Abstract
The cytogenetically normal subtype of acute myeloid leukemia is associated with an intermediate risk which complicates therapeutic options. Lower overall HOX/TALE expression appears to correlate with more favorable prognosis/better response to treatment in some leukemias and solid cancer. The functional significance of the associated gene expression and response to chemotherapy is not known. Three independent microarray datasets obtained from large cohorts of patients along with quantitative polymerase chain reaction validation were used to identify a four-gene HOXA/TALE signature capable of prognostic stratification. Biochemical analysis was used to identify interactions between the four encoded proteins and targeted knockdown used to examine the functional importance of sustained expression of the signature in leukemia maintenance and response to chemotherapy. An 11 HOXA/TALE code identified in an intermediate-risk group of patients (n=315) compared to a group with a favorable risk (n=105) was reduced to a four-gene signature of HOXA6, HOXA9, PBX3 and MEIS1 by iterative analysis of independent platforms. This signature maintained the favorable/intermediate risk partition and where applicable, correlated with overall survival in cytogenetically normal acute myeloid leukemia. We further showed that cell growth and function are dependent on maintained levels of these core genes and that direct targeting of HOXA/PBX3 sensitizes cytogenetically normal acute myeloid leukemia cells to standard chemotherapy. Together the data support a key role for HOXA/TALE in cytogenetically normal acute myeloid leukemia and demonstrate that targeting of clinically significant HOXA/PBX3 elements may provide therapeutic benefit to patients with this subtype of leukemia.
Introduction
Leukemia is associated with genomic aberrations including chromosomal translocations, inversions and deletions.1 Cytogenetic analysis remains the primary method for prognostic sub-classification of acute myeloid leukemia (AML) at presentation.2,3 However, the majority (~50%) of patients presenting with AML are cytogenetically normal (CN-AML) with an intermediate prognosis and marked variation in treatment outcome.1,4 Chemotherapy can induce complete remission in approximately 85% of younger AML patients (aged 16 to 60 years)5 but many relapse with chemo-resistant disease.6 Myeloablative conditioning followed by allogeneic stem-cell transplantation is the currently preferred treatment option, although procedural mortality and graft-versus-host disease severely affect the long-term benefits in many groups of patients.3,7 Accurate predictors of clinical outcome and novel targeted therapies are, therefore, needed to improve treatment for patients.
Molecular analysis, including whole exome and targeted sequencing, has mapped the landscape and frequency of CN-AML-associated mutations that correlate with prognosis to define “more-favorable” or “less-favorable” subgroups. Over 50% of CN-AML cases harbor an NPM1 mutation which, in the absence of FLT3/ITD mutations,8 acts as an independent prognosticator for achieving complete remission and increased overall survival.9–11DNMT3A mutations are reported to be independently associated with a “less-favorable” outcome in CN-AML.12 Whatever combination of genetic mutations that emerges as providing the best prognostic value, it is their downstream targets and associated factors that may provide the basis for therapeutic intervention.
Class I homeobox (HOX) and three-amino loop extension (TALE) homeobox co-factor genes are associated with prognostic subtypes in AML.13–15HOXA cluster genes are normally expressed an order of magnitude higher than the other HOX genes in hematopoietic stem and progenitor cells indicating a particular functional role in hematopoiesis.16 Ectopic expression of individual HoxA genes, in several model systems, resulted in altered hematopoiesis ranging from potentiation of cell proliferation and extended self-renewal for HoxA617 to the onset of an AML phenotype e.g. HoxA9.18,19 Haploinsufficient HoxA cluster mice develop to adulthood but have impaired hematopoietic stem cell function.16HoxA gene deficiency is also associated with defective myeloid, erythroid and lymphoid development.20
The Meis1 TALE gene is a collaborating oncogene to many Hox genes.19,21,22 In several transduction and transplantation models Pbx1 was used as a control TALE gene and was shown to have a limited or negative impact on leukemia initiation.21,22 Direct functions of other Pbx or Meis proteins or isoforms in hematopoiesis are less well-defined, although complementary roles in multiple HOX:TALE complexes are expected, based on their structural similarity.23,24 Deletion of TALE genes is also associated with impaired hematopoiesis. Both Meis1 and Pbx1 are essential for definitive hematopoiesis whereas Pbx2 is reported to be dispensable for normal hematopoiesis25 and Pbx3 deficiency is associated with early death due to central respiratory failure.26
We provide evidence that defined HOX/TALE gene expression signatures may have specific prognostic value and that targeting clinically relevant HOXA/PBX3 axes may provide therapeutic benefit to patients with CN-AML.
Design and Methods
Samples from patients with acute myeloid leukemia
Diagnostic CN-AML peripheral blood or mononuclear cells were obtained following informed consent and anonymized. All studies adhered to the tenets of the Declaration of Helsinki and had local ethical approval.
Gene expression profiling and validation
Expression profiles were generated for patients with favorable cytogenetic risk (MILE Classes 9–11, n=105) and intermediate risk (MILE Class 13, n=315) according to the reported classification (NCBI Gene Expression Omnibus Accession number: GSE13204).27 Two independent cohorts were also examined: de novo CN-AML samples (n=235) with known NPM1-mutation, FLT3-ITD and CEBPα status28; and a custom cDNA array (n=114 patients; Bullinger et al., unpublished). Affymetrix CEL files were imported into the Partek Genomics Suite (St. Louis, MO, USA) as previously reported.29,30HOXA/TALE genes were examined using TaqMan™ chemistry as previously described.14,31 Differentially expressed genes were validated in independent cohorts of patients for the HOXA/PBX3/MEIS1 (n=21) and the HOXA6/HOXA9/PBX3/MEIS1 core signature (n=37). Relative expression values are represented as copy number equivalents, fold-change or percentage of appropriate controls.
Acute myeloid leukemia cell lines and primary acute myeloid leukemia cultures
Two well-characterized human cell lines representative of CN-AML, namely U937 and OCI-AML3, were maintained as described elsewhere.32,33 Low passage numbers of DSMZ-authenticated stocks were used throughout. Primary AML cells were rapidly thawed in the presence of DNase and cultured directly in RPMI 10% fetal calf serum (Invitrogen, Paisley, UK).
Targeted knockdown of HOXA/TALE
AML cell lines were nucleofected using the manufacturer’s dedicated protocol (Amaxa™ GmbH, Cologne, Germany). Primary cells were nucleofected using the dedicated protocol for CD34+ cells. Target short hairpin (sh)RNA and non-targeting control sequences (Online Supplementary Table S1) were obtained commercially or selected following sequential optimization of plasmid delivery and measurable knockdown. Multiple shRNA moieties, specific for widely separated regions along the mRNA sequence targeted, were used throughout. Targeted knockdown was established by quantitative real-time polymerase chain reaction (RQ-PCR) and confirmed by western blot analysis.
Cell cycle, growth, proliferation and caspase 3/7 assays
Samples for cell cycle analysis were processed as previously reported.17 DNA content was measured with an LSR II flow cytometer and analyzed using FACS Diva™ software (BD Biosciences, Franklin Lakes, NJ, USA). Cell growth and viability were determined by trypan blue exclusion and cell proliferation by CellTiter-Glo®. Caspase activity was quantified using Caspase-Glo® 3/7 (Promega, Madison, WI, USA).
Colony-forming unit assays
AML cells and cell lines were cultured for up to 14 days in MethoCult® Classic (Stem Cell Technologies, Vancouver, Canada). Colony staining with 1 mM 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyltetrazolium chloride (INT, Sigma-Aldrich, Gillingham, UK) enabled visualization of colonies and determination of metabolic activity.
Drug treatment
AML cells, pre-treated with or without shRNA, were plated in liquid culture or methylcellulose with 1 μM cytarabine (cytosine arabinoside)34 or 100 ng/mL Mylotarg (gemtuzumab ozogamicin).35
Statistical analysis
ANOVA and Kaplan-Meier analysis of microarray experiments was implemented using Partek Genomics Suite. Two-way ANOVA or Student t-tests were performed on non-microarray-derived data by Graph-Pad Prism software (GraphPad software, Version 5.0, La Jolla, CA, USA) or the SPSS software package (IBM, Portsmouth, UK).
Results
HOXA/TALE expression in favorable/intermediate risk acute myeloid leukemia
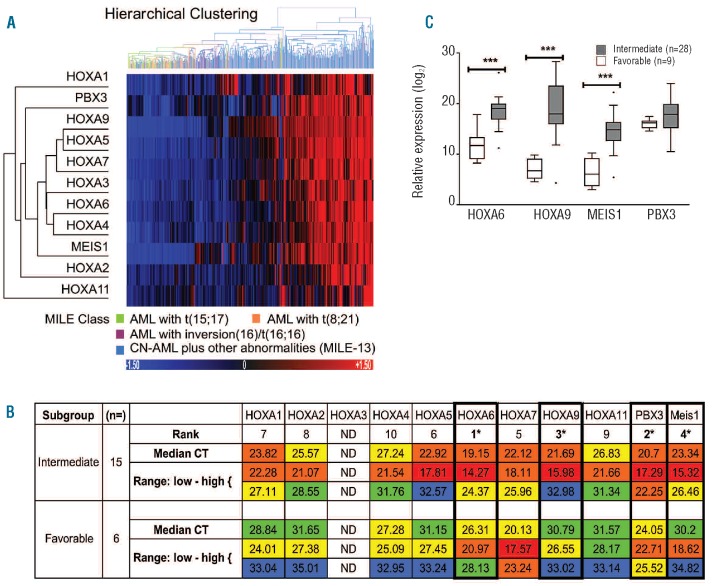
Microarray analysis of a cohort of AML samples (n=420) obtained as part of the MILE study was interrogated by probe-sets for the HOXA cluster and TALE genes. The cohort was made up of a combined favorable cytogenetic risk group consisting of 40 class 9 [t(8;21)], 37 class 10 [t(15;17)] and 28 class 11 [inv(16)/t(16;16)] samples referred to as MILE Classes 9–11 (n=105) and an intermediate risk group classified as CN-AML plus fewer than three structural abnormalities, excluding 11q23, referred to as MILE-13 (n=315). Favorable/intermediate risk AML was partitioned by unsupervised hierarchical clustering on the basis of nine HOXA genes plus PBX3 and MEIS1 (Figure 1A); these genes demonstrated significant differences between MILE-13 and combined MILE Classes 9–11 with a false discovery rate-corrected P-value of less than 0.0000005 (Online Supplementary Table S2). The differential expression of HOXA cluster genes (HOXA1-HOXA11) with the exception of HOXA3 was validated in an independent cohort of patients (n=21) by RQ-PCR. The most consistent and highly ranked genes within the Class 13 equivalent, based on corrected CT values, were HOXA6, PBX3, HOXA9 and MEIS1 (Figure 1B). Differential expression of the four genes was also observed in a more cytogenetically defined independent cohort of patients (n=37) by RQ-PCR (Figure 1C and Online Supplementary Table S3).
Figure 1.
The HOXA/TALE signature stratifies favorable and Intermediate risk AML. (A) An unsupervised hierarchical cluster and heatmap depicting partitioning of a cohort of AML patients (n=420) into those with favorable or Intermediate risk by HOXA/TALE expression using a Euclidean distance metric with average linkage calculation. The favorable risk and intermediate risk groups comprised 105 cases and 315 cases, respectively. Probe-sets for the 11 genes included in the cluster were significantly differentially expressed between Class 13 versus Classes 9–11 with a false discovery rate-corrected P-value < 0.0000005. (B) Tabulated ranked gene expression levels, median and range, represented as CT values corrected for 18S rRNA loading. Color coding indicates overall expression levels: very high expression CT <20 (red); high expression 20 <CT <24 (orange); moderate expression 24 <CT <30 (yellow); low expression 30< CT <35 (green). The numbers of patients within prognostic subgroups are indicated where appropriate. The highest ranked genes in intermediate AML (HOXA6, HOXA9, MEIS1 and PBX3) are highlighted (*). ND not determined; HOXA3 primers not validated. (C) The HOXA/TALE signature was validated in an independent cohort of patients (n=37), by specific RQ-PCR analysis The favorable group (n=9) comprised five t(8;21) and four t(15;17) patients. Relative expression is displayed as copy number equivalents (log2 scale) from standard curve values corrected for 18S rRNA loading.31 Significant differences between risk groups are denoted by ***P≤0.001.
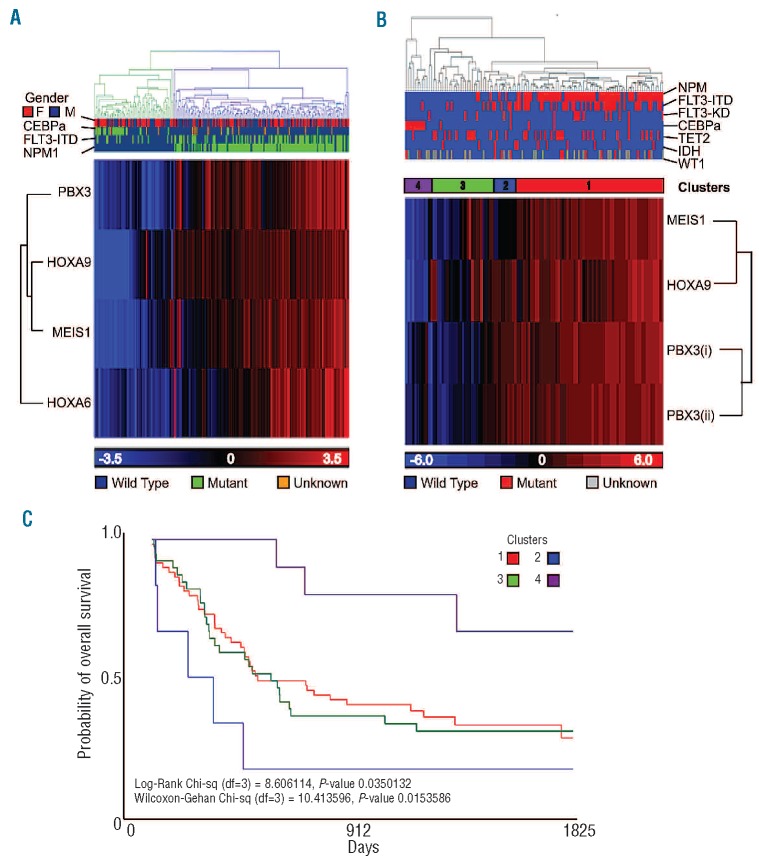
Notably, the four-gene code was sufficient to partition an independent cohort (n=235) of de novo CN-AML patients (Figure 2A) demonstrating that lower expression of the signature correlated in part with favorable cytogenetic risk as defined by NPM1mut/FLT3-ITD/CEBPα status. To investigate whether the HOXA/TALE signature merely reflected known prognostic indicators, data from a custom array applied to a further cohort of CN-AML patients (n=114) with long-term follow up and extensive molecular profiling (Bullinger, personal communication) were examined. Four probes from the custom array corresponding to the HOX/TALE code were again capable of partitioning the patients’ samples by unsupervised hierarchical clustering (Figure 2B). In this case, four distinct clusters were observed (1–4 right to left in the figure) which did not completely align with NPM1, FLT3-ITD, FLT3-TKD, CEBPα, TET2, IDH or WT1 status. In addition, the clusters representing four different states namely: (i) HOXHi/PBXHi/MEISHi; (ii) HOXHi/PBXHi/MEISLo; (iii) HOXint/MEISint/PBXLo; and (iv) HOXLo/MEISLo/PBXLo correlated with overall survival (P=0.015). Furthermore, HOXHi/PBXHi/MEISLo (Cluster 2) may be an indicator of poorer prognosis within this intermediate risk subgroup and is distinct both from patients with NPM1 mutations (mainly located in Cluster 1) and those with CEBPα mutations (mainly Cluster 4) (Figure 2C). Together these data indicate an important role for HOXA6, HOXA9, PBX3 and MEIS1 in intermediate risk/CN-AML.
Figure 2.
HOXA/TALE code partitions CN-AML patients and correlates with overall survival. (A) An unsupervised hierarchical cluster and heatmap depicting partitioning of a cohort of de novo CN-AML patients (n=235) by HOXA6, HOXA9, PBX3 and MEIS1 expression. (B) Probe-sets for the four genes were significantly differentially expressed against NPM-1 mut status with a false discovery rate-corrected P-value < 0.0000005. Unsupervised hierarchical clustering and heatmap generation showed partitioning of a cohort of CN-AML patients’ samples (n=114) with well-defined mutation status based on the probe-sets for HOXA9, PBX3(i) and (ii) and MEIS1. Four distinct gene expression-based clusters 1–4 (right to left) were identified that were not strictly dependent on cytogenetic status. (C) A Kaplan-Meier plot demonstrating correlation between the probe-sets clusters (1–4) and survival (P=0.015), with HOXAHi, PBX3Hi/MEIS1Lo (cluster 2) correlating with worst overall survival and HOXALo/PBX3Lo/MEIS1Lo (cluster 4) associated with best overall survival.
Functional targeting of HOXA/TALE interactions in acute myeloid leukemia cell lines
A targeted knockdown approach (shRNA) was taken to disrupt the HOX/TALE axis at the molecular level. We first wished to investigate the potential targetable HOXA/TALE interactions. A series of co-immunoprecipitation and western blot analyses confirmed known HOXA9:TALE interactions36 and demonstrated that HOXA6 can also interact with PBX and MEIS proteins (Online Supplementary Figure S1). Endogenous proteins were enriched for by immunopreciptation in two AML cell lines, U937 and OCI-AML3. High affinity interactions between HOXA6 and PBX proteins were confirmed irrespective of NPM1 status. Interestingly HOXA6 was also found to interact with MEIS in both AML cell lines, providing additional evidence that HOXA:MEIS interactions are not limited to the Abdominal B-like proteins.
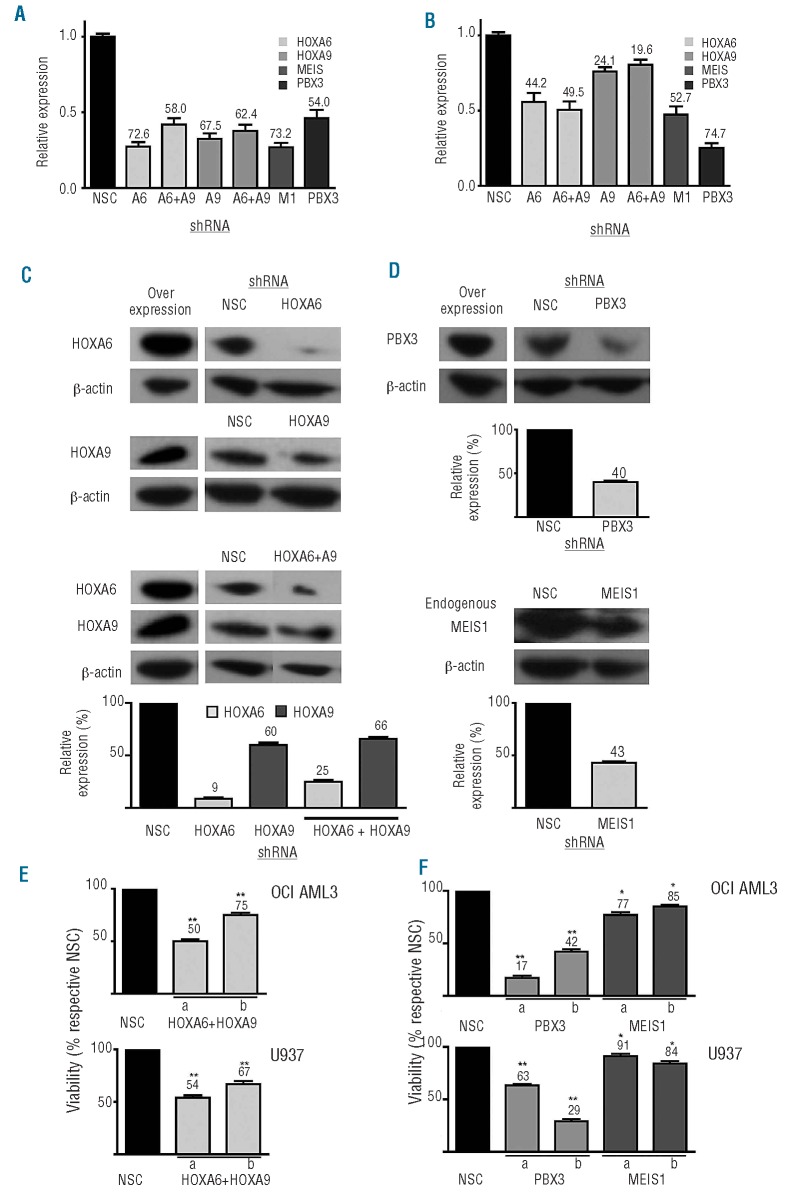
Target shRNA were delivered with high efficiency (>45%) to both cell lines (data not shown). Cell lines displayed comparable HOX/TALE expression to that of primary CN-AML samples (Online Supplementary Table S4). Knockdown of gene expression was demonstrated by RQ-PCR (Figure 3A,B) and specific reduced protein expression was confirmed by western blot analysis of either 293FT cells overexpressing tagged HOXA/PBX3 cDNA or U937 cells for endogenous MEIS1 (Figure 3C,D). Multiple shRNA moieties for each target were used to demonstrate specific effects. Differentially expressed genes associated with single HOXA6 or HOXA9 knockdown in the AML cell lines were obtained using AmpliChip-Leukemia arrays containing 1480 leukemia-associated probe-sets (Online Supplementary Figure S2A). Although knockdown of HOXA6 or HOXA9 resulted in altered expression of a small subset of common genes, associated with cell viability and differentiation in each cell line, significant differences were largely target gene-specific (Online Supplementary Table S5). This suggested that combined knockdown of HOXA6 and HOXA9 would have a more potent effect on AML cells than either alone. Targeted HOXA6/A9 knockdown resulted in minimal compensation by HOXA10, but no reduced expression of other HOX genes, in a similar manner to TALE family members following knockdown of PBX3 or MEIS1 (Online Supplementary Figure S2B).
Figure 3.
Histograms depicting expression of HOXA6, HOXA9, PBX3 and MEIS1 in (A) OCI-AML3 and (B) U937 cell lines following target gene knockdown, compared to non-silenced controls (NSC). Percentage knockdown values at 72 h of either single genes or combinations as stated are labeled. Mean and standard deviation of three experiments are shown where relative expression was calculated using the 2−ΔΔCT method with 18S rRNA values as the calibrator. Western blot analysis shows reduced protein levels of overexpressed HOXA/PBX3 constructs or MEIS1 following incubation with specific shRNA for 48 h in 293FT or U937 cells. Representative data from two or three shRNA sequences per target are shown normalized using β-actin as a loading control and relative to NSC levels (C, D). Reduced viability of (E) HOXA6+HOXA9, PBX3 or (F) MEIS1 knockdown cells (two constructs per gene target a & b) expressed as percentages of respective NSC, measured by trypan blue exclusion with cell counting 24 h post-transfection. Data are representative of n≥4 with mean and standard deviation plotted, P≤ 0.01/0.001 are denoted by */**, respectively.
HOXA/TALE knockdown alters viability and growth of acute myeloid leukemia cell lines
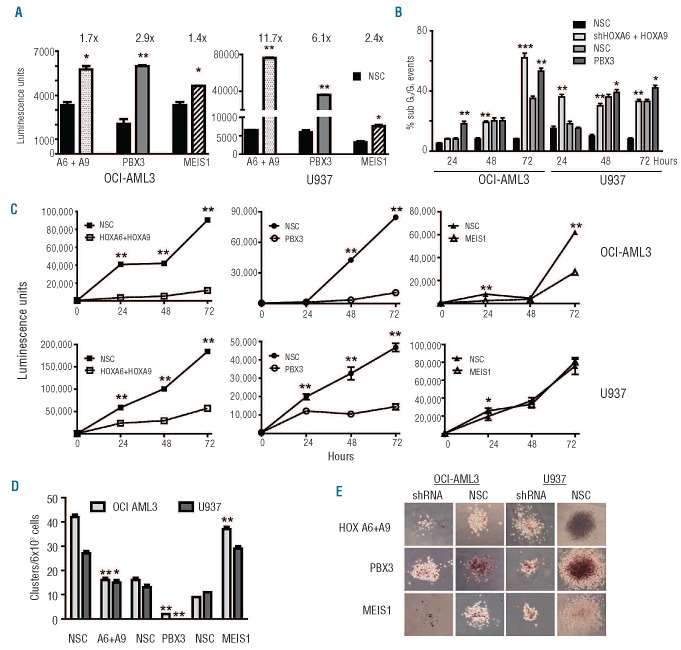
Combined HOXA6+A9 or individual TALE knockdown resulted in reduced viability and altered morphology in both AML cell lines compared to non-silencing controls (Figure 3E-F and Online Supplementary Figure S3). Targeted PBX3 knockdown demonstrated a greater effect than MEIS1. These data were supported by significantly enhanced caspase 3/7 activities, indicative of cellular apoptosis, within 24 h (Figure 4A). Reduced caspase 3/7 activity at later time points reflected poor cell state (Online Supplementary Figure S4). Furthermore, sub G0/G1 events were notably increased following HOXA6+A9 or PBX3 knockdown in both cell lines compared to respective controls (Figure 4B) but not for MEIS1 (data not shown). Consistent with these findings, overall cell growth was significantly reduced following HOXA6+A9 or PBX3 knockdown but less so after MEIS1 knockdown in either cell line (Figure 4C).
Figure 4.
Targeted HOXA/PBX3 knockdown impairs AML cell growth and function. (A) Histogram plots showing increased caspase 3/7 activity (luminescence) 24 h following shRNA knockdown in U937 and OCI-AML3 cells. (B) Targeted knockdown of HOXA/PBX3 resulted in altered cell cycle dynamics and accumulation of sub G0/G1 events measured by propidium iodide staining and flow cytometry analysis. (C) HOXA/PBX3 shRNA reduced overall growth in liquid culture (24–72 h), determined by ATP levels and (D) impaired cluster/colony forming potential following 7 days culture in methylcellulose whereas shMEIS1 resulted in increased clusters compared to respective non-silenced controls (NSC). (E) Reduced cellularity and metabolic activity (INT staining) of colonies following HOXA/TALE knockdown in OCI-AML3 or U937 cells cultured in methylcellulose for 10 days. Graphed data are representative of n≥4 experiments with mean and standard deviation plotted, P≤ 0.01/0.001 are denoted by */**, respectively. Images acquired at 200X and presented at 50X are representative of at least two constructs per gene target by lentiviral or retroviral delivery in each cell line.
The liquid culture assays were extended into methylcellulose to investigate the effect of reduced HOXA/TALE levels on hematopoietic cluster and colony formation. Significant reductions in cluster formation were observed for both cell lines following knockdown of HOXA6+A9 (~2-fold) or PBX3 (~8-fold) (Figure 4D). Notably, reduced MEIS1 levels significantly increased the number of clusters in OCI-AML3 and in U937 cells (>3-fold). Reduced HOXA, PBX3 or MEIS1 levels resulted in reduced cell metabolism and overall colony size in both cell lines as indicated by INT uptake and microscopy (Figure 4E).
HOXA/TALE knockdown impairs growth of primary cytogenetically normal acute myeloid leukemia cells
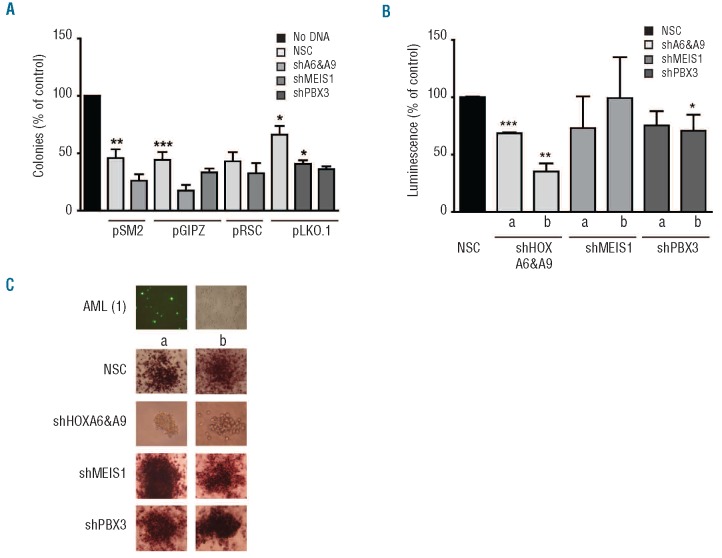
Peripheral blood or bone marrow mononuclear cells from anonymized patients with de novo CN-AML (n=5), were directly nucleofected with individual or combined shRNA (two per target) or respective non-silencing control constructs. RQ-PCR analysis of the patients’ samples showed comparable levels of expression for the four HOXA/TALE genes (Online Supplementary Table S4). Transient knockdown of HOXA6+A9 and PBX3 with specific shRNA constructs (n=2 per target) significantly and consistently reduced overall colony number (Figure 5A) and demonstrated reduced overall cellularity as determined by ATP quantification assays (Figure 5B). Transient transfection (~50%) was determined by positivity for green fluorescent protein and knockdown of HOXA6+A9, using different shRNA combinations, resulted in reduced cellularity and metabolic activity of colonies compared to non-silenced controls as supported by microscopy with INT staining. Although PBX3 knockdown resulted in a significant reduction in colony number, TALE knockdown did not result in a significant change in colony size or INT staining compared to respective controls (Figure 5C).
Figure 5.
Primary CN-AML cell colony formation is sensitive to HOXA/PBX3 levels. (A) Histogram plots demonstrating reduced colony formation of CN-AML samples (n=5) nucleo porated with PBX3, MEIS1 or combined HOXA6 + HOXA9 shRNA (n=2 per target) compared to non-silenced controls (NSC) and cultured in methylcellulose for up to 14 days. (B) Colony cellularity was evaluated by relative ATP fluorescence and plotted as percentage of NSC. Data are plotted as the mean ± S.E.M of duplicate transfection wells for each sample. P≤ 0.05/0.01/0.001 are denoted by */**/***, respectively. (C) Transfection efficiency (pmaxGFP®), and cellularity/metabolic activity were evaluated by fluorescence microscopy and INT staining, respectively. Representative colony images from two different shRNA per target captured at 200X using an inverted microscope (CKX41) with attached digital camera (E620) (both from Olympus, Essex, UK) are shown.
HOXA/TALE knockdown sensitizes acute myeloid leukemia cells to chemotherapy
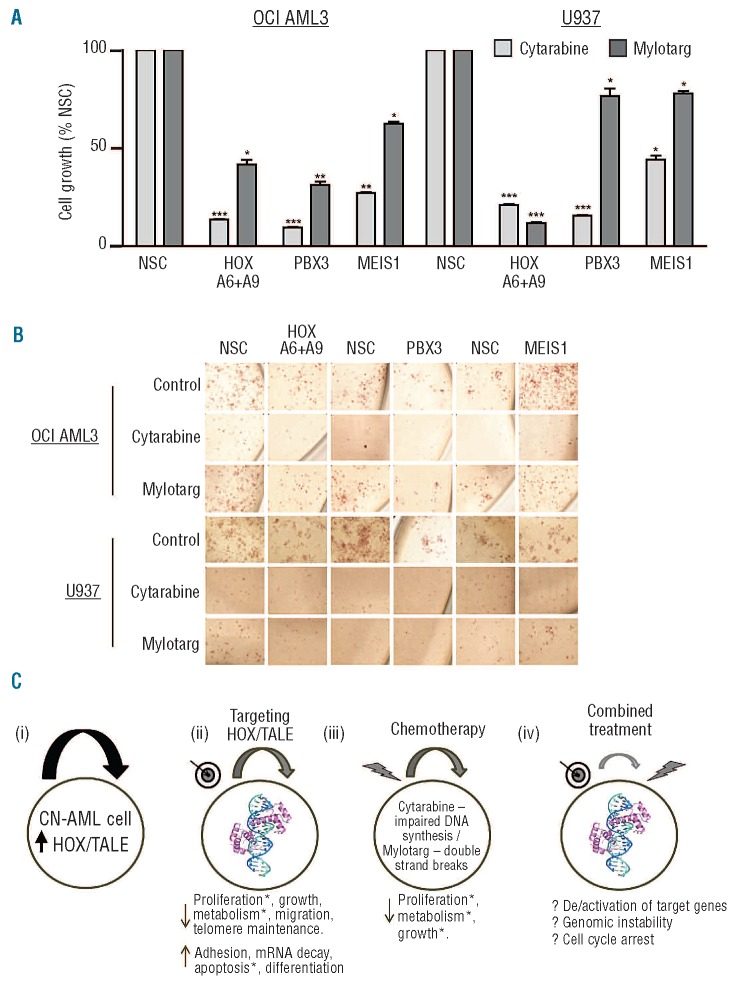
We next decided to investigate whether reduced HOXA/TALE could sensitize AML cell lines to standard-of-care chemotherapy. Initially, OCI-AML3 and U937 cells were shown to be more sensitive to cytarabine-treatment than Mylotarg using published IC50 dosages34,35 (Online Supplementary Figure S5). HOXA/TALE knockdown cells demonstrated significantly less growth in liquid culture and a measurable degree of sensitization compared to non-silencing and non-drug-treated controls (Figure 6A and Online Supplementary Figure S6A). MEIS1 knockdown plus cytarabine resulted in a significant reduction in growth of both cell lines (up to 4-fold) compared to non-silenced controls. Cytarabine treatment in combination with HOXA6+A9 or PBX3 knockdown resulted in a marked reduction in growth of both OCI-AML3 cells (up to 11-fold) and U937 cells (up to 7-fold) after 24 h of treatment. In colony formation assays cells pre-treated with shRNA for 48 h then exposed to cytarabine or Mylotarg showed dramatically impaired colony-forming capacity (Online Supplementary Figure S6B) and cellular metabolism as indicated by reduced INT uptake (Figure 6B) compared to non-silenced controls. Co-treatment with Mylotarg was generally not as cytotoxic as cytarabine. However, HOXA6+A9 or PBX3 knockdown in the presence of Mylotarg consistently reduced AML cell growth. Notably, MEIS1 knockdown plus Mylotarg increased colony formation in both cell lines under these conditions (Figure 6B and Online Supplementary Figure S6B). In all shRNA and drug treatment combinations, HOXA6+A9 or PBX3 knockdown resulted in impaired cluster/colony formation compared to that of their non-silenced control counterparts.
Figure 6.
HOXA/PBX3 knockdown sensitizes AML cells to chemotherapy. (A) OCI-AML3 and U937 cell growth was quantified in liquid culture by ATP levels following pre-conditioning knockdown with the indicated shRNA (48 h) ± cytarabine or Mylotarg treatment for a further 24 h. Data are presented as percentages of the respective non-silenced control cells (NSC). P≤ 0.05/0.01/ 0.001 are denoted by */**/***, respectively. (B) Pre-conditioned and treated cells were also seeded in methylcellulose ± drug, incubated for 10 days and stained with INT for 16 h prior to microscopy. Representative colony images captured at 200X using an inverted microscope (CKX41) with attached digital camera (E620) (both from Olympus, Essex, UK) are shown. (C) HOXA6, HOXA9, PBX3 and MEIS1 are a core part of the HOXA/TALE overexpression signature in CN-AML and may contribute to maintenance of leukemia clones (i). We present evidence indicating that targeting specific HOX/TALE complex components reduces growth of primary CN-AML and cell lines via intrinsic processes and cell–cell / matrix interactions identified by altered gene expression and phenotypic assays* (ii). Standard-of-care chemotherapeutics inhibit DNA synthesis and repair pathways (iii) and combined treatment further reduces CN-AML cell survival by mechanisms which may include (epi)genetic differences, altered cell cycle, destabilization of key cellular complexes or a combination of action (iv).
Discussion
Molecular classification of the heterogeneous CN-AML subgroup is aimed at improving prediction of outcome and response to therapy.3,4,37 With greater availability of next-generation-sequencing data the list of genetic markers38 is likely to increase. Accumulation and integration of these data will ultimately lead to greater understanding of complex genetic landscapes that may stratify patients’ treatment. However, some candidate genes with no demonstrable lesion are dysregulated in AML. Here we investigate whether a subset of homeobox genes can also be used to stratify subgroups of patients and, more importantly, could be candidates for targeted therapy.
A highly statistically significant core HOXA/TALE gene signature (HOXA6, HOXA9, PBX3, MEIS1) that partitions favorable risk from intermediate risk AML was identified in a large cohort of patients (n=420) and validated by RQ-PCR (Figure 1). The candidate signature was further validated in an independent AML dataset.28 CN-AML patients (n=114) were clustered based on HOX/TALE expression, not dependent on known mutations, and HOXAHi/PBX3Hi/MEIS1Lo was shown to be associated with poorer overall survival using a custom array (Figure 2). The identification of a four-gene signature with the ability to stratify CN-AML patients and indeed act further as a prognostic marker, irrespective of current molecular markers, may provide for higher throughput screening in a clinical setting in which large scale analysis is prohibitive.
Knockdown of HOXA6 or HOXA9 resulted in minimal compensation from the HOX network, with only a moderate increase in HOXA10 and decrease in HOXB2 expression noted (Online Supplementary Table S5). Combined HOXA6 plus HOXA9 knockdown, or targeted PBX3 knockdown alone, activated apoptotic pathways and resulted in markedly reduced AML cell growth and colony formation independently of NPM1 or DNMT3A mutations (Figures 3, 4, and 5 and Online Supplementary Figure S4). Notably, targeted knockdown of MEIS1 in primary cells or cell lines enhanced colony formation, indicating a dose-dependent role in AML colony formation or maintenance congruent with the HOXHi/PBX3Hi/MEIS1Lo phenotype. MEIS1 is known to have a role in colony formation22,39 and is essential for the induction and maintenance of MLL leukemogenesis. The data presented here suggest that further investigations will be required to understand the complex role of MEIS1 in CN-AML.
AML cell lines primed by HOXA/PBX3 knockdown demonstrated greater sensitivity to standard chemotherapies than mock-treated lines or those primed with non-silencing controls (Figure 6A,B and Online Supplementary Figure S6). In methylcellulose models, directly evaluating stem/progenitor cell colony-forming potential, the combination of HOXA/PBX3 knockdown and Mylotarg treatment proved highly effective in reducing cluster formation, particularly in OCI-AML3 cells. The difference in response may be due to mutation status or the level of expression of CD33, the primary route for Mylotarg internalization. The NPM1-mut and DNMT3A-mut OCI-AML3 cells are reported to have elevated levels of CD33-antigen compared to U937 cells.40,41
In a recent study Li et al. reported that mir181b overexpression inhibited an MLL model of leukemia in part by decreasing HOXA7, A9, A11 and PBX3 levels.42 A follow-up study indicated that the HOXA/PBX3 interaction is critical for MLL-induced leukemia43. The data presented herein demonstrate that CN-AML cells are also sensitive to HOXA/PBX3 levels and indicate that clinically practical approaches resulting in defined targeting of the HOXA-PBX3-MEIS1 axis alone, as next generation therapy, or in combination with standard chemotherapy (Figure 6C), may provide benefit to intermediate risk, CN-AML patients.
Acknowledgments
The authors would like to thank Dr Hilary Colyer for the processing and cytogenetic analysis of patients’ samples, and staff in the Biological Resource Unit, Bioinformatics and Flow Cytometry Cores, Queen’s University, Belfast for technical support.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
GJD was the recipient of a Yamagiwa-Yoshida Fellowship and AT an American Cancer Society Beginning Investigator Award (both UICC). This work was supported by grants from Leukaemia Lymphoma Research-UK (07016 and 09035) the Northern Ireland Leukaemia Research Fund and the Department for Education and Learning, Northern Ireland and facilitated in part by the COST (European Cooperation in Science and Technology) network (AT, KIM and DG).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29(5):475–86 [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Mrozek K. Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am. 2011;25:(6)1135–61, vii [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–65 [DOI] [PubMed] [Google Scholar]
- 4.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–16 [DOI] [PubMed] [Google Scholar]
- 5.Burnett AK, Hills RK, Milligan DW, Goldstone AH, Prentice AG, McMullin MF, et al. Attempts to optimize induction and consolidation treatment in acute myeloid leukemia: results of the MRC AML12 trial. J Clin Oncol. 2010;28(4):586–95 [DOI] [PubMed] [Google Scholar]
- 6.Balleisen S, Kuendgen A, Hildebrandt B, Haas R, Germing U. Prognostic relevance of achieving cytogenetic remission in patients with acute myelogenous leukemia or high-risk myelodysplastic syndrome following induction chemotherapy. Leuk Res. 2009;33(9):1189–93 [DOI] [PubMed] [Google Scholar]
- 7.Burnett AK, Wheatley K, Goldstone AH, Stevens RF, Hann IM, Rees JH, et al. The value of allogeneic bone marrow transplant in patients with acute myeloid leukaemia at differing risk of relapse: results of the UK MRC AML 10 trial. Br J Haematol. 2002;118(2):385–400 [DOI] [PubMed] [Google Scholar]
- 8.Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118(4):1069–76 [DOI] [PubMed] [Google Scholar]
- 9.Dohner K, Schlenk RF, Habdank M, Scholl C, Rucker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–6 [DOI] [PubMed] [Google Scholar]
- 10.Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106(12):3733–9 [DOI] [PubMed] [Google Scholar]
- 11.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106(12):3747–54 [DOI] [PubMed] [Google Scholar]
- 12.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16(2):186–95 [DOI] [PubMed] [Google Scholar]
- 14.Thompson A, Quinn MF, Grimwade D, O’Neill CM, Ahmed MR, Grimes S, et al. Global down-regulation of HOX gene expression in PML-RARalpha + acute promyelocytic leukemia identified by small-array real-time PCR. Blood. 2003;101(4): 1558–65 [DOI] [PubMed] [Google Scholar]
- 15.Debernardi S, Lillington DM, Chaplin T, Tomlinson S, Amess J, Rohatiner A, et al. Genome-wide analysis of acute myeloid leukemia with normal karyotype reveals a unique pattern of homeobox gene expression distinct from those with translocation-mediated fusion events. Genes Chromosomes Cancer. 2003;37(2):149–58 [DOI] [PubMed] [Google Scholar]
- 16.Lebert-Ghali CE, Fournier M, Dickson GJ, Thompson A, Sauvageau G, Bijl JJ. HoxA cluster is haploinsufficient for activity of hematopoietic stem and progenitor cells. Exp Hematol. 2010;38:(11)1074–86 e1–5 [DOI] [PubMed] [Google Scholar]
- 17.Dickson GJ, Kwasniewska A, Mills KI, Lappin TR, Thompson A. Hoxa6 potentiates short-term hemopoietic cell proliferation and extended self-renewal. Exp Hematol. 2009;37:(3)322–33 e3 [DOI] [PubMed] [Google Scholar]
- 18.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99(1):121–9 [DOI] [PubMed] [Google Scholar]
- 19.Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115(14):2910–8 [DOI] [PubMed] [Google Scholar]
- 20.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89(6):1922–30 [PubMed] [Google Scholar]
- 21.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21(1):224–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Jenkins NA, Copeland NG. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13(10):2235–42 [PubMed] [Google Scholar]
- 24.Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11(12):6149–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selleri L, DiMartino J, van Deursen J, Brendolan A, Sanyal M, Boon E, et al. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol Cell Biol. 2004;24(12):5324–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee JW, Arata A, Selleri L, Jacobs Y, Arata S, Onimaru H, et al. Pbx3 deficiency results in central hypoventilation. Am J Pathol. 2004;165(4):1343–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28(15):2529–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein HU, Ruckert C, Kohlmann A, Bullinger L, Thiede C, Haferlach T, et al. Quantitative comparison of microarray experiments with published leukemia related gene expression signatures. BMC Bioinformatics. 2009;10: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, et al. Novel definition files for human GeneChips based on GeneAnnot. BMC Bioinformatics. 2007;8: 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson GJ, Lappin TR, Thompson A. Complete array of HOX gene expression by RQ-PCR. Methods Mol Biol. 2009;538:369–93 [DOI] [PubMed] [Google Scholar]
- 32.Drexler HG, MacLeod RAF, Nagel S, Dirks WG, Uphoff CC, Steube KG, et al. Guide to leukemia-lymphoma cell lines on CD. Blood. 2005;106(11):165 [Google Scholar]
- 33.Tiacci E, Spanhol-Rosseto A, Martelli MP, Pasqualucci L, Quentmeier H, Grossmann V, et al. The NPM1 wild-type OCI-AML2 and the NPM1-mutated OCI-AML3 cell lines carry DNMT3A mutations. Leukemia. 2012;26(3):554–7 [DOI] [PubMed] [Google Scholar]
- 34.Ohta T, Hori H, Ogawa M, Miyahara M, Kawasaki H, Taniguchi N, et al. Impact of cytidine deaminase activity on intrinsic resistance to cytarabine in carcinoma cells. Oncol Rep. 2004;12(5):1115–20 [PubMed] [Google Scholar]
- 35.Amico D, Barbui AM, Erba E, Rambaldi A, Introna M, Golay J. Differential response of human acute myeloid leukemia cells to gemtuzumab ozogamicin in vitro: role of Chk1 and Chk2 phosphorylation and caspase 3. Blood. 2003;101(11):4589–97 [DOI] [PubMed] [Google Scholar]
- 36.Shen WF, Rozenfeld S, Kwong A, Kom ves LG, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19(4):3051–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–74 [DOI] [PubMed] [Google Scholar]
- 38.Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F, et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood. 2012;120(15):2963–72 [DOI] [PubMed] [Google Scholar]
- 39.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21(21):2762–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Propris MS, Raponi S, Diverio D, Milani ML, Meloni G, Falini B, et al. High CD33 expression levels in acute myeloid leukemia cells carrying the nucleophosmin (NPM1) mutation. Haematologica. 2011;96(10): 1548–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen P, Wang J, Hope K, Jin L, Dick J, Cameron R, et al. Nuclear localizing sequences promote nuclear translocation and enhance the radiotoxicity of the anti-CD33 monoclonal antibody HuM195 labeled with 111In in human myeloid leukemia cells. J Nucl Med. 2006;47(5):827–36 [PubMed] [Google Scholar]
- 42.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119(10):2314–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Huang H, Li Y, Arnovitz S, Chen P, Huang H, et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121(8):1422–31 [DOI] [PMC free article] [PubMed] [Google Scholar]