Abstract
Kallmann syndrome (KS) is a developmental disease that expresses in patients as hypogonadotropic hypogonadism and anosmia. KS is commonly associated with mutations in the extracellular D2 domain of the fibroblast growth factor receptor (FGFR). In this study, for the first time, the molecular basis for the FGFR associated KS mutation (A168S) is elucidated using a variety of biophysical experiments, including multidimensional NMR spectroscopy. Secondary and tertiary structural analysis using far UV circular dichroism, fluorescence and limited trypsin digestion assays suggest that the KS mutation induces subtle tertiary structure change in the D2 domain of FGFR. Results of isothermal titration calorimetry experiments show the KS mutation causes a 10-fold decrease in heparin binding affinity and also a complete loss in ligand (FGF-1) binding. 1H-15N chemical perturbation data suggest that complete loss in the ligand (FGF) binding affinity is triggered by a subtle conformational change that disrupts crucial structural interactions in both the heparin and the FGF binding sites in the D2 domain of FGFR. The novel findings reported in this study are expected to provide valuable clues toward a complete understanding of the other genetic diseases linked to mutations in the FGFR.
Keywords: Fibroblast Growth Factor Receptor, Fibroblast growth factor, Kallmann's syndrome, Nuclear Magnetic Resonance spectroscopy
Introduction
Kallmann syndrome (KS) is a form of idiopathic hypogonadotropic hypogonadism (IHH) that expresses phenotypically with anosmia [1]. Most commonly, KS is expressed with synkinesia, hearing loss, labial or palentine cleft, dental anomalies and inhibited puberty development. An estimated 10% of KS cases are attributed to mutations in the fibroblast growth factor receptor 1 (FGFR1) [2-5].
Fibroblast growth factors (FGFs) are a family of heparin-binding proteins which regulate key cellular processes such as angiogenesis, cell differentiation, morphogenesis, wound healing and tumor growth [6-8]. FGF receptors consist of three extracellular ligand-binding domains (D1, D2, and D3), a single transmembrane helix, and a cytoplasmic tyrosine kinase domain. Cell surface-bound heparan sulfate proteoglycans (HSPGs) that support dimerization or oligomerization of the FGF receptors (FGFRs) are believed to be required for the activation of the FGF signaling pathway [9]. The FGF signaling is shown to be modulated by anosmin-1, a KAL-1 gene product underlying X-linked KS [10]. Anosmin-1 is a 680-amino acid glycosylated protein, which directly binds to HSPG and FGFR and regulates the assembly and activity of the FGF signaling complex [11].
The extracelluar D2 domain of FGFRs is suggested to bind with both HSPGs and FGFs to form a ternary FGF signaling complex. Mutations in the D2 domain of FGFR1 have been associated with families carrying Kallmann syndrome phenotypes [12-16]. The A168S mutation in the D2 domain of FGFR is one of the common KS mutations. Knockout mutational studies in mice have established A168S as a loss-of-function mutation resulting in the inactivation of FGFR. Inactivation of FGFR in mice telencephalon tissue results in the failure of olfactory bulb formation- a phenotype manifestation commonly observed in KS [17]. The molecular basis for the observed loss-of-function associated with KS mutations is still unclear. It is believed that KS mutations either disrupt the anosmin-1/FGFR interaction(s) or cause a drastic structural change in FGFR leading to loss in the binding affinity to the ligand (FGF) [11]. However, very little experimental evidence exists in support or in contradiction of this proposal. In this study for the first time the structural basis for the A168S associated KS is elucidated. The results of the present study are expected to trigger intensive studies leading to a comprehensive understanding of the molecular mechanism underlying the different FGFR mutation related diseases.
Materials and Methods
Mutagenesis, overexpression and protein purification
The A168S mutation on the FGFR2 D2 domain (residues, 145-255) was generated using the Quikchange kit (Stratagene). The mutation was confirmed by automated DNA sequencing. Wild type and the A168S mutant of the D2 domain were overexpressed and purified from Escherichia coli [BL21 (pLysS) strain] according to the method reported previously [18].
Far UV Circular Dichrosim (CD)
Data on the wild type D2 and the A168S mutant were acquired using a Jasco J-710 spectropolarimeter. Protein solutions (90-100 μM) were prepared in 10 mM phosphate buffer containing 100 mM NaCl and 50 mM ammonium sulfate (pH 6.5). Wavelength scans were set to 250-190 nm. CD measurements were made using a 0.1 cm pathlength cuvette. A total of 10 scans were averaged for each sample.
Fluorescence spectroscopy
Measurements were made using a Hitachi F-2500 fluorimeter. Fluorescence measurements were made using a 1.0 cm pathlength and the concentration of each protein sample was adjusted to ∼50 μM. Excitation wavelength was set to 280 nm and intrinsic tryptophan emission was measured from 300 nm to 450 nm. 8-anilino-1-napthalene sulphonate (ANS) binding experiments were performed using an excitation wavelength of 390 nm and an emission wavelength range of 450 nm- 600 nm. In the ANS titration experiments, protein samples were titrated with 3 μl aliquots of 10 mM ANS in 10 mM phosphate buffer containing 100 mM NaCl and 50 mM ammonium sulfate at pH 6.5.
Differential scanning calorimetry (DSC)
DSC measurements were performed using a DASM-1M calorimeter. Protein concentration used was in the range of 1.0 – 1.5 mg/ml in 10 mM phosphate buffer containing 50 mM ammonium sulfate and 100 mM NaCl (pH ∼6.5). The scan rate was set to 1 °C/min and protein samples were heated from 10 °C - 90°C. Data were analyzed using the Origin DSC software provided by Microcal Inc.
Isothermal titration caolorimetry (ITC)
ITC measurements were performed using Microcal VP titration calorimeter (Northhampton, MA, USA). Titrations consisted of 5-10 μl injections of 0.5 mM sucrose octasulfate (a heparin analog) delivered into 0.050 mM concentration of the D2 domain. Injections were delayed for 3.5 minutes to allow the titration peak to return to the baseline prior to additional injections. The titrations curves were analyzed using Origin software supplied by Microcal Inc.
NMR spectroscopy
1H-15N HSQC data were acquired at 298 °K using 0.5 mM 15N labeled D2 domain in 10 mM phosphate buffer (prepared in 95% H2O + 5 %D2O, pH 6.5) containing 50 mM ammonium sulfate and 100 mM NaCl. All experiments were performed using Bruker Avance 700 MHz and 500 MHz NMR spectrometers equipped with cryoprobes. NMR data were referenced to the 1H resonance frequency of 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS).
Results
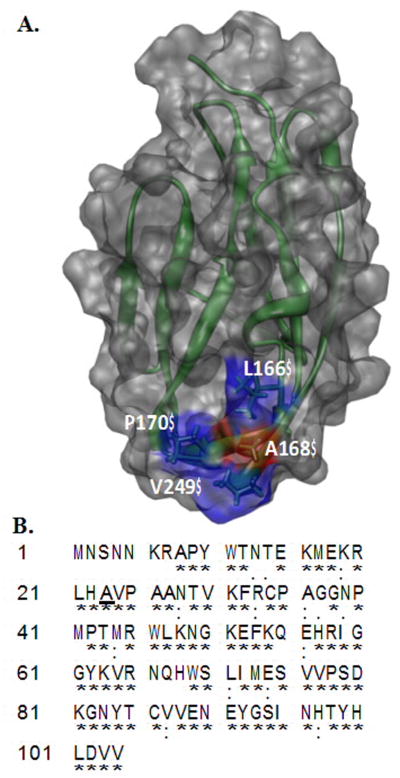
The secondary structure of the D2 domain of FGFR includes 12 antiparallel β-strands arranged in a β-sandwich fold. A168 is well conserved in all four isoforms of the human FGF receptor. Ala168 is located in the middle of a hydrophobic core of residues consisting of L165, P170, V248 and V249 (Fig. 1). In addition, A168 along with L165 and V248 has been shown to contribute to the binding of the ligand (FGF) [19].
Fig. 1.
Panel-A, Depiction of the three-dimensional structure of the D2 domain Worldwide Protein Data Bank (PDB ID: 1WVZ) of FGFR2. The residues forming hydrophobic interactions with FGF include A168 (red), L166 (blue), P170 (blue) and V249 (blue). A168 of the D2 domain contacts Tyr-24 and Met- 142 of FGF. Panel-B Amino acid sequence of the D2 domain of FGFR2. The conserved residues in the D2 domain of FGFRs are indicated by an asterix. Ala168 is conserved in FGFRs and is highlighted in bold.
A168S mutation does not significantly perturb the backbone conformation of the D2 domain
Far UV CD spectrum of the wild type D2 domain shows a strongly negative ellipticity band around 220 nm and a positive ellipticity peak centered at 205 nm. These features of the far UV CD spectrum are typical of β-sheets arranged in a β-sandwich structure [20]. Interestingly, the far UV CD spectrum of the A168S mutant superimposes quite well with that of the wild type of the D2 domain suggesting that the A168S mutation causes no or minimal perturbations in the secondary structure of the receptor domain (Supplementary Fig. S1). These observations are quite surprising because introduction of a polar serine residue in a hydrophobic core can be expected to significantly perturb the backbone conformation of the protein.
A168S mutation causes a subtle but crucial tertiary structural change in the receptor domain
Intrinsic fluorescence serves as a useful and sensitive spectral probe to monitor tertiary structural changes in proteins. D2 domain has three well-conserved tryptophan residues that are partially buried in the interior of the protein [18] The 339 nm emission maximum suggests that the tryptophan residues in the wild type D2 domain are partially exposed to the solvent (Supplementary Fig. S1). The emission maximum of the A168S mutant shows a modest red shift of 3 nm (339 nm to 342 nm) indicating that the mutation causes a subtle tertiary conformational change (Supplementary Fig. S1) resulting in greater solvent exposure of the indole rings of tryptophan residues in the receptor domain.
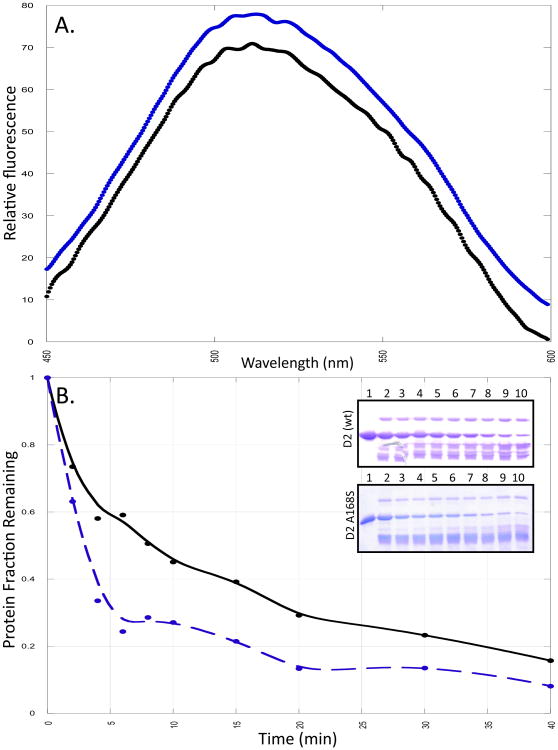
ANS is a popular fluorescent hydrophobic dye, which is commonly used to monitor solvent-exposed hydrophobic surfaces in proteins [12]. ANS in the presence of the wild type D2 domain shows weak emission with an emission maximum centered at 508 nm. (Fig. 2A) However, the emission maximum of the dye when bound to the A168S mutant shows a red shift of 4 nm (from 508 nm to 512 nm) with a very moderate increase in the emission intensity relative to the wild type protein. These fluorescence characteristics suggest that substitution of alanine with serine (A168S) causes a subtle conformational change resulting in increased solvent exposure of hydrophobic surface(s) in the protein.
Fig. 2.
Panel A represents the ANS spectra in the presence of the wild type (black) and the A168S mutant of the D2 domain (blue). The concentration of ANS is 150 μM. Panel B shows the densitometric plot of the limited-trypsin digestion of wild type (black) and the A168S (blue) mutant of D2 domain. Panel B insert shows the 15% SDS PAGE of the limited-trypsin digestion of the wild type and A168S mutant of the D2 domain after 0, 2, 4, 6, 8, 10, 15, 20, 30 and 40 minutes of incubation of the protein domain with trypsin.
The A168S mutation does not significantly affect the thermodynamic stability but increases the conformational flexibility of the D2 domain
Differential scanning calorimetry is a versatile technique that can be used to directly probe the themodynamic stability of proteins [21]. The melting thermogram of the wild type D2 domian shows a relatively sharp melting transition (Tm∼52 °C), from the folded to the unfolded state (Supplementary Fig. S1). The Tm value does not appear to change due to the A168S mutation (Supplementary Fig. S1; Tm∼51.5 °C). These results suggest that the thermodynamic stability of the receptor domain is not significantly affected by the mutation.
Limited trypsin digestion is a useful technique to obtain information on the backbone flexibility of proteins [22, 23]. Lysine and arginine residues located in the flexible and solvent-accessible regions of the protein are more susceptible to trypsin cleavage than when they are located in the interior or the rigid portions of proteins. Therefore, differences in the susceptibility to trypsin cleavage can be expected to provide valuable information on the changes in the backbone flexibility caused by subtle conformational changes induced in the protein due to the A168S mutation.
The D2 domain contains nine lysine residues and six arginine residues. The wild type protein is rapidly cleaved by trypsin in the first few minutes of initiation of the cleavage reaction (Fig. 2). About 40% of the band (Mr ∼13.7 kDa) corresponding to the intact D2 domain remains after 10 minutes of initiation of cleavage (Fig. 2). Interestingly more than 80% of the A168S mutant protein is cleaved within the same time period (Fig. 2). These results in conjunction with those obtained from the fluorescence data suggest that the alanine to serine substitution at position 168 causes subtle conformational change, which significantly increases the backbone flexibility and solvent-accesibility of the hydrophobic surface(s) in the protein.
A168S mutation decreases heparin-binding affinity and causes a complete loss of FGF binding
D2 domain is the most important structural module of the fibroblast growth factor receptor as it provides binding surface to both heparin and the ligand (FGF). Isothermal titration calorimetry (ITC) experiments were performed to examine the effect(s) of the Kallmann mutation (A168S) on the heparin and FGF binding affinities of the D2 domain. ITC is a useful technique to characterize the protein-protein/ligand interactions as it relies on the heat evolved or absorbed during binding interactions.
Heparin is polydisperse and therefore it is challenging to obtain an accurate estimation of its binding affinity to proteins. In this context, we used polyanionic sucrose octasulfate (SOS) to represent heparin binding interactions with the D2 domain. Sucrose octasulfate, unlike heparin, is monodisperse and can be obtained in the pure form (>99% purity). In addition, SOS has been demonstrated to be a good structural and functional mimic for heparin.
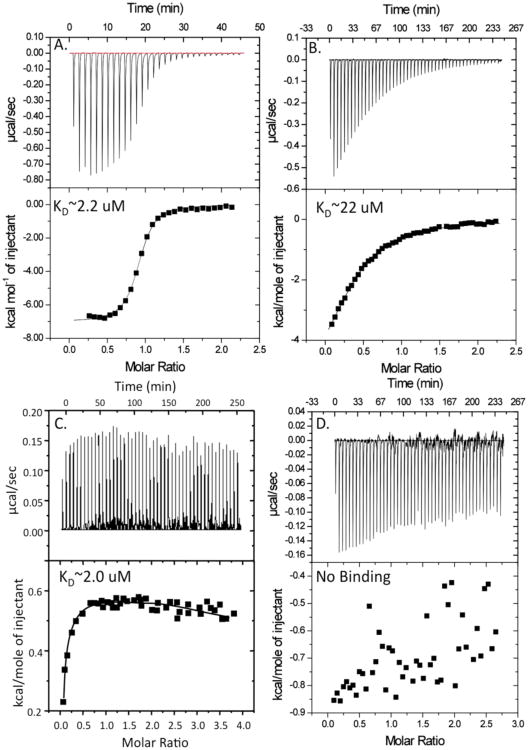
The isothermogram representing the binding of wild type D2 domain to SOS is sigmoidal and the binding constant (Kd) characterizing the SOS-D2 domain interaction(s) is ∼ 2.2 μM (Fig. 3A). SOS and D2 domain bind to each other in a 1:1 stoichiometry. Interestingly, in marked contrast, the A168S versus SOS titration curve is hyperbolic and the binding affinity between A168S mutant and SOS is about 10 times (Kd ∼22 μM) weaker than that of the wild type D2 domain (Fig. 3).
Fig. 3.
Binding isotherm representing the titration of the wild type and the A168S mutant of the D2 domain with SOS and FGF. The top panel shows the raw data from the titration. The bottom panel shows the integrated data derived from the raw data. Panels A and B show the titration of the wild type and the A168S mutant of the D2 domain with SOS. Panels C and D show the isothermogram of the wild type and A168S mutant of the D2 domain titrated with FGF.
The lowering of the heparin binding affinity is unexpected because the mutation site (A168S) is spatially remote from the heparin-binding site in the D2 domain.
Wild type D2 domain binds strongly (Kd∼ 2 μM) to FGF. The binding affinity (Kd) of the isolated D2 domain is in the same range as that of the intact extracellular portion of FGFR suggesting that D2 domain is critical for FGF binding. Interestingly, the A168S mutant of the D2 domain exhibits no or very insignificant binding to FGF. These results suggest that loss-of-function associated with the KS (A168S) mutation is due to complete loss of interaction between the ligand and the receptor.
Structural basis for Kallmann syndrome
A clear picture of the molecular basis for Kallmann syndrome would only emerge when the effects of the mutation on the conformation are understood at an atomic level. In this context, multidimensional NMR experiments were performed to assess the plausible structural changes induced by the KS mutation and also understand the consequences of the conformational change on both heparin and FGF binding.
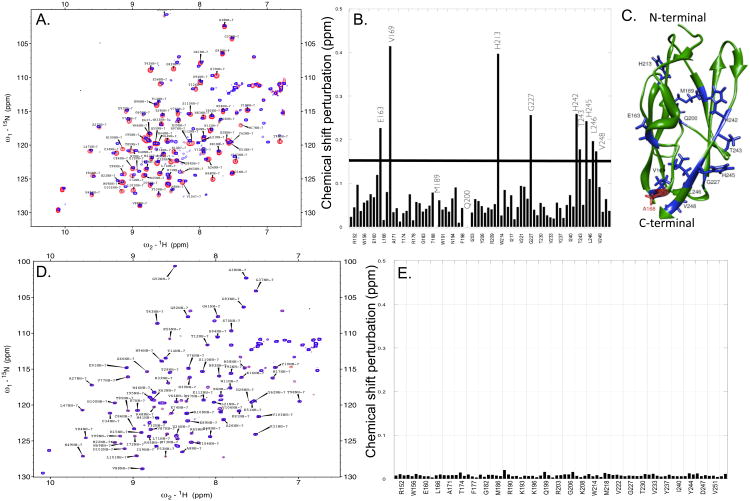
Two-dimensional 1H-15N HSQC spectrum is a finger-print of the backbone conformation of proteins. Each crosspeak in the 1H-15N HSQC spectrum represents an amino acid in a particular backbone conformation of the protein. In this context, we monitored the backbone conformational changes induced due to the Kallmann mutation (A168S) using the 1H-15N chemical shift perturbation (δ) observed in the HSQC spectra (Fig. 4A).
Fig. 4.
Structural changes induced due to the Kallman (A168S) mutation. Panel A shows an overlay of the 1H-15N HSQC spectra of the wild type (red) and the A168S (blue) mutant of the D2 domain of FGFR. Panel B shows the composite 1H-15N chemical shift perturbation (8) calculated from the data shown in Panel A. The horizontal bar represents an arbitrarily set threshold to identify the residues, which show the most significant 1H-15N chemical shift perturbation. Panel C depicts a structural of the D2 domain (PDB ID: 1WVZ) highlighting the A168S mutation site (red) and the most perturbed residues (blue).[18] 1H-15N HSQC spectra of the A168S mutant of the D2 domain in the presence (red) and absence (blue) of FGF is shown in Panel D. Composite 1H-15N chemical perturbation of the A168S mutant calculated from the 1H-15N HSQC data (Panel E). The data suggest that the A168S mutant of the D2 domain does not bind to FGF.
Most of the perturbed residues are located in the vicinity of the mutated residue (A168). Interestingly, residues located in the β-strand G (residues, H242 to V248) located at the C-terminal end of the D2 domain are also significantly perturbed [18]. The observed 1H-15N chemical shift perturbation of residues in β-strand G is plausibly because of their spatial proximity to mutation site (A168) in the three-dimensional structure of the D2 doma in [18]. It appears that the Kallmann mutation causes a subtle conformational change resulting in the disruption of the structural topology of the FGF binding pocket which consequently leads to complete loss in ligand (FGF) binding. Although several residues in the D2 domain are involved in the stabilization of the FGF-D2 domain complex, the intramolecular and intermolecular structural interactions contributed by A168 appear to be quite critical for the formation and stabilization of the ligand-receptor complex. The complete loss in ligand binding affinity of the Kallmann mutant is evident from the insignifi cant 1H-15N chemical shift perturbation [composite 1H-15N chemical shift (δ <0.02 ppm)] of residues observed when 15N enriched A168S mutant of the D2 domain is titrated with unlabeled FGF (Fig. 4D). The lack of binding affinity of the A168S mutant to the ligand is also corroborated by the ITC results discussed earlier (Figs. 3C & 3D).
ITC results, discussed earlier, revealed that the A168S mutant exhibits 10-fold lower affinity to bind to heparin than wild type D2 domain (Figs. 3A & 3B). In this context, the structural basis for the decrease in heparin binding of the Kallmann mutant was assessed by comparing the composite 1H-15N chemical shift perturbation (δ) of the A168S mutant and wild type D2 domain on binding to heparin/SOS. Comparison of the composite 1H-15N chemical shift perturbation plots of the wild type and A168S mutant in the presence of SOS show that residues such as, K161, K174, K196, K208 and R210 in the wild type D2 domain show significant 1H-15N chemical shift perturbation (Supplementary Fig. S3). 3D structures of the FGF-heparin-receptor complex show that these residues located in the D2 domain constitute the heparin-binding pocket [24]. Interestingly, the extent of the composite 1H-15N chemical shift perturbation for residues in the heparin binding pocket are significantly less in the A168S mutant than observed in the wild type D2 domain (Supplementary Fig. S3). These results suggest that the subtle conformational change caused due to the A168S mutation alters the orientation of side-chains of residues involved in electrostatic interaction with heparin and consequently causes the decrease (∼ 10-fold) in affinity to the polysulfated proteoglycan.
Discussion
Kallmann syndrome mutations have been detected in all major structural modules located in the extracellular portion of the FGFR [11]. KS mutations invariably cause a loss-of-function. Several proposals are made to account for the loss-of-function observed in KS. Anosmin-1, a glycosylated protein, encoded by the KAL-1 gene modulates FGF signaling by directly interacting with FGFR [11]. It is believed that the KS mutations cause a disruption of the putative anosmin-1/FGFR interaction and consequently inhibits the FGF signaling process. Alternatively, it is also believed that the loss-of-function of the KS mutations is due to a drastic conformational change leading to the misfolding and plausibly aggregation of the receptor. Contrary to these proposals, the results of the present study clearly suggest that the loss-of-function of the KS (A168S) mutation is due to a subtle conformational change that results in complete disruption of binding interactions between FGF and the receptor. We believe that structure-function studies in the future with other KS mutations can be expected to provide valuable information to understand the general molecular mechanisms underlying FGFR-linked genetic diseases.
Supplementary Material
Highlights.
The structural basis of the Kallmann Syndrome is elucidated.
Kallmann syndrome mutation (A168S) induces a subtle conformational change(s)
Structural interactions mediated by beta-sheet G are most perturbed.
Ligand (FGF)-receptor interaction(s) is completely abolished by Kallmann mutation.
Kallmann mutation directly affects the FGF signaling process.
Acknowledgments
This publication was made possible by Grant Number P30 GM103450-03 from the National Institute of General Medical Sciences, a component of the National Institutes of Health (NIH). Financial support from Department of Energy (DE-02-01ER15161) and the Arkansas Bioscience Institute are also gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Naftolin F, Harris G, Bobrow M. Effect of purified luteinizing hormone releasing factor on normal and hypogonadotrophic anosmic men. Nature. 1971;232:496–497. doi: 10.1038/232496a0. [DOI] [PubMed] [Google Scholar]
- 2.Cadman SM, Kim SH, Hu Y, Gonzalez-Martinez D, Bouloux PM. Molecular pathogenesis of kallmann's syndrome. Horm Res. 2007;67:231–242. doi: 10.1159/000098156. [DOI] [PubMed] [Google Scholar]
- 3.Albuisson J, Pecheux C, Carel JC, Lacombe D, Leheup B, Lapuzina P, Legius E, Matthijs G, Wasniewska M, Dwlpech M, Young J, Hardelin JP, Dode C. Kallmann syndrome: 14 novel mutations in KAL1 and FGFR1 (KAL2) Hum Mutat. 2005;25:98–99. doi: 10.1002/humu.9298. [DOI] [PubMed] [Google Scholar]
- 4.Gill JC, Moenter SM, Tsai PS. Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology. 2004;145:3830–3839. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 5.Raivio T, Sidis Y, Plummer L, Chen H, Ma J, Mukherjee A, Jacobson-Dickman E, Quinton R, Van Vliet G, Lavoie H, Hughes VA, Dwyer A, Hayes FJ, Xu S, Sparks S, Kaiser UB, Mohammadi M, Pitteloud N. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2009;94:4380–4390. doi: 10.1210/jc.2009-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitteloud N, Acierno JS, Jr, Meysing AU, Dwyer AA, Hayes FJ, Crowley WF., Jr Reversible kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the fibroblast growth factor receptor 1 gene. J Clin Endocrinol Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 7.Colvin J, White A, Pratt S, Ornitz D. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 8.Kathir KM, Kumar TK, Yu C. Understanding the mechanism of the antimitogenic activity of suramin. Biochemistry. 2006;45:899–906. doi: 10.1021/bi051389b. [DOI] [PubMed] [Google Scholar]
- 9.Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structure of FGF2 in complex with the extracellular lignad binding domain of FGF receptor 2 (FGFR2) Cell. 2000;101:413–424. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, Guimond SE, Travers P, Cadman S, Hohenester E, Turnbull JE, Kim SH, Bouloux PM. Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1. J Biol Chem. 2009;284:29905–29920. doi: 10.1074/jbc.M109.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Bouloux PM. Novel insights in FGFR1 regulation: Lessons from kallmann syndrome Trends Endocrinol. Metab. 2010;21:385–393. doi: 10.1016/j.tem.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen-White JR, Chorich LP, Bick DP, Sherins RJ, Layman LC. The prevalence of intragenic deletions in patients with idiopathic hypogonadotropic hypogonadism and kallmann syndrome. Mol Hum Reprod. 2008;14:367–370. doi: 10.1093/molehr/gan027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitteloud N, Quinton R, Pearce S, Raivio T, Acerno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng Y, Li W, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi O, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley WJ. Digenic mutations account for viable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato N, Hasegawa T, Hori N, Fukami M, Yoshimura Y, Ogata T. Gonadotrophin therapy in kallmann syndrome caused by heterozygous mutations of the gene for fibroblast growth factor receptor 1: Report of three families: Case report. Hum Reprod. 2005;20:2173–2178. doi: 10.1093/humrep/dei052. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell AL, Dwyer A, Pitteloud N, Quinton R. Genetic basis and variable phenotypic expression of kallmann syndrome: Towards a unifying theory. Trends Endocrinol Metab. 2011;22:249–258. doi: 10.1016/j.tem.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 17.Hebert JM, Lin M, Partanen J, Rossant J, McConnell SK. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130:1101–1111. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- 18.Hung KW, Kumar TK, Kathir KM, Xu P, Ni F, Ji HH, Chen MC, Yang CC, Lin FP, Chiu IM, Yu C. Solution structure of the ligand binding domain of the fibroblast growth factor receptor: Role of heparin in the activation of the receptor. Biochemistry. 2005;44:15787–15798. doi: 10.1021/bi051030n. [DOI] [PubMed] [Google Scholar]
- 19.Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell. 2000;101:413–424. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 20.Kathir KM, Kumar TK, Rajalingam D, Yu C. Time-dependent changes in the denatured state(s) influence the folding mechanism of an all beta-sheet protein. J Biol Chem. 2005;280:29682–29688. doi: 10.1074/jbc.M504389200. [DOI] [PubMed] [Google Scholar]
- 21.Lepock JR. Measurement of protein stability and protein denaturation in cells using differential scanning calorimetry. Methods. 2005;35:117–125. doi: 10.1016/j.ymeth.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 23.Fontana A, Polverino de Laureto P, De Filippis V, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 1997;2:R17–26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 24.Yeh BK, Eliseenkova AV, Plotnikov AN, Green D, Pinnell J, Polat T, Gritli-Linde A, Linhardt RJ, Mohammadi M. Structural basis for activation of fibroblast growth factor signaling by sucrose octasulfate. Mol Cell Biol. 2002;22:7184–7192. doi: 10.1128/MCB.22.20.7184-7192.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.