Abstract
Deregulation of DNA repair enzymes occurs in cancers and may create a susceptibility to chemotherapy. Expression levels of DNA repair enzymes have been shown to predict the responsiveness of cancers to certain chemotherapeutic agents. The RECQ helicases repair damaged DNA including damage caused by topoisomerase I inhibitors, such as irinotecan. Altered expression levels of these enzymes in colorectal cancer (CRC) may influence the response of the cancers to irinotecan. Thus, we assessed RECQ helicase (WRN, BLM, RECQL, RECQL4, and RECQL5) expression in primary CRCs, matched normal colon, and CRC cell lines. We found that BLM and RECQL4 mRNA levels are significantly increased in CRC (P = .0011 and P < .0001, respectively), whereas RECQL and RECQL5 are significantly decreased (P = .0103 and P = .0029, respectively). RECQ helicase expression patterns varied between specific molecular subtypes of CRCs. The mRNA and protein expression of the majority of the RECQ helicases was closely correlated, suggesting that altered mRNA expression is the predominant mechanism for deregulated RECQ helicase expression. Immunohistochemistry localized the RECQ helicases to the nucleus. RECQ helicase expression is altered in CRC, suggesting that RECQ helicase expression has potential to identify CRCs that are susceptible to specific chemotherapeutic agents.
Introduction
Colorectal cancer (CRC) is a leading cause of cancer deaths in the United States, affecting more than 140,000 people annually [1]. The majority of people with CRC are diagnosed with advanced stage disease that will require adjuvant chemotherapy in addition to surgical resection of the primary cancer [2]. Currently, pathologic stage is the best prognostic factor available and is used to identify patients who are most likely to benefit from adjuvant chemotherapy and/or radiation therapy [3]. Although pathologic stage is the best predictive indicator for determining response to adjuvant therapy, 30% to 40% of presumably cured patients develop recurrent disease [3]. Therefore, there is a need for markers that more accurately identify patients who would benefit from more aggressive adjuvant therapy and for markers that identify the most effective agents for treating a specific patient's cancer.
Molecular alterations in CRC, such as altered gene expression levels, have the potential to be more accurate than pathologic stage for predicting response to treatment. Molecular alterations are likely more accurate predictive markers than TNM stage, because they often directly affect biologic processes that mediate the manner in which cancer cells respond to specific chemotherapeutic agents. These alterations often affect processes central to the mechanisms of action of chemotherapeutic agents, such as the regulation of DNA repair and genomic stability, thereby affecting the cancer's response to chemotherapy [4,5].
Genomic instability is one of the hallmarks of cancer and results from the loss of the ability to maintain the integrity of the genome. Loss of genomic stability results in increased mutation rates in cancer cells and appears to play a prominent role in the response of cancers to chemotherapy [4,6]. DNA damage, if left unrepaired, contributes to genomic instability. DNA damage occurs on a regular basis in cells as a consequence of exposure to environmental mutagens and cellular metabolites. The integrity of the genome is normally maintained by DNA repair enzymes, which can be grouped into functional categories depending on the type of DNA damage they repair. Their dysfunction can contribute to altered DNA processing, genomic instability, and increased susceptibility to aberrant genetic alterations [7,8]. Importantly, recent in vitro studies have shown that cancers with loss of function of DNA repair enzymes have an increased susceptibility to chemotherapeutic agents that induce the type of damage normally repaired by those enzymes [7,9]. These studies as well as the successful proof-of-principle studies involving poly (ADP-ribose) polymerase (PARP) inhibitors in BRCA1-deficient tumors [10,11] have led to an intense investigation of DNA repair enzymes as biomarkers in cancers.
From a clinical vantage, the altered expression of DNA repair enzymes has the potential to be used for predicting the response of a cancer to specific chemotherapeutic agents. The differential response of CRCs to DNA-damaging chemotherapeutic agents may reflect the specific DNA repair capabilities of the cancers. Studies have shown that the currently recognized molecular subtypes of CRC can have different clinical behaviors with regard to treatment outcomes. CRCs demonstrating microsatellite instability (MSI) secondary to DNA mismatch repair deficiency have reduced response to 5-fluorouracil [12,13], whereas this same subtype may be more responsive to irinotecancontaining regimens [14]. It is not yet fully understood what drives this variability in clinical response to chemotherapy; however, it is likely rooted in the molecular heterogeneity of each cancer. The variability in clinical response to DNA-damaging chemotherapeutic agents may be driven by heterogeneity in the expression or function of key DNA repair enzymes.
The RECQ helicases are a family of DNA repair enzymes that unwind double-stranded nucleic acids and have a central role in maintaining genomic integrity [15]. The RECQ helicase family includes five ATP-dependentenzymes (WRN, BLM, RECQL, RECQL4, and RECQL5) that have a conserved 3′ to 5′ helicase domain and two additional conserved regions, the RQC and HRDC regions [15]. They modulate complexes involved in homologous recombination, replication fork migration, translocation, and the structure and function of nucleoprotein filaments. They also process topoisomerase I-mediated DNA damage induced by camptothecin derivatives, such as irinotecan [16]. Autosomal recessive loss of function of three of these DNA-modulating enzymes, WRN, BLM and RECQL4, results in human syndromes characterized by constitutional genomic instability and premature aging [17]. These syndromes have also been linked to increased cancer susceptibility, including CRC, making the function of these helicases a point of interest in the pathogenesis of CRC. Furthermore, deficiencies in RECQ helicases have also been shown to result in a cancer-prone phenotype in mice [18,19].
Because the RECQ helicases play a significant role in the maintenance of genomic stability and DNA repair, altered expression of the RECQ helicases may play a role in the pathogenesis of CRC. Furthermore, RECQ helicase expression may be useful for identifying CRCs that are susceptible to specific DNA-damaging chemotherapeutic agents, including those drugs commonly used for the treatment of CRC, 5-fluorouracil, oxaliplatin, and irinotecan [20–22]. Both WRN-and BLM-depleted cells have impaired survival after treatment with the DNA-damaging agents cisplatinum and 5-fluorouracil as well as camptothecin [23]. RECQL5-deficient cell lines show increased sensitivity to camptothecin [20]. Overexpression of one or more of these helicases may mediate resistance to DNA-damaging chemotherapeutic agents, whereas underexpression may identify cancers that are particularly susceptible to these agents. We hypothesized that the expression of the RECQ helicases would differ in primary CRCs compared to matched normal colonic mucosa. Furthermore, we also hypothesized that the expression of the RECQ helicases may differ among the molecular subtypes of CRC. We used quantitative reverse transcription-polymerase chain reaction (qRT-PCR) as well as Western blot analysis and immunohistochemistry (IHC) to assess our hypothesis and found that BLM and RECQL4 expression are increased in CRC, whereas RECQL and RECQL5 expression are decreased. We also show that there are characteristic RECQ helicase expression patterns for different molecular subtypes of CRC. The RECQ helicases are plausible candidates for the development of biomarkers that may predict the therapeutic response of CRCs to chemotherapeutic agents and may be important in CRC pathogenesis.
Materials and Methods
CRC Cases
A total of 46 sporadic primary CRC cases with matched normal colon was assayed. Twenty-six primary CRC cases with matched normal colon tissue were obtained from the Cooperative Human Tissue Network (CHTN) as fresh frozen tissue. An additional 20 primary CRC cases with matched normal colon tissue were obtained in the form of first-strand cDNA (TissueScan Colon Cancer Tissue qPCR Panel III) from OriGene (Rockville, MD). All cases were composed of >60% tumor epithelium.
Human CRC Tissue Microarrays
CRC tissue microarrays (TMAs) were purchased from OriGene (Catalog No. CT565900). TMAs contained 50 x 1 mm cores from formalin-fixed paraffin-embedded (FFPE) samples (40 tumors and 10 normal tissues). Data for individual cases including age, gender, pathology, grade, stage, and so on can be found at http://www.origene.com/Tissue/getTissueMicroArray.aspx?id=TMA002.
Quantitative Real-Time PCR
RNA extraction from fresh frozen tissue was conducted using TRIzol as recommended by the manufacturer, followed by purification using the RNeasy Kit (Qiagen, Foster City, CA). cDNA was made using SuperScript II Reverse Transcriptase (Invitrogen, Grand Island, NY), 1x PCR buffer, MgCl2 (8 mM), RNase out, and 1 to 4 µg of the appropriate total RNA. Real-time quantitative PCR assays were conducted using TaqMan probe and primers (Applied Biosystems, Grand Island, NY) for WRN (Hs00172155_m1), BLM (Hs00172060_m1), RecQL (Hs00262956_m1), RecQL4 (Hs00171627_m1), RecQL5 (Hs00188633_m1), GusB (Hs99999908_m1), and PCNA (Hs00427214_g1) along with TaqMan Universal PCR Mix (Applied Biosystems) as recommended by the manufacturer. GUSB expression was used as a loading control, and PCNA expression was used to normalize for proliferation [24–28]. The relative standard curve method was used in determining RECQ helicase expression levels. Control RNA was obtained from the following CRC cell lines: LS174T for WRN and RECQL and SW480 for BLM, RECQL4, RECQL5, and PCNA.
CpG Island Methylator Phenotype Analysis
Genomic DNA extraction from fresh frozen tissue was conducted using the DNeasy Kit (Qiagen) according to the protocol recommended by the manufacturer. Subsequently, the DNA was bisulfite converted using the Zymo Research EZ DNA methylation kit according to the manufacturer's protocol. CpG island methylator phenotype (CIMP) analysis was conducted using Methylight assays according to the protocol published by Weisenberger et al. [29], using probe primer sets and ALU-based normalization (Table W1).
The percent methylation ratio for each gene was determined for each tumor sample using the following equation: [(gene x mean value for sample)/(ALUC4 mean value for sample)]/[(gene x mean value for the methylated control)/(ALUC4 mean value for the methylated control)] * 100. A sample is scored as being CIMP positive if greater than or equal to three of five genes has a percent methylation ratio of >10.
MSI Analysis
Genomic DNA was extracted from the tissues as described. MSI status of the CHTN samples was determined from genomic DNA according to a previously published protocol [30].
RECQ Helicase Expression and Localization Using IHC
TMAs were deparaffinized and rehydrated in distilled water. Endogenous peroxidase activity was blocked using 0.5% H2O2.The following antibodies and conditions were used for RECQ IHC. For WRN IHC, antigen retrieval was performed using heat-induced epitope retrieval in Epitope Retrieval 2 (Leica Biosystems, Buffalo Grove, IL). Mouse monoclonal 195C to Werner's syndrome helicase (ab62747; Abcam, Cambridge, MA) was diluted 1:100. For BLM IHC, antigen retrieval was as described for WRN. Anti-BLM antibody produced in rabbit (HPA005689; Sigma, St Louis, MO) was diluted 1:100. RECQL antigen retrieval was also as described for WRN. RECQL rabbit polyclonal antibody (sc-25547; Santa Cruz Biotechnology, Dallas, TX) was diluted 1:50. RECQL4 antigen retrieval was performed using heat-induced epitope retrieval in Epitope Retrieval 1 (Leica Biosystems). RECQL4 monoclonal antibody (M09), clone 2G8 (H00009401-M09; Abnova, Taipei City, Taiwan), was diluted 1:50. RECQL5 antigen retrieval was performed as described for WRN. Anti-RECQL5 rabbit polyclonal antibody (Sigma; HPA029971) was diluted 1:150. TMAs were incubated with primary antibody for 20 minutes in a Leica Bond XX. 3,3-Diaminobenzidine substrate was used to visualize RECQ-positive cells. TMAs were counterstained with hematoxylin. Cores were scored using the Allred scoring system, which combines two staining categories (proportion and intensity) to yield a single numerical score [31]. The Allred score ranges from 0 to 8. It is arrived upon by the addition of two scores, a proportion score and an intensity score, to reflect both the number of tumor cells stained as well as how strongly they stain. The proportion score ranges from 0 to 5 (0 = no cells stained, 1 = 1/100 cells stained, 2 = 1/10 cells stained, 3 = 1/3 cells stained, 4 = 2/3 cells stained, 5 = all cells stained), and an intensity score ranges from 0 to 3 (0 = negative, 1 = weak, 2 = intermediate, 3 = strong). Any non-epithelial cells that stained with the antibodies used were not included in this assessment.
Protein Expression Using Western Blot
Cell lysates were prepared in RIPA buffer, and the proteins were resolved using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to polyvinylidene difluoride (PVDF) membranes and subsequently blocked in 5% non-fat dry milk in tris-buffered saline with Tween 20 (TBST). The membranes were incubated overnight with individual primary antibodies diluted 1:1000 at 4°C and then incubated for 1 hour with secondary HRP-conjugated antibodies diluted 1:5000. β-Actin was used as a protein loading control. The following antibodies were used for Western blot analysis experiments: WRN—clone 195C (Sigma); BLM—HPA005689 (Sigma); RECQL—A1107 (Santa Cruz Biotechnology); RECQL4—2814 (Cell Signaling Technology, Danvers, MA); RECQL5—HPA029971 (Sigma); PCNA—sc-7907 (Santa Cruz Biotechnology); β-actin—sc-1616 (Santa Cruz Biotechnology).
Methylation-specific PCR
Genomic DNA was extracted and bisulfite converted, and methylation-specific PCR was conducted as previously described [32]. The primer sequences used are given as follows: RECQL methyl-specific forward primer, 5′-GCGGTCCCAAAAGGGTCAGTTCGGATATCGGATAGTTAAATATCG-3′; RECQL methyl-specific reverse primer, 5′-GCGGTCCCAAAAGGGTCAGTCCGATCAACAAACGAACGTA-3′;RECQL non-methyl-specific forward primer, 5′-GCGGTCCCAAAAGGGTCAGTTGGATATTGGATAGTTAAATATTGG-3′;RECQL non-methyl-specific reverse primer, 5′-GCGGTCCCAAAAGGGTCAGTAAAAACCAATCAACAAACAAACATA-3′; RECQL5 methyl-specific forward primer, 5′-AATTAAAGGTTGTTGGTTGGTTTC-3′; RECQL5 methyl-specific reverse primer, 5′-ACTACGCGACGAATATAAAATTACG-3′;RECQL5 non-methyl-specific forward primer, 5′-AATTAAAGGTTGTTGGTTGGTTTT-3; RECQL5 non-methyl-specific reverse primer, 5′-CTACACAACAAATATAAAATTACATA-3′.
The methylation-specific PCR products were visualized using electrophoresis on a 1.5% agarose gel with ethidium bromide staining with subsequent UV transillumination using an Eagle Eye Imaging System (Stratagene, La Jolla, CA).
Statistical Analysis
As the fold change of RECQ helicase mRNA expression in tumor compared to the matched normal mucosa was determined to be non-Gaussian (KS normality and the D'Agostino and Pearson omnibus normality tests), the significance of the fold change in expression was determined using the Wilcoxon signed rank test with a theoretical median of 1. All subsequent analyses were conducted using non-parametric tests. When comparing the fold change expression of the RECQ helicases in CRC tumor to the normal colonic mucosa, a Wilcoxon signed rank test was used. Subgroup analyses of the expression of each RECQ helicase by stage were conducted using the Kruskal-Wallis test, with subsequent all-pairwise post-hoc comparisons using Mann-Whitney tests. Analysis of RECQ helicase expression by early versus advanced stage (stage I/II vs stage III/IV) was conducted using the Mann-Whitney test. Mann-Whitney and Wilcoxon signed rank tests were used to determined the statistical significance in the IHC studies as well as mRNA expression studies in CRC cell lines compared to normal colonic mucosa. All statistical tests performed were two-tailed analyses.
The fold change of RECQL5 expression was used to divide the tumor samples into two groups, those that have higher RECQL5 expression in normal mucosa compared to tumor and those that have higher RECQL5 expression in tumor compared to normal mucosa. Subsequently, the expression of the other RECQ helicases were analyzed in each of the two groups and their means compared using a Mann-Whitney test. This analysis was also conducted using the expression level of each of the RECQ helicases.
Results
Primary CRCs Have Significantly Altered Expression of RECQ Helicases
We assessed the expression of the five RECQ helicase family members in sporadic primary human CRC samples and the matched normal colon tissue. The expression of some members of the RECQ helicase family has been shown to vary throughout the cell cycle with the highest expression observed predominantly during S-phase [24,25]. As we were concerned that alterations in RECQ helicase expression in specific tumors could be a surrogate marker for proliferation rather than a cell cycle-independent occurrence in the tumor, we normalized the RECQ helicase mRNA expression data for proliferation using the expression levels of PCNA. PCNA, a cell cycle-dependent protein that accumulates in the nucleus during S-phase, can be used to normalize differences that are secondary to proliferation rates rather than basal expression [26–28]. The data without normalization for proliferation are shown in Figure W1.
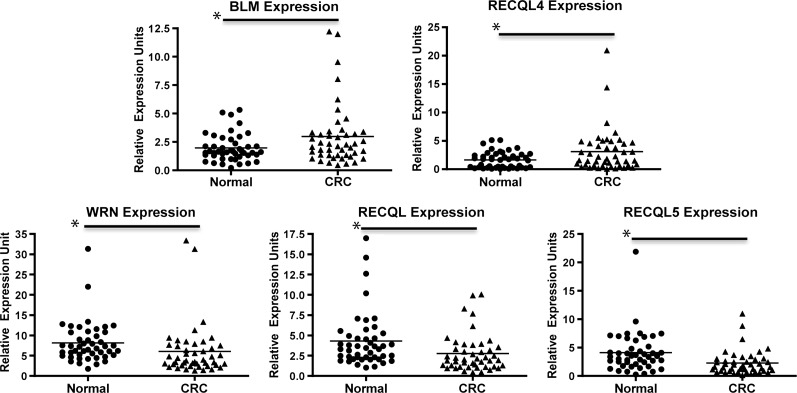
Our results show that, as a group, the primary CRCs display significantly higher BLM and RECQL4 mRNA expression levels compared to normal colonic mucosa (P < .05; Figure 1). In contrast, RECQL and RECQL5 mRNA expression are lower in primary tumors compared to normal colonic mucosa (P < .05). There was also a significant decrease in WRN expression between the CRCs compared to the normal colon (Figure 1). In normal colon, expression of the RECQ helicases showed a similar degree of inter-case variability to that seen in the cancers (Figure 1). Receiver operating characteristic (ROC) analysis also showed non-identity between the expression of WRN, RECQL, RECQL4, and RECQL5 in tumor versus normal colonic mucosa (Figure W2).
Figure 1.
Expression of RECQ helicase mRNA in primary CRCs and normal colonic epithelium. The primary CRCs have higher BLM (P = .0049) and RECQL4 (P = .0002) expression compared to normal colonic mucosa, whereas there is lower expression of RECQL (P = .0046), RECQL5 (P = .0004), and WRN (P = .0049) in primary tumors compared to normal colonic mucosa. Asterisks indicate statistical significance between the two groups identified.
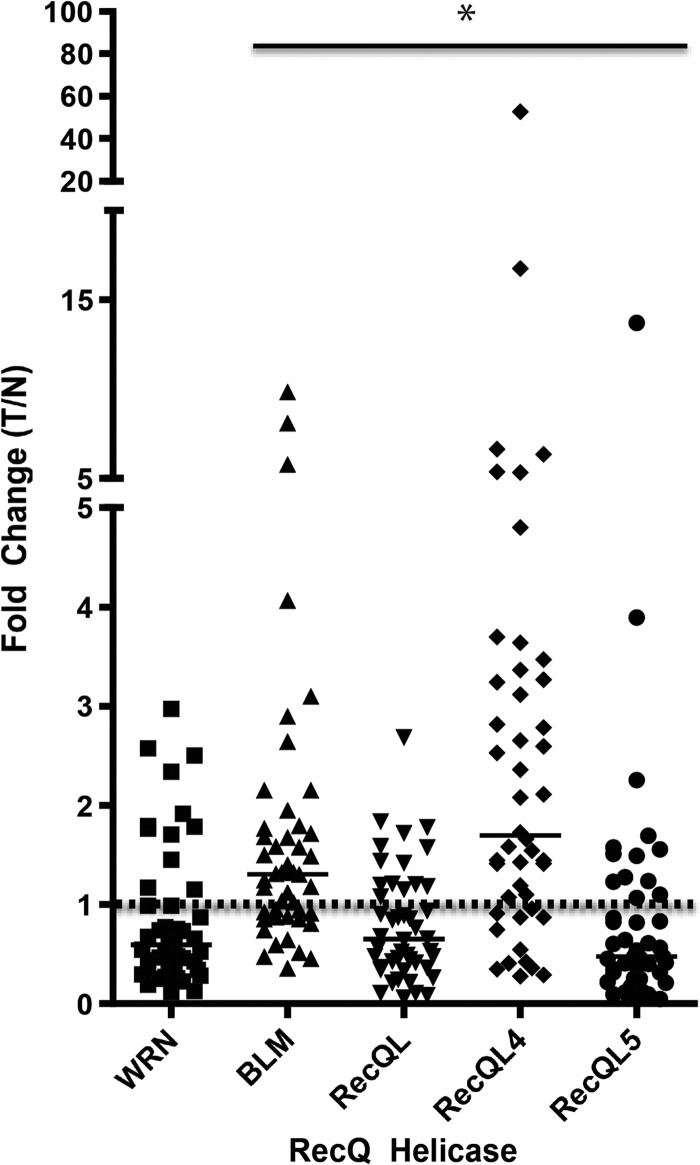
When we compared the fold change of RECQ expression in tumor versus normal colonic mucosa matched pairs, we observed a significantly higher level of expression of BLM and RECQL4 compared to the normal mucosa (Figure 2). We also found a significantly lower level of expression of RECQL and RECQL5 in the CRCs compared to matched normal mucosa (Figure 2). The tumor/normal ratios of expression are given as follows: WRN median = 0.5876, mean = 0.8627, SD = 0.7267 (P = .0881); BLM median = 1.303, mean = 1.795, SD = 1.836 (P = .0011); RECQL median = 0.6496, mean = 0.7973, SD = 0.568 (P = .0103); RECQL4 median = 1.694, mean = 3.604, SD = 7.878 (P < .0001); RECQL5 median = 0.4731, mean = 1.015, SD = 2.036 (P = .0029; Figure 2).
Figure 2.
Fold change of RECQ helicase mRNA expression (CRC/normal colon). Solid horizontal lines indicate the median fold change for each RECQ helicase. The dashed horizontal line indicates a fold change level of 1, where the expression level of the RECQ helicase in the tumor is equal to the expression level in normal colonic mucosa. BLM and RECQL4 expression is significantly higher in the CRCs compared to the matched normal colon (P = .0011 and P < .0001, respectively). RECQL (P = .0103) and RECQL5 (P = .0029) expression is significantly lower in the primary CRCs compared to the matched normal colon mucosa. There is no significant difference in WRN mRNA expression in tumor compared to normal colonic mucosa. Asterisks indicate statistical significance in the fold change compared to the theoretical mean of 1 for the indicated helicases.
Furthermore, we observed substantial heterogeneity in the expression of BLM and RECQL4 in the primary CRCs. The expression varied by 10-fold (BLM) and 50-fold (RECQL4) between CRCs (Figure 2). Our results, before normalization for proliferation, are consistent with data available on RECQ helicase expression in the Oncomine database (for RECQ helicase mRNA expression in CRC versus normal colonic tissue), showing an increase in the expression of all RECQ helicases. Of interest, after normalization for proliferation, we found a decrease in WRN and RECQL5 expression in tumor compared to normal mucosa.
We next assessed the CRCs for correlation of expression among the RECQ helicases in CRC. We found that 3 of 46 (6.5%) cases had increased expression of all the RECQ helicases, whereas 4 of 46 (8.7%) cases had decreased expression of all five RECQ helicases compared to the respective matched normal colonic mucosa; however, this did not correlate with stage.
Characteristic Expression of RECQ Helicases in CIMP and MSI Subtypes of CRC
We also assessed RECQ helicase expression in the conventionally accepted CRC molecular subtypes (MSI and CIMP). We correlated RECQ expression level with the CIMP status of the tumors (CHTN, N = 26) and found that CIMP-positive tumors had high levels of BLM expression (tumor > normal) compared to non-CIMP tumors (Table 1). The percentage of BLM high tumors that displayed CIMP was 26.6%, which is greater than expected by chance. These data suggest that increased BLM expression in CRC may be correlated with CIMP-positive tumor status. There was no correlation between CIMP status and expression of any other RECQ helicase.
Table 1.
CIMP Status of CRC Corresponds with Increased BLM Expression.
| BLM Expression | Non-CIMP | CIMP | Total Cases | Percent CIMP | Group Mean | SD |
| T < N | 11 | 0 | 11 | 0 | 0.75 | 0.22 |
| T > N | 11 | 4 | 15 | 26.6 | 2.62 | 2.59 |
| All cases | 22 | 4 | 26 | 15.3 |
When BLM expression is used to cluster cases into CRCs with BLM expression less in tumor compared to normal and CRCs with BLM expression greater in tumor compared to normal, we identified two distinct subgroups of CRCs (P < .05). Although clustering of cases by BLM expression does not predict differences in expression of any of the other four RecQ helicases, it does correlate with CIMP status. CIMP-positive tumors were observed only in cases where the expression of BLM was greater in the tumors compared to the normal colon. These results suggest that CIMP CRCs tend to correlate with higher BLM expression in the tumor compared to matched normal mucosa and not cases where BLM expression is decreased in the tumor compared to matched normal mucosa.
When we analyzed RECQ helicase expression level and MSI status of the primary sporadic tumors, we found that MSI CRCs (CHTN, N =26) had lower RECQL5 and RECQL expression than the matched normal colonic tissue (Table 2). There was no correlation between MSI status and expression of any other RECQ helicase. There was no statistically significant difference in the expression of any of the RECQ helicases between MSI and microsatellite stable (MSS) tumors.
Table 2.
MSI Tumors Have Low Expression of RECQL and RECQL5.
| Gene Expression | MSS | MSI | Total Cases | Percent MSI | Group Mean | SD |
| RECQL, T < N | 12 | 6 | 18 | 33.3 | 0.50 | 0.30 |
| RECQL, T > N | 8 | 0 | 8 | 0 | 1.71 | 0.45 |
| RECQL5, T < N | 11 | 6 | 17 | 35.3 | 0.38 | 0.25 |
| RECQL5, T > N | 9 | 0 | 9 | 0 | 2.84 | 4.09 |
| All cases | 20 | 6 | 26 | 23.1 |
When either RECQL or RECQL5 expression was used to cluster cases into those that expressed the gene of interest less than the matched normal mucosa or those that expressed the gene more than the matched normal mucosa, we were able to identify two distinct subgroups (P < .05). MSI CRCs were observed only in cases where the expression of RECQL and RECQL5 were less in the CRC compared to the normal colon. These results suggest that MSI cases correlate with low RECQL and RECQL5 expression in the tumor compared to the matched normal mucosa. MSS, microsatellite stable (MSI negative).
Comparison of CRCs that Express High Levels of RECQ Helicases versus CRCs that Express Low Levels of RECQ Helicases
It has been shown in vitro that certain members of the RECQ helicase family can interact during DNA repair. Thus, we conducted an analysis of RECQ helicase expression in CRCs by dividing tumors into the following two groups: 1) those CRCs with RECQ helicase expression greater than normal mucosa and 2) those CRCs with RECQ helicase expression less than normal mucosa. This was done to determine if distinct populations of CRCs with high or low expression of the RECQ helicases could be identified on the basis of expression of any one of the RECQ helicases. To determine if we could identify an association in the expression level of specific RECQ helicases, we assessed the correlation between the expression of RECQ helicases in a pairwise fashion.
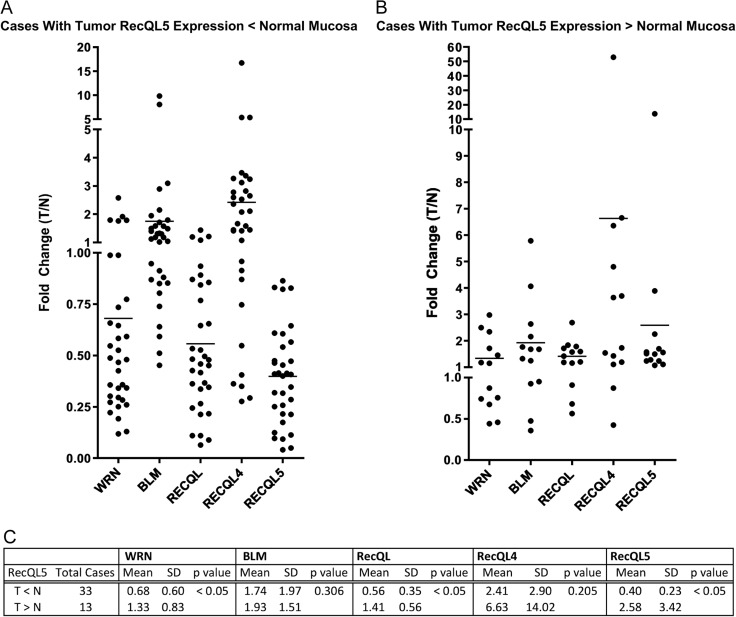
We found that grouping cases by the expression level of RECQL5 as either high (tumor > normal) or low expression (tumor < normal) allowed us to predict similarly high or low expression levels of WRN and RECQL. Grouping of cases by RECQL5 high or low expression identified distinct populations of CRCs with regard to RECQL5, WRN, and RECQL expression (P < .05; Figure 3). Of the tumors that expressed RECQL5 < normal mucosa, 88% also had low levels (T < N) of RECQL expression, and 85% of those tumors had low levels (T < N) of WRN expression (Figure 3).
Figure 3.
Classifying CRC cases by RECQL5 expression can predict the expression levels of WRN and RECQL. (A) Of the tumors that expressed RECQL5 < normal mucosa, 88% also had low levels (T < N) of RECQL expression. (B) Of tumors that expressed RECQL5 > normal mucosa, 85% had low levels (T < N) of WRN expression. (C) Grouping cases by RECQL5 expression T < N or T > N showed statistical significance in expression of RECQL5, RECQL, and WRN (P < .05).
We also found that grouping cases by low expression levels of either RECQL or WRN was able to predict low levels of RECQL5, confirming the relationship seen when the cases were grouped using RECQL5 expression. Grouping cases by expression of RECQL4, however, did not predict differences in expression of the other RECQ helicases.
When BLM expression was used to group tumors into high (tumor > normal) and low (normal < tumor) BLM-expressing groups, we were not able to predict expression of any of the other RECQ helicases. However, CIMP-positive cases were only found among the BLM high tumors and were not found in BLM low tumors (Table 1).
RECQ Expression Analysis by Stage
Subgroup analyses of the mRNA expression of each RECQ helicase by stage showed a statistically significant difference in the expression of WRN in the CRCs compared to normal mucosa by stage (P = .0498), with the highest average value seen in stage II CRCs (Figure W3). However, after all-pairwise post-hoc comparisons, there was not enough power to detect differences in WRN expression among the stages. No significant differences in the expression of BLM (P = .2193), RECQL (P = .5992), RECQL4 (P = .4077), and RECQL5 (P = .3889) among CRC stages I to IV were observed. The significance of these results is unclear given the low level of expression of WRN in the colon and the poor correlation of WRN mRNA expression with WRN protein expression that we have observed (see below).
When we compared the expression levels in localized disease versus advanced disease (stages I and II vs stages III and IV), there were no significant differences in RECQ helicase expression (P > .05). As described in the next section, the same analysis was conducted using Allred scores determined from IHC staining of RECQ helicases in archival CRC cases. The results from the IHC analysis also show no significant difference in protein expression for any of the RECQ helicases across stage, WRN (P = .5709), BLM (P = .3644), RECQL4 (P = .5451), RECQL (P = .1922), or RECQL5 (P = .5604; Figure W3).
Nuclear Localization of RECQ Helicase in Primary CRC Tumors
IHC studies were conducted to determine the localization of the RECQ helicases in CRC tumor tissue and normal colonic mucosa. For all RECQ helicases, IHC analysis consistently localized the helicases to the nucleus (Figure 4). However, RECQL5 was also observed in the cytoplasm as well as in the nucleus. Analysis of Allred scores for each of the RECQ helicases showed decreased expression of RECQL5 in tumors compared to normal mucosa (P < .05), consistent with the RECQL5 mRNA levels we observed (Figure 4). WRN IHC revealed an increase in expression in tumors compared to normal colon mucosa (P < .05), suggesting that protein expression levels and mRNA expression levels for WRN do not correlate well, consistent with data obtained from CRC cell lines (Figure 5). The other RECQ helicases also show statistically significant differences in expression between tumor and normal tissue (P < .05; Figure 5); however, given the small differences in the All red score between tumor and normal tissue, we believe that this is unlikely to be clinically significant. When the intensity score component of the Allred staining score was analyzed separately from the proportion score component, we saw a significant difference in the reduction of RECQL staining intensity and a significant increase in RECQL4 proportion of staining in tumor compared to normal mucosa (P = .0226 and P = .0002, respectively), consistent with the results obtained for RECQL and RECQL4 mRNA expression.
Figure 4.
IHC staining shows localization of RECQ helicases in normal colonic mucosa and CRC. All of the RECQ helicases were detected in the nucleus in both normal colonic mucosa and CRCs. Allred scores for each of the RECQ helicases from IHC of TMAs are also presented. Low expression of RECQL5 in tumors compared to normal mucosa can be seen (P = .0031, Wilcoxon signed rank test), which is consistent with the results obtained from the RECQL5 qRT-PCR assays. WRN showed higher expression in tumors compared to normal colon mucosa (P = .0128, Mann-Whitney), suggesting that immunostaining scores and mRNA expression levels of WRN do not correlate. The other RECQ helicases also show statistically significant differences in expression between tumor and normal tissue (BLM, P = .0046, Wilcoxon signed rank test; RECQL, P = .0287, Wilcoxon signed rank test; RECQL4, P = .0103, Wilcoxon signed rank test); however, this difference likely does not translate to clinical significance. Allred scores for the selected matched tumor normal images chosen to show localization are given as follows: WRN: N =7, T =0; BLM: N =7.75, T =7; RECQL: N = 7.25, T =6; RECQL4: N =7, T =8; RECQL5: N = 6 (cytoplasmic 2), T = 0 (cytoplasmic 2). All images were photographed at x20. Asterisks indicate statistical significance between the two groups identified.
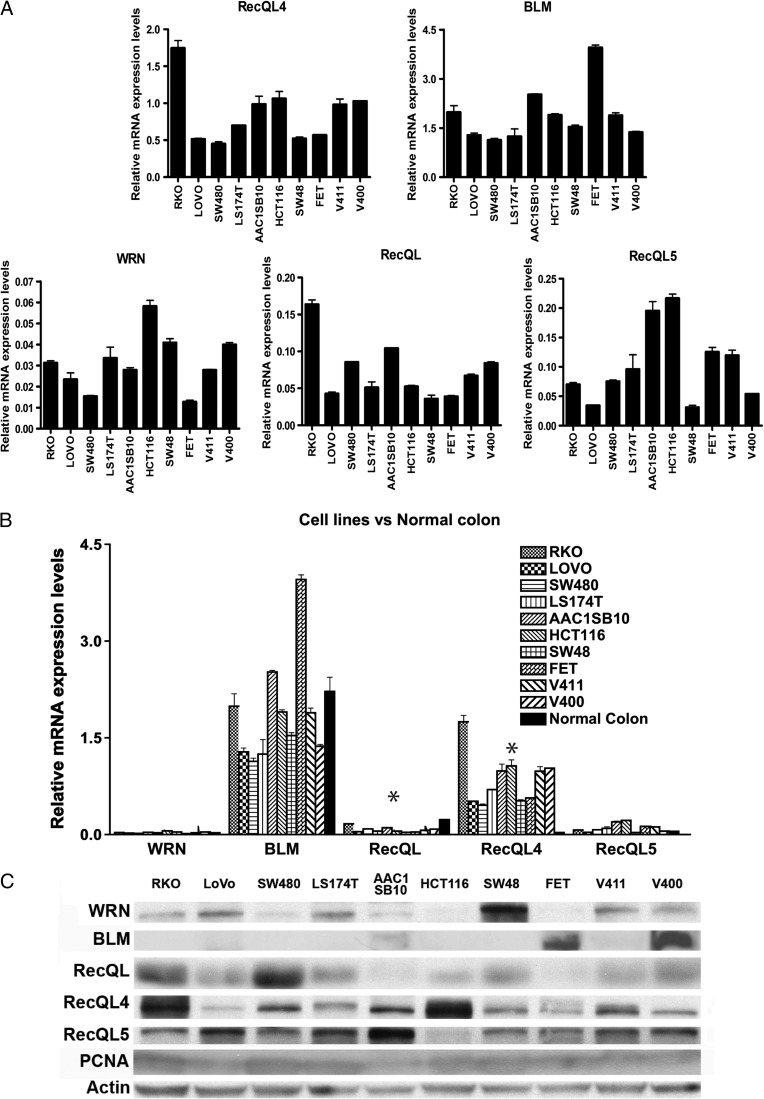
Figure 5.
RECQ helicase expression in CRC cell lines. (A) RECQ helicase mRNA expression was assessed in CRC lines representing the three major molecular subtypes of CRC, MSS (or CIN) cancers (SW480, V400, V411, AAC1/SB10, and HT29), microsatellite unstable (MSI) cancers (HCT116, LoVo, and LS174T), and CIMP cancers (RKO, SW48, and FET). Expression levels of BLM and RECQL4 mRNA were higher than those of RECQL, RECQL5, and WRN, regardless of molecular subgroup. (B) RECQ helicase mRNA expression compared to normal colonic mucosa for CRC cell lines. RECQL4 expression is significantly higher in the cell lines compared to the normal colonic mucosa (P = .002), whereas RECQL expression is significantly decreased (P < .002). Asterisks indicate statistical significance between the average expression of the indicated helicase in CRC cell line compared to normal colonic mucosa. (C) RECQ helicase Western blot results for CRC cell lines. PCNA and actin were both run as normalization controls in light of the known correlation of RECQ helicase expression with the phase of the cell cycle.
CRC Cell Lines Show Expression of RECQ Helicases in a Pattern Similar to Primary CRCs
Given the significant alterations in RECQ helicase expression we observed in primary CRCs, we assessed the expression of RECQ helicases in a panel of CRC cell lines to determine the feasibility of using cell lines to study the functional consequences of alterations in the expression levels of the RECQ helicases. We studied CRC cell lines derived from the three major molecular subtypes of CRC, MSS (or CIN) cancers (SW480, V400, V411, AAC1/SB10, and HT29), MSI cancers (HCT116, LoVo, and LS174T), and CIMP cancers (RKO, SW48, and FET). Similar to the pattern of RECQ helicase expression seen in primary CRCs, the cell lines had higher mRNA expression levels of BLM and RECQL4 than RECQL, RECQL5, and WRN. This was true regardless of the molecular subgroup of the CRC cell line. WRN had the lowest average mRNA expression level of all the RECQ helicase family members (Figure 5A). We observed low expression of RECQL (P < .001) and significantly high expression of RECQL4 (P = .017) in CRC cell lines when compared to normal colonic mucosa (Figure5B), which is consistent with the expression levels we observed in the primary CRCs.
We subsequently assessed RECQ helicase protein expression by immunoblotting protein lysates from these lines (Figure 5C). We found that WRN and RECQL5 protein levels correlated poorly with mRNA levels but that BLM, RECQL, and RECQL4 showed similar patterns of mRNA and protein expression. Interestingly, we found considerable variability among the CRC cell lines, even after normalization for proliferation, of both mRNA and protein expression for BLM and RECQL4. There was up to approximately three-fold difference in expression levels across cell lines (Figure 5, B and C).
Promoter Methylation Status of RECQL and RECQL5
To gain further understanding about the decreased expression of RECQL and RECQL5 seen in CRCs, we assessed the promoter methylation status of both RECQL and RECQL5 in normal colonic mucosa as well as CRC tumors. Our results show the absence of methylation in both the RECQL (N =0/7) and RECQL5 (N =0/8) promoters in normal colonic mucosa. We also found that 14% of CRCs have an aberrantly methylated RECQL5 promoter (N = 3/21), whereas 0% of CRCs have a methylated RECQL promoter (N = 0/16). Thus, the aberrant methylation of the promoters of RECQL and RECL5 does not seem to be a significant mechanism for silencing the expression of these genes.
Discussion
The RECQ helicases are a family of enzymes that maintain genomic stability through their function in DNA processing as helicases and exonucleases. They participate in DNA repair as members of DNA repair complexes [33]. They have been shown to be involved in single-strand break and double-strand break repair [34] as well as in the repair of topoisomerase I-mediated DNA damage induced by camptothecin derivatives such as irinotecan [16]. The loss of function of three of the RECQ helicases, WRN, BLM, and RECQL4, results in human syndromes characterized by a predisposition to cancer, including CRC [33]. In addition, in vitro studies demonstrate that these helicases are important in protecting genomic DNA from genotoxic stress [15,18,24,34–37]. Therefore, we postulated that the expression of the RECQ helicases is altered in CRC and that the aberrant expression of these RECQ helicases may have a role in CRC pathogenesis and in mediating the response of CRC cells to DNA-damaging chemotherapeutic agents. Our results confirm that the expression of RECQ helicases in primary CRC tumors is altered compared to normal colonic mucosa. It is plausible that these changes may be biologically relevant in light of the fact that most RECQ helicases act in protein complexes and even small changes in expression level could disrupt the stoichiometry of these complexes and their function. However, the biologic significance of these changes has yet to be determined. Our current findings support conducting further studies of these helicases to determine if they could be predictive biomarkers or therapeutic targets in CRC.
Our results are consistent with previous findings that RECQL5 may have tumor suppressor effects in CRC [18,19]. We found that RECQL and RECQL5 are expressed at statistically significant decreased levels in CRC when compared to matched normal colonic mucosa (Figure 2). Although loss of function of RECQL and RECQL5 have not been clearly linked to human disease, there is evidence from studies of mouse models as well as in vitro systems that loss of function of these helicases would likely lead to disease given their role in the maintenance of genomic stability [35,36,38–41]. RECQL5 is implicated in DNA replication, transcription, and repair processes, and RECQL5 deficiency is thought to contribute to genomic instability [18,42]. Our data also show that MSI tumors uniformly have decreased RECQL and RECQL5 expression (Table 2). These data suggest that RECQL5 and RECQL may have a role in mediating the degree of MSI seen in MSI CRC warranting further investigation. Furthermore, RECQL5-deficient ApcMin/wt mice have been shown to have an increase in the incidence of tumors formed in the colon. As RECQL5 is highly conserved between mice and humans, these results are consistent with a role for RECQL5 as a tumor suppressor in human CRC [19]. Our data establish the overall decreased expression of both RECQL and RECQL5 in CRCs and are consistent with previous studies suggesting that a decrease in RECQL5 expression in tumors would contribute to the pathogenesis of CRC [18,19,36,37].
We also found that grouping CRC tumors by RECQL5 expression status (high or low) results in two classes of CRC that are distinct not only in their expression of RECQL5 but also in their expression of WRN and RECQL (Figure 3). RECQL5 low (tumor < normal) CRCs tend to also have low WRN and RECQL expression. These data suggest that there may be a biologic link among these three RECQ helicases. Recently, it has been shown that RECQL5 is important for cell survival after treatment with camptothecin, a topoisomerase inhibitor [20,42]. Therefore, tumors with decreased expression of RECQL5 compared to normal colonic mucosa, with similarly decreased expression of WRN and RECQL, may represent a population of cells that are particularly susceptible to topoisomerase I inhibitors such as irinotecan. Further studies will help provide insight into this possibility.
In contrast to RECQL and RECQL5, we found that RECQL4 and BLM expression are significantly increased in CRCs when compared to matched normal colonic mucosa (Figure 2). An increase in the expression of these two RECQ helicases may confer a survival advantage to CRC cells and thereby contribute to the pathogenesis of CRC. RECQL4 and BLM have been shown to have coordinated activities and to cooperate in repairing DNA damage caused by ionizing radiation [24]. Our data are consistent with the possibility that increased expression of BLM and RECQL4 promote the ability of CRC cells to tolerate genotoxic stress. Functional studies are now needed to assess this possibility.
We found increased expression of BLM in primary CRCs despite its role as a tumor suppressor gene in Bloom syndrome and mouse intestinal cancer [43,44]. Increased BLM expression in established CRCs may provide a survival advantage for tumor cells, as it has been shown that BLM can facilitate telomere replication [45,46]. BLM has been co-localized with POT1, TRF1, and TRF2 (proteins that regulate telomere stability and length) in telomerase-deficient, alternative lengthening of telomeres-positive cells. When BLM is depleted in these cells, rapid telomere shortening was shown to occur [45,47]. Given previous data showing that loss of BLM leads to increased susceptibility of cells to killing with DNA-damaging agents [23,34], CRCs that showed an increase in BLM expression compared to normal colonic mucosa may be a subset of cancers that have decreased sensitivity to DNA-damaging agents. Our data showing that CIMP CRCs all have high (tumor > normal) BLM expression may have implications with regard to the effect of chemotherapy on CIMP CRCs compared to non-CIMP CRCs.
We also observed significant overexpression of RECQL4 in the CRCs. Recent studies show that RECQL4 expression is also increased in metastatic prostate cancer cell lines [48] as well as in sporadic osteoblastomas [49]. Loss of RECQL4 activity leads to an accumulation of double-strand breaks, and knockdown of RECQL4 decreases both tumorigenicity and invasiveness [48]. Therefore, increased expression of RECQL4 may likewise be important in the pathogenesis of CRC, conferring a survival advantage to the tumor. The data show that BLM and RECQL4 are the only two RECQ helicases that have increased expression. This is consistent with the co-localization of RECQL4 and BLM during S-phase and their reported coordinated activity [24]. Consistent with previous cell culture studies, our IHC studies show that BLM and RECQL4 are indeed localized to the nucleus (Figure 4). The expression of both BLM and RECQL4 widely range in fold change values, making them good candidates to investigate for a role as predictive biomarkers in CRC.
Our studies showing abnormal expression of the RECQ helicases in CRC suggest that the RECQ helicase genes may also be targets for mutations. Therefore, as an adjunct, we assessed the status of reported RECQ helicase mutations in CRCs in The Cancer Genome Atlas (TCGA) database (cancergenome.nih.gov/; 20/224 cases) and determined the predicted functional consequence of these mutations (Tables W2–W5). We found that there are no reported mutations in RECQL4, whereas there are reported mutations in BLM (4.5%), WRN (4.0%), RECQL (1.8%), and RECQL5 (2.7%). Of the reported mutations, approximately half are predicted to disrupt the function of the gene. Thus, mutational inactivation of the RECQ helicases appears to be a second mechanism through which potential tumor suppressor functions of the RECQ helicases may be disrupted. Furthermore, the lack of reported mutations in RECQL4 and the increased expression of RECQL4 seen in CRCs suggest that this enzyme may be important for tumorigenesis.
Potential mechanisms that control RECQ helicase expression include epigenetic alterations, such as promoter methylation. Our results show that aberrant promoter methylation of RECQL5 occurs in 14% of CRC tumors, suggesting that promoter methylation is responsible for the transcriptional repression of RECQL5 of only a small subset of CRCs. However, aberrant DNA methylation does not appear to be the mechanism responsible for the down-regulation of RECQL in CRCs, as we did not detect RECQL promoter methylation in CRCs. Other possible regulatory mechanisms for RECQ helicases include alterations in transcription factors that enhance or suppress gene expression and post-transcriptional processing of the mRNA. There is evidence that RECQL5 is regulated in part by transcription factor binding with a zinc-binding motif, which is highly conserved among RECQ helicases [50]. There are also data showing that RECQL5 interacts with RAD51 and that this interaction is critical for specific RECQL5 function [51]. Furthermore, E2F binding sites have been identified in RECQ helicase promoters to which both E2F and Rb can bind and recruit an Rb family repressor complex [52]. The precise mechanism by which the RECQ helicases are regulated is not fully understood; however, it is clear that this family of proteins have altered expression in CRCs.
An important technical issue related to our studies is the discrepancy we observed between the mRNA and protein expression of certain RECQ helicases in the primary tumors. In our hands, IHC stains for RECQ expression proved to be less precise in the quantitation of RECQ expression differences compared to qRT-PCR, with many of the Allred scores clustering at high levels for RecQL, RecQL4, and BLM. This is likely because of the more precise quantitative nature of the qRT-PCR assay compared to the more qualitative nature of IHC. The lack of correlation in WRN IHC when compared to mRNA expression in primary tissues was less surprising, as this was consistent with WRN protein expression compared to mRNA expression in CRC cell lines assayed by Western blot analysis. The data suggest that WRN mRNA levels indeed correlate poorly with protein levels. The differences seen in expression of RECQL5 in tumor compared to normal colonic mucosa were consistent at both the mRNA and protein levels, suggesting that these differences are the most robust and the results from the RT-PCR assays are comparable to the IHC studies.
In conclusion, we have shown that there are significant differences among CRCs with regard to the expression of the RecQ helicases as well as differences in the expression of RecQ helicases between CRCs and normal colonic tissues. On the basis of the known function of these helicases, the altered expression of RECQL, RECQL5, RECQL4, and BLM may affect the formation of CRC as well as the response of CRCs to chemotherapy. We also observed characteristic expression patterns of RECQ helicases in the different molecular subtypes of CRC, suggesting that the expression of these DNA repair enzymes may prove to help classify tumor behavior with regard to response to therapy. The studies presented here establish the altered expression profile of RECQ helicases in primary CRCs and provide a strong rationale for further studies of these helicases as predictive biomarkers or therapeutic targets in CRC.
Supplementary Material
Footnotes
Support for these studies was provided by the National Institutes of Health (NIH; RO1CA115513, P30CA15704, UO1CA152756, U54CA143862, and P01CA077852; W.M.G.), Burroughs Wellcome Fund Translational Research Award for Clinician Scientist (W.M.G.), ACS Fellowship PF-11-086-01-TBG (V.V.L.), 2T32DK007742-16 (V.V.L.), American Society of Colon and Rectal Surgeons, General Surgery Resident Research Initiation Grant (V.V.L.), and NIH National Cancer Institute F32CA1591555-01 (V.V.L.). The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W5 and Figures W1 to W3 and are available online at www.transonc.com.
References
- 1.Siegel RL, Ward EM, Jemal A. Trends in colorectal cancer incidence rates in the United States by tumor location and stage, 1992–2008. Cancer Epidemiol Biomarkers Prev. 2012;21:411–416. doi: 10.1158/1055-9965.EPI-11-1020. [DOI] [PubMed] [Google Scholar]
- 2.Saltz LB. Adjuvant therapy for colon cancer. Surg Oncol Clin N Am. 2010;19:819–827. doi: 10.1016/j.soc.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134:1296–1310. doi: 10.1053/j.gastro.2008.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA. Mutator phenotype in cancer: origin and consequences. Semin Cancer Biol. 2010;20:279–280. doi: 10.1016/j.semcancer.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 8.Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol. 2012;9:144–155. doi: 10.1038/nrclinonc.2012.3. [DOI] [PubMed] [Google Scholar]
- 9.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 10.Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, Orr N, Parton M, Smith IE, Reis-Filho JS, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res. 2010;16:6159–6168. doi: 10.1158/1078-0432.CCR-10-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 12.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barratt PL, Seymour MT, Stenning SP, Georgiades I, Walker C, Birbeck K, Quirke P. DNA markers predicting benefit from adjuvant fluorouracil in patients with colon cancer: a molecular study. Lancet. 2002;360:1381–1391. doi: 10.1016/s0140-6736(02)11402-4. [DOI] [PubMed] [Google Scholar]
- 14.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, Jewell SD, Mayer RJ, Goldberg RM, Saltz LB, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Lu X, Luo G. Effect of Recql5 deficiency on the intestinal tumor susceptibility of Apcmin mice. World J Gastroenterol. 2010;16:1482–1486. doi: 10.3748/wjg.v16.i12.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Lu X, Zhou G, Lou H, Luo G. RECQL5 is an important determinant for camptothecin tolerance in human colorectal cancer cells. Biosci Rep. 2011;31:363–369. doi: 10.1042/BSR20100108. [DOI] [PubMed] [Google Scholar]
- 21.Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst) 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70:6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh DK, Popuri V, Kulikowicz T, Shevelev I, Ghosh AK, Ramamoorthy M, Rossi ML, Janscak P, Croteau DL, Bohr VA. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012;40:6632–6648. doi: 10.1093/nar/gks349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botticelli AR, Casali AM, Botticelli L, Zaffe D. Immunohistochemical detection of cell-cycle associated markers on paraffin embedded and formalin fixed needle biopsies of prostate cancer: correlation of p120 protein expression with AgNOR, PCNA/cyclin, Ki-67/MIB1 proliferation-scores and Gleason gradings. Eur J Histochem. 1998;42:41–48. [PubMed] [Google Scholar]
- 27.Bolton WE, Freeman JW, Mikulka WR, Healy CG, Schmittling RJ, Kenyon NS. Expression of proliferation-associated antigens (PCNA, p120, p145) during the reentry of G0 cells into the cell cycle. Cytometry. 1994;17:66–74. doi: 10.1002/cyto.990170109. [DOI] [PubMed] [Google Scholar]
- 28.Turner HE, Wass JA. Are markers of proliferation valuable in the histological assessment of pituitary tumours? Pituitary. 1999;1:147–151. doi: 10.1023/a:1009979128608. [DOI] [PubMed] [Google Scholar]
- 29.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 30.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, et al. Mutational inactivation of transforming growth factor β receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 31.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 32.Luo Y, Tsuchiya KD, II, Park D, Fausel R, Kanngurn S, Welcsh P, Dzieciatkowski S, Wang J, Grady WM. RET is a potential tumor suppressor gene in colorectal cancer. Oncogene. 2012;32:2037–2047. doi: 10.1038/onc.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monnat RJ., Jr Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Semin Cancer Biol. 2010;20:329–339. doi: 10.1016/j.semcancer.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh DK, Ghosh AK, Croteau DL, Bohr VA. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat Res. 2012;736:15–24. doi: 10.1016/j.mrfmmm.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tadokoro T, Ramamoorthy M, Popuri V, May A, Tian J, Sykora P, Rybanska I, Wilson DM, III, Croteau DL, Bohr VA. Human RECQL5 participates in the removal of endogenous DNA damage. Mol Biol Cell. 2012;23:4273–4285. doi: 10.1091/mbc.E12-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X, Lou H, Luo G. A Blm-Recql5 partnership in replication stress response. J Mol Cell Biol. 2011;3:31–38. doi: 10.1093/jmcb/mjq056. [DOI] [PubMed] [Google Scholar]
- 38.Aygun O, Svejstrup JQ. RECQL5 helicase: connections to DNA recombination and RNA polymerase II transcription. DNA Repair (Amst) 2010;9:345–353. doi: 10.1016/j.dnarep.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Brosh RM., Jr Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair (Amst) 2010;9:315–324. doi: 10.1016/j.dnarep.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popuri V, Croteau DL, Brosh RM, Jr, Bohr VA. RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle. 2012;11:4252–4265. doi: 10.4161/cc.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst) 2012;11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Lu X, Zhou G, Barnes EL, Luo G. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol Biol Cell. 2009;20:114–123. doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goss KH, Risinger MA, Kordich JJ, Sanz MM, Straughen JE, Slovek LE, Capobianco AJ, German J, Boivin GP, Groden J. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297:2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama H. RecQ family helicases: roles as tumor suppressor proteins. Oncogene. 2002;21:9008–9021. doi: 10.1038/sj.onc.1205959. [DOI] [PubMed] [Google Scholar]
- 45.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opresko PL. Telomere ResQue and preservation—roles for the Werner syndrome protein and other RecQ helicases. Mech Ageing Dev. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Bhattacharyya S, Sandy A, Groden J. Unwinding protein complexes in ALTernative telomere maintenance. J Cell Biochem. 2010;109:7–15. doi: 10.1002/jcb.22388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Y, Meador JA, Calaf GM, Proietti De-Santis L, Zhao Y, Bohr VA, Balajee AS. Human RecQL4 helicase plays critical roles in prostate carcinogenesis. Cancer Res. 2010;70:9207–9217. doi: 10.1158/0008-5472.CAN-10-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maire G, Yoshimoto M, Chilton-MacNeill S, Thorner PS, Zielenska M, Squire JA. Recurrent RECQL4 imbalance and increased gene expression levels are associated with structural chromosomal instability in sporadic osteosarcoma. Neoplasia. 2009;11:260–268. doi: 10.1593/neo.81384. 263 pp. following 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren H, Dou SX, Zhang XD, Wang PY, Kanagaraj R, Liu JL, Janscak P, Hu JS, Xi XG. The zinc-binding motif of human RECQ5β suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem J. 2008;412:425–433. doi: 10.1042/BJ20071150. [DOI] [PubMed] [Google Scholar]
- 51.Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, El-Naggar S, Clem B, Chesney J, Dean DC. The Rb/E2F pathway and Ras activation regulate RecQ helicase gene expression. Biochem J. 2008;412:299–306. doi: 10.1042/BJ20070975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.