissi C, Godzik A, Hallbook F, Hirose D, Hosomichi K, Ikuta T, Inoko H, Kasahara M, Kasamatsu J, Kawashima T, Kimura A, Kobayashi M, Kozmik Z, Kubokawa K, Laudet V, Litman GW, McHardy AC, Meulemans D, Nonaka M, Olinski RP, Pancer Z, Pennacchio LA, Pestarino M, Rast JP, Rigoutsos I, Robinson-Rechavi M, Roch G, Saiga H, Sasakura Y, Satake M, Satou Y, Schubert M, Sherwood N, Shiina T, Takatori N, Tello J, Vopalensky P, Wada S, Xu A, Ye Y, Yoshida K, Yoshizaki F, Yu JK, Zhang Q, Zmasek CM, de Jong PJ, Osoegawa K, Putnam NH, Rokhsar DS, Satoh N, Holland PW. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome research. 2008;18(7):1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Holland ND. Evolution of neural crest and placodes: amphioxus as a model for the ancestral vertebrate? Journal of anatomy. 2001;199(Pt 1-2):85–98. doi: 10.1046/j.1469-7580.2001.19910085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. Sox proteins and neural crest development. Seminars in cell & developmental biology. 2005;16(6):694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Horigome N, Myojin M, Ueki T, Hirano S, Aizawa S, Kuratani S. Development of cephalic neural crest cells in embryos of Lampetra japonica, with special reference to the evolution of the jaw. Dev Biol. 1999;207(2):287–308. doi: 10.1006/dbio.1998.9175. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Dutton K, Medlin J, Eisen JS. Expression of zebrafish fkd6 in neural crest-derived glia. Mechanisms of development. 2000;93(1-2):161–164. doi: 10.1016/s0925-4773(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38(1):17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18(1):237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman J, Eisen JS. Genetic screen for mutations affecting development and function of the enteric nervous system. Dev Dyn. 2007;236(1):118–127. doi: 10.1002/dvdy.21033. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Fraser SE. Neural crest cell dynamics revealed by time-lapse video microscopy of whole embryo chick explant cultures. Dev Biol. 1998;204(2):327–344. doi: 10.1006/dbio.1998.9082. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Gammill LS. Neural crest migration: patterns, phases and signals. Dev Biol. 2010;344(2):566–568. doi: 10.1016/j.ydbio.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani S. Cephalic neural crest cells and the evolution of craniofacial structures in vertebrates: morphological and embryological significance of the premandibular-mandibular boundary. Zoology (Jena, Germany) 2005;108(1):13–25. doi: 10.1016/j.zool.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Kuratani S, Horigome N. Developmental morphology of branchiomeric nerves in a cat shark, Scyliorhinus torazame, with special reference to rhombomeres, cephalic mesoderm, and distribution patterns of cephalic crest cells. Zoological Society of Japan. 2000;17(7):893–909. [Google Scholar]
- Lakiza O, Miller S, Bunce A, Lee EM, McCauley DW. SoxE gene duplication and development of the lamprey branchial skeleton: Insights into development and evolution of the neural crest. Dev Biol. 2011;359(1):149–161. doi: 10.1016/j.ydbio.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. The avian embryo as a model to study the development of the neural crest: a long and still ongoing story. Mechanisms of development. 2004;121(9):1089–1102. doi: 10.1016/j.mod.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain research reviews. 2007;55(2):237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Teillet MA. The migration of neural crest cells to the wall of the digestive tract in avian embryo. Journal of embryology and experimental morphology. 1973;30(1):31–48. [PubMed] [Google Scholar]
- Li M, Zhao C, Wang Y, Zhao Z, Meng A. Zebrafish sox9b is an early neural crest marker. Development genes and evolution. 2002;212(4):203–206. doi: 10.1007/s00427-002-0235-2. [DOI] [PubMed] [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006;290(1):92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales JR, Henrich T, Ramialison M, Wittbrodt J. New genes in the evolution of the neural crest differentiation program. Genome biology. 2007;8(3):R36. doi: 10.1186/gb-2007-8-3-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley DW. SoxE, Type II collagen, and evolution of the chondrogenic neural crest. Zoological science. 2008;25(10):982–989. doi: 10.2108/zsj.25.982. [DOI] [PubMed] [Google Scholar]
- McCauley DW, Bronner-Fraser M. Neural crest contributions to the lamprey head. Development (Cambridge, England) 2003;130(11):2317–2327. doi: 10.1242/dev.00451. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Developmental cell. 2004;7(3):291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS ONE. 2007;2(8):e787. doi: 10.1371/journal.pone.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development (Cambridge, England) 2010;137(16):2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgreen D, Young HM. Enteric nervous system: development and developmental disturbances--part 1. Pediatr Dev Pathol. 2002;5(3):224–247. doi: 10.1007/s10024-001-0142-y. [DOI] [PubMed] [Google Scholar]
- Northcutt RG. The new head hypothesis revisited. Journal of experimental zoology Part B. 2005;304(4):274–297. doi: 10.1002/jez.b.21063. [DOI] [PubMed] [Google Scholar]
- Northcutt RG, Gans C. The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. The Quarterly review of biology. 1983;58(1):1–28. doi: 10.1086/413055. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Hong CS, Huang X, Delnicki RJ, Saint-Jeannet JP. Functional analysis of Sox8 during neural crest development in Xenopus. Development (Cambridge, England) 2006;133(19):3817–3826. doi: 10.1242/dev.02558. [DOI] [PubMed] [Google Scholar]
- O'Neill P, McCole RB, Baker CV. A molecular analysis of neurogenic placode and cranial sensory ganglion development in the shark, Scyliorhinus canicula. Dev Biol. 2007;304(1):156–181. doi: 10.1016/j.ydbio.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota KG, Kuraku S, Kuratani S. Hagfish embryology with reference to the evolution of the neural crest. Nature. 2007;446(7136):672–675. doi: 10.1038/nature05633. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development (Cambridge, England) 1994;120(3):495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- Reyes M, Zandberg K, Desmawati I, de Bellard ME. Emergence and migration of trunk neural crest cells in a snake, the California Kingsnake (Lampropeltis getula californiae). BMC developmental biology. 2010;10:52. doi: 10.1186/1471-213X-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenstein L, Milanes A, Juarez M, Reyes M, de Bellard ME. Embryonic development of glial cells and myelin in the shark, Chiloscyllium punctatum. Gene Expr Patterns. 2009;9(8):572–585. doi: 10.1016/j.gep.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Baratte B, Shi L, Mazan S. Structure and expression of an Otx5-related gene in the dogfish Scyliorhinus canicula: evidence for a conserved role of Otx5 and Crxgenes in the specification of photoreceptors. Dev Genes Evol. 2001;211(11):533–544. doi: 10.1007/s00427-001-0191-2. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008a;46(11):673–682. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Insights from a sea lamprey into the evolution of neural crest gene regulatory network. The Biological bulletin. 2008b;214(3):303–314. doi: 10.2307/25470671. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Patel K. Wnts and the neural crest. Anatomy and embryology. 2005;209(5):349–355. doi: 10.1007/s00429-005-0459-9. [DOI] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J. SOX9 induces and maintains neural stem cells. Nature neuroscience. 2010;13(10):1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development (Cambridge, England) 1992;116(2):297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic biology. 2002;51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17(12):1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28(6):1102, 1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Uy BR, Simoes-Costa M, Sauka-Spengler T, Bronner ME. Expression of Sox family genes in early lamprey development. The International journal of developmental biology. 2012;56(5):377–383. doi: 10.1387/ijdb.113416bu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu Y, Nakamura N, Lee JA, Cole GJ, Osumi N. Transitin, a nestin-like intermediate filament protein, mediates cortical localization and the lateral transport of Numb in mitotic avian neuroepithelial cells. Development (Cambridge, England) 2007;134(13):2425–2433. doi: 10.1242/dev.02862. [DOI] [PubMed] [Google Scholar]
- Wotton KR, Mazet F, Shimeld SM. Expression of FoxC, FoxF, FoxL1, and FoxQ1 genes in the dogfish Scyliorhinus canicula defines ancient and derived roles for Fox genes in vertebrate development. Dev Dyn. 2008;237(6):1590–1603. doi: 10.1002/dvdy.21553. [DOI] [PubMed] [Google Scholar]
- Young HM, Newgreen D. Enteric neural crest-derived cells: origin, identification, migration, and differentiation. The Anatomical record. 2001;262(1):1–15. doi: 10.1002/1097-0185(20010101)262:1<1::AID-AR1006>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome research. 2008;18(7):1127–1132. doi: 10.1101/gr.076208.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf D, Butland SL, Swanson MI, Bolotin E, Ticoll A, Cheung WA, Zhang XY, Dickman CT, Fulton DL, Lim JS, Schnabl JM, Ramos OH, Vasseur-Cognet M, de Leeuw CN, Simpson EM, Ryffel GU, Lam EW, Kist R, Wilson MS, Marco-Ferreres R, Brosens JJ, Beccari LL, Bovolenta P, Benayoun BA, Monteiro LJ, Schwenen HD, Grontved L, Wederell E, Mandrup S, Veitia RA, Chakravarthy H, Hoodless PA, Mancarelli MM, Torbett BE, Banham AH, Reddy SP, Cullum RL, Liedtke M, Tschan MP, Vaz M, Rizzino A, Zannini M, Frietze S, Farnham PJ, Eijkelenboom A, Brown PJ, Laperriere D, Leprince D, de Cristofaro T, Prince KL, Putker M, del Peso L, Camenisch G, Wenger RH, Mikula M, Rozendaal M, Mader S, Ostrowski J, Rhodes SJ, Van Rechem C, Boulay G, Olechnowicz SW, Breslin MB, Lan MS, Nanan KK, Wegner M, Hou J, Mullen RD, Colvin SC, Noy PJ, Webb CF, Witek ME, Ferrell S, Daniel JM, Park J, Waldman SA, Peet DJ, Taggart M, Jayaraman PS, Karrich JJ, Blom B, Vesuna F, O'Geen H, Sun Y, Gronostajski RM, Woodcroft MW, Hough MR, Chen E, Europe-Finner GN, Karolczak-Bayatti M, Bailey J, Hankinson O, Raman V, LeBrun DP, Biswal S, Harvey CJ, DeBruyne JP, Hogenesch JB, Hevner RF, Heligon C, Luo XM, Blank MC, Millen KJ, Sharlin DS, Forrest D, Dahlman-Wright K, Zhao C, Mishima Y, Sinha S, Chakrabarti R, Portales-Casamar E, Sladek FM, Bradley PH, Wasserman WW. The transcription factor encyclopedia. Genome biology. 2012;13(3):R24. doi: 10.1186/gb-2012-13-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Colored MSA for Sox 8, Sox9 and Sox10 HMG domains.
The sequences are aligned accordingly to nucleotide base similarity with complete HMG domains. The protein sequences were generated through Gblocks (Castresana, 2000). The symbol “#” represents highly conserved amino acids found in the Multiple Sequence Alignment (MSA). Chiloscyllium punctatum; Cp (name colored red), Scyliorhinus canicula; Sc (name colored blue). The outgroup used was Sox4, which is part of the SoxC sub-family (name colored green): Mus musculus Sox4; MmSox4, Xenopus laevis Sox4; XlSox4, Danio rerio Sox4; DrSox4, Takifugu rubripes; TrSox4.
Since the HMG domain is highly conserved in the Sox8, Sox9 and Sox10 subfamilies, the alignment within the MSA had very few gaps and insertions upstream (Bowles et al., 2000). The number symbol “#” in represents homology with the Sox8, Sox9, Sox10 and Sox4 consensus sequences (Castresana, 2000).