Abstract
For several decades, the regulation of casein gene expression by the lactogenic hormones, prolactin and glucocorticoids, has provided an excellent model system in which to study how steroid and peptide hormones regulate gene expression. Early studies of casein gene regulation defined conserved sequence elements in the 5′ flanking region of these genes, including one of which was identified as a γ-interferon activation sequence (GAS). Although this site was thought to interact with a mammary gland-specific factor, purification and cloning of this factor by Bernd Groner and his colleagues revealed it was instead a new member of the signal transducers and activators of transcription family, Stat5, which was expressed in many tissues. The exquisite tissue-specific expression of the casein genes was subsequently shown to depend not on a single transcription factor but on composite response elements that interacted with a number of ubiquitous transcription factors in response to the combinatorial effects of peptide and steroid hormone signaling. More recent studies have defined cooperative effects of prolactin and glucocorticoids as well as antagonistic effects of progesterone on the chromatin structure of both the casein gene proximal promoter region as well as a distal enhancer. Local chromatin modifications as well as long-range interactions facilitated by DNA looping are required for the hormonal regulation of β-casein gene expression. The casein genes are part of a large gene cluster, and the chromatin landscape of the entire cluster is regulated in a tissue-specific and developmental manner. Finally, newly discovered large non coding RNAs, such as the pregnancy-induced non coding RNA (PINC) may play an important role in the epigenetic regulation of mammary gland differentiation.
Keywords: casein genes, chromatin conformation, glucocorticoids, prolactin, Stat5
Introduction
The rapid (within 1 h) prolactin induction of casein mRNA was first demonstrated using rat mammary organ explant cultures in a serum-free, chemically defined medium [1]. In these studies, the presence of hydrocortisone was required for maximal prolactin induction of casein mRNA, which was inhibited in a dose-dependent manner by progesterone. Although this system facilitated the investigation of the mechanisms by which a peptide hormone regulates rapid accumulation of a specific mRNA, it was not amenable to genetic manipulation. The availability of genomic constructs encoding the individual casein genes provided the alternative methods needed to identify those sequences required for tissue-specific and hormonal induction. The first approach was to use comparative genomics to identify several conserved sequence elements in the 5′ flanking DNA of several rat and bovine casein genes [2]. In particular, this identified a conserved sequence element centered at approximately –90 bp 5′ to the start site of transcription, which was later shown to be similar to a γ-interferon activation sequence (GAS) site identified in interferon-regulated genes [3]. These conserved 5′ flanking sequences were hypothesized to contain potential cis-regulatory elements, which might be responsible for the coordinate expression of the functionally related casein genes during mammary gland development. To test this hypothesis, lines of transgenic mice were developed, which contained approximately 500 bp of the 5′ flanking region along with the first exon and 0.5 kb of the first intron linked to a chloramphenicol acetyltransferase (Cat) reporter gene [4]. These sequences proved to be sufficient to specifically target reporter gene expression to the mammary gland of lactating mice.
To further dissect the regulatory sequences responsible for hormonal induction of casein gene expression, however, a cell-based assay in which to test wild-type and mutated constructs was required. Unfortunately, it was not possible to demonstrate appropriate hormonal regulation of milk protein gene-reporter constructs in existing breast cancer cell lines despite the presence of prolactin and glucocorticoid receptors (GRs) in these cell lines. A major advance came with the development of the cloned mammary epithelial HC11 cell line [5] derived from the Comma-D cell line [6]. Prolactin regulation of β-casein gene expression was demonstrated in HC11 cells cultured on a plastic substratum instead of floating collagen gels or matrigel. Most importantly, appropriate hormonal regulation was observed using HC11 cells stably transfected with the β-casein-Cat reporter constructs [7]. These studies demonstrated that β-casein gene promoter activity was regulated by the hormone-induced relief of transcriptional repression and what was thought at the time to be a mammary-specific factor (MGF) [7]. In particular, site-directed mutagenesis was used to identify the importance of the GAS site that interacted with MGF and was required for prolactin induction. In these seminal studies, five nuclear complexes including MGF were shown to interact with the conserved sequences identified in the promoters of several casein genes. MGF was subsequently shown to be a novel member of the cytokine regulated transcription factor family, later designated as signal transducers and activators of gene transcription 5 (Stat5) and conferred the prolactin response. Furthermore, the nuclear factor Yin and Yang 1 (YY1) was shown to prevent the formation of a lactation-associated complex on the β-casein promoter [8, 9]. One major conclusion from these and other subsequent studies is that the hormonal and developmental-specific regulation of casein gene expression was mediated by composite response elements [10, 11]. Although these early studies were focused on the proximal casein promoter region, subsequent studies identified a more distal β-casein gene enhancer region in several different mammalian genes [12–14]. Furthermore, the casein genes were shown to exist in a mammalian-specific gene cluster [14]. Studies in transgenic mice using large cosmid clones suggested that there might even be distal-regulatory elements required for the developmental regulation of the entire cluster [15]. With this background, our laboratory initiated studies to answer the question how these hormonal and developmental cues were integrated not only at the level of the linear DNA sequence but rather in the context of the chromatin architecture of the casein genes.
Integration of prolactin and glucocorticoid signaling by ordered recruitment of specific transcription factors and chromatin modifiers
The availability of several technologies, such as the chromatin immunoprecipitation (ChIP) assay [16] followed by either real-time quantitative PCR or more recently DNA sequencing (ChIP-seq) [17] has revolutionized our ability to analyze transcription factor interactions and chromatin modifications at specific genomic sites as well as globally. Development of the chromosome conformation capture (3C) assay has also provided a quantitative method to allow the identification of physical interactions between chromatin segments of up to several hundred kilobases apart [18]. Using ChIP analysis, we examined the dynamics of recruitment of different transcription factors at the hormonally activated β-casein proximal promoter, as well as the more distal mouse β-casein enhancer, the latter located >6 kb upstream of the transcription start site, using the HC11 cell model [19]. Both glucocorticoids and prolactin were required under the culture conditions used in these experiments, i.e., the use of charcoal-stripped horse serum to remove both steroid hormones and any prolactin capable of interacting with the mouse prolactin receptor. Hormonal stimulation of HC11 cells with Prl alone resulted in a rapid recruitment of Stat5 and histone deacetylases (HDAC) 1 to the β-casein promoter and enhancer and reciprocally the dissociation of YY-1 and HDAC3 from the proximal promoter, but this was not sufficient to promote β-casein transcription. β-Casein gene transcription, however, required treatment with both prolactin and glucocorticoids and the synergistic interaction of the GR and the LAP (liver-enriched activating protein) isoform of CCAAT/enhancer-binding protein β (C/EBPβ) followed by stable association of p300 and phosphorylated RNA polymerase (Pol) II at the promoter and enhancer. Glucocorticoids in the absence of prolactin resulted in a rapid acetylation of histone H3 at both the promoter and the enhancer despite the lack of known glucocorticoid response elements in the enhancer region. This suggested that that the GR may exert multiple functions: it may initiate chromatin remodeling required for transcription initiation and at the same time it may play a bridging role in β-casein activation through interactions with Stat5 and C/EBPβ as well as binding to different coactivators, comodulators, and/or corepressors. Both extracellular matrix and prolactin-regulated transcription of the casein genes also require the concerted action of chromatin remodeling enzymes, such as SWI/SNF, as well as transcription factors and coactivators [20, 21]. ChIP analyses demonstrated that the ATPase activity of SWI/SNF is necessary for recruitment of RNA transcriptional machinery, but not for binding of transcription factors or for histone acetylation [21]. Coimmunoprecipitation analyses also showed that the SWI/SNF complex is associated with STAT5, C/EBPβ, and GR.
Lactogenic hormonal induction of long-distance interactions between β-casein gene regulatory elements
The similar kinetics of assembly of transcription factors, the coactivator p300, and RNA Pol II as well as histone acetylation at the proximal promoter and the distal enhancer suggested that these two regulatory regions might communicate with each other through protein:protein interactions. To test this hypothesis, 3C assays were performed in the presence of both prolactin and glucocorticoids and each hormone alone [22]. Stimulation with both prolactin and hydrocortisone was required for the induction of these long-range interactions between the promoter and the enhancer, and no DNA looping was observed in non-treated cells or cells treated with each of the hormones separately. The lactogenic hormone-induced interaction between the proximal promoter and distal enhancer also was confirmed in hormone-treated primary 3D mammary acini cultures and mammary gland tissue from lactating and non-lactating mice. This long-range interaction was more prevalent in the mammary gland from lactating mice than in MECs isolated from virgin mice. Furthermore, β-casein mRNA induction and long-range interactions between these regulatory regions were inhibited in a progestin-dependent manner after stimulation with prolactin and hydrocortisone in HC11 cells expressing the human progesterone receptor B isoform. In the absence of lactogenic hormones, both β-casein regulatory regions exhibited a constitutive dimethylation of lysine 9 of histone H3 (H3K9me2) [23], a modification usually associated with repressive chromatin structure. Progestins prevented dissociation of the corepressors YY1 and HDAC3 from the promoter as well as the demethylation of lysine 9 of histone 3 induced by Prl and glucocorticoids.
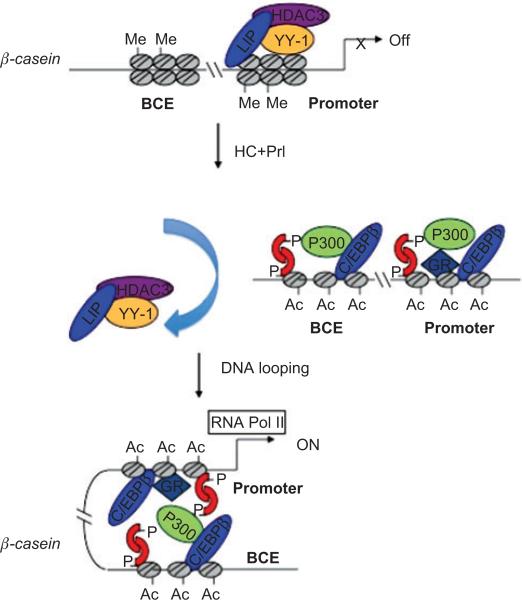
A model to explain the sequential formation of complexes leading to the activation of β-casein gene expression after hormonal stimulation
To summarize these studies, we proposed the following model reproduced in Figure 1. In the absence of lactogenic hormones, YY-1 binds to the β-casein promoter presumably interacting with the LIP isoform of C/EBPβ and HDAC3, which in turn results in the formation of H3K9me2 associated with a repressive chromatin structure. PrlR activation of Stat5 dimerization via Jak 2 phosphorylation followed by nuclear translocation and DNA binding promotes the rapid displacement of YY-1 and HDAC3 from the β-casein promoter. Activated Stat5 then binds to sites adjacent to the C/EBPβ binding sites at both the promoter and enhancer regulatory regions and recruits HDAC1. Consequently, HDAC1 may deacetylate C/EBPβ LAP, and once deacetylated at specific sites, LAP-LAP homodimers and/or LAP-LIP heterodimers can bind with a high affinity to their cognate DNA binding sites as well as interact with other transcription factors, such as GR. After hormonal stimulation with Prl and HC, the transcription factors Stat5, GR, and C/EBPβ rapidly bind to their respective response elements within β-casein regulatory regions and recruit p300 through protein:protein interactions. The recruitment of the coactivator p300 facilitates histone acetylation, which in turn modifies chromatin organization. Interactions between open chromatin structures at the promoter and enhancer mediated through these transcription factors and coactivators enable direct protein-protein contacts facilitated by DNA looping. The formation of the active chromatin loop between distant regulatory elements facilitates binding of the basal transcriptional machinery to the DNA template and initiates transcription. Prolactin alone recruits Stat5 to a complex that is competent to recruit Pol II and stimulates a low level of transcription after 24 h. However, GR recruitment is essential to increase histone acetylation, resulting in an open chromatin structure at both regulatory regions. Thus, the combined treatment with both hormones is required for the formation of an active chromatin loop between the proximal promoter and distal enhancer to achieve maximal β-casein gene transcription. For simplicity, potential GR:Stat5 and GR:C/EBPβ interactions with the distal enhancer are not shown in this figure but may help facilitate long-range looping.
Figure 1.
Hormonal regulation of β-casein gene long-range chromatin interactions.
Developmental changes in the chromatin architecture of the casein gene locus
The casein genes comprise a mammalian-specific gene cluster containing three or four evolutionarily related casein genes and one physically linked gene (κ-casein) with a functional association. To investigate the developmental stage- and tissue-specific changes in the chromatin architecture of the casein gene locus, ChIP-seq was performed on tissues using a variety of different antibodies to identify different histone marks, which differentiate open from closed chromatin (M. Rijnkels et al., Unpublished observations). For example, dimethylated lysine 4 of histone H3 (H3K4me2) is associated with open chromatin and marks active and poised gene promoters as well as active distal regulatory elements. ChIP-seq analysis using an antibody against H3K4me2 showed that the entire casein gene cluster is enriched for this specific histone mark in lactating mammary gland tissue, but this modification is lacking in the liver. Neighboring genomic regions containing genes that are expressed in both tissues show enrichment for H3K4me2 in both tissues. To determine if this open chromatin structure is a result of chromatin reorganization during development and functional differentiation or is an intrinsic property of the mammary epithelium, ChIP-seq analysis was performed in epithelial cells in mammary organoids isolated at different stages of development. A lack of enrichment of H3K4me2 at the promoters and throughout most of the casein gene region was observed in the virgin mammary epithelial cells, which is suggestive of a more closed chromatin organization relative to lactation. Evolutionary conservation of DNA sequences outside of the coding regions of a gene is thought to imply function – a potential role as distal regulatory elements. Previous analysis of the mammalian casein gene loci identified several evolutionarily conserved elements that were not within known promoters or coding regions [14]. Enrichment of H3K4me2 at some of these evolutionary conserved regions in the virgin MEC indicate that changes in epigenetic marks occurred much earlier in these regions than at the promoter and gene regions itself. These evolutionary conserved regions may therefore represent distal regulatory elements that establish a chromatin structure that facilitates 3D interactions leading to casein gene expression.
Long noncoding RNAs may play a role in mammary gland differentiation
Previously, our laboratory identified pregnancy-induced non-coding RNA (PINC) as a gene whose expression was elevated in the involuted rat mammary compared with an age-matched virgin gland [24]. PINC is a mammalian-specific, evolutionary conserved, alternatively spliced, and polyadenylated long non coding RNA (lncRNA) whose expression is induced during pregnancy and then decreases at lactation [25, A.N. Shore et al., Unpublished observations]. We originally hypothesized that PINC might be involved in pregnancy-induced cell fate changes in the mammary gland presumably by affecting epigenetic reprogramming [26]. One feature of an emerging class of lncRNAs including PINC is an association with chromatin-modifying complexes. In fact, as many as 38 % of large intergenic non coding RNAs have been shown to interact with various chromatin-modifying complexes and 24 % specifically interact with polycomb repressive complex 2 (PRC2) [27]. Interestingly, PINC expression is significantly decreased in response to hormonally induced differentiation of HC11 cells (A.N. Shore et al., Unpublished observations). Overexpression of PINC blocks lactogenic differentiation, whereas knockdown enhances markers of differentiation (A.N. Shore et al., Unpublished observations). Finally, PINC interacts with members of the PRC2 complex in both HC11 cells and mammary epithelial cells isolated from mice at day 16 of pregnancy, suggesting that it may be associated with repressive chromatin marks, such as H3K27me3 (A.N. Shore et al., Unpublished observations). Although the precise gene targets of PINC as well as other novel lncRNAs recently shown to be regulated during mammary gland development [28] remain to be established, these studies illustrate a whole new level of complexity of gene regulation. Thus, the message from these studies is that both hormonal and developmental regulation of casein gene expression require more than the binding of a single transcription factor, such as Stat5. Cooperative interactions among several transcription factors and possibly non coding RNAs may be required, and these often result in both local and more distal changes in chromatin architecture.
Acknowledgments
These studies were supported by grants R37CA16303 (J.M.R.), 1R21HD05376, 1R03HD05609, and USDA_6200-51000-048 (M.R.) and a DAMD predoctoral fellowship W81XWH-06-1-0717 (A.S.).
References
- 1.Matusik RJ, Rosen JM. Prolactin induction of casein mRNA in organ culture. A model system for studying peptide hormone regulation of gene expression. J Biol Chem. 1978;253:2343–7. [PubMed] [Google Scholar]
- 2.Yu-Lee LY, Richter-Mann L, Couch CH, Stewart AF, Mackinlay AG, Rosen JM. Evolution of the casein multigene family: conserved sequences in the 5′ flanking and exon regions. Nucleic Acids Res. 1986;14:1883–902. doi: 10.1093/nar/14.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dale TC, Rosen JM, Guille MJ, Lewin AR, Porter AG, Kerr IM, Stark GR. Overlapping sites for constitutive and induced DNA binding factors involved in interferon-stimulated transcription. EMBO J. 1989;8:831–9. doi: 10.1002/j.1460-2075.1989.tb03444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KF, Atiee SH, Rosen JM. Differential regulation of rat beta-casein-chloramphenicol acetyltransferase fusion gene expression in transgenic mice. Mol Cell Biol. 1989;9:560–5. doi: 10.1128/mcb.9.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 1988;7:2089–95. doi: 10.1002/j.1460-2075.1988.tb03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danielson KG, Oborn CJ, Durban EM, Butel JS, Medina D. Epithelial mouse mammary cell line exhibiting normal morpho-genesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci USA. 1984;81:3756–60. doi: 10.1073/pnas.81.12.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt-Ney M, Doppler W, Ball RK, Groner B. Beta-casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Mol Cell Biol. 1991;11:3745–55. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier VS, Groner B. The nuclear factor YY1 participates in repression of the beta-casein gene promoter in mammary epithelial cells and is counteracted by mammary gland factor during lactogenic hormone induction. Mol Cell Biol. 1994;14:128–37. doi: 10.1128/mcb.14.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raught B, Khursheed B, Kazansky A, Rosen J. YY1 represses beta-casein gene expression by preventing the formation of a lactation-associated complex. Mol Cell Biol. 1994;14:1752–63. doi: 10.1128/mcb.14.3.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosen JM, Wyszomierski SL, Hadsell D. Regulation of milk protein gene expression. Annu Rev Nutr. 1999;19:407–36. doi: 10.1146/annurev.nutr.19.1.407. [DOI] [PubMed] [Google Scholar]
- 11.Rosen JM, Zahnow C, Kazansky A, Raught B. Composite response elements mediate hormonal and developmental regulation of milk protein gene expression. Biochem Soc Symp. 1998;63:101–13. [PubMed] [Google Scholar]
- 12.Schmidhauser C, Casperson GF, Myers CA, Sanzo KT, Bolten S, Bissell MJ. A novel transcriptional enhancer is involved in the prolactin- and extracellular matrix-dependent regulation of beta-casein gene expression. Mol Biol Cell. 1992;3:699–709. doi: 10.1091/mbc.3.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winklehner-Jennewein P, Geymayer S, Lechner J, Welte T, Hansson L, Geley S, Doppler W. A distal enhancer region in the human beta-casein gene mediates the response to prolactin and glucocorticoid hormones. Gene. 1998;217:127–39. doi: 10.1016/s0378-1119(98)00380-1. [DOI] [PubMed] [Google Scholar]
- 14.Rijnkels M, Elnitski L, Miller W, Rosen JM. Multispecies comparative analysis of a mammalian-specific genomic domain encoding secretory proteins. Genomics. 2003;82:417–32. doi: 10.1016/s0888-7543(03)00114-9. [DOI] [PubMed] [Google Scholar]
- 15.Rijnkels M, Kooiman PM, Platenburg GJ, van Dixhoorn M, Nuijens JH, de Boer HA, Pieper FR. High-level expression of bovine alpha s1-casein in milk of transgenic mice. Transgenic Res. 1998;7:5–14. doi: 10.1023/a:1008892720466. [DOI] [PubMed] [Google Scholar]
- 16.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 19.Kabotyanski EB, Huetter M, Xian W, Rijnkels M, Rosen JM. Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers. Mol Endocrinol. 2006;20:2355–68. doi: 10.1210/me.2006-0160. [DOI] [PubMed] [Google Scholar]
- 20.Xu R, Nelson CM, Muschler JL, Veiseh M, Vonderhaar BK, Bissell MJ. Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function. J Cell Biol. 2009;184:57–66. doi: 10.1083/jcb.200807021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Spencer VA, Bissell MJ. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J Biol Chem. 2007;282:14992–9. doi: 10.1074/jbc.M610316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabotyanski EB, Rijnkels M, Freeman-Zadrowski C, Buser AC, Edwards DP, Rosen JM. Lactogenic hormonal induction of long distance interactions between beta-casein gene regulatory elements. J Biol Chem. 2009;284:22815–24. doi: 10.1074/jbc.M109.032490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buser AC, Obr AE, Kabotyanski EB, Grimm SL, Rosen JM, Edwards DP. Progesterone receptor directly inhibits beta-casein gene transcription in mammary epithelial cells through promoting promoter and enhancer repressive chromatin modifications. Mol Endocrinol. 2011;25:955–68. doi: 10.1210/me.2011-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginger MR, Gonzalez-Rimbau MF, Gay JP, Rosen JM. Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol Endocrinol. 2001;15:1993–2009. doi: 10.1210/mend.15.11.0724. [DOI] [PubMed] [Google Scholar]
- 25.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci USA. 2006;103:5781–6. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginger MR, Rosen JM. Pregnancy-induced changes in cell-fate in the mammary gland. Breast Cancer Res. 2003;5:192–7. doi: 10.1186/bcr603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, Vargas AC, Campbell IG, Brown MA, Dinger ME, Mattick JS. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]