Abstract
In plants, genes involved in photosynthesis are encoded separately in nuclei and plastids, and tight cooperation between these two genomes is therefore required for the development of functional chloroplasts. Golden2-like (GLK) transcription factors are involved in chloroplast development, directly targeting photosynthesis-associated nuclear genes for up-regulation. Although overexpression of GLKs leads to chloroplast development in non-photosynthetic organs, the mechanisms of coordination between the nuclear gene expression influenced by GLKs and the photosynthetic processes inside chloroplasts are largely unknown. To elucidate the impact of GLK-induced expression of photosynthesis-associated nuclear genes on the construction of photosynthetic systems, chloroplast morphology and photosynthetic characteristics in greenish roots of Arabidopsis thaliana lines overexpressing GLKs were compared with those in wild-type roots and leaves. Overexpression of GLKs caused up-regulation of not only their direct targets but also non-target nuclear and plastid genes, leading to global induction of chloroplast biogenesis in the root. Large antennae relative to reaction centers were observed in wild-type roots and were further enhanced by GLK overexpression due to the increased expression of target genes associated with peripheral light-harvesting antennae. Photochemical efficiency was lower in the root chloroplasts than in leaf chloroplasts, suggesting that the imbalance in the photosynthetic machinery decreases the efficiency of light utilization in root chloroplasts. Despite the low photochemical efficiency, root photosynthesis contributed to carbon assimilation in Arabidopsis. Moreover, GLK overexpression increased CO2 fixation and promoted phototrophic performance of the root, showing the potential of root photosynthesis to improve effective carbon utilization in plants.
Keywords: Arabidopsis root, Chloroplast development, Construction of photosynthetic systems, GLK, Photosynthesis
Introduction
Photosynthetic electron transfer reactions in plants take place in thylakoid membranes inside chloroplasts, in which light energy drives electron transport between a series of multisubunit protein complexes including PSII and PSI. Core reaction centers of PSII and PSI are surrounded by peripheral light-harvesting complexes, LHCII and LHCI, respectively, that capture light energy and transfer it to Chls in the reaction centers. Electrons excited in PSII are transferred stepwise to the plastoquinone pool, Cyt b6f complex, plastocyanin and PSI, where another charge separation creates a strong reductant capable of reducing NADP+ (Rochaix 2011). Whereas photochemical reactions are essential to produce ATP and NADPH required for carbon fixation, excess light energy over the capacity for electron transport leads to the formation of harmful reactive oxygen species and eventually damages cells (Murchie and Niyogi 2011). Therefore, plants strictly regulate construction of the photosynthetic machinery and biogenesis of chloroplasts during photosynthetic organ development.
In plants, the genetic contribution to photosynthesis is shared between the nuclear and plastid genomes (Martin et al. 2002). Whereas genes for LHC proteins and pigment biosynthesis pathways reside in the nuclear genome, those encoding PS core subunits are mainly in the plastid genome. Therefore, chloroplast biogenesis depends on close cooperation between the nuclear and plastid genomes. We recently reported that nuclear-encoded photosynthesis genes form a tight co-expression network with key Chl biosynthetic genes, and suggested that there is a central transcriptional regulation system governing construction of Chl–protein complexes (Masuda and Fujita 2008, Kobayashi et al. 2012b). Golden2-like (GLK) transcription factors are proposed to be involved in the positive regulation of these photosynthesis-associated nuclear genes based on the evidence that Arabidopsis thaliana GLKs (GLK1 and GLK2) directly up-regulate many of those genes (Waters et al. 2009). Whereas loss of GLK activity leads to reduced Chl accumulation in photosynthetic tissues of Arabidopsis (Fitter et al. 2002), rice (Oryza sativa; Wang et al. 2013), tomato (Solanum lycopersicum; Powell et al. 2012) and Physcomitrella patens (Yasumura et al. 2005), increased expression of GLKs induces Chl accumulation and chloroplast biogenesis in non-foliar tissues such as Arabidopsis roots (Kobayashi et al. 2012a), rice calluses (Nakamura et al. 2009) and tomato fruits (Powell et al. 2012), suggesting that GLKs play a crucial role in chloroplast biogenesis during organ development.
Plant roots usually grow underground as heterotrophic organs and depend on aerial leaves for energy, although roots of some epiphytic plants turn green and perform active photosynthesis (Aschan and Pfanz 2003). In Arabidopsis roots, chloroplast development is essentially suppressed even under light conditions, and Chl accumulation is observed only in the upper part of the primary root near the hypocotyl junction (Kobayashi et al. 2012a). Recently we revealed that an auxin/cytokinin signaling pathway is involved in regulation of chloroplast development in the root through GLKs and another transcription factor, LONG HYPOCOTYL5 (HY5) (Kobayashi et al. 2012a). Consistent with the lack of Chl, the expression levels of GLK1 and GLK2 are very low in roots compared with those in leaves (Fitter et al. 2002). Overexpression of GLK genes results in remarkable accumulation of Chl in roots, reaching nearly 10% of the amounts of Chl observed in wild-type leaves (Kobayashi et al. 2012a). This suggests that overexpression of GLK genes derepresses chloroplast development in roots via transcriptional activation of photosynthesis-associated nuclear genes. However, given that GLKs are nuclear transcription factors and various essential proteins in the electron transport chain are encoded in the plastid genome, how the overall photosynthetic machinery is organized in the roots of the wild type and GLK overexpressors (GLKOX) is largely unknown.
In this study, we investigated chloroplast development and photosynthetic functions in the roots of wild-type and GLKOX Arabidopsis. Considering that photosynthesis in the roots contributes to the carbon economy in some species, our findings not only help to elucidate the global regulation of chloroplast biogenesis but also could lead to enhancement of crop photosynthesis.
Results
Chloroplast development in roots of GLK overexpressors
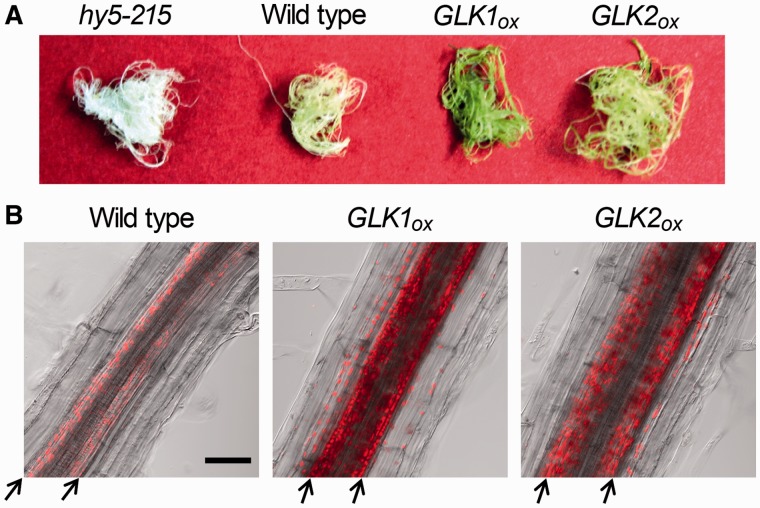
Consistent with our previous report that overexpression of GLK enhances Chl accumulation in roots (Kobayashi et al. 2012a), both GLK overexpressors, GLK1OX and GLK2OX, had visibly green roots compared with the yellowish roots of the wild type (Fig. 1A). A HY5-deficient mutant (hy5-215), which accumulates no Chl in roots (Usami et al. 2004, Kobayashi et al. 2012a), had albino roots. As reported previously (Kobayashi et al. 2012a), wild-type roots showed Chl accumulation only in the stele of the mature primary root (Fig. 1B). In both GLK1OX and GLK2OX roots, the stele of the primary root was the major site of Chl accumulation; however, Chl accumulation was also detected in the outer cell layers, endodermis, cortex and epidermis (Fig. 1B). Moreover, GLKOX plants accumulated Chl in lateral roots, where Chl accumulation was hardly detected in the wild type (Supplementary Fig. S1). These data suggest that overexpression of GLKs triggers ectopic development of chloroplasts in the root.
Fig. 1.
Chl accumulation in GLKOX roots. (A) Color phenotype of roots of 21-day-old plants. GLKOX plants have greenish roots compared with the yellowish wild-type and albino hy5-215 roots. (B) Confocal microscopy of Chl fluorescence in the primary root. Chl autofluorescence micrographs were merged with differential interference contrast images. Arrows indicate boundaries between the stele and endodermis. Bar = 50 µm.
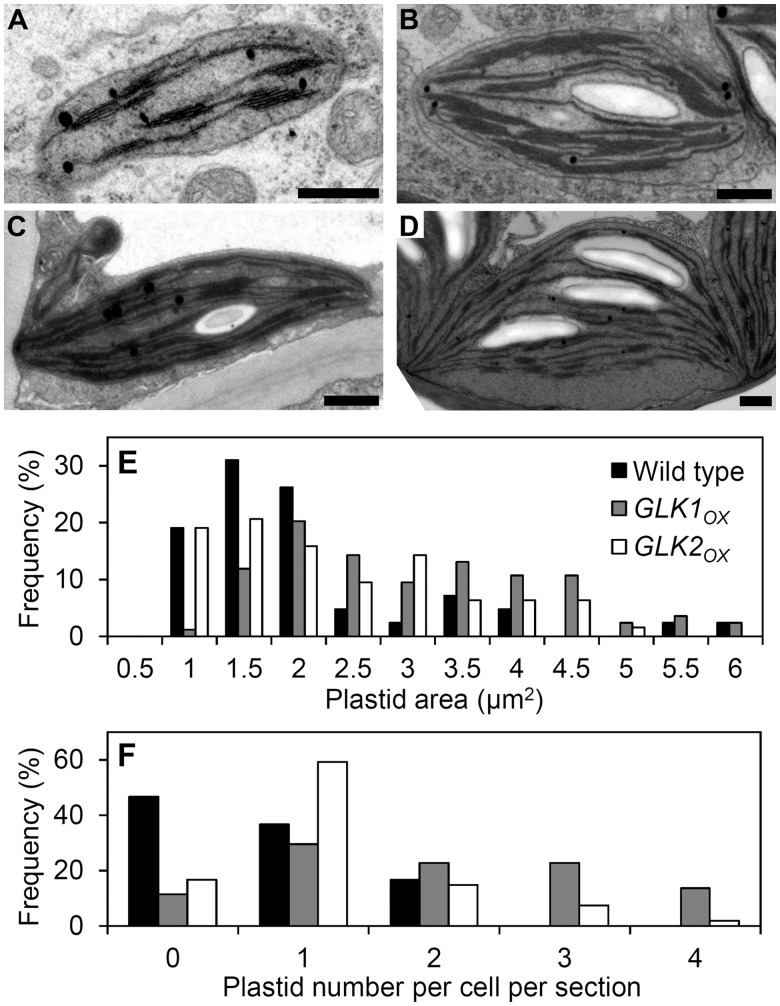
Next, we compared plastid morphology between wild-type and GLKOX plants by observing plastids of primary roots at approximately 4.0 cm from the root–hypocotyl junction with transmission electron microscopy (TEM) (Fig. 2). In the wild type, although the thylakoid membrane networks were poorly formed in the root chloroplasts as compared with those in the leaf chloroplasts, grana stacks were relatively well developed (Fig. 2A, D). The development of thylakoid membrane networks with a highly stacked grana structure was remarkably enhanced in root chloroplasts of both GLKOX lines (Fig. 2B, C). Particularly in the GLK1OX root chloroplasts, thylakoid membranes were extensively stacked and formed very thick and wide grana structures. In addition, chloroplasts of GLKOX roots accumulated starch grains, which were hardly detected in wild-type roots, indicating elevated photosynthetic activity of root chloroplasts in these overexpressors. We also found that chloroplasts in GLK1OX roots were larger than those in wild-type roots (Fig. 2E), presumably due to the pronounced development of thylakoid membrane structures. Furthermore, in GLK1OX and GLK2OX roots, the number of chloroplasts per cell was increased and cells containing multiple chloroplasts were observed more frequently than in wild-type roots (Fig. 2F). These results suggest that GLK overexpression not only enhances chloroplast development via activating biogenesis of thylakoid membranes but also induces chloroplast division in the root. It should be noted that the visible phenotype of root gravitropism which is regulated through amyloplast sedimentation was not affected in GLK1OX roots, suggesting that certain plastids retained their roles in these root tissues.
Fig. 2.
Plastid development in GLKOX roots. (A–C) Plastid ultrastructure in the primary root cells of wild-type (WT) (A), GLK1OX (B) and GLK2OX seedlings (C). (D) A typical chloroplast in a mature Arabidopsis leaf cell. Bars = 0.5 µm. (E) Histogram of plastid size in the outer cells of the stele determined from primary root cross-sections from WT and GLKOX seedlings (n = 42, 87 and 64 for the WT, GLK1OX, and GLK2OX, respectively). (F) Histogram of plastid number in the outer cells of the stele determined from primary root cross-sections (n = 60, 45, and 54 for the WT, GLK1OX, and GLK2OX, respectively).
Gene expression analysis in roots of GLK overexpressors
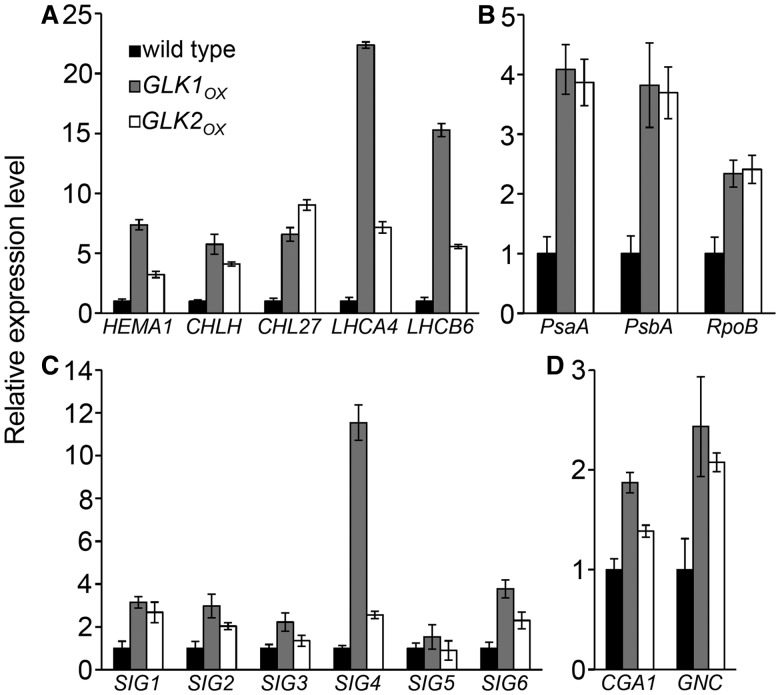
In aerial organs, GLK acts as a transcriptional activator by directly binding to the promoters of nuclear LHC and Chl biosynthetic genes (Waters et al. 2009). Consistent with this, quantitative reverse transcription–PCR (qRT–PCR) analysis revealed that these target genes were highly up-regulated in roots of both GLKOX lines (Fig. 3A). In general, transcripts responded more strongly to GLK1OX than to GLK2OX in roots, as expected from the relatively greener pigmentation in GLK1OX roots (Fig. 1). In GLK1OX, transcript levels of LHC genes (LHCA4 and LHCB6) were increased >10-fold, while those of Chl biosynthetic genes (HEMA1, CHLH and CHL27) were increased >5-fold (Fig. 3A).
Fig. 3.
Quantitative RT–PCR analysis of chloroplast biogenesis-associated gene expression in roots of wild-type and GLKOX seedlings. Expression levels of nuclear-encoded genes for Chl biosynthesis and light harvesting (A), plastid-encoded genes (B), nuclear-encoded genes for sigma factors (C) and GATA-type nuclear transcription factor genes (D). Data are presented as the fold difference from wild-type root samples after normalization to the reference gene ACTIN8. Values are the means ± SE from three independent experiments.
To examine whether GLK overexpression also affects plastid gene expression, we investigated the expression levels of PsaA, PsbA and RpoB in roots of both GLKOX lines (Fig. 3B). The PsaA and PsbA genes, which encode the core proteins of PSI and PSII, respectively, are transcribed by plastid-encoded RNA polymerase (PEP), whereas RpoB, which itself encodes the β-chain of PEP, is transcribed by nuclear-encoded plastid RNA polymerase (NEP) (De Santis-MacIossek et al. 1999). In both GLK1OX and GLK2OX roots, transcript levels of PsaA and PsbA increased 4-fold compared with those in the wild-type roots. The transcript abundance of RpoB also increased in both GLKOX lines, but was lower than those of PsaA and PsbA.
We then examined the expression of genes for sigma factors (SIGs), which are nuclear-encoded transcriptional initiation factors required for the binding of PEP to specific promoters of plastid genes (Fig. 3C). The Arabidopsis genome encodes six SIGs that are localized in plastids and activate subsets of plastid gene promoters in a partly redundant manner (Schweer et al. 2010). As observed for nuclear- and plastid-encoded photosynthetic genes, the expression of all SIG genes, except for SIG5, was up-regulated in both GLKOX lines. Among them, SIG4 in GLK1OX showed the most prominent up-regulation. Because two GATA transcription factors, GNC and CGA1, were recently reported to induce chloroplast development in non-photosynthetic tissues (Chiang et al. 2012), we also analyzed the expression of GNC and CGA1 in GLKOX roots (Fig. 3D). Although GNC and CGA1 have not been identified as direct targets of GLK factors, expression of these genes was increased in GLKOX roots. Our results suggest that GLK overexpression up-regulates not only their primary target genes involved in Chl biosynthesis and light harvesting but also GLK-non-targeted genes associated with chloroplast development in the root.
Pronounced accumulation of peripheral PS proteins in roots of GLK overexpressors
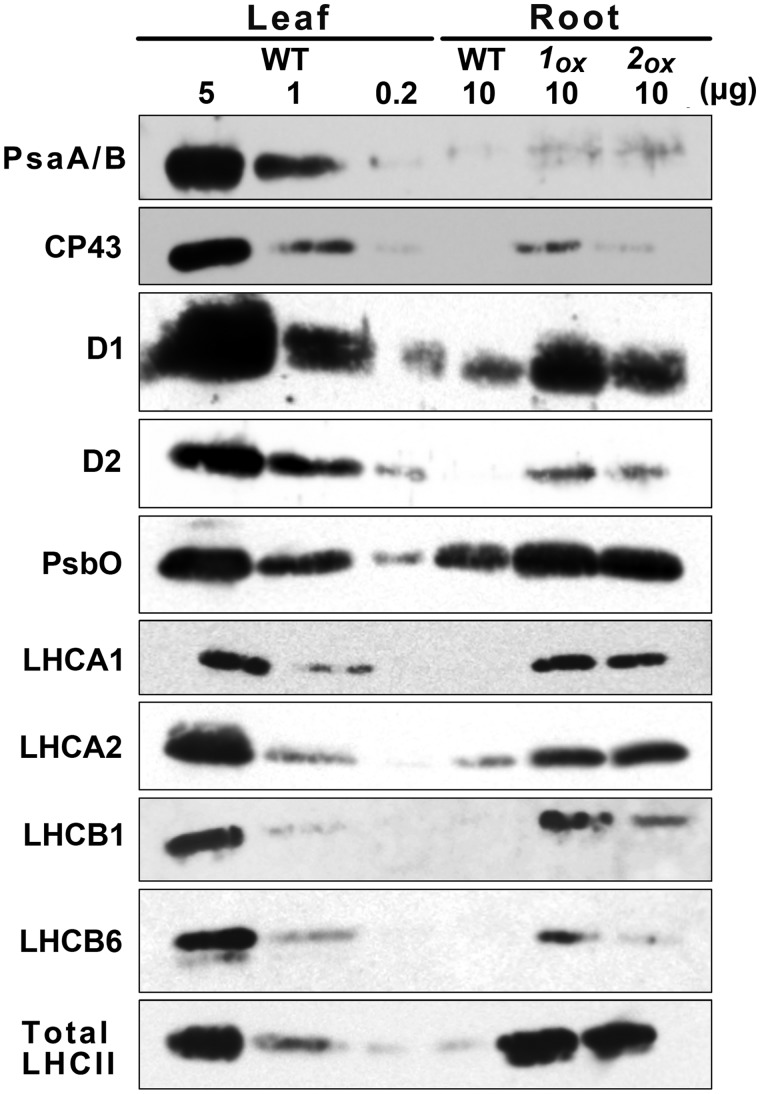
To clarify further the effects of GLK overexpression on chloroplast biogenesis in roots, we examined the levels of photosynthetic proteins in GLKOX roots by immunoblot analysis. A 10 µg aliquot of total membrane protein from roots of wild-type and GLKOX plants was analyzed together with a dilution series (0.2, 1 and 5 µg) of membrane proteins from wild-type leaves (Fig. 4). Since GLK1OX roots contain roughly 10% of the Chl present in wild-type leaves on a fresh weight basis (Kobayashi et al. 2012a), comparison between 10 µg of membrane protein from GLKOX roots and 1 µg from wild-type leaves is appropriate. As shown in Fig. 4, the levels of all membrane photosynthetic proteins were higher in GLKOX roots than in wild-type roots. On the other hand, differences in the protein levels were small between GLK1OX and GLK2OX roots, except in CP43 and LHCB6. Compared with the wild-type leaf samples containing 1 µg of protein, the levels of PsbO and LHC proteins (LHCA1, LHCA2, LHCB1, LHCB6 and total LHCII) in GLKOX root samples containing 10 µg of protein were higher, while those of D1, D2 and CP43 were comparable or lower. Moreover, the amount of PsaA/PsaB, the core reaction center proteins of PSI, was very low even in GLKOX roots. These results show that GLK overexpression induces the accumulation of nuclear-encoded peripheral photosynthetic proteins more strongly than that of plastid-encoded reaction center proteins in PSI and PSII.
Fig. 4.
Differential accumulation of membrane photosynthetic proteins in roots of GLK1OX and GLK2OX. Immunoblot analysis of photosynthetic proteins in 10 µg of total membrane protein from root samples of wild type (WT), GLK1OX (1OX) and GLK2OX (2OX) compared with those in a dilution series (5, 1 and 0.2 µg) of total membrane protein from WT leaves.
Pigment composition in roots of GLK overexpressors
To evaluate the balance among PSI, PSII and their antenna complexes in root chloroplasts, we compared pigment compositions in root samples with those in wild-type leaves (Table 1). As reported previously (Kobayashi et al. 2012a), Chl contents were increased in roots of both GLKOX lines, particularly GLK1OX, compared with those in wild-type roots. In parallel with the Chl accumulation, the amount of carotenoids was also substantially increased in both GLKOX lines. In all root samples, the proportions of Chl b and carotenoids, which are pigments in LHC antennae, relative to Chl a were higher than those in wild-type leaves (Table 1), confirming that there is a higher LHC antenna/reaction center ratio in root chloroplasts.
Table 1.
Pigment composition in roots of the wild type, GLK1OX and GLK2OX, and leaves of the wild type
| Chl a (nmol g−1 FW) | Chl b (nmol g−1 FW) | Chl a/b | Carotenoids |
||
|---|---|---|---|---|---|
| µg g−1 FW | µg µmol−1 Chl a | ||||
| Wild-type leaf | 2,252.7 ± 173.0 | 715.7 ± 59.5 | 3.16 ± 0.04 | 367.7 ± 29.6 | 163.4 ± 1.2 |
| Wild-type root | 19.5 ± 1.6 | 7.5 ± 0.6 | 2.67 ± 0.15 | 8.2 ± 0.6 | 423.6 ± 13.5 |
| GLK1OX root | 134.4 ± 7.9 | 53.9 ± 2.6 | 2.49 ± 0.03 | 58.3 ± 2.7 | 435.6 ± 6.3 |
| GLK2OX root | 62.4 ± 4.5 | 24.0 ± 1.3 | 2.60 ± 0.09 | 24.5 ± 1.1 | 396.1 ± 11.2 |
Values are means ± SE (n = 3 for wild-type leaves, 10 for wild-type roots and 6 for GLK1OX and GLK2OX roots).
To characterize further the PS–LHC complexes in GLK1OX roots, precise photosynthetic pigment analysis was performed by HPLC. In GLK1OX roots, the amount of xanthophylls relative to Chl a was higher than that in wild-type leaves, whereas the proportion of β-carotene was almost equivalent between these two samples (Table 2), consistent with the data showing that LHC antennae are actively formed in root chloroplasts (Fig. 4, Table 1). To estimate the PSI/PSII ratio, we examined the amount of Chl a′ and pheophytin (Phe) a, which are specific to PSI (Kobayashi et al. 1988, Jordan et al. 2001) and PSII (Klimov et al. 1977a, Klimov et al. 1977b, Zouni et al. 2001), respectively. The PSI/PSII ratio estimated from proportions of Chl a′ and Phe a to Chl a was lower in GLK1OX roots than in wild-type leaves, supporting the result obtained by immunoblot analyses that PSI is accumulated less in GLK1OX roots (Fig. 4).
Table 2.
Molar ratios of photosynthetic pigments in wild-type leaves and GLK1OX roots
| Chl a/Chl a′ | Chl a/Phe a | PSI/PSII | β-Carotene/Chl a | Total xanthophyll/Chl a | |
|---|---|---|---|---|---|
| Wild-type leaf | 546 ± 88 | 99 ± 19 | 0.37 ± 0.08 | 0.14 ± 0.04 | 0.68 ± 0.06 |
| GLK1OX root | 565 ± 110 | 70 ± 9 | 0.26 ± 0.06 | 0.12 ± 0.01 | 1.29 ± 0.10 |
Values are means ± SD from five independent samples.
PSs formed in root chloroplasts are different from those in leaf chloroplasts
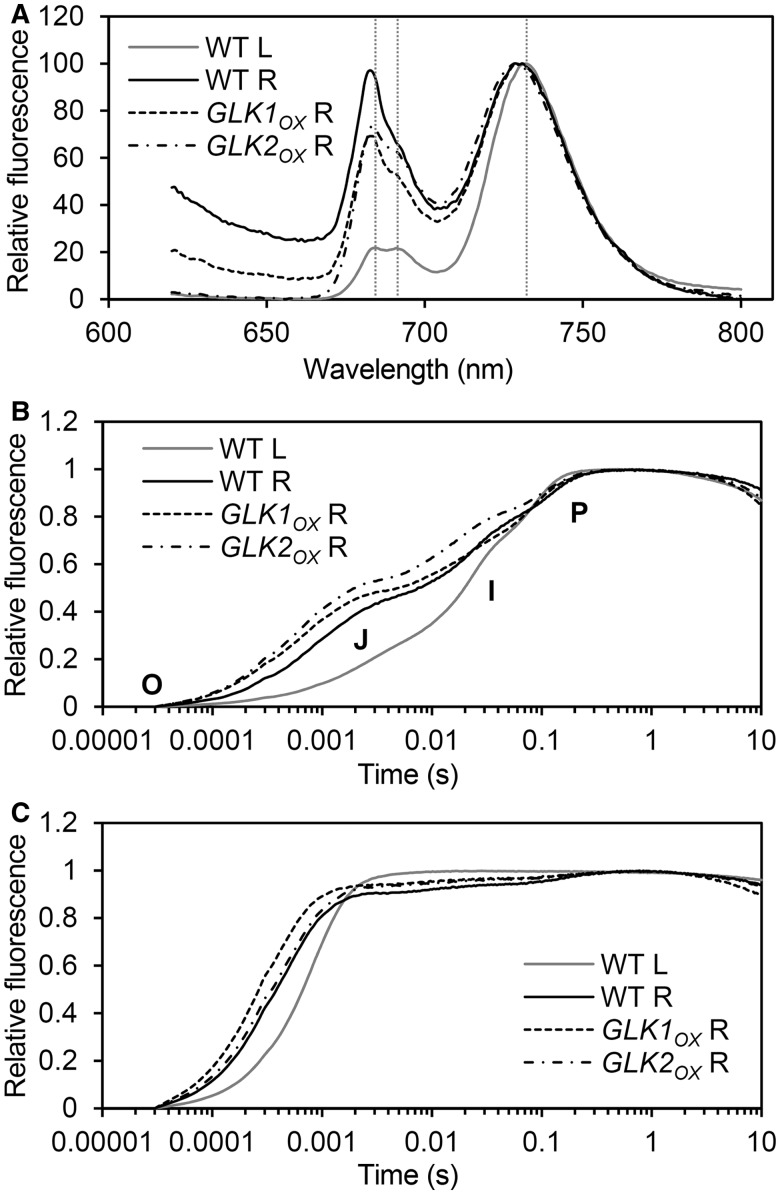
To examine the state of Chl–protein complexes in the roots of wild-type and GLKOX plants, Chl fluorescence spectra were measured at 77K (Fig. 5A). In the emission spectra of wild-type leaves, there were shoulder bands at 684 and 691 nm, which primarily originate from CP43 and CP47 in PSII, respectively (Govindjee 1995). In addition, there was an emission peak at 732 nm, which can be attributed to the PSI–LHCI complex. Since the peaks originating from the PSI–LHCI complex were also detected in all root samples, the spectra were normalized to the fluorescence emission maximum of PSI–LHCI (Fig. 5A). In all root samples, the two shoulder bands originating from PSII were present, but they were slightly shifted to 683 and 689 nm. This shift indicates the presence of LHCII complexes that are excitonically uncoupled from PSII (Hölzl et al. 2009). For PSI, the emission maximum of root samples was also shifted to lower wavelengths (729–731 nm), indicating the presence of LHCI proteins that were either weakly or not coupled to PSI reaction centers, an effect that is usually observed in plants suffering from a decrease in PSI reaction center (Stöckel et al. 2006).
Fig. 5.
Comparison of PS complexes in roots of the wild type and GLKOX lines with those in wild-type leaves. Characteristics of PS complexes were examined in samples from roots of wild type (WT R), GLK1OX (GLK1OX R) and GLK2OX (GLK2OX R) and from wild-type leaves (WT L). (A) Chl fluorescence emission spectra at 77K. Vertical dashed lines represent emission peaks at 684, 691 and 732 nm observed in WT L samples. Slight blue shifts in these peaks were observed in all root samples. Representative data from multiple independent experiments are shown (n > 3). (B and C) Transient fluorescence induction kinetics of Chl in the absence (B) or presence (C) of 40 µM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). Values are means from three independent experiments. Two inflections, labeled J and I, are observed between the levels O (origin) and P (peak) only in the absence of DCMU (B).
To evaluate the functionality of the photosynthetic apparatus in root chloroplasts of GLKOX plants, we analyzed the transient kinetics of Chl fluorescence using a logarithmic timing series (Fig. 5B). Wild-type leaves displayed a typical polyphasic fluorescence rise exhibiting the origin-inflection-intermediary peak–peak (O-J-I-P) transient (Govindjee 1995). Similar kinetics was observed in leaves of both GLKOX lines (Supplementary Fig. S2), demonstrating that GLK overexpression does not affect electron transport of PSII in leaves. The O-J-I-P transient was also observed in all root samples, indicative of functional electron transport in the root PSII. However, as reported previously (Kobayashi et al. 2012a), the O-J transition, which constitutes the photochemical phase of the Chl a fluorescence rise, occurred with a higher fluorescence yield in the wild-type roots than in the leaves, suggesting that electron transport from QA to QB is insufficient in the root chloroplasts. Moreover, such high Chl fluorescence at the O-J transition was also observed in roots of GLKOX plants, indicating that GLK overexpression does not improve the efficiency of photosynthetic electron transport in the root chloroplasts.
We next measured Chl fluorescence kinetics in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) (Fig. 5C). DCMU inhibits electron transfer from QA to QB, which results in a rapid reduction of total QA (Govindjee 1995). In wild-type roots, the fluorescence rise was much faster than that in leaves, indicating that QA reduction occurs more rapidly in root chloroplasts than in leaf chloroplasts. This phenomenon was more pronounced in the GLK1OX roots, consistent with the finding that GLK1OX enhances the formation of antennae relative to reaction centers in root chloroplasts.
Photosynthetic electron flow in root chloroplasts
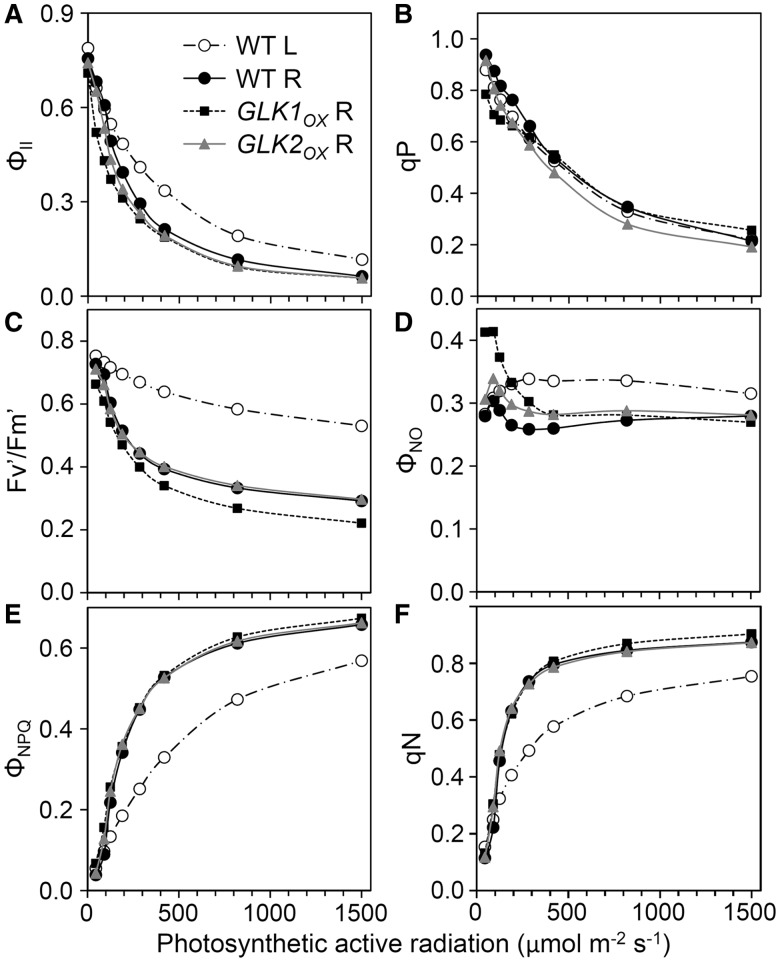
To evaluate photosynthetic activity in root chloroplasts, we analyzed Chl fluorescence using pulse amplitude modulation (PAM) techniques (Maxwell and Johnson 2000). First we determined the maximum quantum efficiency of PSII (Fv/Fm) in the dark (Fig. 6), which represents the intrinsic photosynthetic efficiency of PSII. As observed previously (Kobayashi et al. 2012a), Fv/Fm in dark-adapted roots of the wild type was slightly lower than that in leaves, suggesting a slight decrease in maximal efficiency of light utilization in the root PSII. In GLKOX plants, particularly GLK1OX, Fv/Fm levels were further decreased in roots, although those in leaves were unchanged. Next we analyzed the light–response curves of Chl fluorescence from PSII. In all root samples, the photochemical quantum yield of PSII (ΦII) was lower than that in wild-type leaves under middle to high photosynthetically active radiation (PAR) (Fig. 7A). In addition, GLK1OX roots had lower ΦII even under low PAR (Supplementary Fig. S3A). The ΦII can be viewed as a product of two components, the coefficient of photochemical quenching (qP) and the maximum quantum efficiency of open PSII (Fv′/Fm′). Based on the ‘puddle’ model (Kramer et al. 2004), qP represents the QA redox status and thus the openness of PSII. In all root samples, qP was not very different from that in the wild-type leaves (Fig. 7B), except in the GLKOX roots under low PAR (Supplementary Fig. S3B). When the fraction of open PSII was estimated using another coefficient (qL) on the basis of the ‘lake’ model (Kramer et al. 2004), PSII in the roots was found to be in an even more oxidized state than that in the leaves (Supplementary Fig. S3F). On the other hand, Fv′/Fm′ was substantially decreased in all root samples compared with that in wild-type leaves (Fig. 7C; Supplemental Fig. S3C). These data suggest that the low ΦII in roots resulted mainly from low PSII quantum efficiency and not PSII acceptor-side limitation.
Fig. 6.
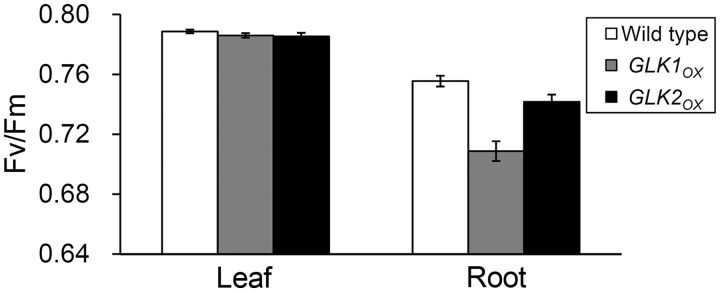
Maximum quantum yield of PSII (Fv/Fm). Fv/Fm levels were compared between leaves and roots from the wild type, GLK1OX and GLK2OX. Values are means ± SE (n = 5 for leaves and 7 for roots in each line).
Fig. 7.
Light–response curves of Chl fluorescence parameters. Wild-type leaves (WT L) and roots of the wild type (WT R), GLK1OX (GLK1OX R) and GLK2OX (GLK2OX R) were dark adapted for 5 min prior to the measurements and exposed for 3 min to each light intensity. (A) Effective quantum yield of PSII (ΦII). (B) Coefficient of photochemical quenching (qP), a measure of the redox state of the PSII acceptor side. (C) Maximum PSII quantum yield under light conditions (Fv′/Fm′). (D) Quantum yield of non-regulated energy dissipation (ΦNO). (E) Quantum yield of regulated energy dissipation (ΦNPQ). (F) Coefficient of non-photochemical quenching (qN). Data are means of multiple experiments (n = 5 for wild-type leaves and 7 for each root sample).
Absorbed light energy by LHCII–PSII can be divided into ΦII, quantum yield of light-induced non-photochemical quenching (ΦNPQ) and quantum yield of non-light-induced non-photochemical quenching (ΦNO) (Kramer et al. 2004). Whereas ΦNO levels were similar in all samples (Fig. 7D), except for GLK1OX roots under low PAR (Supplementary Fig. S3D), ΦNPQ levels in all root samples were higher than those in wild-type leaves (Fig. 7E; Supplementary Fig.S3E). The ΦNPQ component represents the fraction of regulated heat dissipation. In fact, in all root samples, the coefficient of non-photochemical quenching (qN) was also higher than that in wild-type leaves (Fig. 7F).
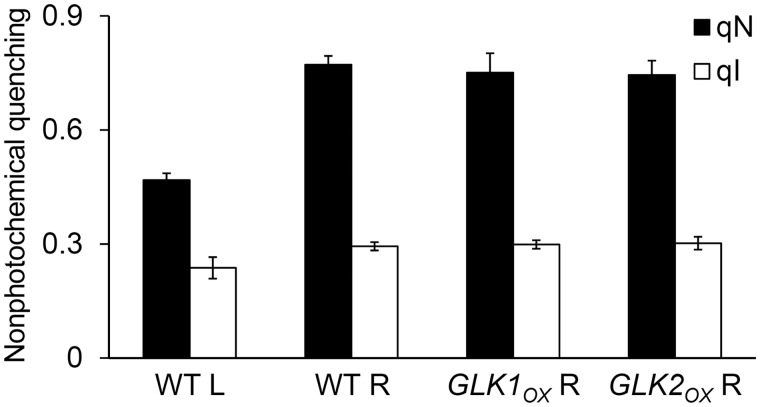
To explore this further, we evaluated the photoinhibitory component (qI) of qN, which is characterized by its very slow relaxation kinetics in the range of hours (Murchie and Niyogi 2011). Total qN was determined in plant samples after a 10 min exposure to actinic light (420 µmol photons m−2 s−1). After relaxation for 15 min in the dark, the remaining qN components, corresponding to qI, were determined. The contribution of qI to the total qN in root samples was similar to that in wild-type leaves (Fig. 8), suggesting that the rapidly reversible component related to the xanthophyll cycle is the main contributor to the increase in non-photochemical quenching in roots. In the leaves, there were no large differences in the light–response curves of Chl fluorescence between wild-type and GLKOX plants (Supplementary Fig. S4).
Fig. 8.
Contribution of photoinhibition to total non-photochemical quenching (qN). Wild-type leaves (WT L) and roots of the wild type (WT R), GLK1OX (GLK1OX R) and GLK2OX (GLK2OX R) were dark adapted for 15 min and then exposed to light stress (420 µmol photons m−2 s−1) for 10 min. First, total qN was measured at the end of the light stress. After relaxation of the rapidly reversible qN component with additional dark treatment for 15 min, the remaining qN component was determined as the photoinhibition-related qN component (qI). Values are the means ± SE from three independent experiments.
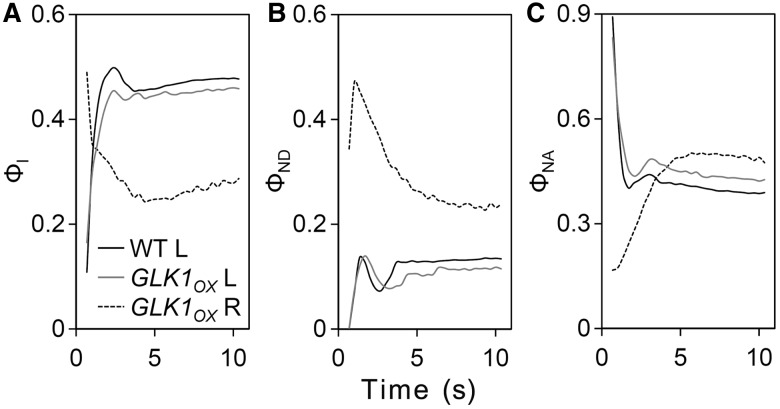
We also evaluated the redox state of PSI by analyzing changes in P700+ levels during moderate light (126 µmol photons m−2 s−1) exposure (Klughammer and Schreiber 2008). Although the P700 signal from wild-type roots was too low to evaluate, clear P700+ signal was detected in GLK1OX roots (Fig. 9) and used for comparison with those from leaves of the wild type and GLK1OX. In both wild-type and GLK1OX leaves, the photochemical quantum yield of PSI (ΦI), which represents the fraction of open P700, increased rapidly after light exposure and maintained high steady-state levels. In contrast, ΦI in GLK1OX roots decreased after illumination and remained at lower levels. We then analyzed the quantum yield of non-photochemical energy dissipation in PSI due to donor-side limitations (ΦND) or acceptor-side limitations (ΦNA). ΦND represents the fraction of P700+, whereas ΦNA represents the fraction of P700 that cannot be oxidized in a given state (Pfundel et al. 2008). In GLK1OX roots, ΦND levels stayed higher than those in wild-type and GLK1OX leaves. Meanwhile, steady-state levels of ΦNA were not markedly different among all samples, although inverse profiles of ΦNA were detected between GLK1OX roots and leaf samples during the first few seconds, as observed for ΦI. These data indicate that the efficiency of light utilization in PSI decreased in GLK1OX roots mainly due to donor-side limitation as compared with that in leaves.
Fig. 9.
Quantum yield of PSI. Slow induction kinetics of the PSI quantum yields in wild-type leaves (WT L), and GLK1OX leaves (GLK1OX L) and roots (GLK1OX R) were measured for 10 min under actinic light (126 µmol photons m−2 s−1). (A) Photochemical quantum yield of PSI (ΦI). (B and C) Non-photochemical quantum yield of energy dissipation due to PSI donor-side limitation (ΦND) (B) and PSI acceptor-side limitation (ΦNA) (C). Data are means of two independent measurements.
Photosynthetic activity and photoautotrophic growth of roots in GLK overexpressors
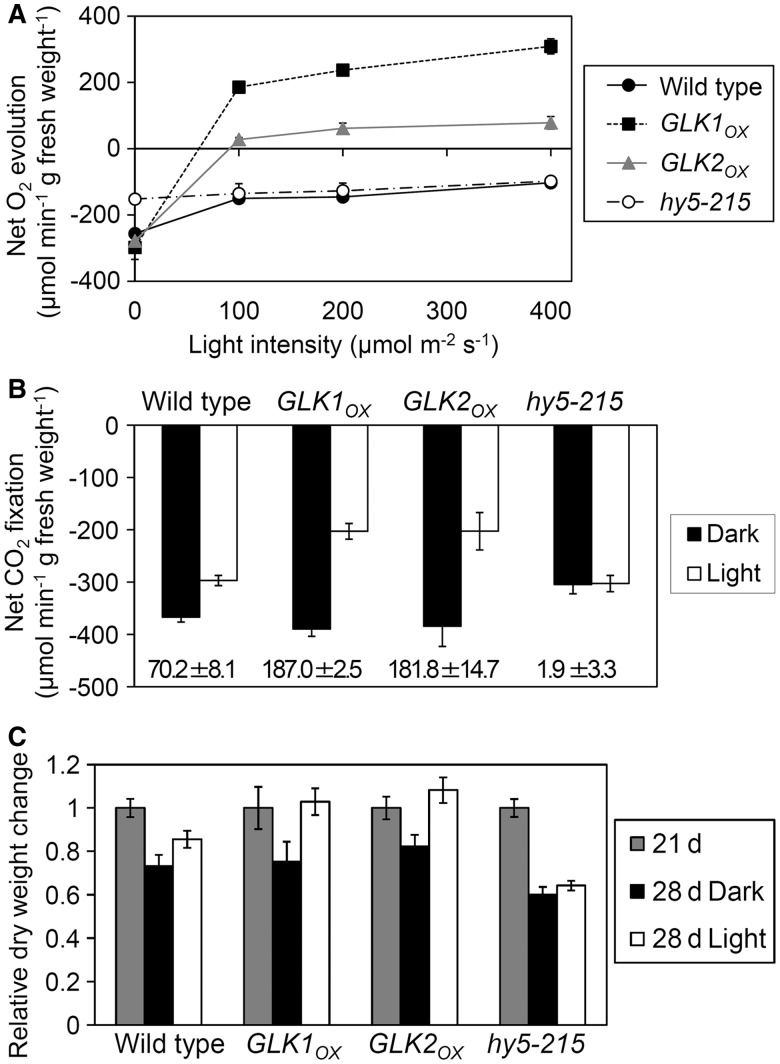
Finally, we evaluated the effects of GLK overexpression on overall photosynthetic performance in the root. For this experiment, roots were excised from 21-day-old seedlings and their photosynthetic activities were measured. Fig. 10A shows the light–response curve of net O2 evolution under a high CO2 (5%) condition. In both GLKOX lines, O2 evolution activity overcame the light compensation point at <100 µmol photons m−2 s−1, whereas wild-type roots did not achieve net O2 production even at the highest light condition (400 µmol photons m−2 s−1). Furthermore, roots of GLKOX plants exhibited light-dependent CO2 fixation activity under an atmospheric CO2 (0.039%) condition (Fig. 10B). Both GLKOX lines had much higher CO2 fixation activity than the wild type at 200 µmol photons m−2 s−1. We also tested the hy5-215 mutant, which cannot accumulate Chl in the root (Fig. 1A) (Usami et al. 2004, Kobayashi et al. 2012a), as a negative control. As expected, neither O2 evolution nor CO2 fixation activities were detected in the hy5-215 roots.
Fig. 10.
Photosynthetic activity of roots. (A) Light–response curve of net O2 evolution activity in roots. The zero level represents the light compensation point. (B) Net CO2 fixation activity in roots under dark or light (200 µmol photons m−2 s−1) conditions. Numbers in the graph indicate the gross CO2 fixation rate (µmol CO2 min−1 g FW−1) in each root sample calculated by subtracting the CO2 fixation rate in the dark from that in the light. (C) Change in dry weight of detached roots. Roots detached from 21-day-old seedlings were further incubated for 7 d without a carbon source under dark or light (60 µmol photons m−2 s−1) conditions. The dry weight of the 28-day-old detached roots was normalized to the initial weight of the 21-day-old root. Values are means ± SE from three independent experiments for A and B, and from seven independent experiments for C.
To investigate photoautotrophic growth activity, excised roots from 21-day-old seedlings were further incubated on Murashige and Skoog (MS) medium in the absence of any available carbohydrates for 7 d in the dark or light (60 µmol photons m−2 s−1). After a 7 d incubation, dry weights of the excised roots were compared with the initial root weights of 21-day-old seedlings (Fig. 10C). The change in dry weight after incubation without carbon sources represents the balance between photosynthesis and respiration activities. In the hy5-215 mutant, dry weight decreased to the same extent in dark- and light-treated roots, consistent with the absence of photosynthetic activity in hy5-215 roots (Fig. 10A, B). A decrease in dry weight compared with the initial weight was also observed in wild-type roots under both light and dark conditions, although light treatment slightly reduced the decrease. In both GLKOX lines, in contrast, dry weights of light-treated roots were much larger than those of dark-treated roots and comparable with the initial levels, suggesting that carbon fixation by photosynthesis entirely compensates for the loss due to the respiration in these roots. These results show that the roots of GLKOX plants have the ability to perform photosynthesis that can maintain their weights without sugar. It should be noted, however, that the growth of whole GLKOX plants was the same or less than that of the wild type, probably due to excessive Chl accumulation in leaves in these plants (Waters et al. 2008) which was not advantageous for regulated photosynthetic growth.
Discussion
Coordinated up-regulation of genes for chloroplast biogenesis in the nucleus and plastids
Consistent with the previous report that nuclear genes for enzymes involved in Chl biosynthesis and LHC proteins are direct targets of GLK factors in Arabidopsis (Waters et al. 2009), the expression of these genes was strongly up-regulated in GLKOX roots (Fig. 3A). In addition, genes that were not identified as direct targets of GLKs such as GNC, CGA1 and most genes for SIGs were also up-regulated in GLKOX roots (Fig. 3C, D). Although we cannot exclude the possibility that these genes are directly targeted by GLKs in the root, it is likely that the induction of the primary GLK targets secondarily influences the expression of GLK-non-targeted genes related to chloroplast biogenesis. Similar results were obtained in rice GLK1OX callus, in which counterparts of Arabidopsis GLK-non-targeted genes, such as most nuclear-encoded PS genes, were globally up-regulated (Nakamura et al. 2009). Such global up-regulation of chloroplast-related genes would contribute to chloroplast development in the GLKOX roots. As an example, GNC and CGA1, which were up-regulated in GLKOX roots, induce development and division of chloroplasts in the hypocotyl and the root (Chiang et al. 2012), consistent with our data in Figs. 1 and 2. Because GLKOX roots substantially accumulated the gene products of not only GLK direct targets (LHCA1, LHCB1 and LHCB6) but also non-direct targets (LHCA2 and PSBO) (Fig. 4), an intact peripheral LHC antenna system nearly equivalent to that in wild-type leaves could be constructed in GLKOX roots. Furthermore, carotenoids substantially accumulated in GLKOX roots together with Chl (Tables 1, 2), although the genes involved in carotenoid biosynthesis have not been identified as GLK targets (Waters et al. 2009). These results suggest a global influence of GLK-induced Chl–LHC antenna formation in roots.
In addition to the induction of photosynthesis-associated nuclear genes, the expression of PEP-dependent photosynthetic genes (PsaA and PsbA) was up-regulated in roots of both GLKOX lines (Fig. 3B). As proposed previously (Nakamura et al. 2009), it is likely that the increased gene expression of SIGs, which trigger PEP-dependent transcription (Schweer et al. 2010), leads to the up-regulation of the PEP-dependent genes in GLKOX roots. Moreover, expression of the NEP-dependent RpoB gene, which encodes the β-chain of PEP, was increased in GLKOX roots, highlighting the possibility that overall PEP activities are elevated in these roots. Our recent study suggests that thylakoid membrane biogenesis changes nucleoid morphology and globally activates plastid gene expression independently of photosynthesis (Kobayashi et al. 2013). Furthermore, one of the nuclear-encoded plastid RNA polymerases, RPOTmp, is reported to be tightly associated with thylakoid membranes (Azevedo et al. 2008). Thus, the thylakoid membrane biogenesis in GLKOX root plastids may also positively influence global plastid gene expression. In addition, the increased plastid number per cell in GLKOX roots (Fig. 2F) would contribute to the increased transcripts of plastid-encoded genes.
Reduced light utilization in the imbalanced PSs in root plastids
Protein analyses clearly revealed high abundance of LHC antenna complexes relative to the reaction centers in GLKOX roots (Fig. 4). A similar tendency was observed in wild-type roots although the amount of each photosynthetic protein was much lower than that in GLKOX roots. Moreover, low Chl a/b and high xanthophyll/Chl a ratios were detected not only in GLKOX roots but also in the wild-type roots (Table 1), suggesting that chloroplasts developed in the root have relatively large antenna complexes per reaction center. Consistent with these data, enhanced grana formation was observed in root chloroplasts (Fig. 2). In particular, GLK1OX root chloroplasts had substantially enlarged grana thylakoids (Fig. 2B). Based on the model that attractive forces between trimeric LHCs on closely appressed thylakoids cause membrane adhesion during grana formation (Standfuss et al. 2005), the excessive accumulation of LHC antenna systems would cause the hyperstacking of grana thylakoids in GLKOX root chloroplasts. In support of this idea, hyperstacked grana structures were also observed in the rice non-yellow coloring 1 mutant, in which LHC–Chl b complexes are selectively accumulated during senescence due to the loss of Chl b-degrading activity (Kusaba et al. 2007). Because QA reduction in the presence of DCMU occurred more rapidly in all root samples than in wild-type leaves (Fig. 5C), the imbalance between the antennae and the reaction centers in root chloroplasts may lead to excessive input of light energy from antenna complexes to the PSII reaction centers, which may result in the inefficient electron transport to the quinone pool in root chloroplasts (Fig. 5B).
The PAM analyses revealed that ΦII levels were considerably decreased in all root samples particularly at middle to high PAR (Fig. 7A). These changes accompanied remarkable increases in thermal dissipation of light energy (Fig. 7E), which occurs in the LHCII antenna (Murchie and Niyogi 2011). Thus, it is likely that the high antenna/reaction center ratio causes excess excitation of root PSII, thereby increasing thermal dissipation of light energy within the antenna and decreasing the PSII photochemical efficiency. This idea is supported by the substantial accumulation of carotenoids relative to Chl a in root chloroplasts (Table 1). Xanthophylls, in particular, showed a marked increase in GLK1OX roots (Table 2), consistent with the high degree of non-photochemical quenching observed in the roots. It should be noted that the intrinsic PSII efficiency represented by Fv/Fm was also lower in all root samples than in wild-type leaves (Fig. 6), which may be caused by the partial uncoupling of LHCII from the PSII reaction center (Fig. 5A), and/or the inefficient electron transport from QA to QB (Fig. 5B). The decrease in intrinsic PSII efficiency could be partly responsible for the decrease in the actual PSII efficiency under the light. In addition, GLK1OX roots showed a decrease in ΦII even under low PAR (Fig. 7A; Supplementary Fig. S3A). As represented by the lower qP (Fig. 7B; Supplementary Fig. S3B) and qL (Supplementary Fig. S3F), PSII in GLK1OX roots was in a more reduced state than in wild-type leaves and other root samples under low PAR, suggesting that electron transport downstream of PSII is retarded in GLK1OX root chloroplasts. In the GLK1OX root, ΦNO levels, which represent the proportion of non-regulated energy dissipation by heat and fluorescence (Kramer et al. 2004), increased under low PAR (Fig. 7D; Supplementary Fig. S3D). Because regulated thermal energy dissipation is less functional under low PAR (Fig. 7E; Supplementary Fig. S3E), the excess input of light energy from enlarged antenna systems into the reaction centers may cause over-reduction of PSII and increase the energy lost in a non-regulated manner in GLK1OX root chloroplasts.
Despite the similar up-regulation of PsaA and PsbA in GLKOX roots (Fig. 3B), accumulation of the PSI reaction center (PsaA/PsaB) was much lower than that of PSII reaction center proteins D1 (PsbA) and D2 (PsbD) and core antenna CP43 (PsbC) (Fig. 4). This result is confirmed by the pigment analyses in GLK1OX roots (Table 2). Considering the similar accumulation of LHCA and LHCB proteins in GLKOX roots, the reason that the PSI reaction center failed to accumulate is not clear. The assembly of PSI involves assembly-dependent regulation of biosynthesis of the major chloroplast-encoded subunits (Wostrikoff et al. 2004). In fact, several factors are involved in assembly, stability and regulation of PSI complexes (Ozawa et al. 2009). Thus, it is possible that such essential factors for PSI accumulation may be deficient in roots even under GLK overexpression. P700 measurements showed that PSI in GLK1OX roots is in a more oxidized state than in leaf chloroplasts due to donor-side limitations (Fig. 9), suggesting that LHCI–PSI in GLK1OX root chloroplasts functions effectively and electron transport to PSI is rather retarded. In GLK1OX roots, the low photochemical efficiency of PSII (Fig. 7A, C) may lead to the donor-side limitation in PSI even though PSII is more abundant than PSI (Fig. 4, Table 2). Alternatively, it is possible that the electron transport chain downstream of PSII, such as Cyt b6f complexes or plastocyanin, is impaired in the GLK1OX root as observed in the lower qP level (Supplementary Fig. S3B).
GLK factors improve phototrophic growth in roots
Although the overexpression of GLKs could not improve photochemical efficiency in root chloroplasts, total photosynthetic activities measured in terms of O2 evolution and CO2 fixation greatly increased in those roots (Fig. 10A, B). Furthermore, GLKOX roots showed the ability to perform photosynthesis that can maintain their weights without sugar (Fig. 10C), demonstrating that the mass production of photosynthetic machinery increases total CO2 fixation in those roots. It is interesting to note that wild-type roots also showed gross O2 evolution and CO2 fixation activities although the level was much lower than in GLKOX roots, demonstrating that root photosynthesis can contribute to carbon assimilation in Arabidopsis. Not only leaves but also other so-called non-photosynthetic organs such as stem, root, fruit and flower have the potential to perform photosynthetic CO2 fixation (Aschan and Pfanz 2003). With the exception of aerial roots of some orchid species, which can serve as primary photosynthetic organs, most green photosynthesizing roots are considered to contribute to internal CO2 recycling using respiratory-released CO2 (Aschan and Pfanz 2003). Because Arabidopsis roots, which lack stomata, accumulate Chls inside the stele (Kobayashi et al. 2012a), the recycling of internally respired CO2 may be the main role of photosynthesis there. As we reported previously, Chl accumulation in the wild-type Arabidopsis root occurs predominantly in the basal area near the hypocotyl junction and decreases toward the tip even when the light is equally irradiated to the whole root (Kobayashi et al. 2012a). Considering that only the upper parts of the root in the soil can receive light, the root base may have an inherent ability to develop chloroplasts and perform photosynthesis. As proposed in the case of photosynthesis in seeds and fruits (Kalachanis and Manetas 2010, Tschiersch et al. 2011), it is possible that the high antenna/reaction center ratio in root chloroplasts is of advantage in the low-light environments of roots growing in the soil.
In this study, we demonstrated that induction of Chl biosynthesis and chloroplast development by GLK overexpression increases photosynthesis in roots. Further increases in photosynthetic activity of roots achieved by improving the balance between antennae and reaction centers and the photochemical efficiency could promote overall biomass production in plants.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana wild type (Columbia), hy5-215 (Oyama et al. 1997), GLK1OX and GLK2OX (Waters et al. 2008) were grown vertically on solid medium containing 1× MS plant salt mixture (Wako), 1% (w/v) sucrose and 0.8% (w/v) gelrite (Wako), pH 5.7 at 23°C under continuous white light (60 µmol photons m−2 s−1) for 21 d unless stated otherwise.
Microscopic analyses
For Fig. 1B, primary roots at approximately 4.0 cm from the root–hypocotyl junction were examined using a confocal laser scanning microscope (LSM700; Carl Zeiss). Chl autofluorescence between 660 and 700 nm was detected under 488 nm laser excitation and merged with differential interference contrast images.
For ultrastructure analysis of root plastids, primary roots at approximately 4.0 cm from the root–hypocotyl junction were analyzed by TEM according to Toyooka et al. (2000) with modification (Kobayashi et al. 2012a). For quantification of the number and size of plastids, high-resolution TEM images prepared as described above were merged in a whole cross-section image of the stele of the primary root. All plastids observed in the outer three cell layers of the cross-section were counted and used for area quantification with ImageJ software (National Institutes of Health).
qRT–PCR analysis
Total RNA was extracted from roots of 21-day-old seedlings using the RNeasy Plant Mini kit (Qiagen). Genomic DNA digestion and reverse transcription were performed using the PrimeScript RT reagent Kit with gDNA Eraser (TAKARA BIO INC.) according to the manufacturer’s instructions. cDNA amplification was performed using the Thunderbird PreMix kit (Toyobo) and 200 nM gene-specific primers, listed in Supplementary Table S1. Thermal cycling consisted of an initial denaturation step at 95°C for 10 s, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. Signal detection and quantification were performed in duplicate using MiniOpticon (Bio-Rad). The relative abundance of all transcripts amplified was normalized to the constitutive expression level of ACTIN8 (Pfaffl 2001). Three independent biological experiments were performed for each root sample.
Immunoblot analysis
The membrane protein fraction was prepared from roots or leaves of 21-day-old seedlings as described previously (Kobayashi et al. 2013). A 10 µg aliquot of total membrane protein from each root sample was electrophoresed together with a dilution series (0.2–5 µg) of total membrane protein from wild-type leaves and electrotransferred onto PROTRAN nitrocellulose membranes (Schleicher & Schuell) as described (Kobayashi et al. 2007). Protein bands that reacted with primary antibodies were secondarily labeled with goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase (Thermo Scientific) and detected using a chemiluminescence reagent (ImmunoStar LD, Wako) and an imager (ImageQuant LAS 4000 mini, GE Healthcare). Antibodies against PsaA/PsaB and total LHCII were kindly provided by R. Tanaka, Hokkaido University, Sapporo, Japan, and those against D1 and D2 were kindly provided by M. Ikeuchi, The University of Tokyo, Tokyo, Japan. Antibodies against PsbO, CP43, LHCA1, LHCA2, LHCB1 and LHCB6 were from AgriSera.
Spectroscopic pigment determination
Plant tissues crushed in liquid nitrogen were homogenized in 80% acetone, and debris was removed by centrifugation at 10,000×g for 5 min. The absorbance of the supernatant at 720, 663, 647, 645 and 470 nm was measured with an Ultrospec 2100 pro spectrophotometer (GE Healthcare Biosciences). The Chl (a and b) and carotenoid contents of the samples were determined according to Melis et al. (1987) and Lichtenthaler (1987), respectively.
HPLC pigment analysis
The leaf or root tissue was ground in a glass mortar for 1 min with approximately 30 ml of anhydrous Na2HPO4 (–4°C) as a desiccant. The ground material was transferred to a glass beaker, to which approximately 20 ml of chloroform (–20°C) was added, and the mixture was sonicated for 2 min at 4°C. The extract was then filtered and dried under a vacuum. The green solid material obtained by the above procedure was immediately redissolved in approximately 20 µl of chloroform. For carotene, Phe a, Chl a′ and Chl a analyses, a 3–5 µl aliquot of the extract was injected into a silica HPLC column (YMC-pak SIL, 250×4.6 mm i.d.) cooled to approximately 4°C in an ice–water bath. The sample was eluted isocratically with degassed hexane/2-propanol/methanol (100/0.7/0.2, v/v/v) at a flow rate of 0.9 ml min−1, and was monitored with a JASCO UV-970 detector (λ = 425 nm) and a JASCO Multiwavelength MD-915 detector (λ = 300–800 nm) in series. For xanthophyll and Chl a analyses, an aliquot of 3–5 µl of extract was injected onto a reversed-phase HPLC column (Kaseisorb LC ODS 2000-3, 250×4.6 mm i.d.) cooled to 4°C in an ice–water bath. The pigments were eluted isocratically with degassed water/ethanol/methanol/2-propanol (3/86/13/1, v/v/v/v) at a flow rate of 0.3 ml min−1, and monitored with a JASCO UV-2070 detector (λ = 425 nm) and a SHIMADZU Multiwavelength SPD-M10A VP photodiode array detector (λ = 300–800 nm) in series.
Chl fluorescence measurement
Fluorescence emission spectra of Chl proteins at 77K were obtained directly from plant tissues in liquid nitrogen using a spectrofluorometer under 435 nm excitation (RF-5300PC, Shimadzu).
For Chl fluorescence induction experiments (Fig. 5B, C), leaves or 1 cm segments of primary roots from the hypocotyl junction were excised and dark-incubated for 5 min before initiation of the experiments. Five segments of primary roots were used for an experiment in a batch. When required, the tissues were infiltrated with 40 µM DCMU and 150 mM sorbitol by depression before dark incubation. Chl fluorescence transients were measured in a logarithmic time series between 30 µs and 10 s after the onset of strong actinic light (1,650 µmol photons m−2 s−1) with an LED pump-probe spectrometer (JTS-10, BioLogic).
Photochemical efficiency analysis was performed using a PAM fluorometer (Junior-PAM, Walz) at room temperature. Plants were pre-incubated under dim light (∼5 µmol photons m−2 s−1) for >30 min before experiments. Then, single leaves or batches of 1 cm segments of the primary roots from the hypotocyl junction were dark-incubated for 5 min on the leaf clip with the MS medium. After measuring minimum Chl fluorescence (Fo) in the dark, maximal Chl fluorescence (Fm) was determined with a saturating pulse. After actinic light treatment for 3 min, stationary fluorescence (F) and maximum fluorescence under the actinic light (Fm′) were determined followed by the measurement of minimal fluorescence of illuminated samples (Fo′) after far-red light treatment. From these fluorescence yields, photosynthetic parameters were calculated according to the following equations (Van Kooten and Snel 1990, Maxwell and Johnson 2000): Fv/Fm = (Fm – Fo)/Fm, Fv′/Fm′ = (Fm′ – Fo′)/Fm′, ΦII = (Fm′ – F)/Fm′, qP = (Fm′ – F)/(Fm′ – Fo′), qN = 1 – (Fm′– Fo′)/(Fm – Fo). The actual PSII efficiency ΦII can be transformed into a product of the PSII openness (qP) and the quantum efficiency of open PSII (Fv′/Fm′): ΦII = (Fm′ – F)/Fm′ = qP×Fv′/Fm′. The qL, ΦNPQ and ΦNO were determined according to the method of Kramer et al. (2004).
P700 absorbance measurement
The redox state of P700 was determined in a batch of leaves or roots from 21-day-old seedlings by measuring the absorbance change at 830–875 nm reference beams using Dual-PAM-100 (Waltz) at room temperature (Klughammer and Schreiber 2008). The measurements were performed using the automated induction and recovery program provided by the Dual-PAM software (Pfundel et al. 2008). The maximal P700 signal (Pm) was determined by application of a saturating pulse (15,000 µmol photons m−2 s−1) after far-red pre-illumination. Then actinic light (126 µmol photons m−2 s−1) was supplied for 10 min and saturating pulses were given every 20 s to determine the maximum P700 signal under the actinic light (Pm′). Each saturating pulse was followed by a 1 s dark interval to determine the minimum level of the P700+ signal (Po). Quantum yields in PSI were calculated according to Pfundel et al. (2008) although the correct interpretation of these parameters remains debatable.
Oxygen evolution and CO2 fixation analysis
For O2 evolution and CO2 fixation analysis, roots (approximately 0.2 g FW) excised from 21-day-old seedlings were used. The O2 evolution rate was determined with a Clark-type oxygen electrode (LD2, Hansatech Instruments). Roots wetted with liquid MS medium were incubated in an air chamber with 5% CO2 at 23°C. Changes in O2 concentration in the chamber were monitored with the electrode under several light conditions (0–400 µmol photons m−2 s−1).
The CO2 fixation rate was measured with a portable infrared gas analyzer (LI-6400, LI-COR) and a 6400-17 whole plant Arabidopsis chamber. During the measurement, temperature and CO2 concentration in the chamber were kept at 23°C and 390 µl CO2 l−1, respectively.
Dry weight measurement
Roots detached from 21-day-old seedlings at the root–hypocotyl junction were collected to determine the initial weight or further incubated for 7 d on vertical MS medium without sucrose under continuous light (60 µmol photons m−2 s−1) or in the dark. Collected samples were dehydrated completely at 70°C for 48 h with dry silica gel and immediately weighed to avoid water absorption from the air.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Education, Sports, Science and Culture in Japan [Grants-in-Aid for Scientific Research on Priority Areas (Nos. 24570042, 22370016 and 24770055) and the Global Center of Excellence Program (K03)]; the Japan Society for the Promotion of Science [research fellowships of for young scientists] and a RIKEN post-doctoral fellowship (to K.K.); the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation [grant GBMF3070 to K.K.N.].
Supplementary Material
Acknowledgments
We thank Mayumi Wakazaki (RIKEN, Yokohama, Japan) for performing electron microscopy and Chika Ikeda (RIKEN, Yokohama, Japan) for her technical assistance.
Glossary
Abbreviations
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- Fv/Fm
maximum quantum efficiency of PSII
- Fv′/Fm′
maximum quantum efficiency of open PSII
- GLK
golden2-like
- GLKox
GLK overexpressor
- HY5
long hypocotyl 5
- LHC
light-harvesting complex
- MS
Murashige and Skoog
- NEP
nuclear-encoded plastid RNA polymerase
- O-J-I-P
origin-inflection-intermediary peak–peak
- PAM
pulse amplitude modulation
- PAR
photosynthetically active radiation
- PEP
plastid-encoded RNA polymerase
- Phe
pheophytin
- qI
photoinhibitory component of qN
- qL
coefficient of open PSII on the basis of the ‘lake’ model
- qN
coefficient of non-photochemical quenching
- qP
coefficient of photochemical quenching
- qRT–PCR
quantitative reverse transcription–PCR
- SIGs
sigma factors
- TEM
transmission electron microscopy
- ΦI
photochemical quantum yield of PSI
- ΦII
quantum yield of PSII
- ΦNA
quantum yield of non-photochemical energy dissipation in PSI due to acceptor-side limitations
- ΦND
quantum yield of non-photochemical energy dissipation in PSI due to donor-side limitations
- ΦNO
quantum yield of non-light induced non-photochemical quenching
- ΦNPQ
quantum yield of light-induced non-photochemical quenching
Disclosures
The authors have no conflicts of interest to declare.
References
- Aschan G, Pfanz H. Non-foliar photosynthesis—a strategy of additional carbon acquisition. Flora. 2003;198:81–97. [Google Scholar]
- Azevedo J, Courtois F, Hakimi MA, Demarsy E, Lagrange T, Alcaraz JP, et al. Intraplastidial trafficking of a phage-type RNA polymerase is mediated by a thylakoid RING-H2 protein. Proc. Natl Acad. Sci. USA. 2008;105:9123–9128. doi: 10.1073/pnas.0800909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012;160:332–348. doi: 10.1104/pp.112.198705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, et al. Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J. 1999;18:477–489. doi: 10.1046/j.1365-313x.1999.00473.x. [DOI] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Govindjee Sixty-three years since Kautsky: chlorophyll a fluorescence. Aust. J. Plant Physiol. 1995;22:131–160. [Google Scholar]
- Hölzl G, Witt S, Gaude N, Melzer M, Schöttler MA, Dörmann P. The role of diglycosyl lipids in photosynthesis and membrane lipid homeostasis in Arabidopsis. Plant Physiol. 2009;150:1147–1159. doi: 10.1104/pp.109.139758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauß N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- Kalachanis D, Manetas Y. Analysis of fast chlorophyll fluorescence rise (O-K-J-I-P) curves in green fruits indicates electron flow limitations at the donor side of PSII and the acceptor sides of both photosystems. Physiol. Plant. 2010;139:313–323. doi: 10.1111/j.1399-3054.2010.01362.x. [DOI] [PubMed] [Google Scholar]
- Klimov VV, Allkhverdiev SI, Demeter S, Krasnovsky AA. Photoreduction of pheophytin in photosystem 2 of chloroplasts with respect to the redox potential of the medium. Dokl. Akad. Nauk SSSR. 1977a;249:227–230. [Google Scholar]
- Klimov VV, Klevanik AV, Shuvalov VA, Krasnovsky AA. Reduction of pheophytin in the primary light reaction of photosystem II. FEBS Lett. 1977b;82:183–186. doi: 10.1016/0014-5793(77)80580-2. [DOI] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U. Saturation pulse method for assessment of energy conversion in PS I. PAM Appl. Notes. 2008;1:11–14. [Google Scholar]
- Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keränen M, et al. Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell. 2012a;24:1081–1095. doi: 10.1105/tpc.111.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl Acad. Sci. USA. 2007;104:17216–17221. doi: 10.1073/pnas.0704680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Narise T, Sonoike K, Hashimoto H, Sato N, Kondo M, et al. Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 2013;73:250–261. doi: 10.1111/tpj.12028. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Obayashi T, Masuda T. Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal Behav. 2012b;7:922–926. doi: 10.4161/psb.20760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Watanabe T, Nakazato M, Ikegami I, Hiyama T, Matsunaga T, et al. Chlorophyll a′/P700 and pheophytin a/P680 stoichiometries in higher plants and cyanobacteria determined by HPLC analysis. Biochim. Biophys. Acta. 1988;936:81–89. [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth. Res. 2004;79:209–218. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell. 2007;19:1362–1375. doi: 10.1105/tpc.106.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophyll and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:349–382. [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Fujita Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008;7:1131–1149. doi: 10.1039/b807210h. [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence—a practical guide. J. Exp. Bot. 2000;51:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- Melis A, Spangfort M, Andersson B. Light-absorption and electron transport balance between photosystem-II and photosystem-I in spinach chloroplasts. Photochem. Photobiol. 1987;45:129–136. [Google Scholar]
- Murchie EH, Niyogi KK. Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 2011;155:86–92. doi: 10.1104/pp.110.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Muramatsu M, Hakata M, Ueno O, Nagamura Y, Hirochika H, et al. Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol. 2009;50:1933–1949. doi: 10.1093/pcp/pcp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S, Nield J, Terao A, Stauber EJ, Hippler M, Koike H, et al. Biochemical and structural studies of the large Ycf4–photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell. 2009;21:2424–2442. doi: 10.1105/tpc.108.063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfundel E, Klughammer C, Schreiber U. Monitoring the effects of reduced PS II antenna size on quantum yields of photosystems I and II using the Dual-PAM-100 measuring system. PAM Appl. Notes. 2008;1:21–24. [Google Scholar]
- Powell AL, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science. 2012;336:1711–1715. doi: 10.1126/science.1222218. [DOI] [PubMed] [Google Scholar]
- Rochaix JD. Assembly of the photosynthetic apparatus. Plant Physiol. 2011;155:1493–1500. doi: 10.1104/pp.110.169839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G. Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription—recent lessons from Arabidopsis thaliana. Eur. J. Cell Biol. 2010;89:940–946. doi: 10.1016/j.ejcb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Standfuss J, Terwisscha van Scheltinga AC, Lamborghini M, Kühlbrandt W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 2005;24:919–928. doi: 10.1038/sj.emboj.7600585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J, Bennewitz S, Hein P, Oelmüller R. The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 2006;141:870–878. doi: 10.1104/pp.106.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch H, Borisjuk L, Rutten T, Rolletschek H. Gradients of seed photosynthesis and its role for oxygen balancing. Biosystems. 2011;103:302–308. doi: 10.1016/j.biosystems.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A. Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol. 2004;45:1798–1808. doi: 10.1093/pcp/pch205. [DOI] [PubMed] [Google Scholar]
- Van Kooten O, Snel JFH. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 1990;25:147–150. doi: 10.1007/BF00033156. [DOI] [PubMed] [Google Scholar]
- Wang P, Fouracre J, Kelly S, Karki S, Gowik U, Aubry S, et al. Evolution of GOLDEN2-LIKE gene function in C(3) and C (4) plants. Planta. 2013;237:481–495. doi: 10.1007/s00425-012-1754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008;56:432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Girard-Bascou J, Wollman FA, Choquet Y. Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 2004;23:2696–2705. doi: 10.1038/sj.emboj.7600266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell. 2005;17:1894–1907. doi: 10.1105/tpc.105.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauß N, Saenger W, et al. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.