Abstract
Benthic algae are associated with coral death in the form of stress and disease. It's been proposed that they release exudates, which facilitate invasion of potentially pathogenic microbes at the coral-algal interface, resulting in coral disease. However, the original source of these pathogens remains unknown. This study examined the ability of benthic algae to act as reservoirs of coral pathogens by characterizing surface associated microbes associated with major Caribbean and Indo-Pacific algal species/types and by comparing them to potential pathogens of two dominant coral diseases: White Syndrome (WS) in the Indo-Pacific and Yellow Band Disease (YBD) in the Caribbean. Coral and algal sampling was conducted simultaneously at the same sites to avoid spatial effects. Potential pathogens were defined as those absent or rare in healthy corals, increasing in abundance in healthy tissues adjacent to a disease lesion, and dominant in disease lesions. Potentially pathogenic bacteria were detected in both WS and YBD and were also present within the majority of algal species/types (54 and 100% for WS and YBD respectively). Pathogenic ciliates were associated only with WS and not YBD lesions and these were also present in 36% of the Indo-Pacific algal species. Although potential pathogens were associated with many algal species, their presence was inconsistent among replicate algal samples and detection rates were relatively low, suggestive of low density and occurrence. At the community level, coral-associated microbes irrespective of the health of their host differed from algal-associated microbes, supporting that algae and corals have distinctive microbial communities associated with their tissue. We conclude that benthic algae are common reservoirs for a variety of different potential coral pathogens. However, algal-associated microbes alone are unlikely to cause coral death. Initial damage or stress to the coral via other competitive mechanisms is most likely a prerequisite to potential transmission of these pathogens.
Introduction
Coral diseases have contributed to the regional collapse of important reef-building species worldwide [1]–[3]. Despite a concerted global initiative to characterise coral diseases over the past two decades, the causes of disease outbreaks remain largely unknown [4]. Their impacts are continuing to increase in number and spatial extent, raising hypotheses that they could be associated with large-scale environmental stressors such as rising ocean temperature or acidification [5], [6]. Additionally, nutrient pollution has been shown to increase the spread of several coral diseases and thus could exacerbate disease outbreaks locally [7], [8]. A recent but still largely unexplored hypothesis is that coral diseases may be enhanced by the increased abundance of benthic algae which reefs are experiencing on regional and global scales [9]–[12].
Competition between corals and benthic algae is frequent on reefs especially under conditions of reduced grazing and increased nutrients, a change which favors the growth of macroalgae and turf algae [13], [14]. The incidence of coral disease has been found to be positively correlated with increasing algal cover [15]–[18], and the experimental link between direct algal contact and coral disease has been established in the field [19], [20]. However, there is no clear mechanistic link between algae and coral diseases. Contact between corals and algae commonly cause a suite of stresses for corals, such as localized tissue death, reductions in tissue thickness and zooxanthellae density and lower photosynthetic efficiency often resulting in bleaching [21], [22], all of which could increase coral susceptibility to disease pathogens. Current evidence suggests that algae have the ability to directly harm corals in three principal ways; 1) shading, 2) abrasion resulting in a physical injury, and 3) the release of poisonous allelochemicals (reviewed by [23]). There are also numerous indirect ways the algae have been shown to have an effect, notably: 4) via the release of primary metabolites, which has been shown to stimulate microbial activity at the coral/algal interface [24]–[26], 5) via the release of secondary metabolites which alter the coral associated microbial community [27]–[30], 6) by attracting corallivores, which cause increased coral mortality [31], and lastly 7) by acting as a vector for coral disease pathogens [19]. In contrast to the above mentioned studies which illustrate numerous direct and indirect effects of algae on coral [32], showed that the presence of macro algae adjacent to corals had no observable effect on coral health and disease prevalence at least in relation to Yellow Band Disease within the Caribbean. However, in their experiments, macroalgae were not directly touching the coral, which suggests that water borne algal exudates alone are not sufficient to illicit a stress response on the coral.
Regardless of the main trigger for coral death by algae, with regard do disease onset the source of coral pathogens remains unknown. A possible mechanism for this, is that algae may act as vectors of coral pathogens, transmitting them from their surfaces to corals via direct contact [19]. showed that direct contact of the green algae Halimeda opuntia to apparently healthy corals could trigger white plague disease and found the bacterial pathogen, Aurantimonas coralicida (the proposed causal agent of WP Type II disease, [33]) to be associated with H. opuntia. Furthermore, more recently [20], showed that, under experimental conditions, contact between the coral Acropora pulchra and the green filamentous macroalgae Chlorodesmis fastigiata often resulted in a ciliate infection on the coral. However, in both cases, it has not yet been demonstrated whether the pathogens were directly transmitted by the algae.
As a first step, it seems relevant to study the availability of potential coral pathogens associated with algal surfaces and to determine which of these are also associated with specific coral diseases. Many species of bacteria have been accredited to causing specific coral diseases [33]–[39], in particular members from the genus Vibrio such as V. shiloi, V. coralliilyticus and V. harveyi, along with others such as Aurantimonas corallicida [40]. recently characterized the bacterial communities on the surface of benthic algae and found that some matched with sequences related to bacteria associated with coral disease states highlighted by other studies. However, no study has simultaneously investigated the dominant bacterial communities associated with both algae and healthy/diseased corals from the same reefs using the same methodology. As the microbial communities of the coral holobiont vary both spatially [41] and temporally [42], it is important that samples are collected at the same time, extracted in the same manor and analysed using the same technique. In addition to potential bacterial pathogens, other microorganisms such as ciliates have been shown to be associated with many coral diseases [43]–[45]. Three species, two from the genus Philaster and a Varistrombidium, have been shown to ingest coral algal symbionts and it has been hypothesized that they cause the pathology of the diseases known collectively as White Syndrome [45]. Despite their importance in many ecosystems, ciliates have received considerably little attention and their abundance on algal surfaces has not been studied to date.
Here we examine the bacterial and ciliate communities associated with various macroalgal species and turf algae using culture independent techniques (16S and 18S rRNA genes) and compare them to potential pathogens of two diseases dominating the Indo-Pacific and Caribbean regions: White Syndrome (WS) and Yellow Band Disease (YBD) respectively. Several Vibrios have been proposed as WS pathogens [37], [39]. However, a recent study suggest that ciliates are largely responsible for the macroscopic signs of the disease [45]. YBD has no named causal agent to date, however the Vibrio ‘core group’, similar to species reported to cause WS in the Indo-Pacific, has been shown to play a role in the degeneration and deformation of the coral algal symbionts in the disease [35], [46]. ‘Potential pathogens’ in this study were defined as those absent or rare in healthy corals, increasing in abundance in apparently healthy tissues (those adjacent to the disease lesion), and dominant in the disease lesion itself [45]. These potential pathogens were identified by examining coral disease-associated microbes on the same reefs, at the same time and using the same procedures as those associated with various algae. We were interested in detecting only those microbes associated with the surface of the algal species sampled as these will be the microbes which will most likely come into contact with the coral interface and therefore most likely pass from algae to coral, hence we utilized a modified DGGE-based technique known to only detect the dominant members of the microbial communities allowing for a larger sample size to be studied at relatively lower cost techniques such as high throughput sequencing.
Methods
Sample collection
Samples of corals and algae were collected at Heron Island, on the Great Barrier Reef in the Indo-Pacific and at Los Roques, Venezuela in the Caribbean in accordance with collection permits issued by the Great Barrier Reef Marine Park Authority (GBRMPA) and the Instituto Nacional de Parques (INPAQUES), respectively. Coral collection procedure follows that of [45]. At Heron Island, colonies of Acropora muricata, suffering from signs of White Syndrome, were tagged and monitored for 4 days to follow the progression of disease lesions. Samples were collected from actively progressing disease lesions (DL; n = 3) and apparently healthy tissue (AH; n = 3) ∼1 cm away from the DL interface. Non-diseased coral colonies (ND; n = 3) were sampled as controls. Alongside these coral samples, eleven algal species (n = 3 replicates per species, listed in Fig. 1) were collected. These were common to abundant at Heron Island, were observed to encounter corals and represented a wide range of morphological and taxonomic groups. All samples were collected in situ into sterile 50 ml falcon tubes and transported back to the laboratories. The water was then replaced with 100% EtOH and stored at −20°C until extraction. Coral and algal samples were centrifuged at 20,000 g to concentrate loosely associated microbes and surface mucus layer (of the corals specifically) [47]. Although this technique is a novel approach to study both coral and algal microbial associates, the presence of coral DNA in the extracted samples illustrates that this technique successfully samples both tissue and surface mucus of corals. All samples were then vacuum centrifuged to reduce the volume of EtOH/homogenized tissues to 10 ml in each sample. A subset of the homogenized tissue and mucus was then sampled and weighed to allow for standardization between sample types.
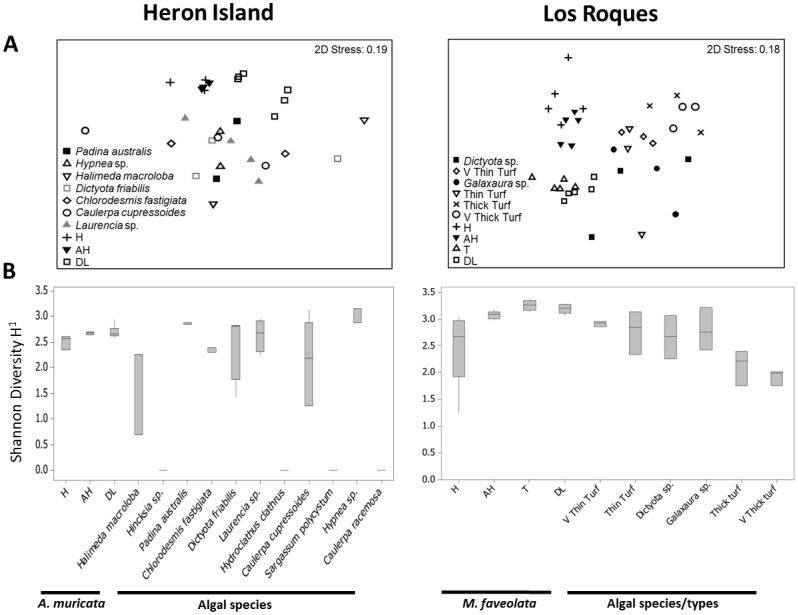
Figure 1. Bacterial 16S rRNA gene communities associated with various coral and algal samples at Heron Island and Los Roques.
A) Multidimensional scaling (MDS) plot showing changes in bacterial communities. B) Shannon-Wiener Diversity Index. The middle bars are medians, boxes show quartiles, whiskers show 95% CI of data. H = non diseased, AH = apparently healthy, DL = disease lesions, T = transition.
In Los Roques, Montastraea faveolata showing signs of YBD were tagged and monitored as above. Coral samples were taken from the same areas, with the addition of a further set of samples at the transition (bleached) zone (T; n = 3) of the disease lesion. Two algal species and four turf algal types (n = 3 replicates per type) were collected alongside the coral samples: Dictyota sp., Galaxaura sp., very thin turf (<2 mm), thin turf (2–<5 mm), thick turf (5–10 mm) and very thick turf (>1 cm). Turf algae consisted of diminutive mixed assemblages of filamentous algae, juvenile macroalgae and cyanobacteria [48]. Turf thickness was measured in situ with calipers and samples were scrapped using a sterile scalpel blade into 50 ml falcon tubes and transported to the surface. Samples were processed and stored similar to those taken from Heron Island. All algal replicates were taken at least 10 meters apart from each other to minimize the chance of individual variation within species.
DNA extraction, DGGE and sequence analysis
All samples were extracted using the Qiagen DNeasy Blood and Tissue Kit, spin column protocol [49]. A portion of the bacterial 16S rRNA gene was amplified using universal eubacterial primers (GC-357F) (5′- CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCAGCACGGGGGG-CCTACGGGAGGCAGCAG-3′) and (518R) (5′-ATTACCGCGGCTGCTGG-3′). All reactions were performed using a Hybaid PCR Express thermal cycler. PCR and DGGE were carried out as described by [49]. To identify the dominant DGGE bands across samples, representative bands (n = 50) were excised and sequenced to account for known DGGE artifacts such as heteroduplexes [50]. Excised bands were left overnight in Sigma molecular grade water, vacuum centrifuged, re-amplified with primers 357F and 518R, labeled using Big Dye (Applied biosystems) transformation sequence kit and sent to Genevision (Newcastle University, UK) for sequencing. Bacterial operational taxonomic units (OTUs) were defined from DGGE band-matching analysis using BioNumerics 3.5 (Applied Maths BVBA) as described by [49]. From the same extracted samples as above, ciliate 18S rRNA genes were amplified directly using the primers CilF (5′-TGGTAGTGTATTGGACWACCA-3′) CilDGGE-r (5′TGAAAACATCCTTGGCAACTG-3′). PCR and DGGEs were carried out as described in [45].
Statistical analysis
DGGE images were analyzed using BioNumerics version 3.5. Quantification of samples is based on the densitometric curves extracted from the banding patterns. Minimum profiling was set at 10% which allows elevation of the band with respect to the background noise of the gel. Any bands specified as ‘uncertain’ were removed at this step. This analysis allows for both the presence/absence of the OTUs and relative intensity to be analyzed. A one-way analysis of similarity (ANOSIM) based on Bray-Curtis similarities of both band intensity patterns and presence/absence only were performed to test for differences between DGGE profiles of the bacterial 16S rRNA and ciliate 18S rRNA gene assemblages associated with different coral health states and the algal species using PRIMER v6 [51]. Pairwise comparisons within ANOSIM were used to contrast between specific sample types [52]. Non-metric multidimensional scaling (MDS) was used to visualize variations in bacterial and ciliate communities among the different samples. Shannon-Weiner diversity (H') was calculated as an estimate of diversity and analysed using PERMANOVA followed by pairwise tests.
Results
16S rRNA gene bacterial community
In both study sites, 16S rRNA gene bacterial communities differed among sample types (ANOSIM, Heron Island: R = 0.371, p<0.001; Los Roques: R = 0.743, p = 0.001; Figs. 1a). Coral samples regardless of host health consistently differed from algal samples regardless of species/types (Fig. 1; Tables S1 & S2). Healthy coral tissue differed more with diseased and/or transition tissue (in the case of YBD) than with apparently healthy tissue. Variation among algal samples was less (Fig. 1; Fig. 2). Bacterial communities did not differ among algal species which hosted bacteria at Heron Island (Fig. 1; Fig. 2; Table S1). At Los Roques, thick turf and very thick turf were significantly different than the other algal types (Fig. 1; Fig. 3; Table S2). Shannon-Wiener diversity varied significantly between sample types for both Heron Island and Los Roques (PERMANOVA F = 24.264, p = 0.001; F = 26.342, p = 0.001 respectively) (Tables S3 & S4). Coral-associated bacterial diversity increased with reduced health of the host (Fig. 1b). Bacterial diversity was generally lower in the algal samples when compared to the coral with the exception of Padina australis, Laurencia sp. and Hypnea sp. from Heron Island (Fig. 1b). However, there was no significant differences in bacterial diversity between and within both the coral and algal samples at Heron Island (Fig. 1; Table S3), and only the bacterial diversity of the transition band in corals showing signs of YBD and the lesion interface itself were significantly different to healthy coral samples at Los Roques (Fig. 1; Table S4). At Heron Island, four out of the eleven algal species (Hincksia sp., Hydroclathrus clathrus, Sargassum polycystum and Caulerpa racemosa) showed no bacterial associates, and only a few bacterial ribotypes were detected on Halimeda macroloba (Fig. 2). In contrast, surface-associated bacteria were present in all algal species/types at Los Roques (Fig. 3). Fewer bacteria were found on thick and very thick turf compared to thinner turf types and the two macro-algal species.
Figure 2. Heatmap summarizing the relative density of dominant DGGE bands according to BioNumerics software for 16S rRNA gene bacterial profiles of coral and algal samples collected in Heron Island.
White blocks signifies that that ribotype was not detected in the sample. All replicates (N = 3 per sample type) have been included in this figure. ND = non diseased, AH = apparently healthy, DL = disease lesions, T = transition. For full algal species names see Fig. 1.
Figure 3. Heatmap summarizing the relative density of dominant DGGE bands according to BioNumerics for 16S rRNA gene bacterial profiles of coral (M.faveolata) and algal samples collected in Los Roques.
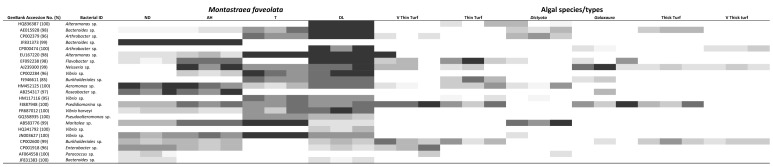
All replicates (N = 3 per sample type) have been included in this table. Abbreviations as in Fig. 1 and key as in Fig. 2.
Four potential WS bacterial pathogen ribotypes (NCBI accession numbers JF831360, JN406279, JN406285 and JN406280) identified as an Arcobacter sp., an Aeromonas sp., a Cyanobacterium sp., and a Clostridium sp. respectively, were detected in coral samples at Heron Island. Three of these (Aeromonas sp.; Cyanobacterium sp. and Clostridium sp.) were also present in algal samples, with all three being detected on Padina australis and Halimeda macroloba (Fig. 2). Overall 6 of the 11 algal species (54%) and 18 of the 33 algal samples (54%) included potential bacterial pathogens. Aeromonas sp. was detected in 15 of the 33 algal samples (45%), Cyanobacterium in 11/33 (33%) and Clostridium in 8/11 (24%) (Fig. 2). A further two species, Pseudoalteromonas (JN406297) and Shewanella (JN406298) increased in abundance in the WS diseased lesion samples, but they were also present in healthy tissues. These were only detected in two algal species, Hypnea and Halimeda and only in one of the replicates of each of the algae. Out of the algae in which surface bacteria were detected for, only Caulerpa cupressiodes was free of these potential WS pathogens.
In Los Roques, nine bacterial ribotypes were identified as potential YBD pathogens, these included ribotypes relating to a Burkholderiales sp. (NCBI accession number FJ946611), a Pseudoalteromonas sp. (GQ358935), three Vibrio sp. (CP002284, HM117116 & HQ341792), a Alteromonas sp. (HQ836387), a Neisseria sp. (AJ239300) and two Arthrobacter sp. (CP002379 & CP000474) (Fig. 3). Six of these (the Neisseria sp., the Alteromonas sp., the two Arthrobacter sp., the Burkholderiales sp. and one of the Vibrio sp.) were found to be associated with algae of some kind. All algal species/types and all algal replicate samples harbored potential bacterial pathogens. Neisseria was the most common bacterial ribotype detected in the algal samples and was present in 5 of the 6 algal species/types (83%) and 13 of the 18 algal samples (72%). Out of the two Arthrobacter, one was present in Dictyota and very thin turf, whilst the other was present in Galaxaura and very thick turf.
18S rRNA gene ciliate assemblage
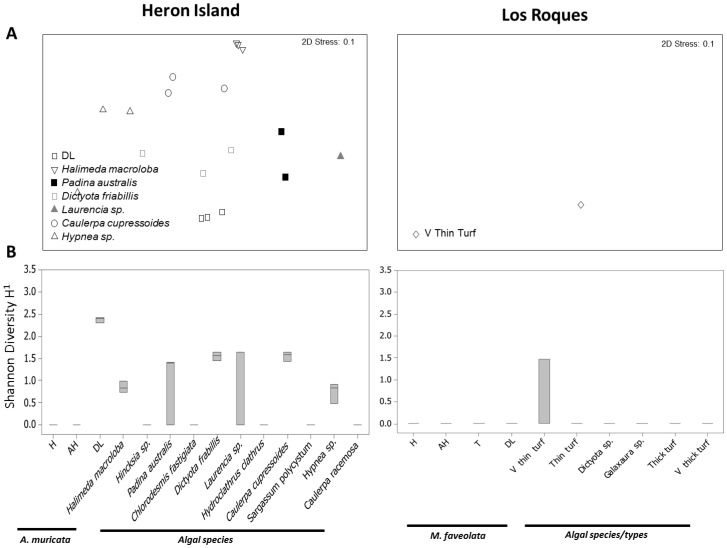
In Heron Island, ciliate assemblage associated with corals showing signs of WS differed from any other (coral or algal) sample (ANOSIM R = 0.67, p = 0.001 and pairwise tests p<0.05) (Fig. 4a; Fig. 5). These ciliates were consistently absent from healthy and apparently healthy tissues. WS associated ciliates included ribotypes similar to Aspidisca sp. (JN406268), Diophrys sp. (JN406270), Philaster sp. (JN626268 and JN626269), Pseudocarnopsis sp. (HQ013358), Euplotes sp. (JN406271 and HQ013357), Glauconema sp. (JN406267) and Varistrombidium sp. (HQ204551) (Fig. 5). Six out of the eleven algal species (Halimeda macroloba, Padina australis, Dictyota friabilis, Laurencia sp., Caulerpa cupressoides, Hypnea sp.) hosted ciliates (Fig. 5), with eight ciliate species being detected. Five of these ciliate species were associated with WS diseased corals: Aspidisca sp., the two Philaster sp. (JN626268 and JN626269), Varistrombidium sp. and Pseudocarnopsis sp. The other three had ribotypes related to Anteholosticha sp. (FJ156105), Ichthyophthirius sp. (DQ270015) and Dileptus sp. (HM581676). Four of the 11 algal species (36%) and 10 of the 33 algal samples (30%) included WS associated ciliates. In contrast to WS, ciliates were not associated with Yellow Band Disease (YBD) tissues or any other coral samples collected from Los Roques (Fig. 6), however they have been visually observed in some cases of the disease (see Video S1). Only one algae (very thin turf) showed the presence of ciliates in the Los Roques samples (Fig. 4; Fig. 6), however the diversity of ciliates associated with this algae was consistent between all replicates. Three different ciliate ribotypes were detected on very thin turf including ribotypes closely related to Parathspidium Sp. (FJ875140), Sterkiella sp. (AB684399) and a Philaster sp. (JN626268).
Figure 4. Ciliate 18S rRNA gene communities associated with various coral and algal samples at Heron Island and Los Roques.
A) Multidimensional scaling (MDS) plot showing changes in ciliate communities. B) Shannon-Wiener Diversity Index. The middle bars are medians, boxes show quartiles, whiskers show 95% CI of data. Abbreviations as in Fig. 1.
Figure 5. Heatmap summarizing the relative density of dominant DGGE bands according to BioNumerics for 18S rRNA gene ciliate profiles of coral (A. muricata) and algal samples collected in Heron Island.
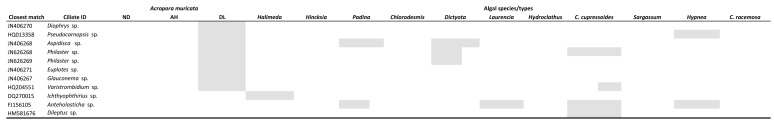
All replicates (N = 3 per sample type) have been included in this table. Abbreviations as in Fig. 1 and key as in Fig. 2. For full algal species names see Fig. 1.
Figure 6. Heatmap summarizing the relative density of dominant DGGE bands according to BioNumerics for 18S rRNA gene ciliate profiles of coral (M. faveolata) and algal samples collected in Los Roques.
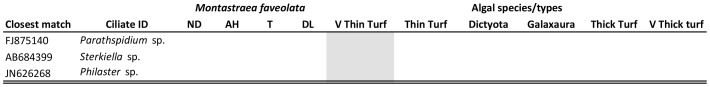
All replicates (N = 3 per sample type) have been included in this table. Abbreviations as in Fig. 1 and key as in Fig. 2.
Discussion
To determine whether benthic algae act as a source of coral pathogens, we examined dominant surface microbes (bacteria and ciliates) associated with a variety of algae species and related them, using the same non culture molecular technique (DGGE), to dominant potentially pathogenic microbes associated with two coral diseases. In the Indo Pacific, several potentially pathogenic bacteria detected in this study have previously been shown to be associated with coral disease states, such as Vibrio sp. (JN406288), Arcobacter sp. (JF831360), Aeromonas sp. (JN406279), Cyanobacterium sp. (JN406285), Clostridium sp. (JN406280), Pseudoalteromonas sp. (JN406297) and Shewanella (JN406298) [39], . Similarly, out of the five ciliates detected within the algal samples, three have been shown to be involved in coral diseases [45], two species from the genus Philaster (JN626268 and JN626269) and a Varistrombidium sp. (HQ204551). A further ciliate detected in the algae samples has been identified as the causal agent of an aquarium fish disease known as Ich (the ciliate Ichthyophthirius sp. DQ270015) [64]. In all cases except for ciliates at Los Roques which were not associated with YBD, potential bacterial and ciliate pathogens were detected in a majority of algal species or types, supporting the hypothesis that benthic algae are a common reservoir for coral disease pathogens.
A meta-analysis study reviewing coral disease associated bacteria conducted by [56], showed that, in diseased corals, the bacterial communities were consistently dominated by an increase in Rhodobacter, Clostridia and Cyanobacteria. In our study, the latter two orders increased in abundance in the WS samples, but were absent in YBD tissues. In contrast, although they were identified in both coral diseases representatives of Rhodobacter decreased in abundance in diseased tissues contrary to that reported by [56] and patterns observed for other diseases such as Black Band Disease [55], White Plague and White Band Disease [57], [58]. These contrasts between studies are likely to be brought about by the molecular profiling techniques utilized and specific primers chosen, as methodological studies have shown that primer choice can dramatically alter the end result of the bacterial assemblages [59]. This was the reasoning, at least in part, why this study analyzed the microbial diversity of both algae and corals at the same time, rather than simply relying on previous studies to highlight similarities and differences within the two community types.
Several Vibrio species such as Vibrio coralliilyticus and V. harveyi have been identified as potential WS pathogens by numerous previous studies [37], [39]. However, although the technique employed here (DGGE) does not bias against vibrios as demonstrated by the dominance of Vibrio sp. in the YBD diseased coral tissue, only one strain of vibrio closely related to V. harveyi (JN406288) was found in the coral samples at Heron Island. This strain was not identified as a potential pathogen since it had similar relative band intensity in both diseased and healthy tissues. This finding agrees with other studies [45], [60], which also used more stringent non culture molecular techniques such as clone libraries to show the same result. Furthermore, this ribotype was not detected on any of the algae sampled in this study.
The YBD bacterial community profiles showed similar results to that of the Indo-Pacific with numerous potential pathogens present in the coral samples. These included ribotypes related to Neisseria sp. (AJ239300), Burkholderiales sp. (FJ946611), Pseudoalteromonas sp. (GQ358935), Alteromonas sp. (HQ836387) and two Arthrobacter sp. (CP002379 & CP000474). All but the Pseudoalteromonas sp. were found to be associated with various algal species, though in relatively low dominance. Although no known causal agent has been identified for YBD to date, the disease lesion has previously been associated with a significant increase in the core Vibrio group at the lesion interface and in the transition band, giving rise to the disease name [35], [46]. This is consistent with results in our study whereby a large number of ribotypes closely related to different Vibrios were found in diseased samples. However, two of these Vibrio ribotypes (FR687012 & JN003627) were found in healthy samples showing that healthy tissues can harbor bacteria normally considered potentially pathogenic, a result increasingly seen in recent studies [45], [46], [58]. Two Vibrios (HM117116 & JN003627) were found associated with algae (Dictyota and thin turf) and only one of these (HM117116) was considered as a potential pathogen of YBD, being absent in healthy tissues and present in the diseased tissues.
[40] expanded the coral holobiont concept to benthic algae on the basis that, like corals, algae hosted abundant and distinctive microbial communities and that these microbes provided benefits to their host. Similarly to their study and in support of this concept, we found that dominant algal surface associated microbes were distinct from those associated with corals and varied among algal species. Interestingly certain species such as Halimeda macroloba exhibited low density bands as opposed to Hypnea sp. or Chlorodesmis fastigiata. Furthermore, four of the eleven algal species (Hincksia sp., Hydroclathrus clathrus, Sargassum polycystum and Caulerpa Racemosa) sampled in Heron Island showed no recoverable surface associated microbes. In our view, it remains unlikely that this result is an extraction error, as all samples were treated individually and no recoverable bacterial or ciliate sequences were detected in any of the replicate samples, however further analysis on these algae should be conducted to confirm this. In support of our results, algae commonly exhibit mucus release and tissue sloughing, they release different primary and secondary compounds and may have resident bacteria deeper within their tissues which have specific antibiotic and antifouling properties [61], [62]. Together these factors may create these distinctive microbial communities which are associated with the different algal species.
[40] also highlighted that algae have a higher diversity of bacterial OTUs than those associated with corals. In contrast, our study showed that the dominant bacterial ribotypes associated with the surface of the algae were in general no more diverse than those present on the surface of corals. In fact in most species, diversity was generally lower than the coral samples as a whole. This difference in results between the two studies may in part be methodological [40]. homogenised the algal tissue giving total bacterial diversity associated with the algal species and utilized deep sequencing techniques which allow insight into the whole bacterial community. In our study, samples of both coral and algae were centrifuged to optimize the detection of surface associated microbes and the non-culture molecular technique, DGGE, was used to characterize the dominant members of the microbial communities associated with the samples, as these will be the microbes which will most likely come into contact with the coral interface and therefore most likely pass from algae to coral. While this vastly reduces the cost of analyses, it has the disadvantage of underestimating total bacterial diversity.
Similar to that observed for bacterial associates, ciliated protozoans were detected within algae as well as diseased lesions, although only in the case of WS [45]. recently showed that four species ingest algal symbionts from corals (two from the genus Philaster, one Euplotes sp. and one Varistrombidium sp.). It was argued that these ciliates, particularly the two Philaster sp., were largely responsible for the macroscopic signs of the disease (WS), namely the sharp demarcation between apparently healthy tissues and the denuded skeleton giving rise to the name white syndrome. In this study, the two main potential agents, identified as Morph 1 and Morph 2 in [45], were found in two algal species (D. friabilis and C. cupressoides) in Heron Island. Although these ciliates were not consistently prevalent in the algae (1–2 out of the three replicates of each algal species), our study suggests that algae can act as a reservoir for ciliates pathogenic to corals and in particular involved in WS.
A recent experimental study by [20] showed that corals which were in direct contact with the algae Chlorodesmis fastigiata often resulted in a ciliate infection similar to Brown Band Disease. They visually investigated tufts of C. fastigiata at the time to see if this algae was a carrier for the ciliate pathogens, but failed to find any signs. In support, using a more rigorous molecular profiling technique, we also found no ciliates associated with the algae. These results suggest that these ciliate infections may be the result of the interaction between the algae and the coral. Such interaction commonly results in damaged coral tissue, which may attract the ciliates, in turn leading to infection. Thus, the algae may not be the source of this specific pathogen. In contrast to that of Heron, at Los Roques, only one algal type (very thin turf) harboured ciliates. Interestingly, one of the three species detected in this type was 98% similar to the same Philaster sp. (JN626268) associated with WS in the Indo-Pacific. Although we only studied pathogens associated with YBD in which ciliates were apparently not pathogenic, it remains to be investigated whether other Caribbean diseases, such as White Plague and White Band Disease, which share similar gross macroscopic signs with WS, could be associated with this species of ciliate.
In conclusion, this study shows that algae do contain certain potential coral pathogens. As reservoirs of these pathogens, algae therefore have the potential to act as vectors of coral diseases, transmitting pathogens to corals by direct contact. However, it has been argued that the increase in coral diseases combined with a decrease in host densities suggests opportunistic infections of compromised hosts rather than the involvement of specific pathogens is more likely [63]. It is highly probable that other factors mediated by algae such as the release of primary and secondary metabolites which cause abnormal stress or trauma to corals [24], [25], [27]–[29], are required to weaken the corals immune defense and subsequently facilitate invasion by any number of opportunistic pathogens. As algae are becoming increasingly abundant globally and as reefs are under the assault of an increasing number of stressors, it is urgent to determine whether algal-surface associated pathogens can migrate to coral surfaces, and whether this transmission is dependent upon coral health.
Supporting Information
Pairwise tests of 16S rRNA gene bacterial communities within separated coral and algal samples at Heron Island. (ND) Healthy coral, (AH) apparently healthy, (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Pairwise tests of 16S rRNA gene bacterial communities within separated coral and algal samples at Los Roques. (ND) Healthy coral, (AH) apparently healthy, (T) transition and (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Pairwise tests of bacterial diversity within separated coral and algal samples at Heron Island. (ND) Healthy coral, (AH) apparently healthy, (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Pairwise tests of bacterial diversity within separated coral and algal samples at Los Roques. (ND) Healthy coral, (AH) apparently healthy, (T) transition and (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Time-lapse image of Yellow band Disease, showing the presence of ciliates at the disease lesion interface, although ciliates were not detected in the samples in this study, this illustrates that they are likely playing some kind of role YBD.
(WMV)
Funding Statement
This work was supported by the Natural Environmental Research Council, UK (NE/E006949). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bythell J, Sheppard C (1993) Mass Mortality of Caribbean Shallow Corals. Marine Pollution Bulletin 26: 296–297. [Google Scholar]
- 2. Bythell JC, Hillis-Starr ZM, Rogers CS (2000) Local variability but landscape stability in coral reef communities following repeated hurricane impacts. Marine Ecology-Progress Series 204: 93–100. [Google Scholar]
- 3. Aronson RB, Precht WF, Macintyre IG (1998) Extrinsic control of species replacement on a Holocene reef in Belize: the role of coral disease. Coral Reefs 17: 223–230. [Google Scholar]
- 4. Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, et al. (2002) Climate warming and disease risks for terrestrial and marine biota. Science 296: 2158–2162. [DOI] [PubMed] [Google Scholar]
- 5. Bruno JF, Selig ER, Casey KS, Page CA, Willis BL, et al. (2007) Thermal stress and coral cover as drivers of coral disease outbreaks. PLOS Biology 5: 1220–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, et al. (2007) Coral reefs under rapid climate change and ocean acidification. Science 318: 1737–1742. [DOI] [PubMed] [Google Scholar]
- 7. Bruno JF, Petes LE, Harvell CD, Hettinger A (2003) Nutrient enrichment can increase the severity of coral diseases. Ecology Letters 6: 1056–1061. [Google Scholar]
- 8. Voss JD, Richardson LL (2006) Nutrient enrichment enhances black band disease progression in corals. Coral Reefs 25: 569–576. [Google Scholar]
- 9. Hughes TP (1994) Catastrophes, phase shifts, and large scale degradation of the Caribbean coral reef. Science 265: 1574–1551. [DOI] [PubMed] [Google Scholar]
- 10. McClanahan T, Polunin N, Done T (2002) Ecological states and the resilience of coral reefs. Conservation Ecology 6. [Google Scholar]
- 11. Bruno JF, Sweatman H, Precht WF, Selig ER, Schutte VGW (2009) Assessing evidence of phase shifts from coral to macroalgal dominance on coral reefs. Ecology 90: 1478–1484. [DOI] [PubMed] [Google Scholar]
- 12. Done TJ, Dayton PK, Dayton AE, Steger R (1991) Regional and local variability in recovery of shallow coral communities - Moorea, French-Polynesia and central. Great Barrier Reef Coral Reefs 9: 183–192. [Google Scholar]
- 13. Burkepile DE, Hay ME (2006) Herbivore vs. nutrient control of marine primary producers: Context-dependent effects. Ecology 87: 3128–3139. [DOI] [PubMed] [Google Scholar]
- 14. Vermeij MJA, van Moorselaar I, Engelhard S, Hornlein C, Vonk SM, et al. (2010) The Effects of Nutrient Enrichment and Herbivore Abundance on the Ability of Turf Algae to Overgrow Coral in the Caribbean. Plos One 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hayes RL, Goreau NI (1998) The significance of emerging diseases in the tropical coral reef ecosystem. Rev Biol Trop 46: 173–185. [Google Scholar]
- 16. Harvell D, Aronson R, Baron N, Connell JH, Dobson AP, et al. (2004) The rising tide of ocean diseases: unsolved problems and research priorities. Frontiers in Ecology and the Environment 2: 375–382. [Google Scholar]
- 17. Goreau TJ, Cervino J, Goreau M, Hayes R, Hayes M, et al. (1998) Rapid spread of diseases in Caribbean coral reefs. Revista De Biologia Tropical 46: 157–171. [Google Scholar]
- 18. Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, et al. (1999) Review: Marine ecology - Emerging marine diseases - Climate links and anthropogenic factors. Science 285: 1505–1510. [DOI] [PubMed] [Google Scholar]
- 19. Nugues MM, Smith GW, Van Hooidonk RJ, Seabra MI, Bak RPM (2004) Algal contact as a trigger for coral disease. Ecology Letters 7: 919–923. [Google Scholar]
- 20. Bender D, Diaz-Pulido G, Dove S (2012) Effects of macroalgae on corals recovering from disturbance. Journal of Experimental Marine Biology and Ecology 429: 15–19. [Google Scholar]
- 21. Quan-Young LI, Espinoza-Avalos J (2006) Reduction of zooxanthellae density, chlorophyll a concentration, and tissue thickness of the coral Montastraea faveolata (Scleractinia) when competing with mixed turf algae. Limnology and Oceanography 51: 1159–1166. [Google Scholar]
- 22. Rasher DB, Hay ME (2010) Chemically rich seaweeds poison corals when not controlled by herbivores. Proceedings of the National Academy of Sciences of the United States of America 107: 9683–9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCook LJ, Jompa J, Diaz-Pulido G (2001) Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs 19: 400–417. [Google Scholar]
- 24. Smith JE, Shaw M, Edwards RA, Obura D, Pantos O, et al. (2006) Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecology Letters 9: 835–845. [DOI] [PubMed] [Google Scholar]
- 25. Haas AF, Nelson CE, Wegley Kelly L, Carlson CA, Rohwer F, et al. (2011) Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. Plos One 6: e27973–e27973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson CE, Goldberg SJ, Kelly LW, Haas AF, Smith JE, et al. (2013) Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. Isme Journal 7: 962–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rasher DB, Stout EP, Engel S, Kubanek J, Hay ME (2011) Macroalgal terpenes function as allelopathic agents against reef corals. Proceedings of the National Academy of Sciences of the United States of America 108: 17726–17731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morrow KM, Paul VJ, Liles MR, Chadwick NE (2011) Allelochemicals produced by Caribbean macroalgae and cyanobacteria have species-specific effects on reef coral microorganisms. Coral Reefs 30: 309–320. [Google Scholar]
- 29. Morrow KM, Ritson-Williams R, Ross C, Liles MR, Paul VJ (2012) Macroalgal Extracts Induce Bacterial Assemblage Shifts and Sublethal Tissue Stress in Caribbean Corals. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thurber RV, Burkepile DE, Correa AMS, Thurber AR, Shantz AA, et al. (2012) Macroalgae Decrease Growth and Alter Microbial Community Structure of the Reef-Building Coral, Porites astreoides. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolf AT, Nugues MM (2013) Synergistic effects of algal overgrowth and corallivory on Caribbean reef-building corals. Ecology doi.org/10.1890/12-0680.1 [DOI] [PubMed] [Google Scholar]
- 32. Vu I, Smelick G, Harris S, Lee SC, Weil E, et al. (2009) Macroalgae Has No Effect on the Severity and Dynamics of Caribbean Yellow Band Disease. Plos One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denner EBM, Smith GW, Busse HJ, Schumann P, Narzt T, et al. (2003) Aurantimonas coralicida gen. nov., sp nov., the causative agent of white plague type II on Caribbean scleractinian corals. International Journal of Systematic and Evolutionary Microbiology 53: 1115–1122. [DOI] [PubMed] [Google Scholar]
- 34. Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, et al. (2003) Vibrio coralliilyticus sp nov., a temperature dependent pathogen of the coral Pocillopora damicornis . International Journal of Systematic and Evolutionary Microbiology 53: 309–315. [DOI] [PubMed] [Google Scholar]
- 35. Cervino JM, Thompson FL, Gomez-Gil B, Lorence EA, Goreau TJ, et al. (2008) The Vibrio core group induces yellow band disease in Caribbean and Indo-Pacific reef-building corals. Journal of Applied Microbiology 105: 1658–1671. [DOI] [PubMed] [Google Scholar]
- 36. Kushmaro A, Banin E, Loya Y, E S, Rosenberg E (2001) Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica . International Journal of Systematic and Evolutionary Microbiology 51: 1383–1388. [DOI] [PubMed] [Google Scholar]
- 37. Luna GM, Bongiorni L, Gili C, Biavasco F, Danovaro R (2010) Vibrio harveyi as a causative agent of the White Syndrome in tropical stony corals. Environmental Microbiology Reports 2: 120–127. [DOI] [PubMed] [Google Scholar]
- 38. Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, et al. (2009) Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. Isme Journal 3: 512–521. [DOI] [PubMed] [Google Scholar]
- 39. Sussman M, Willis BL, Victor S, Bourne DG (2008) Coral Pathogens Identified for White Syndrome (WS) Epizootics in the Indo-Pacific. Plos One 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barott KL, Rodriguez-Brito B, Janouskovec J, Marhaver KL, Smith JE, et al. (2011) Microbial diversity associated with four functional groups of benthic reef algae and the reef building coral Montastraea annularis . Environmental Microbiology 13: 1192–1204. [DOI] [PubMed] [Google Scholar]
- 41. Guppy R, Bythell JC (2006) Environmental effects on bacterial diversity in the surface mucus layer of the reef coral Montastraea faveolata . Marine Ecology-Progress Series 328: 133–142. [Google Scholar]
- 42. Haapkylae J, Unsworth RKF, Seymour AS, Melbourne-Thomas J, Flavell M, et al. (2009) Spatio-temporal coral disease dynamics in the Wakatobi Marine National Park, South-East Sulawesi, Indonesia. Diseases of Aquatic Organisms 87: 105–115. [DOI] [PubMed] [Google Scholar]
- 43. Ainsworth TD, Kvennefors EC, Blackall LL, Fine M, Hoegh-Guldberg O (2007) Disease and cell death in white syndrome of Acroporid corals on the Great Barrier Reef. Marine Biology 151: 19–29. [Google Scholar]
- 44. Work TM, Aeby GS (2011) Pathology of tissue loss (white syndrome) in Acropora sp corals from the Central Pacific. Journal of Invertebrate Pathology 107: 127–131. [DOI] [PubMed] [Google Scholar]
- 45. Sweet MJ, Bythell J (2012) Ciliate and bacterial communities associated with White Syndrome and Brown Band Disease in reef building corals. Environ Microbiol 14: 2184–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Croquer A, Bastidas C, Elliott A, Sweet MJ (2013) Bacterial assemblages shifts from healthy to yellow band disease states in the dominant reef coral Montastraea faveolata. Environmental Microbiology Reports 5: 90–96. [DOI] [PubMed] [Google Scholar]
- 47. Koren O, Rosenberg E (2006) Bacteria Associated with mucus and tissues of the coral Oculina patagonica in the Summer and Winter. Applied and Environmental Microbiology 72: 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steneck RS, Dethier MN (1994) A functional grup approach to the structure of algal dominated communities. Oikos 69: 476–498. [Google Scholar]
- 49. Sweet MJ, Croquer A, Bythell JC (2010) Temporal and spatial patterns in waterborne bacterial communities of an island reef system. Aquatic Microbial Ecology 61: 1–11. [Google Scholar]
- 50. Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Current Opinion in Microbiology 2: 317–322. [DOI] [PubMed] [Google Scholar]
- 51. Clarke KR, Warwick RM (2001) A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Marine Ecology-Progress Series 216: 265–278. [Google Scholar]
- 52. Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecology 26: 32–46. [Google Scholar]
- 53. Casadevall A, Pirofski LA (2003) The damage-response framework of microbial pathogenesis. Nature Reviews Microbiology 1: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mouchka M, Hewson I, Harvell D (2010) Coral-Associated Bacterial Assemblages: Current Knowledge and the Potential for Climate-Driven Impacts. Integrative and comparative biology doi:–10.1093/icb/icq061: p1–13 [DOI] [PubMed] [Google Scholar]
- 55. Cooney RP, Pantos O, Le Tissier MDA, Barer MR, O'Donnell AG, et al. (2002) Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environmental Microbiology 4: 401–413. [DOI] [PubMed] [Google Scholar]
- 56. Mouchka ME, Hewson I, Harvell CD (2010) Coral-Associated Bacterial Assemblages: Current Knowledge and the Potential for Climate-Driven Impacts. Integrative and Comparative Biology 50: 662–674. [DOI] [PubMed] [Google Scholar]
- 57. Pantos O, Bythell JC (2006) Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture-independent 16S rRNA techniques. Diseases of Aquatic Organisms 69: 79–88. [DOI] [PubMed] [Google Scholar]
- 58. Pantos O, Cooney RP, Le Tissier MDA, Barer MR, O'Donnell AG, et al. (2003) The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environmental Microbiology 5: 370–382. [DOI] [PubMed] [Google Scholar]
- 59. Sanchez O, Gasol JM, Massana R, Mas J, Pedros-Alio C (2007) Comparison of different denaturing gradient gel electrophoresis primer sets for the study of marine bacterioplankton communities. Applied and Environmental Microbiology 73: 5962–5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kvennefors ECE, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O (2010) Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site and species-specificity of common bacterial associates. PLoS One 5: e10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sevak HP, Amsler CD, McClintock JB, Maschek JA, Amsler MO, et al. (2012) Algicidal activity and potential antifouling defenses in macroalgae from the western Antarctic Peninsula including probable synergistic effects of multiple compounds. Botanica Marina 55: 311–315. [Google Scholar]
- 62. Paul VJ, Ritson-Williams R (2008) Marine chemical ecology. Natural Product Reports 25: 662–695. [DOI] [PubMed] [Google Scholar]
- 63. Barott KL, Rohwer FL (2012) Unseen players shape benthic competition on coral reefs. Trends in Microbiology 20: 621–628. [DOI] [PubMed] [Google Scholar]
- 64. Xu DH, Klesius PH, Bosworth BG, Chatakondi N (2012) Susceptibility of three strains of blue catfish, Ictalurus furcatus (Valenciennes), to Ichthyophthirius multifiliis. Journal of Fish Diseases 35: 887–895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pairwise tests of 16S rRNA gene bacterial communities within separated coral and algal samples at Heron Island. (ND) Healthy coral, (AH) apparently healthy, (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Pairwise tests of 16S rRNA gene bacterial communities within separated coral and algal samples at Los Roques. (ND) Healthy coral, (AH) apparently healthy, (T) transition and (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Pairwise tests of bacterial diversity within separated coral and algal samples at Heron Island. (ND) Healthy coral, (AH) apparently healthy, (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Pairwise tests of bacterial diversity within separated coral and algal samples at Los Roques. (ND) Healthy coral, (AH) apparently healthy, (T) transition and (DL) disease lesion. * p<0.05; ns: not significant.
(DOCX)
Time-lapse image of Yellow band Disease, showing the presence of ciliates at the disease lesion interface, although ciliates were not detected in the samples in this study, this illustrates that they are likely playing some kind of role YBD.
(WMV)