Abstract
An unexplained increase in the incidence of parapneumonic empyema (PPE) in pneumonia cases has been reported in recent years. The present study investigated the genetic and biological specifications of new isolates of torque teno mini virus (TTMV) detected in pleural effusion samples from children hospitalised for severe pneumonia with PPE.
A pathogen discovery protocol was applied in undiagnosed pleural effusion samples and led to the identification of three new isolates of TTMV (TTMV-LY). Isolated TTMV-LY genomes were transfected into A549 and human embryonic kidney 293T cells and viral replication was assessed by quantitative real-time PCR and full-length genome amplification. A549 cells were further infected with released TTMV-LY virions and the induced-innate immune response was measured by multiplex immunoassays.
Genetic analyses of the three TTMV-LY genomes revealed a classic genomic organisation but a weak identity (<64%) with known sequences. We demonstrated the in vitro replication of TTMV-LY in alveolar epithelial cells and the effective release of infectious viral particles. We also showed a selective production of inflammatory mediators in response to TTMV infection.
This study reports the description of replicative TTMV-LY isolated from parapneumonic effusions of children hospitalised with PPE, suggesting a potential role of the virus in the pathogenesis of pneumonia.
Introduction
About 1.8 million children aged <5 years die from pneumonia every year [1]. A wide range of pathogens are recognised as causative agents of pneumonia, but 14–23% of cases remain aetiologically undiagnosed, despite continuing optimisation of diagnostic tests for the detection of respiratory pathogens [2, 3].
Different methods have been developed for the identification of unsuspected, new and divergent pathogens. In recent years, these techniques have led to the discovery of several viruses in respiratory specimens, including human metapneumovirus, bocavirus, WU and KI polyomaviruses, and torque teno virus (TTV) [4]. Some of these have been confirmed subsequently as causative agents of lower respiratory tract infections (LRTI), but the involvement of most of these viruses remains a widely debated issue. TTV belongs to the recently created viral family Anelloviridae, composed of single-stranded circular DNA viruses, such as TTV (Alphatorquevirus), torque teno midi virus (Gammatorquevirus) and torque teno mini virus (TTMV) (Betatorquevirus) [5].
In this study, we isolated and characterised three new species of TTMV (TTMV-LY), detected in pleural fluids from children hospitalised in France with parapneumonic empyema (PPE) by a method of pathogen discovery. This pathology, with increasing incidence, is associated with a high morbidity and frequently requires prolonged hospitalisation and invasive procedures [6]. As the respiratory tract is thought to be a site of primary infection and continual replication of TTV [7], we conducted further investigations to evaluate to what extent these new viruses may be associated with the pathology. We report here that TTMV-LY isolates actively replicated in alveolar epithelial cells and were able to modulate their innate immune response.
Experimental procedures
Patients and samples
A prospective study was performed of 28 children, aged from 5 months to 14 years (12 males and 16 females), hospitalised for PPE between April 4, 2007 and March 30, 2009 in the paediatric intensive care unit of Hospices Civils de Lyon (Lyon, France). This study was approved by all participating institutional review boards (CE 06–139) and signed informed consent was obtained from all parents. 28 nasopharyngeal aspirates were obtained using disposable mucus extractors within 48 h of admission. 25 pleural effusion samples were collected at the time of chest drain insertion. The samples were kept frozen at -70°C until further study.
Respiratory pathogens nucleic acids detection
The presence of the 25 most common respiratory pathogens was tested in the 28 nasal aspiration and 25 pleural effusion samples using the FTD Respiratory Pathogens 21+ assay (Fast Track Diagnostics, Junglinster, Luxembourg) in combination with the AgPath-ID One-Step RT-PCR Kit (Ambion, Austin, TX, USA) after total nucleic acid extraction using the NucliSENS® easyMAG platform (bioMérieux, Craponne, France) (named easyMAG below), as recommended by the manufacturer.
TTMV isolation by pathogen discovery and genetic characterisation
Five negative pleural effusion samples were subjected to a pathogen discovery protocol adapted from Allander et al. [8] and based on the cloning and sequencing of random RT-PCR products. “Homebrew” sequence analysis software was used to identify new or divergent sequences from those deposited in databases. Three out of five samples harbouring fragments of TTMV sequences were subjected to TTMV full-length genome amplification by inverse PCR with back-to-back TTMV-LY-9F and TTMV-LY-8R primers (table 1). Amplicons were cloned and sequenced through both strands. Using the MEGA5 programme (http://megasoftware.net/mega.php), multiple alignments were generated by ClustalW (www.clustal.org/omega), and phylogenetic trees were constructed by the neighbour-joining method. Predictive secondary structures were modelled with RNA structure 4.5 software (D.H. Mathews, M. Zuker and D.H. Turner). Details of the procedures are given in the online supplementary material. The three full-length genomes obtained in the present study were deposited in the GenBank database under the following accession numbers: JX134044, JX134045 and JX134046.
Table 1– Primers and probes used in this study.
| Primers | Sequence | Nucleotide position | Target | GATC-site position | Amplification conditions | |
| Inverse PCR | ||||||
| TTMV-LY-9F | 5'-GACCACAAACCGTCACTTAGTTCC-3' | 42–65 | Betatorquevirus | NA | 95°C for 1 min 40 cycles: 95°C for 30 s 54°C for 1 min 72°C for 3 min 72°C for 10 min |
|
| TTMV-LY-8R | 5'-GCAATTTAAATTAGGTGGTTTTCCTG-3' | 40–15 | ||||
| TTMV-LY-11F | 5'-CTAGRTCAGTCTGGCGGAACGG-3' | 288–309 | Betatorquevirus | NA | ||
| TTMV-LY-10R | 5'-CCCCTTGACTACGGTGGTTTCAC-3' | 278–256 | ||||
| Real-time PCR | ||||||
| TTMV-LY-F | 5'-ATTCGAATGGCTGAGTTTATGC-3' | 170–191 | Betatorquevirus | 210, 579, 2674 | 95°C for 3 min 35 cycles: 95°C for 10 s 54°C for 30 s 72°C for 34 s |
|
| TTMV-LY-R | 5'-CCTTGACTACGGTGGTTTCAC-3' | 276–256 | ||||
| TTMV-LY-P | 5'-CCAGACGGAGACCGGATCACTTCA-3' (FAM-BHQ1) | 194–217 | ||||
| TTMV-LY-2F | 5'-CACGAATTAGCCAAGACTGGGCAC-3' | 2398–2421# | TTMV-LY2 | 512, 1382, 2456, 2732, 2746# |
||
| TTMV-LY-2R | 5'-TGCAGGCATTCGAGGGCTTGTT-3' | 2494–2473# | ||||
| TTMV-LY-2P | 5'-ACAGAGCCAGATCGACAGCAACTGCT-3' (FAM-BHQ1) | 2445–2470# | ||||
Nucleotide position and GATC-sites are indicated based on TGP96 (AB041962) sequence. TTMV-LY: torque teno mini virus isolates; NA: not applicable; F: forward; R: reverse; P: probe. #: nucleotide positions are based on TTMV-LY2 sequence.
Cell culture and TTMV transfection or infection
Two cell lines were used to study the replication of these new isolates of TTMV: the human embryonic kidney (HEK) 293T cell line and A549 alveolar epithelial cell line (both from CelluloNet, UMS3444/US8 Biosciences Gerland-Lyon Sud, Lyon, France). HEK293T cells were maintained in DMEM, 10% fetal bovine serum, 2 mM glutamine and antibiotics, while A549 cells were maintained in F12K medium, 10% fetal bovine serum, 2 mM glutamine and antibiotics. Cloned TTMV DNA was linearised with EcoRV and BamHI (Roche Diagnostics, Meylan, France). Viral and vector DNAs were cotransfected into cells (1 μg per 105 cells on 12-well plates) with JetPEI reagent (PolyPlus-transfection, Illkirch, France) as recommended by the manufacturer. Controls included transfection with vector alone or cells transfected with JetPEI alone. Transfected cultures were incubated overnight at 37°C and 5% carbon dioxide. After 18 h, cells were washed three times with PBS before adding fresh medium. The transfection efficiencies were optimised with a reporter plasmid. For the infection studies, healthy cells, seeded in 24-well plates, were infected with the supernatants of TTMV DNA-transfected cells that had been harvested 3 days post-transfection, and incubated at 37°C and 5% carbon dioxide for 2 h, 7 h and 24 h. Cells were then washed three times with PBS and incubated with fresh medium for 6 h. All experiments were performed at least twice, in duplicate.
TTMV-LY replication
For all experiments, supernatants were collected at each prescribed time, centrifuged and immediately treated with DNaseI for 2 h at 37°C and frozen at -80°C. Replicated TTMV DNA was extracted using easyMAG and eluted in 50 μL. Cells were harvested in 2 mL of NucliSENS lysis buffer (bioMérieux) for the extraction of nucleic acids with easyMAG and eluted in 100 μL of elution buffer. Total cellular DNA was then digested with DpnI or NdeII, which cut the dam-methylated GATC-sites of input TTMV and the nonmethylated GATC-sites of replicative TTVMV, respectively (table 1). To completely digest DNA and reduce the background level observed in qPCR, exonuclease III (ExoIII; Fermentas, Burlington, Canada) was added, as described elsewhere [9]. Full-length genomes of TTMV were detected by inverse PCR. Briefly, the reaction mix was composed of 5 μL of nucleic acids, 1 μM of each primer (TTMV-LY-11F and TTMV-LY-10R) (table 1), 2 mM of deoxynucleotide triphosphate mix and Expand High Fidelity enzyme mix (Invitrogen, Darmstadt, Germany). Quantification of replicated TTMV was achieved by TaqMan real-time PCR assays (Eurogentec, Seraing, Belgium) with primers targeting specifically a GATC-site. The qPCR assays were optimised as previouly recommended [10]. The amplification step was realised in 25 μL volumes containing 12.5 μL iQ Supermix (Bio-Rad, Hercules, CA, USA), 0.3 μM each of primers and probe (table 1) and 5 μL of DNA. Amplification was detected with a CFX96 detection machine (BioRad). DNA from untransfected cells was included in each reaction. For transfection studies, the replication rate was calculated as the ratio of the quantified amount of replicative TTMV detected after DpnI digestion to the total quantified amount of TTMV (replicative and residual input TTMV genomes).
Measurements of cytokine/chemokine release
Cell-free supernatants collected from the TTMV-infected A549 cell line cultures were assayed for simultaneous quantification of human epidermal growth factor, eotaxin, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon (IFN)-α2, IFN-γ, interleukin (IL)-10, IL-12(p40), IL-12(p70), IL-13, IL-15, IL-17, IL-1ra, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IP-10, monocyte chemoattractant protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β, tumour necrosis factor (TNF)-α, TNF-β and vascular endothelial growth factor (VEGF), using a commercial multiplex bead-based immunoassay kit (Milliplex; Millipore, Billerica, MA, USA).
Results
Prevalence of respiratory pathogens in nasal aspiration and pleural effusion positive samples
The identification of the main respiratory agents in nasal aspiration and pleural effusion samples of children hospitalised for severe pneumonia was performed using a sensitive multiplex real-time molecular assay. In nasal aspiration samples, rhinovirus, parainfluenza viruses and human metapneumovirus were found as the main respiratory pathogens (table 2). Bacterial agents were found in 80% of the pleural effusion samples whereas 20% remained undiagnosed.
Table 2– Prevalence of respiratory pathogens in nasopharyngeal aspirates and pleural effusion samples from 28 children hospitalised with parapneumonic empyema.
| Nasopharyngeal aspirates | Pleural effusion | |
| Subjects | 28 | 25 |
| Bacterial detection | ||
| Streptococcus pneumoniae | 15 | 19 |
| Staphylococcus aureus | 2 | 1 |
| Viral detection | ||
| Rhinovirus | 8 | |
| Bocavirus | 3 | |
| Parainfluenza viruses | 4 | |
| Human metapneumovirus | 4 | |
| Human coronavirus NL63 | 2 | |
| Respiratory syncitial virus | 2 | |
| Influenza A and B | 0 | |
| Adenovirus | 1 | |
| Cases of co-detection | 6 | 0 |
| Undiagnosed | 5 (18) | 5 (20) |
| TTMV detection# | ||
| Positive samples | 8 | 9 |
| Cases of co-detection | 6 (with one or several viruses) | 6 (with S. pneumoniae) |
Data are presented as n or n (%). Clinical criteria were based on respiratory complaints and fever, difficulty in breathing, pulmonary infiltrates compatible with pneumonia, a chest radiograph at admission confirmed by ultrasounds, white blood cell counts >20 G·L−1 or neutrophilis >10 G·L−1 and C-reactive protein level >60 mg·L−1 after 12 h of fever. Undiagnosed nasal aspiration and pleural effusion samples came from different patients, except for one of them. TTMV: torque teno mini virus. TTMV was detected by real-time PCR with primers and probes indicated in table 1. #: Isolation and genetic characterisation of TTMV sequences in undiagnosed pleural effusion samples.
A pathogen discovery protocol based on random RT-PCR was performed on the five negative pleural effusion samples, and several short sequences related to TTMV were obtained for three of them. The characterisation of TTMV required the amplification and sequencing of the full-length virus genome. Betatorquevirus-specific inverse PCR was performed and the corresponding TTMV-LY genomes were amplified. The TTMV-LY genomes were composed of 2912 bp, 2979 bp and 2915 bp for TTMV-LY1, -LY2 and -LY3, respectively. Using PCR, the TTMV-LY genomes found in pleural effusion samples were also identified in the corresponding nasal aspiration sample for each patient. Among the three patients for whom the pleural effusion samples were only positive for TTMV, two were male and one was female. They were aged 48 months, 7 months and 37 months for TTMV-LY1, -LY2 and -LY3, respectively, and presented the same clinical features as the other patients of the cohort. Phylogenetic analyses were carried out on the nucleotide sequence of predictive open reading frame (ORF)1, as recommended by the International Committee on Taxonomy of Viruses [5]. Based on the phylogenetic tree (fig. 1), all three new isolates of TTMV-LY belong to genogroup I and are related to previously identified TGP96 (64% of maximum identity) and Pt-TTV8-II (44% of maximum identity) isolates. TTMV-LY isolates possess at least three ORFs: one major ORF1 composed of 673, 667 and 659 amino acids (aa) for TTMV-LY1, -LY2 and -LY3, respectively, and several overlapping ORFs. The TTMV-LY1 and -LY3 have two overlapping ORFs of 91 and 101 aa, and 97 and 129 aa, respectively, as opposed to TTMV-LY2, which has ORFs of 100 and 109 aa, and an additional ORF3 of 113 aa. One other feature of these viruses is the highly conserved 5′-untranslated region (UTR), as shown in figure 2. Figure 2a shows that TTMV-LY2 possesses a significant insertion in this 5′-UTR, forming a predictive secondary structure of RNA sequence (fig. 2b). This structure appeared to be capable of forming a stable loop-and-dimer structure.
Figure 1–
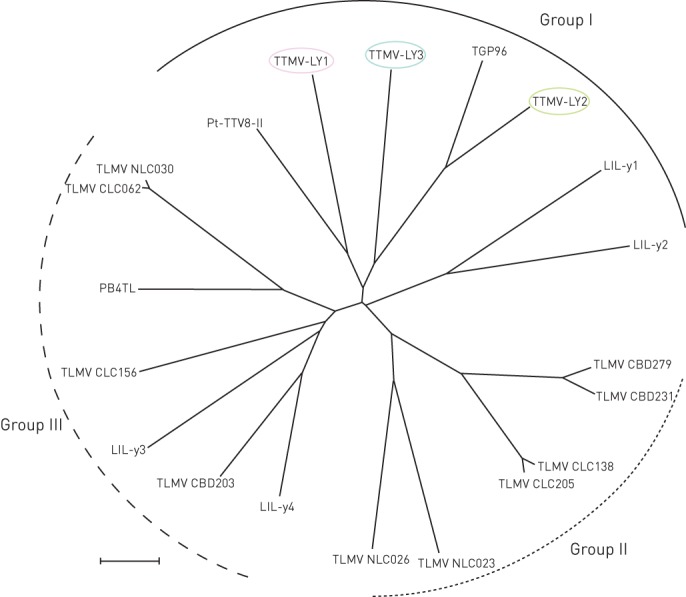
Phylogenetic tree of the open reading frame-1 nucleotide sequence of torque teno mini virus (TTMV) isolates TTMV-LY1, -LY2, -LY3 and Betatorquevirus constructed by the neighbour-joining method. TTMV-LY genomes isolated from pleural effusion samples are circled. Scale bar = 0.1 nucleotide substitutions per position.
Figure 2–
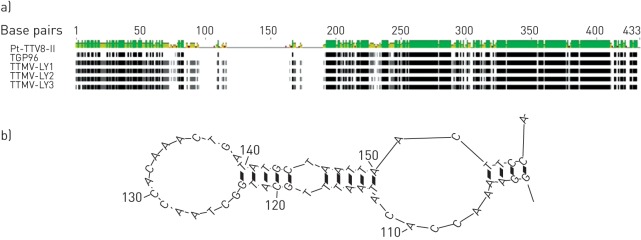
Multiple alignment in the 5′-untranslated region of the newly defined phylogenetic group I nucleotide sequences revealed the presence of a significant insertion in the torque teno mini virus (TTMV) isolates TTMV-LY2 genome (a), which could form a stem-loop pattern in the predictive secondary structure (b). Residues that are identical among the sequences are given a black background. Lines between residues indicate deletions between the sequences. A: adenine; C: cytosine; G: guanine; T: thymine.
Transfection of the TTMV-LY genomes into cells and in vitro replication
After transfection of the linearised virus genomes, we monitored: 1) the replication of the new TTMV isolates in the A549 and HEK293T cells; and 2) the release of viral particles in the supernatants, by quantifying newly synthesised genomes with a specific normalised qPCR assay for 1 week. We initially observed that, as shown in figure 3a, all the TTMV-LY isolates actively replicated in both cell lines. There was no significant difference between the replication mean rates for the three isolates in A549 cells (25%) and in HEK293T (10%). As observed in the kinetic experiments (fig. 3b), the mean rate of replication rose regularly from day 2 to day 4, reaching 35% of the replicative form of TTMV-LY in both cell lines. This replication rate remained relatively constant in A549 cells until day 7, but in HEK293T cells it slightly decreased beginning on day 4. The release of TTMV-LY particles showed a similar pattern as to the replication rate within the cells, with a peak at day 4 post-transfection, with an eight-fold increase for A549 and a 20-fold increase for HEK293T supernatants.
Figure 3–
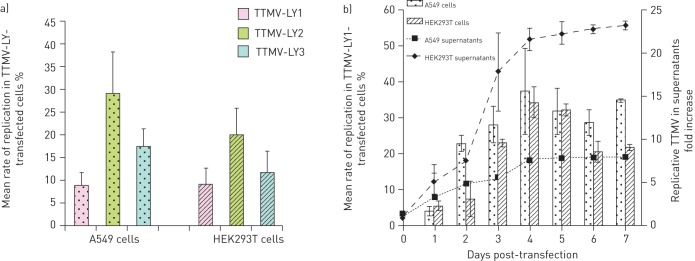
Replication of torque teno mini virus (TTMV) isolates TTMV-LY genomes after transfection produce more replicative TTMV in human embryonic kidney (HEK) 293T cells than in A549 cells as quantified by real-time qPCR. Histograms represent the mean±sd rate of replication of TTMV-LY in the cells transfected with the linearised full-length genomes. Replication of TTMV-LY was a) measured at day 3 post-transfection or b) monitored over a week following transfection. The curves in b) indicate the quantification of TTMV-LY1 in the culture medium of cells at days 0–7 post-transfection after DNaseI (Thermo Scientific, Illkirch, France) treatment and nucleic acid extraction. Results are expressed as the ratio of quantified TTMV-LY1 at day 1–7 post-transfection to quantified TTMV-LY1 at day 0. Data are shown only for TTMV-LY1, because the replication profile among the three viruses is similar.
Furthermore, we ensured that the replicative TTMV-LY detected in figure 3 effectively corresponded to viral particles and not merely to partial fragments of DNA. For this purpose, the complete genome amplification of TTMV-LY in the A549 cells (fig. 4a) and supernatants (fig. 4b) were measured. We observed the presence of a 2.9-kb PCR fragment representing the recircularised replicative TTMV for the three TTMV-LY isolates, verified by sequencing analyses (data not shown). We confirmed that circular full-length genomes were present in cells and were released into the supernatants of A549 cells, indicating the presence of viral particles.
Figure 4–
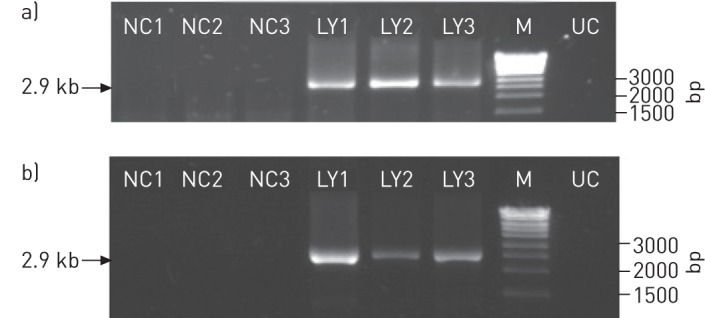
Detection, by PCR with back-to-back primers, of replicative recircularised genomes of torque teno mini virus (TTMV) isolates TTMV-LY1, -LY2 and -LY3 in a) A549 cells and b) in the supernatants of cells at day 3 post-transfection. NC1, NC2, and NC3: negative control for each genome. LY1, LY2 and LY3: cells transfected with the TTMV-LY1, -LY2 and -LY3 genomes, respectively; M: DNA molecular weight marker; UC: uninfected cells.
TTMV-LY infectious particles replicate in alveolar epithelial cells
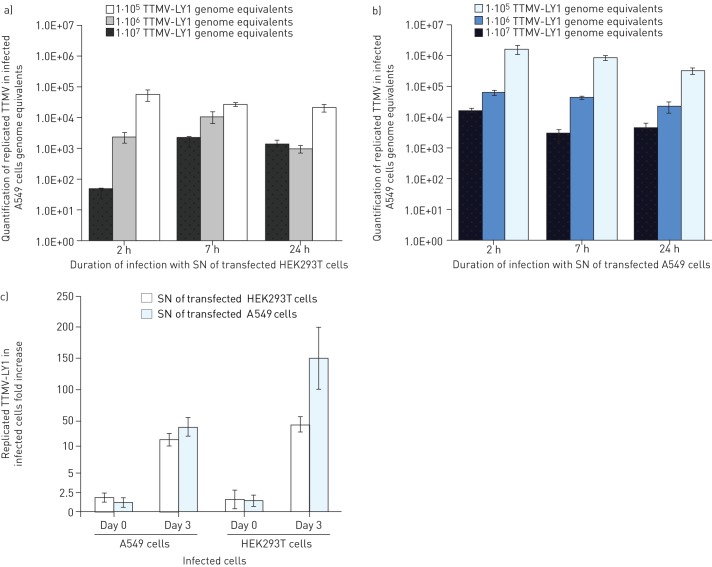
The capacity for producing replicative particles was measured by quantifying the TTMV-LY genomes in A549 cells. We exposed pulmonary cells for 2 h, 7 h and 24 h with the neosynthetised virus particles released at day 3 post-transfection into the supernatants of the transfected A549 and HEK293T cells. We noticed that the optimal condition for A549-infected TTMV-LY was after incubation with the supernatants of A549 and HEK293T transfected cells for 2 h and 7 h, respectively (fig. 5a and b). We also observed that the efficiency of TTMV A549 cell entry was more significant when the viral load was highest (107 genome equivalents). Moreover, the quantity of TTMV-LY within the A549 cells was systematically greater for infections with the supernatants of A549 cells than within HEK293T cells, with 1.7×106 versus 5.6×104 after 2 h of infection with 107 genome equivalents, suggesting that the viral particles produced by pulmonary cells may be more infectious. This result was confirmed by the quantification of replicated TTMV-LY after infection of the two cell lines (fig. 5c). Infected A549 cells systematically showed the highest viral titre with 48- and 167-fold increases for the infections with the HEK293T and A549 supernatants, respectively, versus 22- and 44-fold increases for the HEK293T cells at day 3. This observation was made for all three TTMV-LY genomes (data not shown).
Figure 5–
Torque teno mini virus (TTMV)-LY isolates infect the A549 alveolar cell line preferentially over the human embryonic kidney (HEK)293T cell line, as quantified by real-time PCR. a) and b) A549 cells were incubated with TTMV-LY1 for 2 h, 7 h, and 24 h with three different viral loads of 105, 106, and 107 genome equivalents, harvested from the supernatants (SN) of transfected A549 or HEK293T cells at day 3 post-transfection. The quantity of TTMV-LY measured in culture-medium exposed cells was null. c) The quantity of replicated TTMV-LY was measured at day 0 and day 3 post-infection in the 2-h exposed A549 and HEK293T cells with a viral load of 1×107 genome equivalents. Results are expressed as fold increase compared to cells exposed with the supernatants of cells exposed to the TTMV-LY genomes without transfection reagent. Data are presented as mean±sd.
Each individual TTMV-LY modulates the innate immune response of pulmonary epithelial cells in a specific manner
In this study, we used a 29-plex bead-based immunoassay to measure the levels of a wide range of inflammatory mediators in the supernatant of pulmonary cells infected with TTMV-LY1, -LY2 or LY3. As shown in table 3, at 6 h post-infection TTMV-LY induced the production of several cytokines and chemokines, which are significantly enhanced compared to those produced in uninfected cells. We observed that TTMV-LY1 drives the production of a large number of pro-inflammatory (IFN-γ, RANTES, IL-2 and IL-12), chemotactic (MIP-1b) and anti-inflammatory cytokines (IL-10 and IL-13), as well as growth factors (G-CSF, GM-CSF and IL-7). The two other TTMV-LY genomes induce a weaker response, with the induction of RANTES, IL-12(p40), MCP-1, VEGF and IL-13 for TTMV-LY2, and IL-2, IL-12(p40) and MCP-1 for TTMV-LY3. Only IL-12 was released into supernatants for the three TTMV-LY isolates after infection of the cells.
Table 3– Secretion of inflammatory mediators into the supernatant of 2 h-infected A549 cells at 6 h post-infection.
| Soluble mediators pg·mL−1 | Mock | TTMV-LY1 | TTMV-LY2 | TTMV-LY3 |
| Pro-inflammatory cytokines | ||||
| IFN-γ | 4.5±3.0 | 20.5±4.9 | 7.8±2.7 | 7.2±0.8 |
| IL-2 | 26.6±5.2 | 51.3±6.8 | 32.3±2.4 | 33.4±1.2* |
| IL-12 (p40) | 16.6±4.2 | 36.3±4.9* | 22.2±1.0* | 22.3±1.1* |
| IL-12 (p70) | 10.2±1.8 | 20.9±2.1* | 10.1±0.8 | 10.6±0.6 |
| Anti-inflammatory cytokines | ||||
| IL-10 | 54.8±4.9 | 95.7±5.4* | 60.2±1.2 | 55.3±0.9 |
| IL-13 | 82.0±6.1 | 120.2±1.3* | 94.6±1.2* | 89.2±3.8 |
| Chemotactic cytokines | ||||
| RANTES | 28.1±0.1 | 67.7±12.4* | 71.6±14.2* | 33.1±2.4 |
| MIP-1β | 43.4±1.9 | 67.9±0.4** | 43.5±2.7 | 44.2±2.6 |
| MCP-1 | 868±197 | 1271±48 | 2108±524 | 1610±550 |
| Growth factors | ||||
| G-CSF | 55.5±3.6 | 176.2±38.1* | 58.1±1.7 | 58.5±4.0 |
| GM-CSF | 30.9±7.5 | 56.9±1.0* | 44.1±2.7 | 40.0±5.9 |
| IL-7 | 37.8±6.5 | 77.8±7.1* | 41.1±1.1 | 39.2±1.3 |
| VEGF | 156.5±20.0 | 218.6±13.5 | 254.6±32.8* | 220.9±42.3 |
Data are expressed as mean±sd. Experiments were performed twice in duplicate. TTMV-LY: torque teno mini virus isolates; IFN: interferon; IL: interleukin; MIP: macrophage inflammatory protein; MCP: monocyte chemoattractant protein; G-CSF: granulocyte colony-stimulating factor; GM-CSF: granulocyte-macrophage colony-stimulating factor; VEGF: vascular endothelial growth factor. Bold type indicates a trend towards statistical significance. *: statistically different from mock at p<0.05 (t-test); **: statistically different from mock at p<0.01 (t-test).
Discussion
This study examined the potential involvement of new isolates of TTMV, identified in pleural effusion samples, in the pathogenesis of severe pneumonia in children. The main findings are: 1) TTMV-LY can deeply colonise lungs; 2) alveolar epithelial cells, where efficient replication occurred, were permissive to the TTMV-LY isolates; and 3) TTMV-LY infection modulates the innate immune response of pulmonary cells by inducing the production of inflammatory mediators.
PPEs result from an inflammation of the pleura space as a consequence of infection by bacterial pathogens [11]. Bacteria are frequently described in pleural fluids, and were also confirmed in this study in pleural effusion samples, but the presence of a viral origin remains poorly documented [12, 13]. This study is the first to reveal the presence of an anellovirus in parapneumonic effusions. Anelloviruses are characterised by extreme genetic diversity, a high prevalence in various populations and a wide distribution in body fluids. This ubiquity, together with the absence of suitable in vitro culture systems, has hampered progress in the investigation of this group of viruses [14].
In previous molecular epidemiological studies, unequivocal demonstration that anelloviruses are implicated in the pathogenesis and severity of disease is still lacking or is poorly demonstrated [15, 16]. Here, we found the presence of full-length TTMV genome in nasal aspirates and pleural effusions in children hospitalised with severe pneumonia. In this cohort, as in a Chinese cohort of 216 nasal aspiration samples of children suffering from LRTI (data not shown), no correlation has been observed between the severity of the disease and the presence of TTMV, the age or the sex of the patients. However, the presence of TTMV complete genome linked to the viral replication may indicate its role in the pathogenesis of pneumonia.
This study reports the isolation of three new full-length TTMV genomes (TTMV-LY) in children with severe pneumonia. As a first step, the biological properties of these new isolates of TTMV-LY were investigated. We first transfected the full-length genome of the three TTMV-LY viruses to ascertain whether the viral replication takes place in HEK293T cells, as already demonstrated [17, 18], and subsequently in alveolar epithelial A549 cells, wich act as a model of type II pneumocytes [19]. We described for the first time that TTMV, like TTV, was able to replicate in cell lines after transfection of its genome and thus lead to the production of infectious virions. We observed that the viral infection was more efficient in alveolar cells than in kidney cells, suggesting a better tropism of the TTMV-LY genome in pulmonary cells and a strengthened role in the pathogenesis of LRTI. We hypothesised that, like porcine circoviruses and papillomaviruses, the potential pathogenicity of TTMV could be restricted to some species or a group of species [20, 21]. Indeed, in this study, high genetic TTMV diversity was observed only among different patients, but the same full-length genome was systematically found in the different samples taken from the same patient. All three genomes, moreover, belonged to the same phylogenetic branch of Betatorquevirus (group I). We investigated the conserved 5′UTR genome sequence of the three TTMV-LY genome sequences to determine if specific replication patterns were present. No assumed origin of replication (_AGT_TTACA) was identified, but an insertion of 200 bases forming a stem-loop structure in the conserved 5′-UTR was detected in the TTMV-LY2. As previously described, this secondary structure could be involved in the viral pathogenicity as enhancer and promoter elements [22]. Despite this secondary structure presented by TTMV-LY2, no gain in the replication yield was observed compared to the other TTMV-LYs. However, the effects of such genetic modifications could be expressed in another biological manner, in response to the interaction with the host cell, as observed for many viruses.
As a second step, we investigated the response of the host A549 cells to TTMV-LY infection by measuring the production of soluble mediators implicated in the inflammation process. Alveolar type II epithelial cells maintain alveolar integrity by forming the alveolar barrier, producing surfactants and repairing injured type I epithelium [23]. Moreover, they are an integral part of the lung innate immunity, acting to intensify the function of dendritic cells and alveolar macrophages by cytokine secretion [24, 25]. No cytopathic effect was observed in A549 cells, but infection with TTMV-LY induced a complex innate immune response, differently modulated by the three isolates. Thus, TTMV-LY infection resulted in the production of not only pro-inflammatory cytokines, including IFN-γ as previously described [26], but significant levels of IL-10 or IL-13 anti-inflammatory cytokines were also measured, corroborating that TTMV-LY may modify innate immune balance, as already demonstrated for TTV [27]. The fact that different isolates of TTMV can impact differently on the production of soluble mediators is not surprising in view of what has been observed with other viral infections [28, 29], and may reflect their recognition by different receptors and/or interaction through different molecular pathways. IL-12 seems to play a central role in the response to TTMV-LY infection. This cytokine, enhancing the proliferation and cytotoxicity of activated T-cells and natural killer cells, is thought to be a key factor in the regulation of the host defence against many intracellular pathogens [30]. We may assume that TTMV-LY-mediated lung inflammation may result from the abundant secretion of IL-12, although further studies would be needed to prove this.
Taken together, these results suggest that TTMV-LY isolates can invade the alveolar cells, where they replicate and induce innate immune imbalance in the respiratory tract. Further studies aimed at understanding the physiopathological implications of TTMV as an aetiological agent in respiratory diseases are necessary, as required for every newly discovered pathogen. These investigations would allow to us learn about the presumed need to enlarge the range of researched pathogens to the anelloviruses in the case of PPE, in order to improve the efficiency of diagnosis and treatment of this pathology.
Supplementary Material
Acknowledgments
The authors thank all the patients who participated in this study, and the International S&T Cooperation Programme of China (2010DFB33270), which supported a part of the study in China. We especially thank UMS3444/US8 Biosciences Gerland-Lyon Sud (Lyon, France) for the contribution of their facilities (flow cytometry and CelluloNet) and the availability of their staff.
Footnotes
This article has supplementary material available from www.erj.ersjournals.com
Support statement: This research was supported by the Fondation Mérieux (Lyon, France). The International S&T Cooperation Programme of China (2010DFB33270) supported a part of the study in China. UMS3444/US8 Biosciences Gerland-Lyon Sud (Lyon, France) contributed facilities (flow cytometry and CelluloNet) and staff.
Conflict of interest: None declared.
References
- 1.World Health Organization/The United Nations Children’s Fund. Global Action Plan for the Prevention and Control of Pneumonia (GAPP). Geneva, WHO Press, World Health Organization, 2009.
- 2.Gentile A, Bardach A, Ciapponi A, et al. Epidemiology of community-acquired pneumonia in children of Latin America and the Caribbean: a systematic review and meta-analysis. Int J Infect Dis 2012; 16: e5–e15 [DOI] [PubMed] [Google Scholar]
- 3.Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis 2011; 52: Suppl. 4, S284–S289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lysholm F, Wetterbom A, Lindau C, et al. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS ONE 2012; 7: e30875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Committee on Taxonomy of Viruses. Current Virus Taxonomy. www.ictvonline.org/virusTaxonomy.asp Date last accessed: June 7, 2013. Last updated: 2012. [Google Scholar]
- 6.Girdhar A, Shujaat A, Bajwa A. Management of infectious processes of the pleural space: a review. Pulm Med 2012; 2012: 816502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maggi F, Pifferi M, Fornai C, et al. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. J Virol 2003; 77: 2418–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allander T, Tammi MT, Eriksson M, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA 2005; 102: 12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faurez F, Dory D, Henry A, et al. Replication efficiency of rolling-circle replicon-based plasmids derived from porcine circovirus 2 in eukaryotic cells. J Virol Methods 2010; 165: 27–35 [DOI] [PubMed] [Google Scholar]
- 10.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55: 611–622 [DOI] [PubMed] [Google Scholar]
- 11.Light RW. Parapneumonic effusions and empyema. Proc Am Thorac Soc 2006; 3: 75–80 [DOI] [PubMed] [Google Scholar]
- 12.Falguera M, Carratalà J, Bielsa S, et al. Predictive factors, microbiology and outcome of patients with parapneumonic effusion. Eur Respir J 2011; 38: 1173–1179 [DOI] [PubMed] [Google Scholar]
- 13.Thijsen SFT, Luderer R, Van Gorp JMH, et al. A possible role for Epstein–Barr virus in the pathogenesis of pleural effusion. Eur Respir J 2005; 26: 662–666 [DOI] [PubMed] [Google Scholar]
- 14.Okamoto H. History of discoveries and pathogenicity of TT viruses. Curr Top Microbiol Immunol 2009; 331: 1–20 [DOI] [PubMed] [Google Scholar]
- 15.Maggi F, Bendinelli M. Human anelloviruses and the central nervous system. Rev Med Virol 2010; 20: 392–407 [DOI] [PubMed] [Google Scholar]
- 16.Borkosky SS, Whitley C, Kopp-Schneider A, et al. Epstein–Barr virus stimulates torque teno virus replication: a possible relationship to multiple sclerosis. PLoS ONE 2012; 7: e32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Villiers E-M, Borkosky SS, Kimmel R, et al. The diversity of torque teno viruses: in vitro replication leads to the formation of additional replication-competent subviral molecules. J Virol 2011; 85: 7284–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakkola L, Tommiska J, Boele LCL, et al. Construction and biological activity of a full-length molecular clone of human torque teno virus (TTV) genotype 6. FEBS J 2007; 274: 4719–4730 [DOI] [PubMed] [Google Scholar]
- 19.Foster KA, Oster CG, Mayer MM, et al. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp Cell Res 1998; 243: 359–366 [DOI] [PubMed] [Google Scholar]
- 20.Bernard H-U, Calleja-Macias IE, Dunn ST. Genome variation of human papillomavirus types: phylogenetic and medical implications. Int J Cancer 2006; 118: 1071–1076 [DOI] [PubMed] [Google Scholar]
- 21.Todd D, McNulty MS, Adair BM, et al. Animal circoviruses. Adv Virus Res 2001; 57: 1–70 [DOI] [PubMed] [Google Scholar]
- 22.Suzuki T, Suzuki R, Li J, et al. Identification of basal promoter and enhancer elements in an untranslated region of the TT virus genome. J Virol 2004; 78: 10820–10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason RJ. Biology of alveolar type II cells. Respirology 2006; 11 Suppl. S1:S12–S15 [DOI] [PubMed] [Google Scholar]
- 24.Gereke M, Jung S, Buer J, et al. Alveolar type II epithelial cells present antigen to CD4+ T cells and induce Foxp3+ regulatory T cells. Am J Respir Crit Care Med 2009; 179: 344–355 [DOI] [PubMed] [Google Scholar]
- 25.Kannan S, Huang H, Seeger D, et al. Alveolar epithelial type II cells activate alveolar macrophages and mitigate P. aeruginosa infection. PLoS ONE 2009; 4: e4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocchi J, Ricci V, Albani M, et al. Torque teno virus DNA drives proinflammatory cytokines production and secretion by immune cells via toll-like receptor 9. Virology 2009; 394: 235–242 [DOI] [PubMed] [Google Scholar]
- 27.Maggi F, Pifferi M, Tempestini E, et al. TT virus loads and lymphocyte subpopulations in children with acute respiratory diseases. J Virol 2003; 77: 9081–9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell WC. Adenoviruses: update on structure and function. J Gen Virol 2009; 90: 1–20 [DOI] [PubMed] [Google Scholar]
- 29.Sørensen LN, Reinert LS, Malmgaard L, et al. TLR2 and TLR9 synergistically control herpes simplex virus infection in the brain. J Immunol 2008; 181: 8604–8612 [DOI] [PubMed] [Google Scholar]
- 30.Ruan S, McKinley L, Zheng M, et al. Interleukin-12 and host defense against murine pneumocystis pneumonia. Infect Immun 2008; 76: 2130–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.