Abstract
Tumor necrosis factor receptor 1 (TNFR1)-associated death domain protein (TRADD) is an important adaptor in TNFR1 signaling and has an essential role in nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation and survival signaling. Increased expression of TRADD is sufficient to activate NF-κB. Recent studies have highlighted the importance of NF-κB activation as a key pathogenic mechanism in glioblastoma multiforme (GBM), the most common primary malignant brain tumor in adults.We examined the expression of TRADD by immunohistochemistry (IHC) and find that TRADD is commonly expressed at high levels in GBM and is detected in both cytoplasmic and nuclear distribution. Cytoplasmic IHC TRADD scoring is significantly associated with worse progression-free survival (PFS) both in univariate and multivariate analysis but is not associated with overall survival (n = 43 GBMs). PFS is a marker for responsiveness to treatment. We propose that TRADD-mediated NF-κB activation confers chemoresistance and thus a worse PFS in GBM. Consistent with the effect on PFS, silencing TRADD in glioma cells results in decreased NF-κB activity, decreased proliferation of cells, and increased sensitivity to temozolomide. TRADD expression is common in glioma-initiating cells. Importantly, silencing TRADD in GBM-initiating stem cell cultures results in decreased viability of stem cells, suggesting that TRADD may be required for maintenance of GBM stem cell populations. Thus, our study suggests that increased expression of cytoplasmic TRADD is both an important biomarker and a key driver of NF-κB activation in GBM and supports an oncogenic role for TRADD in GBM.
Introduction
Tumor necrosis factor receptor 1 (TNFR1)-associated death domain protein (TRADD) is an important component of the TNFR1 signaling network [1,2]. It plays a key role in TNFR1-induced nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation and cell survival. Increased expression of TRADD is known to activate NF-κB [1]. In common with other members of the TNF superfamily, TRADD may also induce apoptosis, depending on the cellular context [3]. TRADD also has a role in innate immunity and has been implicated in toll-like receptor 3 and toll-like receptor 4 signaling [4]. TRADD functions as an adaptor and is involved in assembly of a signaling platform at TNFR1 that includes other proteins such as TNF receptor-associated factor 2 and receptor interacting protein 1 (RIP1) [5]. Activation of TNFR1 has been shown to result in the formation of sequential signaling complexes, including a membrane-associated complex that mediates survival and includes TRADD and a subsequent cytosolic death complex composed of TRADD, RIP1, and Fas-associated protein with death domain [6]. Although TRADD has been considered a cytoplasmic protein, it can shuttle between the cytoplasm and the nucleus and has both a putative nuclear localization signal and a putative nuclear export signal [7–9]. In the nucleus, TRADD may interact with key regulatory proteins and/or influence transcription of key genes [8,9].
Although TRADD has been investigated intensively in inflammatory and immune signaling, much less is known about its role in cancer. On the basis of TRADD's role in proinflammatory signaling and the key role of inflammation in the pathogenesis of cancer, an oncogenic role for TRADD is plausible. However, TRADD also has a role in cell death and, furthermore, is located in a chromosomal region (16q22.1) showing frequent loss of heterozygosity in various cancers [8,10], suggesting a tumor suppressive role for TRADD. A recent study has shown that TRADD plays a tumor suppressive role in an experimental model of skin cancer in mice and suggested that low levels of TRADD conferred a worse prognosis [8].
Glioblastoma multiforme (GBM) is the most common primary malignant tumor of the central nervous system in adults [11]. The treatment of GBM remains an intractable problem [12]. The Cancer Genome Atlas has provided a comprehensive picture of genetic abnormalities in GBM [13–17]. The molecular pathogenesis of glioma is thought to involve multiple genetic alterations that result in aberrant activity of pathways involved in proliferation, cell cycle regulation, and apoptosis [16–19]. The genetic changes detected frequently in primary GBM include INK4A loss, epidermal growth factor receptor (EGFR) amplification and mutation, phosphatase and tensin homolog loss, and mouse double minute 2 homolog amplification, among other abnormalities. TRADD is reported to be highly expressed in the mesenchymal subtype of GBM along with other NF-κB genes such as RelB [16].
Aberrant NF-κB activation is widespread in cancer [20,21]. NF-κB has important roles in cell viability and cell cycle progression and influences proliferation of cancer cells and resistance to treatment [22,23]. There is also an extensive cross talk between NF-κB activation and oncogenic and tumor suppressor signaling pathways [24,25]. Recent studies suggest that NF-κB activation is an important contributor to the malignant phenotype in GBM [12]. It has been suggested that two major pathways activate NF-κB in GBM [26]. First, EGFR gene amplification and aberrant EGFR gene amplification are detected in 40% to 50% of GBMs and increased EGFR signaling is well documented to activate NF-κB [27–30]. Second, NF-κB inhibitor α (NFKBIA, which encodes IκBα) is commonly deleted in non-EGFR-amplified GBMs resulting in NF-κB activation [26]. However, additional pathways such as transforming growth factor β also activate NF-κB in GBM [31]. Furthermore, other members of the TNFR/NF-κB superfamily, such as A20, have also been implicated in maintenance of stem cells in GBM [32].
Gliomas are graded I to IV in order of increasing malignancy, and GBM (grade IV) is the most malignant. TRADD is a key component of the NF-κB signaling pathway and functions primarily as an adaptor. Recent studies have demonstrated that NF-κB activation is common in GBM and proposed aberrant EGFR signaling and IκBα deletion as major mechanisms of NF-κB activation in GBM. However, additional mechanisms are likely. For example, increased expression of components of NF-κB pathway may also be sufficient for NF-κB activation. We have previously shown that increased expression in RIP1 is sufficient to induce NF-κB activation in glioma cells. Furthermore, high levels of RIP1 are common in GBM and confer a worse prognosis [25].
In this study, we propose that increased TRADD expression is an important mechanism of NF-κB activation in GBM. We show that TRADD expression is robust in GBMs. Increased levels of cytoplasmic TRADD results in a worse progression-free survival (PFS) in GBM patients. Nuclear presence of TRADD has no effect on prognosis. Consistent with the effect on PFS, silencing TRADD in glioma cells results in decreased NF-κB activity, decreased proliferation of cells, and increased sensitivity to temozolomide. Silencing TRADD in GBM stem cell cultures results in decreased viability of cells, suggesting that TRADD may be required for maintenance of GBM stem cell populations. Thus, our data suggest that cytoplasmic TRADD plays an oncogenic role in GBM.
Materials and Methods
Antibodies, Reagents, and Western Blot
TRADD antibody was purchased from Novus Biologicals (Littleton, CO; Cat. No. NB-100-92112). IκBα and actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Phospho-p65 and p65 antibodies were purchased from Cell Signaling Technology (Danvers, MA). Protein quantitation was performed using a BIO-RAD kit (Hercules, CA). Western blot was performed according to standard protocols.
Protein lysates were prepared for Western blot using a lysis buffer containing 1% NP-40, 150 mM NaCl, 0.25 deoxycholate, 1 mM EGTA, 1 mM NaF, 50 mM Tris-HCl, 1 mM PMSF, 2 mM orthovanadate, and a protease inhibitor cocktail.
RNA Interference
TRADD was silenced using lentiviral shRNA ready-to-use transduction particles from Santa Cruz Biotechnology (sc-36709-V, containing three target-specific constructs) and puromycin selection according to the manufacturer's protocol. As a control, we used control (scrambled, non-sequence) shRNA lentiviral particles also from Santa Cruz Biotechnology. TRADD silencing was confirmed with Western blot.
Luciferase Assays
TRADD-silenced or control shRNA cells were plated onto 24-well dishes followed by transfection with an NF-κB-LUC reporter. A dual-luciferase reporter assay system was used according to the instructions of the manufacturer (Promega, Madison, WI). Firefly luciferase activity was measured in a luminometer and normalized on the basis of Renilla luciferase activity. Experiments were done in triplicate and repeated twice.
Cell Proliferation Assays
5K cells were plated onto 24-well dishes. The cells were trypsinized, analyzed by trypan blue exclusion for dead cells, and counted using an automated cell counter after 72 hours of plating. At least three independent experiments were done in triplicate.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Conversion Assays
U251 control shRNA or TRADD-silenced cells were plated at 5K per well in 96-well dishes. An MTT kit from Roche Applied Science (Indianapolis, IN) was used according to the manufacturer's instructions.
Primary Tumors/Immunohistochemistry
Informed consent was obtained and all studies were conducted according to Institutional Review Board-approved protocols at the University of Texas Southwestern Medical Center.
The TRADD antibody used was from Novus Biosciences (Cat. No. NB100-92112; Novus Biologicals) and was used for both Western blot and immunohistochemistry (IHC). We validated this antibody in TRADD knockdown cells by Western blot. As can be seen in Figure 4A, the TRADD signal is lost in cells when TRADD is knocked down with siRNA. The antibody was developed for IHC in the Neuropathology IHC Core at the University of Texas Southwestern Medical Center. An isotype-matched control antibody was used as a negative control.
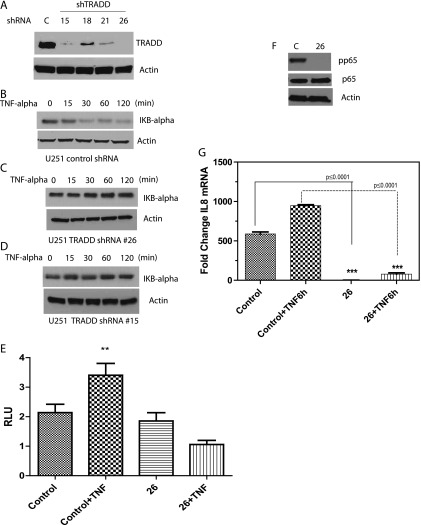
Figure 4.
(A) U251MG cells were infected with control (scrambled) lentivirus or with TRADD shRNA lentivirus, followed by puromycin selection and Western blot of clones infected with TRADD shRNA. Clones 15 and 26 show the best silencing of TRADD. (B) Degradation of IκBα in response to TNF-α (20 ng/ml) can be seen in U251MG control shRNA cells, showing that TNF-α activates NF-κB in these cells. (C and D) Failure of IκBα degradation in response to TNF-α in TRADD-silenced clones 15 and 26, demonstrating that TRADD is essential for NF-κB activation. (E) A reporter assay in clone 26 with an NF-κB responsive luciferase reporter. While NF-κB transcriptional activity increases in response to TNF-α in control shRNA cells, there is no increase in TRADD-silenced clone 26, again confirming the requirement for TRADD in NF-κB activation (one-way ANOVA, P = .0031). (F) Western blot showing that phosphorylation of the p65 subunit of NF-κB is in response to TNF-α is impaired in TRADD-silenced clone 26. (G) Quantitative real-time polymerase chain reaction experiment showing that mRNA levels of the NF-κB target gene IL-8 is decreased in TRADD-silenced clone 26, both at a basal level (P ≤ .0001) and in response to TNF-α (P ≤ .0001) compared to control. Newman-Keuls multiple comparison test was used for statistical analysis.
Cores (1.5 mm) for tissue microarrays (TMAs) were punched using a tissue microarrayer (Beecher Instruments, Sun Prairie, WI). The donor blocks were formalin-fixed, paraffin-embedded tissue blocks of human gliomas. TMA sections of human gliomas were cut at 4-µm thickness and deparaffinized. For antigen retrieval, the sections were heated for 30 minutes at 95°C in CC1 Solution (Ventana Medical Systems, Tucson, AZ). The sections were exposed to a rabbit polyclonal antibody to human and mouse TRADD (NB100-92112; Novus Biologicals) at 1:100 dilution. The signal was developed with the alkaline phosphatase-based ultraView universal detection system (Ventana Medical Systems). The chromogen was ultraView DAB (Ventana Medical Systems). The immunostaining procedure was performed using the Benchmark XT automated stainer (Ventana Medical Systems). The IHC staining intensity for TRADD was scored semiquantitatively. The final IHC score was derived from IHC staining intensity multiplied by the percentage of tumor cells showing positive staining as we have described previously [33].
Primary GBM cultures were generated directly from human tumor specimens. A papain dissociation system was used to dissociate tumors. Cells were then cultured in neurobasal medium supplemented with B27 without vitamin A and with EGF (10 ng/ml) and basic fibroblast growth factor (10 ng/ml), as described previously [34].
Annexin FACS
Annexin assay was done by using Annexin-V-FLUOS Staining Kit (Roche Applied Science). Neurosphere cultures from TRADD-silenced or control shRNA cells (1 x 106) or U251MG cells with control shRNA or TRADD shRNA were plated onto six-well plates. The cells were incubated for 25 minutes at room temperature with propidium iodide and Annexin-V-FLUOS labeling solution in incubation buffer (supplied by the manufacturer). Annexin and or propidium iodide-positive cells were detected by flow cytometry. TRADD silencing was confirmed by Western blot.
Statistical Analysis
Our data set contains 43 GBM patients with detailed clinical information. All patients have received radiotherapy and chemotherapy.
The primary end point was PFS, defined as the time from the completion of surgery to tumor recurrence, death from any cause, or last follow-up contact. We also studied overall survival (OS), defined as the time from the completion of surgery to death or last follow-up contact. To evaluate the association of TRADD levels with survival, patients were divided into two equal-sized groups based on the median of their IHC scorings. Survival curves for PFS and OS were examined using the Kaplan-Meier method and were compared using log-rank test. Univariate and multivariate survival analyses were performed using Cox proportional hazards model. Multivariate models were adjusted for age (in years, as a continuous variable), gender (male or female), Karnofsky performance score (KPS; as a continuous variable), and surgery (partial removal or complete removal).
Error bars represent the means ± SDs of three independent experiments. All data were analyzed for significance using GraphPad Prism 5.0 software, where P < .05 was considered statistically significant. One-way analysis of variance (ANOVA) and two-tailed t test were used to compare groups.
Results
TRADD Expression in GBM
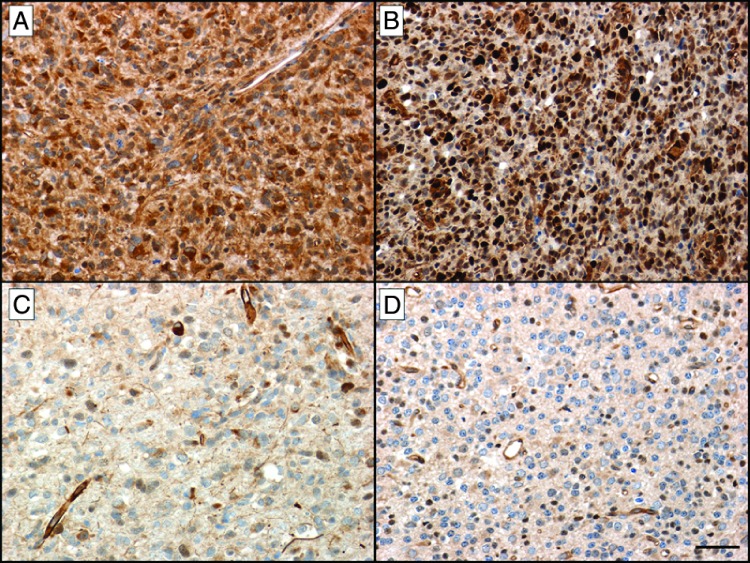
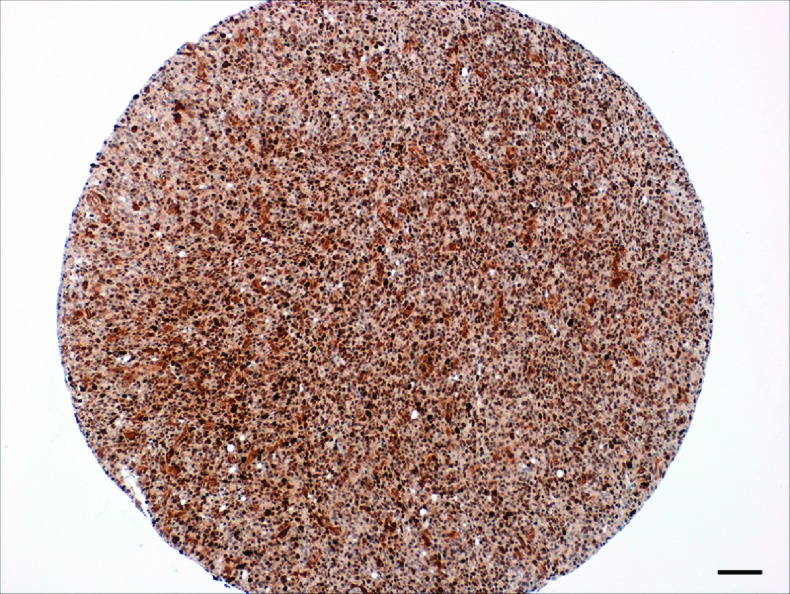
Because TRADD plays a key role in NF-κB activation, we investigated TRADD levels in GBM. TRADD expression was examined in GBM TMAs. GBMs were assessed for the presence of TRADD in nucleus and cytoplasm, and a semiquantitative TRADD score was compiled using the intensity of staining as well as the number of stained cells in each tumor. Nuclear as well as cytoplasmic staining was studied. Figure 1 shows representative photomicrographs of TRADD IHC in human GBMs on a TMA. Figure 1A shows strong cytoplasmic positivity for TRADD in a GBM, whereas Figure 1B shows strong nuclear and moderate cytoplasmic positivity for TRADD in a second case of GBM. Figure 1C shows absent to weak positivity for TRADD in a third case of GBM. Figure 1D shows absent to weak positivity for TRADD in an oligodendroglioma [World Health Organization (WHO) grade II], a type of low-grade glioma. Figure 2 shows a representative low-power photomicrograph of a TMA core from a human GBM showing strong expression of TRADD.
Figure 1.
Representative photomicrographs of TRADD IHC in human gliomas on a TMA. (A) Strong cytoplasmic positivity for TRADD in a glioblastoma. (B) Strong nuclear and moderate cytoplasmic positivity for TRADD in a second case of glioblastoma. (C) Absent to weak positivity for TRADD in a third case of glioblastoma. (D) Absent to weak positivity for TRADD in an oligodendroglioma (WHO grade II), a type of low-grade glioma (IHC for TRADD with hematoxylin counterstain. Size bar, 50 µm. Original magnification, x200).
Figure 2.
A representative low-power photomicrograph of a TMA core from a human glioblastoma showing strong expression of TRADD (IHC for TRADD with hematoxylin counterstain. Scale bar, 100 µm).
Cytoplasmic TRADD Expression Confers a Worse Prognosis in GBM
Next, we investigated whether TRADD expression influences prognosis in GBM. Our data set contains 43 GBM patients with detailed clinical information (Table W1). All patients received radiotherapy and chemotherapy.
The primary end point was PFS, defined as the time from the completion of surgery to tumor recurrence, death from any cause, or last follow-up contact. We also studied OS, defined as the time from the completion of surgery to death or last follow-up contact. To evaluate the association of TRADD levels with survival, patients were divided into two equal-sized groups based on the median of their IHC scorings. Survival curves for PFS and OS were examined using the Kaplan-Meier method and were compared using log-rank test. Univariate and multivariate survival analyses were performed using Cox proportional hazards model [35]. Multivariate models were adjusted for age (in years, as a continuous variable), gender (male or female), KPS (as a continuous variable), and surgery (partial removal or complete removal).
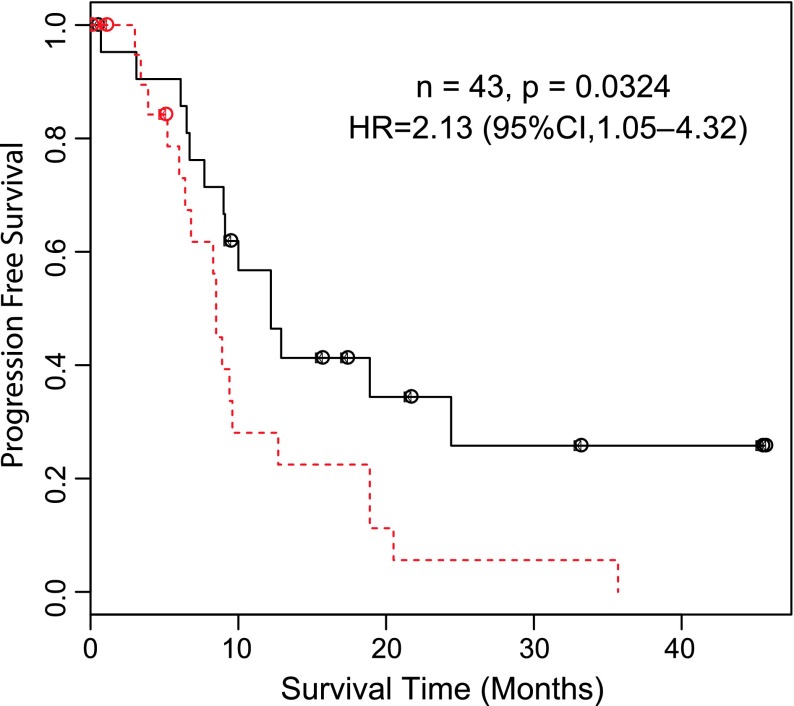
The cytoplasmic TRADD level showed significant association with PFS. The group with high IHC scoring had significantly worse survival outcomes than the low IHC group: hazard ratio (HR) = 2.13, 95% confidence interval (CI) = 1.05 to 4.32, P = .0324 (Figure 3). Multivariate analysis adjusting for preselected significant prognostic factors showed that the IHC scoring is an independent prognostic factor for PFS: HR = 2.471, 95% CI = 1.143 to 5.341, P = .021 (Table 1). When we examined the association between the cytoplasmic TRADD level and patients' OS, we found no significant difference in OS between high and low IHC scoring groups: HR = 1.034, 95% CI = 0.489 to 2.186, P = .884. It should be noted that no association between nuclear TRADD IHC and OS or PFS was detected.
Figure 3.
Association of cytoplasmic TRADD level with PFS. To evaluate the association of TRADD levels with survival, patients were divided into two equal-sized groups based on the median of their IHC scorings. Survival curves for PFS were generated using the Kaplan-Meier method and were compared using log-rank test. Univariate analyses was performed using Cox proportional hazards model (multivariate analysis is described in Table 1). Dashed line represents high cytoplasmic TRADD, while solid line represents low cytoplasmic TRADD.
Table 1.
Multivariate Cox Regression Analysis of the Cytoplasmic TRADD Level and Clinical Variables in GBM Patients.
| HR | 95% CI | P | ||
| IHC levels | ||||
| High versus low | 2.471 | 1.143 | 5.341 | .021 |
| Age | 0.970 | 0.929 | 1.012 | .159 |
| Gender | ||||
| Male versus female | 0.544 | 0.228 | 1.298 | .170 |
| KPS | 0.956 | 0.908 | 1.007 | .090 |
| Surgery | ||||
| PR versus CR | 1.454 | 0.422 | 5.005 | .553 |
CR indicates complete response; PR, partial response.
Silencing TRADD Inhibits NF-κB Activation
To examine the role of TRADD in an experimental cell culture model of glioma, we used the established GBM cell line U251MG and undertook to stably silence TRADD in these cells using lentiviral shRNA. Two clones with stably silenced TRADD were identified (Figure 4A) and designated as clones 15 and 26. As a control, we use scrambled (non-sequence) shRNA. Since TRADD is involved in NF-κB activation, we tested whether silencing TRADD in these cells influenced TNF-α-induced NF-κB activation. NF-κB subunits are normally sequestered in the cytoplasm by IκBα. Addition of TNF-α to cells results in degradation of IκBα allowing NF-κB subunits to translocate to the nucleus and initiate gene transcription. Thus, we examined the degradation of IκBα by Western blot as a readout for NF-κB activation. As can be seen in Figure 4B, while IκBα is degraded in response to TNF-α stimulation in control shRNA cells, we do not detect degradation of IκBα in TRADD-silenced clones (Figure 4, C and D). Furthermore, we examined the effect of TRADD silencing on TNF-α-induced NF-κB transcriptional activity by using an NF-κB responsive luciferase reporter assay. Silencing of TRADD inhibited TNF-α-induced NF-κB transcriptional activity as is shown in Figure 4E. In addition, phosphorylation of the p65 subunit is impaired in TRADD-silenced cells (Figure 4F). Finally, induction of an NF-κB target gene [interleukin-8 (IL-8)] is also impaired in TRADD-silenced cells (Figure 4G).
A Role for TRADD in Proliferation of Glioma Cells and in Resistance to Temozolomide
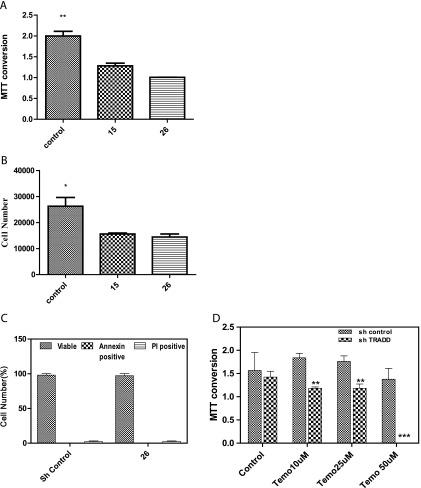
NF-κB has important roles in cell viability and cell cycle progression and influences proliferation of cancer cells and resistance to treatment. Thus, we examined whether silencing TRADD influenced the viability/proliferation of cells. We examined the viability/proliferation of U251 control shRNA and TRADD-silenced clones 15 and 26 in monolayer culture using cell counting and an MTT assay. As can be seen in Figure 5, A and B, the clones with stably silenced TRADD exhibited decreased proliferation of cells compared to control (scrambled) shRNA-expressing cells. TRADD also has been reported to have an effect in promoting cell death. The lack of cell death in TRADD-silenced cells was confirmed by an Annexin-FACS assay (Figure 5C). Thus, the prosurvival role of TRADD appears to dominate in glioma cells.
Figure 5.
(A) An MTT conversion assay was conducted comparing U251 cells with control shRNA or stably silenced TRADD (clones 15 and 26). MTT conversion is higher in control cells compared to TRADD-silenced cells suggesting increased proliferation in control cells (one-way ANOVA, P = .005). Cell death was excluded by trypan blue exclusion in parallel experiments. (B) Analysis of cell proliferation in U251 TRADD-silenced and control shRNA clones. 5K cells were plated onto 24-well dishes for 72 hours followed by trypsinization and counting in an automated cell counter. Cell proliferation is decreased in TRADD-silenced clones 15 and 26 (one-way ANOVA, P = .04). (C) An Annexin-FACS assay was conducted to determine if TRADD silencing leads to cell death. The percentage of viable cells was compared in control and TRADD-silenced cells by unpaired t test. No cell death was detected in TRADD-silenced clone 26. (D) An MTT conversion assay was conducted in U251 cells with control shRNA or TRADD-silenced clone 26 to assess the response to increasing doses of temozolomide. While U251 control shRNA cells are somewhat resistant to temozolomide, silencing TRADD increases the vulnerability of cells to temozolomide at 10 µM (P ≤ .04), 25 µM (P ≤ .04), and 50 µM (P ≤ .0003) in comparison to the control shRNA cells. Data were analyzed by two-tailed unpaired t test.
Temozolomide is a first-line chemotherapeutic drug in GBM and is administered as the standard of care to patients with GBM. We examined whether the presence of TRADD influenced the effect of temozolomide on the viability of U251MG glioma cells in TRADD-silenced cells (clone 26) compared to control shRNA cells. As can be seen in Figure 5D, U251 control shRNA cells are somewhat resistant to the effect to temozolomide. However, silencing TRADD results in a significantly decreased viability of U251 cells in response to temozolomide. These findings are consistent with a prosurvival role of TRADD in glioma cells and also with our finding that increased TRADD levels result in a worse PFS in GBMs, possibly by conferring resistance to treatment with temozolomide.
The Role of TRADD in Glioma-Initiating Stem-Like Cells
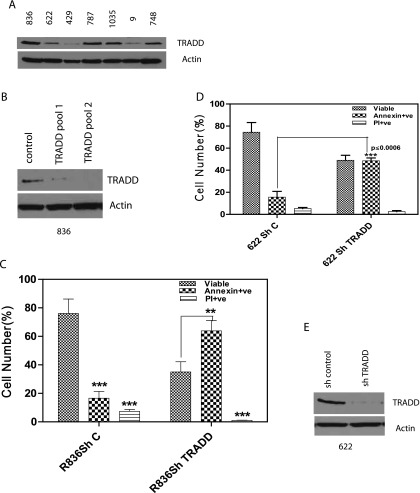
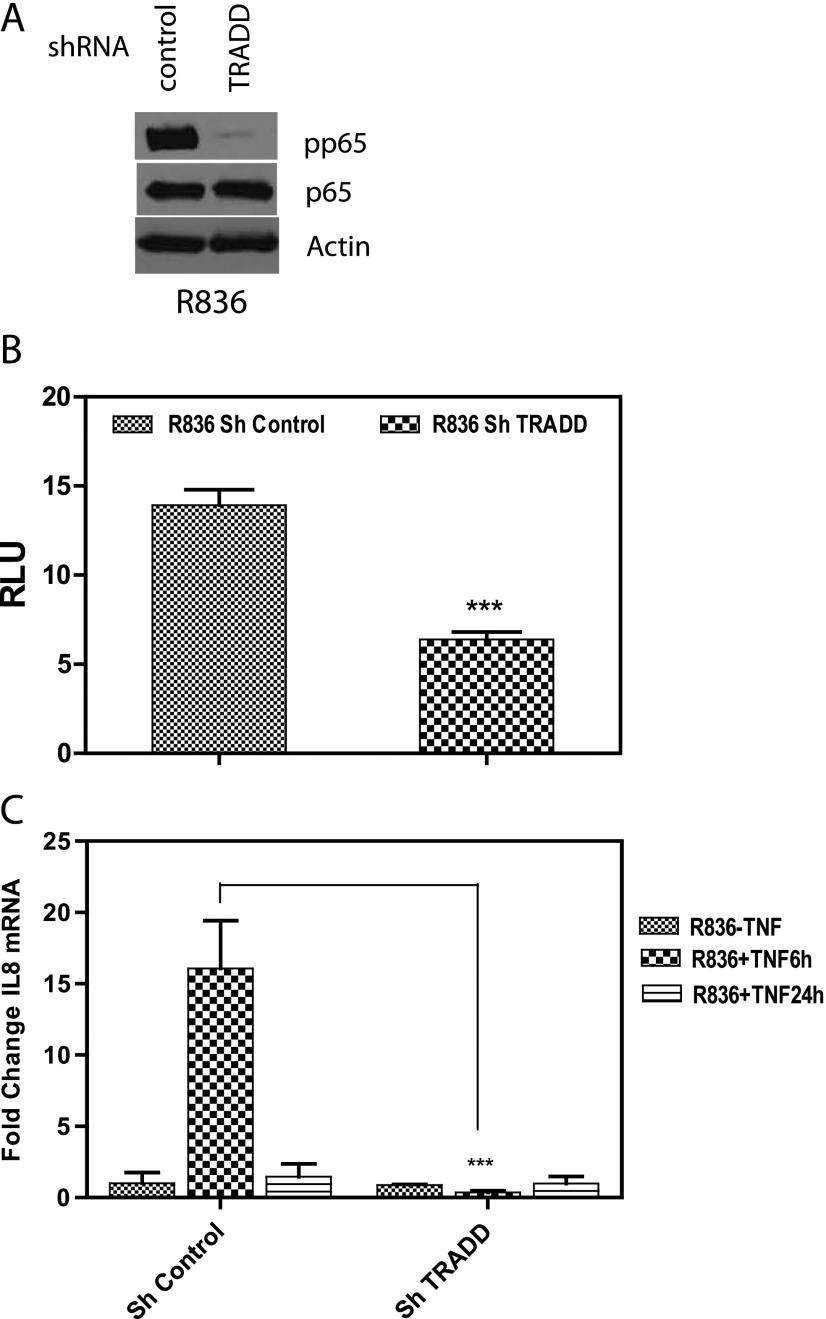
While established GBM cell lines provide an isogenic and widely used experimental model for glioma studies, glioma-initiating stem-like cells derived from human GBM samples and cultured as neurospheres in defined medium resemble primary GBMs more closely compared to established GBM cell lines [36]. Thus, we examined a panel of primary GBM cultures grown as neurospheres for the presence of TRADD. TRADD can be detected in neurosphere cultures derived from all GBM tumors (Figure 6A). Next, we silenced TRADD in glioma stem-like cultures derived from GBM tumor 836. Two TRADD-silenced pools were generated with better silencing of TRADD in pool 2 (Figure 6B). We conducted an Annexin-FACS assay to determine the effect of TRADD silencing in TRADD pool 2 compared to control shRNA cells. Interestingly, silencing TRADD resulted in a sharp drop in viability of these stem-like cultures with a large increase in Annexin-positive cells as shown in Figure 6C. A similar result was obtained in GBM622 as shown in Figure 6D. Silencing of TRADD in GBM622 is shown in Figure 6E. In addition, we confirmed that TRADD silencing results in impaired NF-κB activation by comparing phosphorylation of the p65 subunit of NF-κB, reporter assays, and induction of an NF-κB target gene (IL-8; Figure 7, A–C). These findings demonstrate a requirement for TRADD in NF-κB activation and maintenance of GBM stem cell populations and are consistent with a previous study that revealed a role for NF-κB in the maintenance of glioma stem-like cultures [37].
Figure 6.
(A) A panel of glioma stem-like cultures grown as neurospheres from various GBMs was screened for the presence of TRADD by Western blot. All of the stem cell cultures express TRADD. (B) TRADD was silenced in neurosphere culture derived from GBM836 using lentiviral shRNA followed by puromycin selection, and two pools were examined. Pool 2 has better silencing of TRADD. Control shRNA (scrambled) cells were used as control. (C) While the majority of control shRNA cells are viable, silencing TRADD in glioma stemcell cultures results in cell death with increased Annexin-positive cells (P ≤ .008, two-tailed unpaired t test). (D) A second GBM primary neurosphere culture from GBM622 with silenced TRADD also shows increased Annexin-positive cells in TRADD-silenced cells compared to control shRNA cells (P ≤ .0006, two-tailed unpaired t test). (E) Western blot showing silencing of TRADD in GBM622 neurosphere cultures.
Figure 7.
Impaired NF-κB activation in TRADD-silenced glioma stem cell cultures (836, pool 2). (A) Phosphorylation of the p65 subunit of NF-κB decreased in TRADD-silenced glioma stem cell cultures compared to control shRNA cells. (B) A reporter assay in TRADD-silenced cells with an NF-κB responsive luciferase reporter. NF-κB transcriptional activity is significantly higher in control cells compared to TRADD-silenced cells. Cells were treated with TNF-α for 6 hours. (C) Quantitative real-time polymerase chain reaction experiment showing that mRNA levels of the NF-κB target gene IL-8 - increases after 6 hours of TNF-α exposure in control cells, while it does not increase in TRADD-silenced cells (P ≤ .0001). TRADD silencing is shown in Figure 6B. Newman-Keuls multiple comparison test was used for statistical analysis.
Discussion
While TRADD has been investigated extensively in inflammation and immune signaling, relatively little is known about its role in cancer. TRADD belongs to the TNF receptor superfamily, whose members may influence survival or cell death depending on the cellular context. It is well established that TRADD has a role in NF-κB activation and promotion of cell survival. Aberrant NF-κB activation is widespread in cancer and promotes the malignant phenotype and resistance to treatment. The mechanisms of NF-κB activation in cancer are diverse but may include overexpression of individual components of the NF-κB signaling network. In GBM, NF-κB activation is well documented and a focus of considerable investigation. It has been postulated that the major mechanisms of NF-κB activation in GBM are aberrant EGFR signaling and deletion of NFKBIA (which encodes IκBα) is commonly deleted in non-EGFR-amplified GBMs resulting in NF-κB activation [26–29].
However, data from The Cancer Genome Atlas have documented that increased expression of components of the NF-κB signaling network including TRADD and RelB can be detected in the mesenchymal subtype of GBMs. Since it is known that overexpression of TRADD is sufficient to activate NF-κB in vitro [1], increased expression of TRADD at the protein level may be sufficient to activate NF-κB in GBMs. Furthermore, our findings that increased TRADD expression is common in GBM suggest that TRADD-mediated activation of NF-κB may be an important mechanism in GBM. Furthermore, TRADD overexpression may cooperate with other mechanisms such as deletion of NFKBIA to activate NF-κB.
TRADD can be detected in both cytoplasmic and nuclear compartments. The major function of cytoplasmic TRADD is likely to be NF-κB activation in response to extrinsic stimuli. Our data suggest that high levels of cytoplasmic TRADD confer a worse prognosis, in terms of PFS in GBM, a finding that is valid both in univariate and multivariate analyses, demonstrating that cytoplasmic TRADD confers a worse PFS in GBM. PFS is an indicator of responsiveness to treatment. Thus, it is likely that high levels of cytoplasmic TRADD inhibit sensitivity to chemotherapeutic drugs such as temozolomide. This hypothesis is supported by our experimental data showing that silencing TRADD increases sensitivity to temozolomide in glioma cells. The TRADD-mediated resistance to temozolomide is presumably secondary to increased NF-κB activation driven by increased levels of cytoplasmic TRADD. Increased levels of TRADD did not influence OS, consistent with the devastating and intractable nature of GBM. Less is known about nuclear TRADD, and in our analysis, nuclear TRADD did not influence prognosis. However, nuclear TRADD may play an important role in regulating apoptosis and have a tumor suppressive influence in other cancers. A recent study has proposed that nuclear TRADD promotes p19Arf stability and tumor suppression [8]. TRADD has also been reported to form a complex with STAT-1 in the nucleus [9]. Thus, emerging evidence suggests that TRADD may have complex roles in cancer depending on the tumor type and microenvironment of the tumor. However, our data indicate that increased cytoplasmic TRADD plays an oncogenic role in GBM.
The oncogenic role of TRADD in GBM is likely related to the role of TRADD in NF-κB activation. In addition to its well-known role in inflammation, NF-κB activation also influences cell survival, cell cycle progression, and proliferation. Indeed, there is an extensive cross talk between NF-κB components and growth factor signaling and cell cycle pathways [38–43]. Activation of NF-κB also regulates cell cycle progression and cell proliferation by mechanisms that include the expression of cytokines that promote proliferation of tumor cells [44–46] and by inducing expression of cyclin D1 and c-Myc [47,48]. In our experiments, silencing TRADD resulted in decreased NF-κB activation and decreased proliferation of glioma cells. Importantly, silencing TRADD increased the susceptibility of glioma cells to temozolomide, consistent with the effect of TRADD on PFS. Another intriguing finding of this study is expression of TRADD in glioma stem-like cultures derived from primary GBMs. We detected TRADD in all of the stemlike cultures we tested. Importantly, silencing TRADD in GBM stem cell cultures inhibited NF-κB activation and sharply reduced the viability of cells indicating a requirement for TRADD in maintaining stem cell cultures. Our findings are consistent with a previous study that revealed a role for NF-κB in the maintenance of glioma stem-like cultures [37].
In summary, cytoplasmic expression of TRADD is common in GBM and adversely influences prognosis, presumably by activation of NF-κB and conferring resistance to treatment. Experimental loss of function studies show an important role of TRADD in proliferation of GBM cells and in conferring resistance to temozolomide. In GBM stem cell populations, TRADD is commonly expressed and may be required for maintenance of GBM stem cells. Furthermore, we propose that TRADD overexpression is an important mechanism of NF-κB activation in GBM. Thus, TRADD plays an oncogenic role in GBM and may be an important biomarker as well as an attractive target for therapeutic intervention in this disease.
Supplementary Material
Footnotes
This work was supported in part by National Institutes of Health (NIH) grant R01NS062080 to A.A.H. and by R01 CA139217 to D.A.B. S.B. is supported by grants from the NIH (R01 CA149461), National Aeronautics and Space Administration (NNX13AI13G), and the Cancer Prevention and Research Institute of Texas (RP100644).
This article refers to supplementary material, which is designated by Table W1 and is available online at www.neoplasia.com.
References
- 1.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 2.Pobezinskaya YL, Liu Z. The role of TRADD in death receptor signaling. Cell Cycle. 2012;11:871–876. doi: 10.4161/cc.11.5.19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 4.Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, Huang HL, Pike KA, Hao Z, Su YW, et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc Natl Acad Sci USA. 2008;105:12429–12434. doi: 10.1073/pnas.0806585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 6.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 7.Morgan M, Thorburn J, Pandolfi PP, Thorburn A. Nuclear and cytoplasmic shuttling of TRADD induces apoptosis via different mechanisms. J Cell Biol. 2002;157:975–984. doi: 10.1083/jcb.200204039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chio II, Sasaki M, Ghazarian D, Moreno J, Done S, Ueda T, Inoue S, Chang YL, Chen NJ, Mak TW. TRADD contributes to tumour suppression by regulating ULF-dependent p19Arf ubiquitylation. Nat Cell Biol. 2012;14:625–633. doi: 10.1038/ncb2496. [DOI] [PubMed] [Google Scholar]
- 9.Wesemann DR, Qin H, Kokorina N, Benveniste EN. TRADD interacts with STAT1-α and influences interferon-γ signaling. Nat Immunol. 2004;5:199–207. doi: 10.1038/ni1025. [DOI] [PubMed] [Google Scholar]
- 10.Scheuerpflug CG, Lichter P, Debatin KM, Mincheva A. Assignment of TRADD to human chromosome band 16q22 by in situ hybridization. Cytogenet Cell Genet. 2001;92:347–348. doi: 10.1159/000056927. [DOI] [PubMed] [Google Scholar]
- 11.Surawicz TS, McCarthy BJ, Kupelian V, Jukich PJ, Bruner JM, Davis FG. Descriptive epidemiology of primary brain and CNS tumors: results from the Central Brain Tumor Registry of the United States, 1990–1994. Neuro Oncol. 1999;1:14–25. doi: 10.1093/neuonc/1.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reardon DA, Wen PY. Therapeutic advances in the treatment of glioblastoma: rationale and potential role of targeted agents. Oncologist. 2006;11:152–164. doi: 10.1634/theoncologist.11-2-152. [DOI] [PubMed] [Google Scholar]
- 13.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Mischel PS, Cloughesy TF, Nelson SF. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci. 2004;5:782–792. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 15.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network, author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karin M, Cao Y, Greten FR, Li ZW. NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 21.Pacifico F, Leonardi A. NF-κB in solid tumors. Biochem Pharmacol. 2006;72:1142–1152. doi: 10.1016/j.bcp.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 22.Schauer IG, Zhang J, Xing Z, Guo X, Mercado-Uribe I, Sood AK, Huang P, Liu J. Interleukin-1β promotes ovarian tumorigenesis through a p53/NF-κB-mediated inflammatory response in stromal fibroblasts. Neoplasia. 2013;15:409–420. doi: 10.1593/neo.121228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain G, Voogdt C, Tobias A, Spindler KD, Moller P, Cronauer MV, Marienfeld RB. IκB kinases modulate the activity of the androgen receptor in prostate carcinoma cell lines. Neoplasia. 2012;14:178–189. doi: 10.1593/neo.111444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomasova D, Mulay SR, Bruns H, Anders HJ. p53-independent roles of MDM2 in NF-κB signaling: implications for cancer therapy, wound healing, and autoimmune diseases. Neoplasia. 2012;14:1097–1101. doi: 10.1593/neo.121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S, Hatanpaa KJ, Xie Y, Mickey BE, Madden CJ, Raisanen JM, Ramnarain DB, Xiao G, Saha D, Boothman DA, et al. The receptor interacting protein 1 inhibits p53 induction through NF-κB activation and confers a worse prognosis in glioblastoma. Cancer Res. 2009;69:2809–2816. doi: 10.1158/0008-5472.CAN-08-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Carpenter G. Epidermal growth factor activation of NF-κB is mediated through IκBα degradation and intracellular free calcium. Oncogene. 1998;16:2095–2102. doi: 10.1038/sj.onc.1201731. [DOI] [PubMed] [Google Scholar]
- 28.Habib AA, Chatterjee S, Park SK, Ratan RR, Lefebvre S, Vartanian T. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-κB (NF-κB)-inducing kinase to activate NF-κB. Identification of a novel receptor-tyrosine kinase signalosome. J Biol Chem. 2001;276:8865–8874. doi: 10.1074/jbc.M008458200. [DOI] [PubMed] [Google Scholar]
- 29.Biswas DK, Cruz AP, Gansberger E, Pardee AB. Epidermal growth factor-induced nuclear factor κB activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci USA. 2000;97:8542–8547. doi: 10.1073/pnas.97.15.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor (EGFR) in glioma: signal transduction, neuropathology, imaging and radioresistance. Neoplasia. 2010;12:675–684. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyler CE, Rich JN. Looking in the miR-ror: TGF-β-mediated activation of NF-κB in glioma. J Clin Invest. 2012;122:3473–3475. doi: 10.1172/JCI66058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hjelmeland AB, Wu Q, Wickman S, Eyler C, Heddleston J, Shi Q, Lathia JD, Macswords J, Lee J, McLendon RE, et al. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010;8:e1000319. doi: 10.1371/journal.pbio.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S, Zhao D, Hatanpaa KJ, Mickey BE, Saha D, Boothman DA, Story MD, Wong ET, Burma S, Georgescu MM, et al. RIP1 activates PI3K-Akt via a dual mechanism involving NF-κB-mediated inhibition of the mTOR-S6K-IRS1 negative feedback loop and down-regulation of PTEN. Cancer Res. 2009;69:4107–4111. doi: 10.1158/0008-5472.CAN-09-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 35.Collett D. Modelling Survival Data in Medical Research. New York, NY: Chapman & Hall/CRC; 2003. [Google Scholar]
- 36.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Nogueira L, Ruiz-Ontanon P, Vazquez-Barquero A, Lafarga M, Berciano MT, Aldaz B, Grande L, Casafont I, Segura V, Robles EF, et al. Blockade of the NFκB pathway drives differentiating glioblastoma-initiating cells into senescence both in vitro and in vivo. Oncogene. 2011;30:3537–3548. doi: 10.1038/onc.2011.74. [DOI] [PubMed] [Google Scholar]
- 38.Albanese C, Wu K, D'Amico M, Jarrett C, Joyce D, Hughes J, Hulit J, Sakamaki T, Fu M, Ben-Ze'ev A, et al. IKKα regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell. 2003;14:585–599. doi: 10.1091/mbc.02-06-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y, Bonizzi G, Seagroves TN, Greten FR, Johnson R, Schmidt EV, Karin M. IKKα provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell. 2001;107:763–775. doi: 10.1016/s0092-8674(01)00599-2. [DOI] [PubMed] [Google Scholar]
- 40.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, et al. RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 41.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 43.Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM. IKKα controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 2006;25:3801–3812. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aggarwal BB. Nuclear factor-κB: the enemy within. Cancer Cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 46.Kirillova I, Chaisson M, Fausto N. Tumor necrosis factor induces DNA replication in hepatic cells through nuclear factor κB activation. Cell Growth Differ. 1999;10:819–828. [PubMed] [Google Scholar]
- 47.Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M. NF-κB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol Cell Biol. 1999;19:2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim DW, Gazourian L, Quadri SA, Romieu-Mourez R, Sherr DH, Sonenshein GE. The RelA NF-κB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene. 2000;19:5498–5506. doi: 10.1038/sj.onc.1203945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.