Abstract
We have integrated and analyzed a large number of data sets from a variety of genomic assays using a novel computational pipeline to provide a global view of estrogen receptor 1 (ESR1; a.k.a. ERα) enhancers in MCF-7 human breast cancer cells. Using this approach, we have defined a class of primary transcripts (eRNAs) that are transcribed uni- or bidirectionally from estrogen receptor binding sites (ERBSs) with an average transcription unit length of ∼3–5 kb. The majority are up-regulated by short treatments with estradiol (i.e., 10, 25, or 40 min) with kinetics that precede or match the induction of the target genes. The production of eRNAs at ERBSs is strongly correlated with the enrichment of a number of genomic features that are associated with enhancers (e.g., H3K4me1, H3K27ac, EP300/CREBBP, RNA polymerase II, open chromatin architecture), as well as enhancer looping to target gene promoters. In the absence of eRNA production, strong enrichment of these features is not observed, even though ESR1 binding is evident. We find that flavopiridol, a CDK9 inhibitor that blocks transcription elongation, inhibits eRNA production but does not affect other molecular indicators of enhancer activity, suggesting that eRNA production occurs after the assembly of active enhancers. Finally, we show that an enhancer transcription “signature” based on GRO-seq data can be used for de novo enhancer prediction across cell types. Together, our studies shed new light on the activity of ESR1 at its enhancer sites and provide new insights about enhancer function.
Enhancers are genomic regulatory elements that (1) carry sequence information for transcription factor binding, (2) may be located far from TSSs, (3) regulate gene expression regardless of location and orientation, and (4) play key roles in controlling tissue-specific gene expression (Bulger and Groudine 2011; Ong and Corces 2011). Current models posit that enhancers function by promoting communication with target gene promoters through chromatin loops or by tracking of enhancer-bound transcription factors through intervening chromatin to target gene promoters (Bulger and Groudine 2011; Ong and Corces 2011; Kolovos et al. 2012). Recent studies have focused intense interest on the properties of enhancers, beyond the binding of sequence-specific transcription factors, which might give clues to their mechanisms of action and aid in their identification. In this regard, histone modifications (e.g., H3 lysine 4 monomethyl, H3K4me1; H3 lysine 27 acetyl, H3K27ac), histone variants (e.g., H2A.Z), coactivators (e.g., EP300, CREBBP, Mediator), and an open chromatin architecture (e.g., DNase I hypersensitivity) have been identified as genomic features that mark or identify enhancers (Melgar et al. 2011; Natoli and Andrau 2012). Differential association of these features with enhancers in a given cell may define distinct classes of enhancers that specify distinct gene regulatory mechanisms and biological outcomes (Creyghton et al. 2010; Ghisletti et al. 2010; Rada-Iglesias et al. 2011; Wang et al. 2011; Zentner et al. 2011; Pham et al. 2012; Rada-Iglesias et al. 2012; Shen et al. 2012; Vahedi et al. 2012; Whyte et al. 2012; Xu et al. 2012; Ostuni et al. 2013). Enhancer profiles may even provide useful clinical signatures for cancer diagnosis and prognosis (Akhtar-Zaidi et al. 2012; Ross-Innes et al. 2012).
More recently, several studies have shown that many enhancers overlap with sites of RNA Pol II loading, active RNA Pol II transcription, and the production of enhancer RNAs (“eRNAs”) (De Santa et al. 2010; Kim et al. 2010; Hah et al. 2011; Wang et al. 2011; Djebali et al. 2012). A common signature of enhancer transcription is the production of short (i.e., ∼1 to 2 kb) eRNAs that are transcribed bidirectionally (Kim et al. 2010). We and others have recently shown that the genomic binding sites for the estrogen receptor (ESR1) and other steroid hormone receptors overlap with sites of transcription (Hah et al. 2011; Wang et al. 2011). The role of transcription in enhancer function is unknown, but the act of transcription may help to create an open chromatin environment that promotes enhancer function (Natoli and Andrau 2012). Alternatively, the stable accumulation of eRNAs may play a functional, perhaps even structural, role and may facilitate gene looping (Orom et al. 2010; Orom and Shiekhattar 2011; Natoli and Andrau 2012; Lai et al. 2013; Melo et al. 2013).
In the studies described herein, we used Global Run-On Sequencing (GRO-seq), a method that assays the location and orientation of all active RNA polymerases genome-wide (Core et al. 2008), to generate a global profile of active transcription at ESR1 binding sites (ERBSs) in MCF-7 human breast cancer cells in response to a short time course of E2 treatment. We integrated the data from our GRO-seq assays with data from a variety of other genomic assays (e.g., ChIP-seq, DNase-seq, ChIA-PET) using a novel computational pipeline to provide a comprehensive and global view of ESR1 enhancers and their regulation by E2 in MCF-7 cells. Together, our studies have shed new light on the activity of ESR1 at its enhancer sites and provide new insights about enhancer function in general, including the potential roles of enhancer transcription.
Results
ESR1 enhancers are sites of estrogen-induced transcription
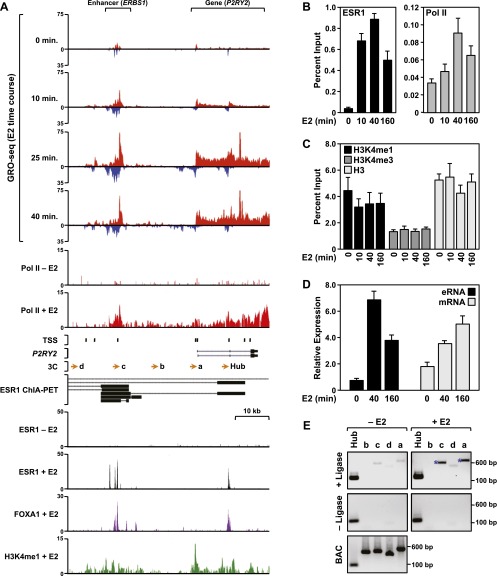
In a previous study using GRO-seq to characterize the estrogen-regulated transcriptome in MCF-7 cells, we identified hundreds of transcribed regions in the genome generating primary transcripts that overlap estrogen receptor binding sites (ERBSs) (Hah et al. 2011). In this study, we have undertaken a comprehensive identification and analysis of ESR1 enhancer transcription in MCF-7 human breast cancer cells by integrating a wide array of genomic data sets with locus-specific molecular analyses. Multiple examples of transcribed ESR1 enhancers located upstream of estrogen-regulated target genes are shown in browser track representation in Figure 1A and Supplemental Figures 1 and 2. These include an ERBS, which we refer to as ERBS1, located ∼20 kb upstream of the promoter of the P2RY2 gene, as well as these additional enhancer/gene pairs: ERBS2/GREB1, ERBS3/SBNO2, ERBS4/SMAD7, and ERBS5/PGR. We also include as a negative control an ERBS (ERBS6) that does not produce enhancer transcripts. As shown in the GRO-seq browser tracks in Figure 1A, transcription of the P2RY2 gene and a region around ERBS1 is up-regulated rapidly in a short time course of treatment with 17β-estradiol. The transcripts from ERBS1 (Fig. 1A), as well as ERBSs 2–5 (Supplemental Fig. 2), are produced bidirectionally from both strands of DNA, reminiscent of the enhancer RNAs (eRNAs) described previously (Kim et al. 2010), and the transcribed regions are associated with RNA Pol II and previously identified transcription start sites (TSSs) (Yamashita et al. 2011).
Figure 1.
The ESR1 enhancer of the estrogen-responsive P2RY2 gene produces bidirectional transcripts in MCF-7 cells. (A) Browser tracks of GRO-seq, ChIP-seq (Pol II, ESR1, FOXA1, and H3K4me1), ChIA-PET, TSS locations, and gene annotation for P2RY2 and its distal ESR1 binding site (ERBS1). The data are from MCF-7 cells treated with a time course of E2 (GRO-seq) or a single time point of E2 (45 or 60 min). TSSs identified previously based on a published data set from MCF-7 cells (Yamashita et al. 2011) are located as indicated. (Orange arrows) The locations of primers used for 3C assays. The black bars shown for the ChIA-PET data indicate the “head” and “tail” making contact in the gene loops, which are indicated by the dotted black lines. Scale bars show the length of the indicated region. A more detailed set of genomic data for P2RY2, as well as data for additional enhancer/gene pairs, can be found in Supplemental Figures 1 and 2. (B,C) ChIP-qPCR analyses showing recruitment of ESR1 and Pol II (B) or levels of H3K4me1, me3, and H3 (C) at ERBS1 in response to a time course of E2 treatment. Each bar represents the mean + the SEM for three or more independent biological replicates. (D) RT-qPCR analyses showing the expression of ERBS1 eRNA and P2RY2 mRNA in response to a time course of E2 treatment. Each bar represents the mean + the SEM for three or more independent biological replicates. (E) 3C-PCR assay showing E2-induced looping between ERBS1 and the P2RY2 gene. The lowercase letters correspond to the primers denoted by orange arrows shown in panel A. The assays were conducted in the presence (experimental) or absence (control) of DNA ligase, as indicated. Digested and ligated bacterial artificial chromosome (BAC) DNA spanning the entire P2YR2 locus was used as a PCR control. The size of the PCR fragments in base pairs is shown. One representative experiment from three conducted is shown.
As expected, these ERBSs are also associated with previously characterized enhancer features, including the pioneer transcription factor FOXA1, histone H3 lysine 4 monomethylation (H3K4me1), the histone acetyltransferases EP300 and CREBBP (a.k.a. p300 and CBP), p160 steroid receptor coactivator proteins (NCOA1, 2, and 3), and an open chromatin architecture as defined by DNase-seq and formaldehyde-assisted isolation of regulatory elements (FAIRE)–seq (Fig. 1A; Supplemental Fig. 2). Like the ERBS eRNAs, many of these enhancer features are induced by treatment with E2. These ERBSs are also involved in chromatin looping events that promote physical interactions with their target genes, as defined by chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) (Fig. 1A; Supplemental Fig. 2; Fullwood et al. 2009). Locus-specific molecular assays, including ChIP-qPCR, RT-qPCR, and 3C-PCR, confirm the localization of RNA Pol II, H3K4me1, and H3K27ac at these ERBSs (Fig. 1B,C; Supplemental Figs. 3A,B, 4A–D, 5), as well as the steady-state production of the eRNAs (Fig. 1D; Supplemental Figs. 3C, 6A–E) and enhancer looping events (Fig. 1E; Supplemental Fig. 3D,E) as the genes are induced by E2. Interestingly, an ERBS not associated with eRNA production (ERBS6) lacks many of the enhancer features described above (Supplemental Figs. 1, 2, 4D, 5, 6F). Collectively, these genomic and locus-specific analyses illustrate clearly the range of features associated with ESR1 enhancers, including the production of eRNAs.
Global identification and characterization of estrogen-regulated ESR1 enhancer transcripts
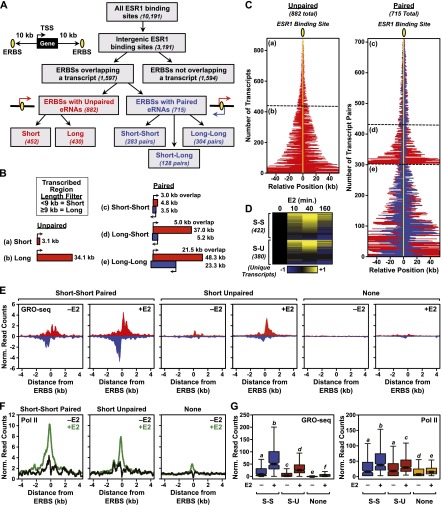
To obtain a global view of ESR1 enhancer transcripts, we developed a computational pipeline to identify eRNAs overlapping with ERBSs using GRO-seq data (Fig. 2A). Starting from a set of all ERBSs (∼10,000) defined previously (Welboren et al. 2009), we narrowed the list to those that are intergenic (i.e., >10 kb away from the beginning or end of an annotated RefSeq gene; ∼3000), which allowed us to avoid ambiguities caused by eRNAs overlapping annotated transcribed regions. We then separated them into those that have (∼1600) and those that do not have (∼1600) an overlapping transcript, as defined by GRO-seq. Next, we classified those with overlapping transcripts based on the production of transcripts from both strands of DNA (“Paired”; 715) or from one strand of DNA (“Unpaired”; 882). Finally, we applied a length filter for the transcription unit/primary transcript, defining those <9 kb as “short” and those ≥9 kb as “long” to exclude those that might overlap with annotated promoters (Fig. 2B,C). For the purposes of the remaining studies shown herein, we focused on two classes of enhancer transcripts: (1) short unpaired (S-U) and (2) short–short paired (S-S), which we call eRNAs. Many of the transcripts in the remaining classes (i.e., long unpaired, long–short paired, and long–long paired) do not resemble eRNAs as previously described (Kim et al. 2010) and are likely to represent other types of noncoding RNAs.
Figure 2.
Genome-wide identification of ESR1 enhancer transcripts in MCF-7 cells using GRO-seq. (A) Flowchart of ERBS classification in MCF-7 cells based on genomic location, eRNA production, and length of the transcribed region based on ChIP-seq and GRO-seq. (B) Schematics of average transcribed regions overlapping ERBSs in MCF-7 cells in five classes: (a) short unpaired, (b) long unpaired, (c) short–short paired, (d) long–short paired, and (e) long–long paired. “Short” and “Long” indicate a transcribed region <9 kb or ≥9 kb, respectively. (Red and blue boxes) Transcription from opposite strands. (C) Graphical representation of the positions and orientations of eRNAs (indicated by red and blue lines) relative to ESR1 binding sites (indicated by yellow oval and line) for unpaired and paired eRNAs in MCF-7 cells. The position relative to the ERBS is indicated in kilobases. (a–e) Correspond to the categories shown in panel B. (Red and blue lines) Transcription from opposite strands. (D) Heat map showing the expression of E2-regulated short unpaired (S-U) and short–short paired (S-S) eRNAs over a time course of E2 treatment in MCF-7 cells based on GRO-seq data. The data were median centered and scaled to the 0-min time point. Yellow and blue indicate up-regulated and down-regulated transcripts, respectively. Only unique transcripts are shown (i.e., those transcripts that overlap more than one ERBS are represented once). (E) Metaplot analyses of GRO-seq reads surrounding ERBSs associated with short–short paired transcripts, short unpaired transcripts, or no transcripts in MCF-7 cells ± E2 treatment. (F) Metaplot analyses of Pol II ChIP-seq reads surrounding ERBSs associated with short–short paired transcripts, short unpaired transcripts, or no transcripts in MCF-7 cells ± E2 treatment. (G) Box plot representation of GRO-seq and Pol II ChIP-seq reads associated with short–short paired transcripts (S-S), short unpaired transcripts (S-U), or no transcripts in MCF-7 cells ± E2 treatment.
Production of the short unpaired and short–short paired ESR1 enhancer transcripts is regulated by E2 over a short time course of treatment (0, 10, 40, and 160 min) (Fig. 2D). About 70% of the transcripts are E2 up-regulated, with maximum effects for most up-regulated transcripts occurring at 40 min and for most down-regulated transcripts occurring at 160 min (Fig. 2D). The up-regulation is evident in metaplots of the GRO-seq data (Fig. 2E) and corresponds to the levels of RNA Pol II at the ERBS, as expected (Fig. 2F,G). For the paired eRNAs, the extent of transcription from each strand is highly correlated with the other (Pearson's R = 0.7470408, P-value < 2.2 × 10−16) (Supplemental Fig. 7), suggesting a common regulatory mechanism. Unbiased motif analyses indicate that ERBS with or without transcripts are highly enriched for predicted estrogen response elements and share some other motifs in common as well (e.g., SP1), which may function as tethering factors for ESR1 (Supplemental Fig. 8).
The production of eRNAs from ERBSs positively correlates with a wide variety of enhancer features
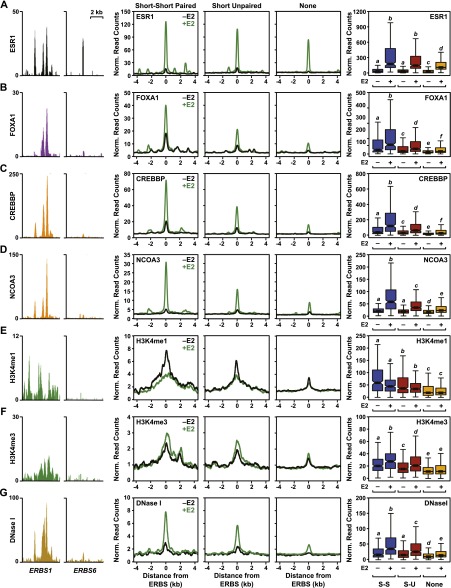
To better understand how the production of eRNAs from ERBSs may relate to enhancer function, we mined a large number of existing genomic data sets from MCF-7 cells (see Supplemental Table 1). Although all three classes of ERBSs that we examined (i.e., those with S-S paired eRNAs, short unpaired eRNAs, and no eRNAs) have similar mean and median levels of ESR1 binding by ChIP-seq (less than twofold difference) (Fig. 3A), considerable differences were observed among these groups with respect to other enhancer properties. For example, ERBSs producing short bidirectional eRNAs (i.e., S-S paired transcripts) have significantly higher mean and median levels of pioneer factors (e.g., FOXA1; TFAP2C, a.k.a. AP2γ), ESR1 coregulators (e.g., CREBBP, EP300, NCOA1, NCOA2, and NCOA3), and enhancer histone modifications (e.g., H3K4me1), as well as the most accessible chromatin structures (defined by DNase-seq and FAIRE-seq), than ERBS producing no transcripts (Wilcoxon rank-sum test, P-value < 0.001) (Fig. 3; Supplemental Fig. 9). Moderate differences in the levels of ESR1 binding across the three classes of ERBSs cannot account for the differences in transcript production, because ERBS without transcripts have disproportionately lower levels of GRO-seq signals than ERBS with transcripts (Supplemental Fig. 10A). Furthermore, ERBSs selected for similar levels of ESR1 binding show significant differences in the enrichment of enhancer features (Wilcoxon rank-sum test, P-value < 0.001) (Supplemental Fig. 10B). Together, these data indicate that the production of eRNAs at ERBS correlates with properties that are generally associated with active enhancers.
Figure 3.
The production of eRNAs from ERBSs positively correlates with the recruitment of coactivators, the levels of histone modifications, and the chromatin state in MCF-7 cells. Browser tracks, metaplots, and boxplots showing a positive correlation between eRNA production at ERBS with known markers of enhancer function. (Left two panels) Browser track representations of coactivator or histone modification ChIP-seq data, or DNase-seq data, as indicated on the y-axis for ERBS1 and ERBS6. (Middle three panels) Metaplot analyses of ChIP-seq or DNase-seq read counts for sets of ERBSs with short–short paired, short unpaired, or no transcripts in the presence (green line) or absence (black line) of E2 treatment. (Right panel) Box plot representations of ChIP-seq or DNase-seq data for sets of ERBSs with short–short paired (blue boxes), short unpaired (maroon boxes), or no transcripts (orange boxes) in the presence (+) or absence (−) of E2 treatment. (A) ESR1 ChIP-seq. (B) FOXA1 ChIP-seq. (C) CREBBP ChIP-seq. (D) NCOA3 ChIP-seq. (E) H3K4me1 ChIP-seq. (F) H3K4me3 ChIP-seq. (G) DNase-seq.
The production of eRNAs from ERBSs positively correlates with enhancer looping to target genes
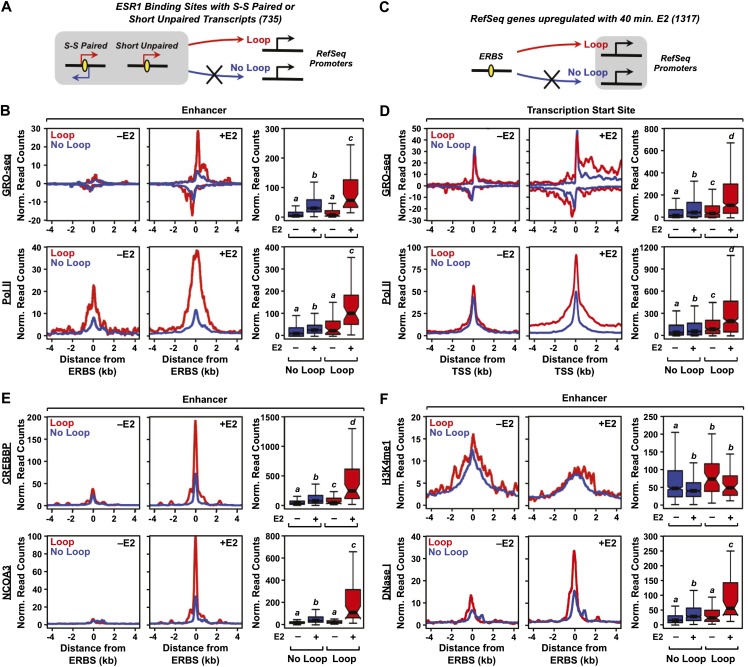
Models of enhancer function implicate looping to target gene promoters as a key mechanistic step controlling gene expression. Indeed, ESR1 enhancers participate in extensive looping interactions (Fullwood et al. 2009), and estrogen-dependent looping from distal ERBSs correlates with estrogen-dependent target gene activation (Fig. 1D,E; Carroll et al. 2005; Pan et al. 2008). To determine how the production of eRNAs from ERBSs might relate to chromatin looping, we mined an existing ESR1 ChIA-PET data set (Fullwood et al. 2009), which maps ESR1-dependent looping on a genome-wide basis, for looping events between an intergenic ERBS and a target gene promoter. Specifically, we assayed for loops originating within a 2-kb window around the peak center of ERBSs with transcripts (i.e., either S-S paired or S-U) and looping to a 10-kb window around the TSSs of target genes (Fig. 4A). We then integrated the looping information with enhancer transcription data from our GRO-seq analyses, producing metaplots of GRO-seq reads for ERBS with or without loops. We observed that ∼40% of ERBSs with an eRNA are involved in looping events versus ∼20% of ERBSs without an eRNA. This represents a twofold enrichment (P < 0.0001), indicating a greater likelihood of looping when the enhancer is transcribed.
Figure 4.
The production of eRNAs from ERBSs positively correlates with enhancer looping to target genes in MCF-7 cells. (A,C) Schematics of the looping analyses based on ESR1 ChIA-PET data. ERBSs (enhancers) were queried to determine if they loop to the promoters of RefSeq genes, based on ESR1 ChIA-PET data. Looping was assayed within a 2-kb (±1 kb) window around ESR1 peak centers and a 10-kb (±5 kb) window around the TSSs of target genes. (B) Metaplots and boxplots for GRO-seq (top) and Pol II ChIP-seq (bottom) data for ERBSs that either loop to (Loop, red lines) or do not loop to (No Loop, blue lines) target gene promoters. (D) Metaplots and boxplots for GRO-seq (top) and Pol II ChIP-seq (bottom) data for target gene promoters that are either looped to (Loop, red lines) or are not looped to (No Loop, blue lines) ERBSs. (E) Metaplots and boxplots for CREBBP ChIP-seq (top) and NCOA3 ChIP-seq (bottom) data for ERBSs that either loop to (Loop, red lines) or do not loop to (No Loop, blue lines) target gene promoters. (F) Metaplots and boxplots for H3K4me1 ChIP-seq ChIP-seq (top) and DNase-seq (bottom) data for ERBSs that either loop to (Loop, red lines) or do not loop to (No Loop, blue lines) target gene promoters.
We also observed that ERBSs looping to target gene promoters have significantly greater mean and median levels of enhancer transcription and RNA Pol II loading than those ERBSs that do not loop to a promoter (Wilcoxon rank-sum test, P-value < 0.001) (Fig. 4B; Supplemental Fig. 11A). In addition, we assayed ∼1300 E2-up-regulated target genes and found that those that are looped to from an intergenic ERBS have elevated levels of transcription, as defined by GRO-seq, compared with those that are not looped to from an intergenic ERBS (Fig. 4C,D; Supplemental Fig. 12). Further integration with additional genomic data indicates that E2-dependent production of eRNAs from ERBSs also positively correlates in a significant manner with coregulator recruitment and an open chromatin structure, but not the levels of histone methylation, at the enhancers (Wilcoxon rank-sum test, P-value < 0.001) (Fig. 4E,F; Supplemental Fig. 11B–G). RAD21, a component of the cohesin complex, which is thought to facilitate gene looping (Kagey et al. 2010), is enriched at ERBSs that loop to target genes (Supplemental Fig. 13A). siRNA-mediated knockdown of RAD21 results in an approximately twofold to threefold increase in basal eRNA production, as well as a modest (∼25%–50%) reduction in E2-induced eRNA production at four ERBSs that we examined in gene-specific assays (Supplemental Fig. 13B,C). These results suggest a functional link between E2-dependent production of eRNAs at ERBSs, enhancer looping, and target gene activation.
Inhibition of eRNA production by flavopiridol does not inhibit enhancer complex assembly or looping to target gene promoters
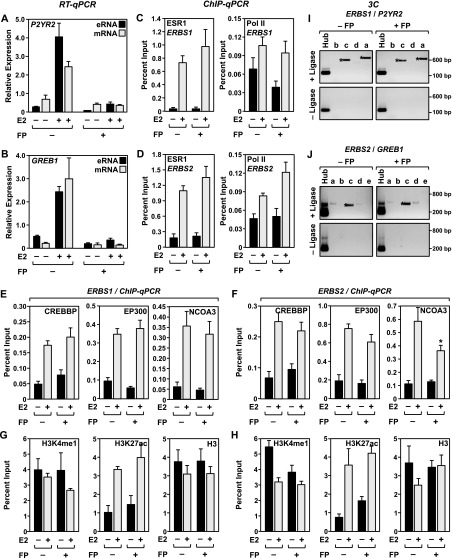
Our results have shown that the production of eRNAs from ERBSs correlates with many indicators of “active” enhancers (e.g., RNA Pol II, coregulators, H3K4me1, looping to target gene promoters), but the precise role of eRNAs in enhancer function is unknown. To address the role of E2-induced eRNAs in ESR1 enhancer function, we used the small-molecule drug flavopiridol (FP), an inhibitor of the CDK9 kinase of the P-TEFb complex (Chao et al. 2000), to block enhancer transcription and the stable, steady-state accumulation of eRNAs originating from ERBSs. MCF-7 cells were pretreated with FP for 1 h prior to treatment with E2. In locus-specific assays using RT-qPCR, FP efficiently blocked the production and accumulation of eRNAs from all five of the ERBSs that we examined, as well as mRNAs from nearby E2-regulated target genes looped to from these enhancers (Fig. 5A,B; Supplemental Fig. 6). This experimental system gave us the opportunity to examine the assembly of ESR1 enhancer complexes in the absence of eRNAs.
Figure 5.
Inhibition of eRNA production by flavopiridol does not inhibit ESR1, Pol II, or coregulator binding, alter H3K4me1 or H3K27ac levels, or prevent enhancer looping at ERBSs in MCF-7 cells. Locus-specific assays for E2-responsive enhancers showing the effects of a 1-h pre-treatment with flavopiridol (FP) on various molecular outcomes in MCF-7 cells. Each bar represents the mean + the SEM for three or more independent biological replicates. (A,B) Treatment with flavopiridol inhibits the E2-dependent production and steady-state accumulation of eRNAs and target gene mRNAs. RT-qPCR analyses for selected eRNAs and mRNAs in response to E2 treatment. (A) ERBS1 eRNA/P2RY2 mRNA and (B) ERBS2 eRNA/GREB1 mRNA. (C,D) ChIP-qPCR analyses for ESR1 (left) and Pol II (right) for ERBS1 (C) and ERBS2 (D) in the absence or presence of E2 and flavopiridol, as indicated. (E,F) ChIP-qPCR analyses for CREBBP (left), EP300 (middle), and Pol II (right) for ERBS1 (E) and ERBS2 (F) in the absence or presence of E2 and flavopiridol, as indicated. (G,H) ChIP-qPCR analyses for H3K4me1 (left), H3K27ac (middle), and H3 (right) for ERBS1 (G) and ERBS2 (H) in the absence or presence of E2 and flavopiridol, as indicated. (I,J) 3C-PCR analyses showing that looping between distal ERBSs and target genes in the presence of E2 is not blocked by flavopiridol (FP). (I) ERBS1/P2RY2 and (J) ERBS2/GREB1. The lowercase letters correspond to the primers denoted by orange arrows shown in Figure 1A. The assays were conducted in the presence (experimental) or absence (control) of DNA ligase, as indicated. The size of the PCR fragments in base pairs is shown. One representative experiment from three conducted is shown.
Treatment of MCF-7 cells with FP did not affect the E2-dependent binding of ESR1 or the loading of RNA Pol II at the ERBSs (Fig. 5C,D; Supplemental Fig. 14A–D), which occurred normally in the presence of the drug. Likewise, treatment with FP did not dramatically affect the recruitment of coregulators to the ERBSs (Fig. 5E,F; Supplemental Fig. 15) or the levels of H3K4me1 and H3K27ac at the ERBSs (Fig. 5G,H; Supplemental Fig. 14E–H). Thus, although the production of eRNAs correlates well with markers of active enhancers, the production and stable accumulation of eRNAs are not required for the assembly of ESR1 enhancer complexes at ERBSs. Furthermore, treatment with FP did not affect E2-dependent looping between ERBSs and E2-regulated target genes (Fig. 5I,J), indicating that the production and stable accumulation of eRNAs are not required for chromatin looping, at least under the conditions that we tested herein.
eRNAs defined by GRO-seq can be used to predict enhancers
As clearly illustrated in the data shown above, the production of eRNAs from ERBSs tracks very well with known features of active enhancers. As such, we reasoned that eRNA production can be used in the absence of any other genomic information to identify active enhancers de novo. We tested this hypothesis in the series of experiments described below.
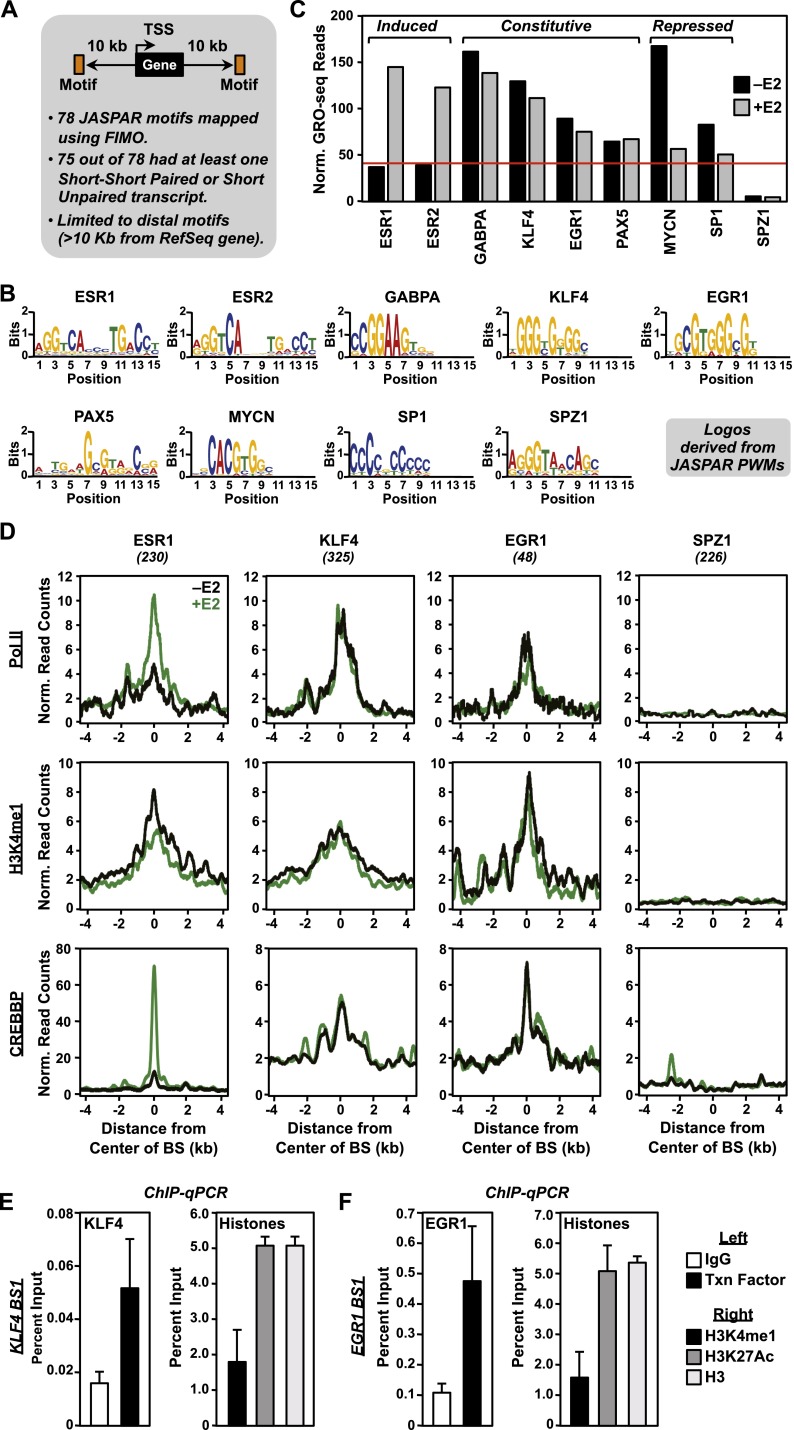
First, we used a directed approach to determine whether any known motifs for sequence-specific DNA binding transcription factors are enriched in GRO-seq reads in MCF-7 cells. To this end, we mapped all occurrences of 78 transcription factor motifs using position weight matrices (PWMs) from the JASPAR database (Bryne et al. 2008) to the human genome using FIMO (Grant et al. 2011), filtering for those located in intergenic regions (i.e., >10 kb away from the start or end of an annotated gene) (Fig. 6A). Seventy-five out of the 78 motifs overlapped at least one S-S paired or S-U transcript originating within a 2-kb window (±1 kb) around the center of the motif. Then, for each occurrence of each motif, we collected all GRO-seq reads within a 1-kb window (±0.5 kb) around the center of the motif and normalized them to the total number of occurrences of the motif. As expected, this analysis identified motifs for estrogen receptors (e.g., ESR1, ESR2) as dramatically enriched (i.e., well above the median) in GRO-seq reads in the E2-treated condition (Fig. 6B,C).
Figure 6.
Directed search for ESR1 and non-ESR1 enhancers in MCF-7 cells using GRO-seq data. (A) Schematic of the directed enhancer search using GRO-seq data. Seventy-eight motifs from the JASPAR database were mapped to the human genome using FIMO. For all intergenic motifs (>10 kb from RefSeq genes) with eRNAs (either short–short paired or short unpaired) originating within a 2-kb window around the center of the motif (i.e., ±1 kb relative to the motif), we collected the GRO-seq reads within a 1-kb window around the center of the motif (i.e., ±0.5 kb relative to the motif) and normalized them to the total number of occurrences of the motif. (B) Bar graph showing the normalized GRO-seq read count density per occurrence for nine selected motifs from the JASPAR database ± E2. (C) Web logos for the nine selected motifs shown in panel B generated using the JASPAR position weight matrices (PWMs). (D) Metaplots of Pol II, H3K4me1, and CREBBP ChIP-seq data from MCF-7 cells treated with (green lines) or without (black lines) E2 for four selected motifs from panel B. (E,F) ChIP-qPCR assays of KLF4 (E) and EGR1 (F) binding (left panels) and enhancer-associated histone modifications (H3K4me1 and H3K27ac; right panels) at cognate predicted binding sites. Each bar represents the mean + SEM for three or more independent biological replicates.
Other mapped motifs (e.g., GABPA, KLF4, EGR1, PAX5, MYCN, and SP1) also showed enrichment in GRO-seq reads in patterns that were either constitutive or repressed by E2 (Fig. 6B,C; Supplemental Fig. 16). In addition, we observed that some mapped motifs (e.g., SPZ1) showed no enrichment of GRO-seq reads, which served as a useful control in this experiment. These motifs show patterns of RNA Pol II, H3K4me1, and CREBBP based on ChIP-seq data that correspond well with the GRO-seq results (Fig. 6D; Supplemental Fig. 17). Furthermore, ChIP-qPCR assays confirm the binding of GABPA, KLF4, and EGR1 at their cognate predicted enhancers, but not control regions lacking the respective motifs (Fig. 6E,F; Supplemental Fig. 18). Together, the results from the directed search suggest that enhancers for a variety of transcription factors are active in MCF-7 cells and that GRO-seq can be used to identify them.
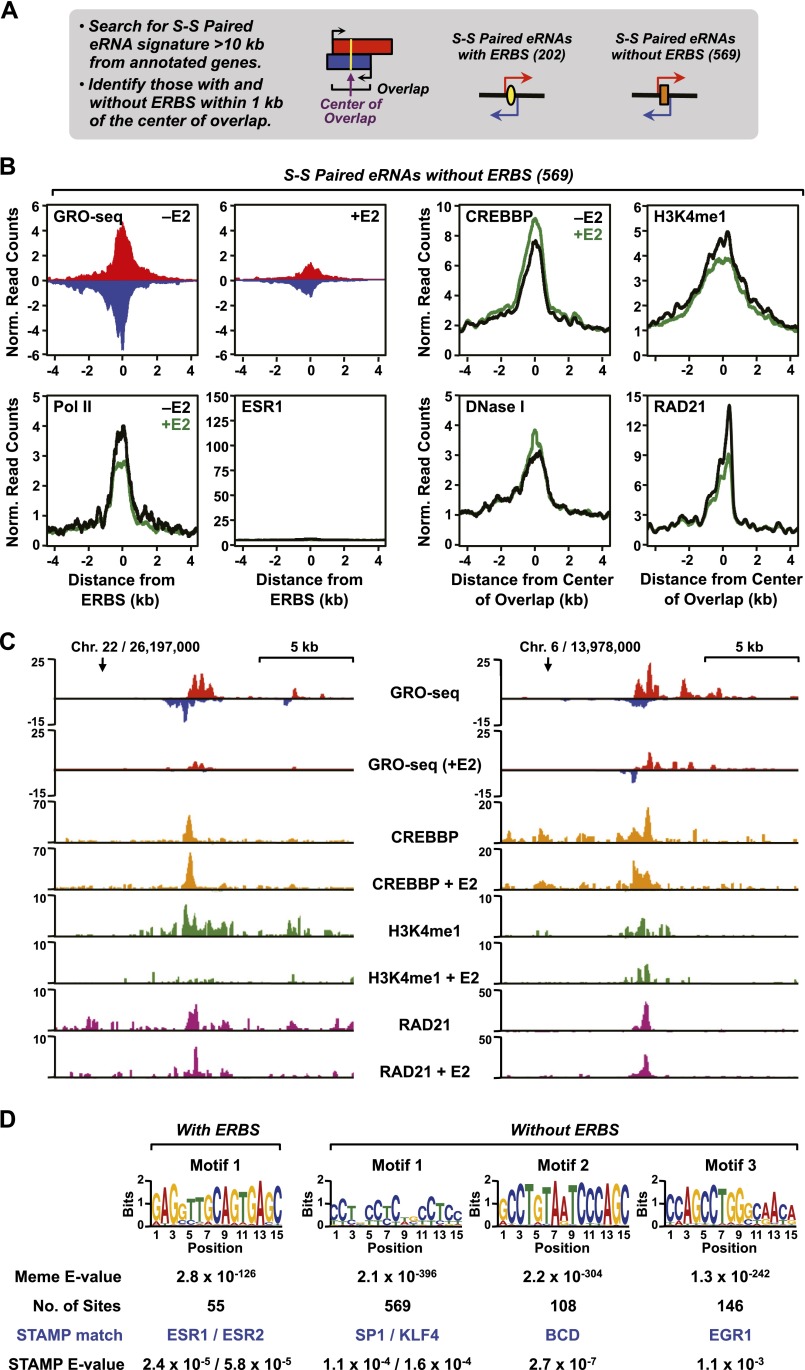
Second, we used an unbiased search for eRNA “signatures” to identify active enhancers in MCF-7 cells in the absence of any transcription factor binding or motif information. Specifically, we searched the called transcripts from our MCF-7 GRO-seq data for short, divergent, overlapping, and intergenic eRNA pairs that fit the average signature obtained from ERBSs [see Fig. 2B(c)], limiting our search to S-S paired transcripts to make it as stringent as possible (Fig. 7A). We identified 771 occurrences that fit the criteria, which we further divided based on the presence of an ERBS within 1 kb of the center of the peak (202 with an ERBS and 569 without an ERBS). Given the stringency of our search criteria, this is unlikely to be a comprehensive set, but it demonstrates proof-of-principle. The putative enhancers that we identified without an ERBS are of particular interest, since they represent an unbiased identification that does not rely on transcription factor binding data. Interestingly, the transcription of many enhancers in this group is regulated by E2 (33% are down-regulated and 13% are up-regulated) as indicated by our GRO-seq data (Fig. 7B, top left; Fig. 7C), even though they lack ESR1 binding, as determined by ChIP-seq. These results suggest that the estrogen signaling pathway can impact enhancers that lack ESR1 binding. The patterns of E2-dependent regulation of transcription for these enhancers mirror those of the nearest-neighbor genes (Supplemental Fig. 19), as might be expected if there is a functional link between distal enhancers and nearby target genes.
Figure 7.
Unbiased search for ESR1 and non-ESR1 enhancers in MCF-7 cells using GRO-seq data. (A) Schematic of the unbiased enhancer search using GRO-seq data. All intergenic (>10 kb away from the start or end of an annotated RefSeq gene) short–short paired eRNAs <9 kb and with an average overlap of 3 kb were identified in the set of called transcripts from MCF-7 cells. (B) GRO-seq data, Pol II, ESR1, CREBBP, H3K4me1, and RAD21 ChIP-seq data, and DNase-seq data (as indicated) from the analysis described in panel A were collected, mapped relative to the center of the plus and minus strand overlap of the short–short paired eRNAs, and expressed as metaplots. (C) Browser tracks of GRO-seq and selected ChIP-seq data for two enhancers without ERBSs identified in the unbiased search described in panel A. (D) Web logos and statistical parameters for the top motifs identified in a search of enhancers identified in panel A. All occurrences of the short–short paired transcripts were collected and subjected to motif analysis. De novo motif searching was performed on a 1-kb region around the center of the plus and minus strand overlap (±500 bp) using MEME. The predicted motifs from MEME were matched to known motifs using STAMP.
We used the set of putative enhancers without an ERBS to align other genomic data to the center of the divergent eRNA overlap, which revealed features that would be expected of active enhancers, including an enrichment of RNA Pol II, CREBBP, and H3K4me1, as well as an open chromatin architecture, as determined by DNase-seq (Fig. 7B). Despite the fact that the transcription of many enhancers in this group is dramatically reduced by E2 treatment (Fig. 7B, top left; Fig. 7C), the other enhancer properties that we examined were largely unaffected (Fig. 7B,C). This result mirrors what we observed with the ESR1 enhancers in the presence of FP, namely, a preservation of enhancer features in spite of inhibited transcription.
To determine which transcription factors might underlie the predicted enhancers identified based on the called transcripts from GRO-seq, we performed de novo motif analyses on a 1-kb region around the center of the eRNA overlap using MEME (Bailey et al. 2009). The predicted motifs from MEME were then matched to known motifs using STAMP (Parks and Beiko 2010). As expected, the predicted enhancers with an ERBS were enriched for EREs, whereas those without an ERBS were not (Fig. 7D; data not shown). The predicted enhancers without an ERBS were enriched in motifs for transcription factors such as SP1, KLF4, Bicoid-related factors, and EGR1 (Fig. 7D). These same motifs were also enriched in our directed search (Fig. 6C; Supplemental Fig. 16), giving more confidence to this approach.
To determine how enhancer prediction using GRO-seq compares with enhancer prediction using H3K4me1 or H3K27ac ChIP-seq, we called enhancers using all three parameters under basal (i.e., −E2) conditions. We found that a greater percentage of enhancers called by GRO-seq have Pol II–dependent promoter looping events than those called by H3K4me1 or H3K27ac (Supplemental Fig. 20A,B). In addition, enhancers called by GRO-seq have a greater enrichment of DNase-seq and CREBBP ChIP-seq signals than those called by H3K4me1 or H3K27ac (Supplemental Fig. 20C). Furthermore, DNase-seq and CREBBP ChIP-seq signals at enhancers called by eRNAs (and to a lesser extent H3K4me1) track better with Pol II looping than those called by H3K27ac (Supplemental Fig. 20D). Collectively, these results indicate that (1) enhancers called by GRO-seq are “active,” and (2) the presence of eRNAs may more accurately predict active enhancers than H3K4me1 or H3K27ac. Finally, application of our GRO-seq-based enhancer prediction pipeline to data from mouse embryonic stem (mES) cells (Supplemental Fig. 21) further demonstrates the power of using GRO-seq to perform unbiased enhance prediction in the absence of other genomic information.
Discussion
In the studies described herein, we integrated and analyzed a large number of genomic data sets using a novel computational pipeline to provide a comprehensive and global view of ESR1 enhancers in the MCF-7 human breast cancer cell line. Our results indicate that intergenic ERBSs produce enhancer transcripts that are similar to eRNAs described previously (Kim et al. 2010) and have the following properties: (1) They originate from one or both strands of DNA, with an average transcription unit (i.e., primary transcript) length of ∼3–5 kb; (2) the majority are up-regulated by short treatments with E2 (i.e., 10, 25, or 40 min) with kinetics that, in many cases, precede or match the induction of the target gene; and (3) they are detectable by RT-qPCR using either random hexamer or oligo(dT) primers for RT, suggesting that they may be polyadenylated, but perhaps minimally so since they do not give strong signals in poly(A) RNA-seq data sets from MCF-7 cells (data not shown).
Relation of eRNA production to features of active enhancers
The enrichment of several genomic features has been proposed to be marks of active enhancers, including H3K4me1, H3K27ac, EP300/CREBBP, RNA Pol II, and an open chromatin architecture (for review, see Maston et al. 2012; Natoli and Andrau 2012). Our results indicate that the production of eRNAs at ERBSs strongly correlates with the enrichment of these features and, in fact, may be a good indicator of active enhancers. In the absence of eRNA production, strong enrichment of these features is not observed, even though ESR1 binding is evident. ERBSs producing eRNAs are also enriched for enhancer–promoter loops. The formation of such loops is thought to play an important role in the communication between signal-dependent enhancers and their regulated target genes (Fullwood et al. 2009; Bulger and Groudine 2011). Indeed, target genes that are looped to by an intergenic ERBS show greater levels of transcription than those that are not looped to by an intergenic ERBS. Interestingly, the enrichment of coregulators (e.g., CREBBP, EP300, NCOAs) correlates well with enhancer looping, whereas the enrichment of H3K4me1, H3K4me2, and H3K4me3 does not. Thus, histone H3K4 methylation at enhancers is not a good predictor of ERBS looping to target gene promoters, even though it clearly marks ERBSs.
Our results indicate that although ESR1 may bind to some enhancer “hot spots” (i.e., genomic locations with pre-opened chromatin where a number of different sequence-specific DNA-binding transcription factors bind) (Li et al. 2011), ESR1 can also bind to genomic locations with closed chromatin, inducing chromatin opening in an E2-dependent manner. This is suggested by the decrease in H3K4me1 levels upon ESR1 binding (Fig. 3E), is evident from our analysis of the DNase-seq data (Fig. 3G), and is consistent with previous reports for ESR1 action at enhancers (He et al. 2012). In this regard, we observed a unimodal enrichment of K3K4me1, 2, and 3 at ERBSs, which colocalizes with the site of ESR1 binding (Fig. 3E,F; Supplemental Fig. 8E), in a region thought to be occupied by a nucleosome that is remodeled upon ESR1 binding (Wang et al. 2011; He et al. 2012). In contrast, studies of other transcription factors have revealed a bimodal enrichment pattern with transcription factor binding in the middle, in a region thought to be nucleosome-free (He et al. 2010, 2012; Kim et al. 2010; Wang et al. 2011). Nucleosome remodeling events at steroid receptor enhancers are a common theme, as illustrated by ESR1, androgen receptor, and glucocorticoid receptor, although the precise mechanisms of binding and nucleosome rearrangements may differ (He et al. 2010, 2012; John et al. 2011).
The assembly of enhancer complexes can be dissociated from eRNA production
The function of enhancer transcription and the stable, steady-state accumulation of eRNAs are unknown. Some have suggested that the act of transcription helps to create an open chromatin environment that promotes enhancer function, while others have suggested that the stable accumulation of eRNAs may play a functional, perhaps even structural, role (Bulger and Groudine 2011; Orom and Shiekhattar 2011; Maston et al. 2012; Natoli and Andrau 2012). Two aspects of our results have shed some light on these questions, as well as the order of operations at signal-regulated enhancers. First, using the drug FP, we showed that many features of enhancers, including the assembly of enhancer complexes and the modification of histones, can be dissociated from eRNA production. FP blocks transcription elongation but not preinitiation complex assembly or transcription initiation, so it targets a specific aspect of enhancer function. As we observed, FP efficiently blocks enhancer transcription and the stable, steady-state accumulation of eRNAs at ERBSs, but had no effect on any of the E2-dependent enhancer features that we examined, including enhancer–promoter looping. Second, in our unbiased search for enhancers, we identified a large set of putative enhancers actively transcribed but lacking ERBSs. Transcription of these enhancers was dramatically reduced by treatment with E2, in many cases being completely abrogated. However, as observed in the experiments with FP, inhibition of the transcription of these enhancers had, in most cases, little or no effect on the enhancer features that we monitored. Together, these results clearly show that the assembly of enhancer complexes can be dissociated from eRNA production, suggesting that eRNA production occurs after the assembly of active enhancers. These results are consistent with previous results showing that eRNAs are not expressed, despite normal levels of RNA Pol II loading at enhancers, when the cognate promoter is removed (Kim et al. 2010). They do not, however, suggest that eRNA production is unnecessary for enhancer function or target gene activation. In fact, FP also inhibits target gene activation, so the potential role of eRNA production in that aspect of enhancer function could not be assayed in our studies. Further studies will be required to resolve these issues.
Unbiased prediction of enhancers based on eRNA production
Our genomic analyses have defined an enhancer transcription “signature” based on GRO-seq data (i.e., short, bidirectional transcription units) that can be used for de novo enhancer prediction. Using GRO-seq-defined transcription and the enhancer transcription signature, one can query any cell type for active enhancers in the absence of any histone modification or transcription factor binding information from ChIP-seq, as we did for MCF-7 cells and mES cells. The power of this approach is the ability to use a single genomic data set (i.e., GRO-seq) for each cell type, which would allow profiling of enhancers across a large collection of cell lines or tissues. Then, using transcription factor binding site motif analyses, one could work backward to determine which transcription factors may underlie those enhancers (see Fig. 7D, for example), which after selection could be assayed by ChIP-seq. Collectively, our results indicate that (1) enhancers called by GRO-seq are “active” and (2) the presence of eRNAs may more accurately predict active enhancers than other commonly used enhancer marks.
Methods
Additional details about all of the methods listed below, information about the antibodies used, and additional methods for the analysis of GRO-seq data and other genomic data can be found in the Supplemental Material.
Cell culture and treatments
MCF-7 cells were plated for experiments in phenol red–free MEM (Sigma-Aldrich) supplemented with 5% charcoal-dextran-treated calf serum (CDCS) prior to treatment. As indicated, the cells were treated with 100 nM E2 for the times specified, with or without a 1-h pre-treatment of 1 μM flavopiridol (Sigma-Aldrich).
Locus-specific molecular assays: ChIP-qPCR, RT-qPCR, and 3C-PCR
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) (Kininis et al. 2007), reverse transcription-quantitative PCR (RT-qPCR) (Hah et al. 2011), and chromosome conformation capture-PCR (3C-PCR) (Pan et al. 2008) were performed essentially as described previously using MCF-7 cells and locus-specific primers designed to detect the end products of interest by qPCR or PCR (see Supplemental Table S2). Each experiment was performed a minimum of three times with independent biological samples to ensure reproducibility.
GRO-seq
Global run-on sequencing (GRO-seq) was performed as described previously (Hah et al. 2011) from two biological replicates of E2-treated (0, 10, and 40 min) MCF-7 cells. The data sets, which were used for all genome-scale analyses, are available from NCBI/GEO using accession number GSE43836. For some gene-specific analyses showing genome browser tracks, we also used data from GRO-seq libraries generated from other E2 treatment time points (e.g., 25 min; GEO accession number GSE41324).
Analysis of GRO-seq data
GRO-seq data were analyzed using software described previously (Hah et al. 2011) and the approaches described below. Software, scripts, and other information can be obtained by contacting W. Lee Kraus. Read alignment, transcript calling, determination of regulation by E2, and the generation of heat maps were performed as described previously (Hah et al. 2011).
Defining classes of ESR1 enhancer transcripts (eRNAs)
The repertoire of genomic ESR1 binding sites (ERBSs) was extracted from ChIP-seq data provided in Welboren et al. (2009) (GEO accession number GSM365926). Those ERBSs >10 kb away from the 5′ or 3′ ends of annotated genes were defined as “Intergenic ERBSs.” They were divided into three classes based on the presence, location, and orientation of GRO-seq-defined transcripts: (1) those overlapping transcripts originating from both strands of DNA, running in opposite directions as a divergent pair; (2) those overlapping a transcript originating from one strand of DNA only (“Unpaired); and (3) those not overlapping a transcript. The transcripts in classes 1 and 2 were further categorized based on the length of the transcript unit/primary transcript as “short” (length < 9 kb; eRNAs, by our definition) and “long” (length > 9 kb; which likely represent other classes of noncoding RNAs, such as lncRNAs).
Analysis of ChIP-seq data
ChIP-seq data sets from MCF-7 cells were obtained from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) online databases (see Supplemental Table 1). The raw files were aligned to hg18 using Bowtie (Langmead et al. 2009). Uniquely mappable reads were converted into bigWig files using BEDTools (Quinlan and Hall 2010) for visualization in the UCSC Genome Browser.
Analysis of ChIA-PET data
We used the ESR1-dependent intrachromosomal loops defined in the ChIA-PET data set from Fullwood et al. (2009) to examine if the ERBSs defined by Welboren et al. (2009) are (1) involved in loops and (2) if they loop to the promoters of 19,008 unique RefSeq genes. We assayed for loops within a 2-kb window (±1 kb) around ESR1 peak centers and a 10-kb window (±5 kb) around the transcription start sites (TSS) of genes. We used GRO-seq data to determine the amount of transcription at the ERBSs and the target genes involved in looping events. Various metaplot representations of GRO-seq and ChIP-seq data were used to compare the properties of ERBSs and genes with or without looping events.
Genomic data analysis and visualization
We used metaplots to illustrate the distribution of GRO-seq, ChIP-seq, and ChIA-PET reads around ESR1 peak maxima (or other genomic features) using the metaplot function in our GRO-seq package (Hah et al. 2011). We also used boxplots to minimize the bias caused by outliers in the data, which can overly influence metaplot representations. The interquartile regions (IQRs) of the boxplots were used to plot metaplots centered on the ERBSs.
Predicting enhancers based on GRO-seq data
We used GRO-seq data combined with DNA-binding transcription factor motif information to predict active enhancers with directed and unbiased approaches.
Directed search for enhancers based on GRO-seq data
Motifs for all curated, nonredundant, vertebrate transcription factors (130 total) in the JASPAR database (Bryne et al. 2008) were searched using FIMO (Grant et al. 2011) with a P-value threshold of 1 × 10−6; 78 out of 130 motifs were identified and mapped in hg18 at this P-value. From the total set of identified motifs, we selected only intergenic motifs (>10 kb from RefSeq genes) that have eRNAs originating within a 2-kb window around the center of the motif (i.e., ±1 kb relative to the motif). For each occurrence of a specific intergenic motif with an overlapping eRNA that we selected, we collected all GRO-seq reads within a 1-kb window around the center of the motif (i.e., ±0.5 kb relative to the motif) and normalized them to the total number of occurrences of the motif.
Unbiased search for enhancers based on GRO-seq data
We searched our MCF-7 GRO-seq data sets for sites in the genome expressing intergenic (>10 kb away from the start or end of an annotated RefSeq gene) short–short paired eRNAs using the following parameters: primary transcript length shorter than 9 kb and an average overlap of 3 kb. All occurrences that fit these criteria were collected and subjected to motif analysis. De novo motif searching was performed on a 1-kb region around the center of the plus and minus strand overlap (±500 bp) using the command-line version of MEME (Bailey and Elkan 1994). The predicted motifs from MEME were matched to known motifs using STAMP (Parks and Beiko 2010).
Data access
The new GRO-seq data sets described herein are available from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE43836.
Acknowledgments
We thank John Lis, Leighton Core, T.K. Kim, Xin Luo, Miao Sun, and Hector Franco for helpful discussions and critical comments on this manuscript. This work was supported by an NIH training award (T32HD052471) and a postdoctoral fellowship from the PhRMA Foundation to C.D., and a grant from the NIH/NIDDK (DK058110) to W.L.K.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.152306.112.
References
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the Forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem 275: 28345–28348 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G 2010. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS 2011. FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS 2012. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res 22: 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27: 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolovos P, Knoch TA, Grosveld FG, Cook PR, Papantonis A 2012. Enhancers and silencers: An integrated and simple model for their function. Epigenetics Chromatin 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R 2013. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol 12: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, Landt SG, Snyder M, Green MR 2012. Characterization of enhancer function from genome-wide analyses. Annu Rev Genomics Hum Genet 13: 29–57 [DOI] [PubMed] [Google Scholar]
- Melgar MF, Collins FS, Sethupathy P 2011. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol 12: R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. 2013. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49: 524–535 [DOI] [PubMed] [Google Scholar]
- Natoli G, Andrau JC 2012. Noncoding transcription at enhancers: General principles and functional models. Annu Rev Genet 46: 1–19 [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R 2011. Noncoding RNAs and enhancers: Complications of a long-distance relationship. Trends Genet 27: 433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171 [DOI] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E 2008. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem 283: 32977–32988 [DOI] [PubMed] [Google Scholar]
- Parks DH, Beiko RG 2010. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26: 715–721 [DOI] [PubMed] [Google Scholar]
- Pham TH, Benner C, Lichtinger M, Schwarzfischer L, Hu Y, Andreesen R, Chen W, Rehli M 2012. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood 119: e161–e171 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J 2012. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11: 633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O'Shea JJ 2012. STATs shape the active enhancer landscape of T cell populations. Cell 151: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG 2009. ChIP-seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, Hou S, Stamatoyannopoulos JA, Mikkola HK, Yuan GC, et al. 2012. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell 23: 796–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K, Suzuki Y 2011. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res 21: 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]