Abstract
Identifying the genetic changes driving adaptive variation in natural populations is key to understanding the origins of biodiversity. The mosaic of mimetic wing patterns in Heliconius butterflies makes an excellent system for exploring adaptive variation using next-generation sequencing. In this study, we use a combination of techniques to annotate the genomic interval modulating red color pattern variation, identify a narrow region responsible for adaptive divergence and convergence in Heliconius wing color patterns, and explore the evolutionary history of these adaptive alleles. We use whole genome resequencing from four hybrid zones between divergent color pattern races of Heliconius erato and two hybrid zones of the co-mimic Heliconius melpomene to examine genetic variation across 2.2 Mb of a partial reference sequence. In the intergenic region near optix, the gene previously shown to be responsible for the complex red pattern variation in Heliconius, population genetic analyses identify a shared 65-kb region of divergence that includes several sites perfectly associated with phenotype within each species. This region likely contains multiple cis-regulatory elements that control discrete expression domains of optix. The parallel signatures of genetic differentiation in H. erato and H. melpomene support a shared genetic architecture between the two distantly related co-mimics; however, phylogenetic analysis suggests mimetic patterns in each species evolved independently. Using a combination of next-generation sequencing analyses, we have refined our understanding of the genetic architecture of wing pattern variation in Heliconius and gained important insights into the evolution of novel adaptive phenotypes in natural populations.
Natural selection acting on heritable genetic variation has generated much of the extraordinary biological diversity we observe in nature. However, in the 150 years since Darwin and Wallace independently posited the theory of natural selection, we still have only a rudimentary understanding of how adaptive variation arises and spreads in natural populations. The molecular basis of adaptive variation in natural populations has been identified in only a handful of traits (Martin and Orgogozo 2013). These examples provide the foundation for our current understanding of the genetic architecture of adaptation and underpin efforts to unite molecular, developmental, and evolutionary biology into a single evolutionary synthesis (Stern and Orgogozo 2009).
The array of adaptive wing color patterns of Heliconius butterflies offers an exceptional opportunity to explore the functional changes that drive complex adaptive traits in natural populations. Heliconius butterflies display bright wing color patterns that are under strong natural selection (Benson 1972; Mallet and Barton 1989; Mallet et al. 1990; Kapan 2001)—they warn potential predators that the butterflies are unpalatable (Chai 1986). Selection favoring regional mimicry among these and other noxious butterflies drives the remarkable diversity in wing color patterns—characterized by extreme divergence within species and striking convergence among distantly related species (Turner 1975). This pattern of convergence and divergence is best exemplified by the Müllerian mimics, H. erato and H. melpomene. Since the divergence of the two species 13–26 million years ago (Pohl et al. 2009), both have undergone parallel radiations, such that the range of each species is composed of an identical patchwork of divergent color pattern races stitched together by a series of narrow hybrid zones. This replicate-rich and highly variable system has become a textbook example of evolution by natural selection and provides a remarkable template to explore the repeatability of evolution.
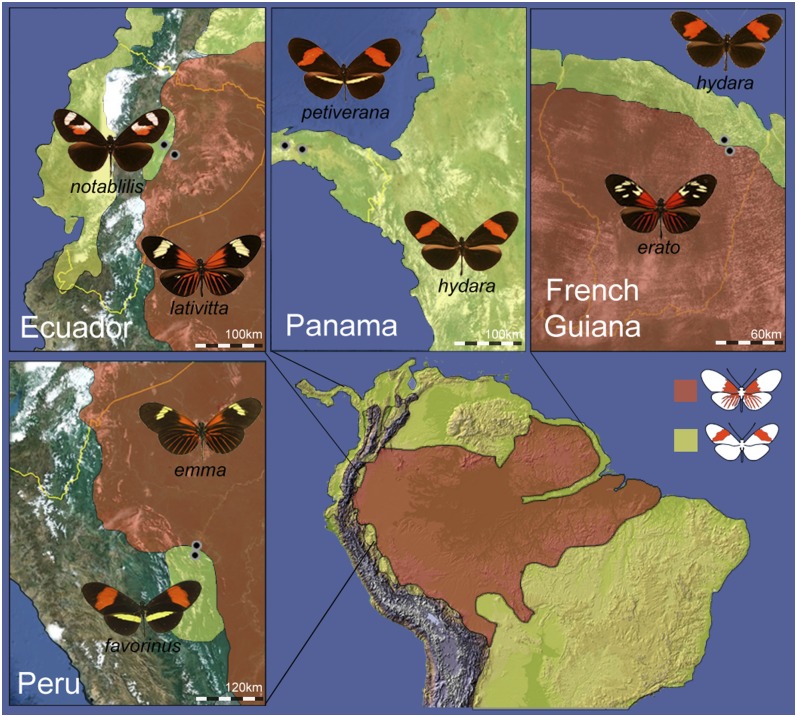
Heliconius show striking variation in complex red color pattern elements across both the forewing and hindwing. Red color patterns in H. erato and H. melpomene consist of three distinct elements—the color of the forewing band, the presence or absence of the red “dennis” patch on the proximal portion of the forewing, and the presence or absence of red hindwing rays (Sheppard et al. 1985; Papa et al. 2008). These elements comprise two major red phenotypes—“postman” and “rayed” (Fig. 1). Postman races are characterized by a red forewing band and absence of both the red dennis patch and hindwing rays and are found in disjunct populations across Central and South America. Rayed races, in contrast, have a yellow forewing band, red dennis patch, and hindwing rays. The rayed races are found throughout the Amazon Basin but not on the Pacific coast of South America or in Central America (Fig. 1).
Figure 1.
Distribution of the H. erato color pattern radiation with approximate sampling locations from four H. erato hybrid zones. Shaded areas are distributions of H. erato rayed (red) and postman (yellow) races (based on Hines et al. 2011; Rosser et al. 2012).
Substantial progress has been made in understanding the genetic basis of red phenotypic variation in Heliconius, but the causative variant remains elusive. The region was initially described as a complex of three tightly linked loci (Sheppard et al. 1985). However, the region modulating red variation was positionally cloned in both H. erato (Counterman et al. 2010) and H. melpomene (Baxter et al. 2010) to a single shared 400-kb genomic interval—referred to as the D interval in H. erato and the B/D interval in H. melpomene. Targeted resequencing identified an ∼150-kb region of divergence between divergent red color pattern races of H. melpomene (Nadeau et al. 2012). Gene expression analyses across this interval supported the transcription factor optix as the only gene with expression patterns consistent with a role in red pattern formation (Reed et al. 2011). Gene expression differences and the highly conserved amino acid sequence variation in optix suggest that variable red patterns are driven by cis-regulatory variation (Reed et al. 2011). Phenotypic recombinants between red pattern elements have been occasionally observed (Mallet 1989), implicating the potential involvement of multiple, tightly linked, cis-regulatory variants in this region that generate differences in the spatial expression of optix and result in diverse red color pattern phenotypes.
Identifying the genomic region responsible for red color pattern variation has allowed reassessment of the history of color pattern diversification in these co-mimics using the region most likely to reflect mimetic history. Phylogenetic reconstruction of the history of color pattern evolution in both H. erato and H. melpomene found that sequences from the gene optix cluster races strongly by color pattern, rather than the clustering by geographic proximity observed with other genomic markers (Hines et al. 2011). This suggests a single origin for geographically disjunct rayed color pattern phenotypes within each species. The evolutionary history between the two co-mimetic species is likely more complex. Heliconius erato and H. melpomene belong to divergent clades and are not known to hybridize, precluding the hypothesis of adaptive introgression that has been shown between more closely related species within the melpomene/cydno/silvaniform clade (The Heliconius Genome Consortium 2012; Pardo-Diaz et al. 2012). However, the question remains as to whether or not the convergent color patterns have a common origin through the use of ancestral variation or if they evolved independently through unique de novo mutations. Inferring the evolutionary history of this striking mimetic phenotype requires high resolution sequence data across a more finely defined genomic region regulating color pattern variation in both species.
In this study, we use a combination of next-generation sequencing technologies to (1) explore the genetic variation that modulates spatially complex adaptive red wing color pattern variation in natural populations of Heliconius erato butterflies; and (2) compare the genomic regions responsible for parallel mimetic color pattern radiations in H. erato and H. melpomene. We start by using transcriptome sequencing to annotate protein coding genes across the H. erato red color pattern interval. We then examine whole genome resequencing data from divergent color pattern races across four geographically distinct H. erato hybrid zones. In these admixture zones, gene flow between divergent races homogenizes the genomes, while strong selection on color pattern creates narrow regions of genetic divergence around these genomic targets of selection. We use these hybrid zone data to localize signatures of selection across 1 Mb of the red color pattern locus and identify a noncoding region likely responsible for regulating expression of optix and ultimately driving spatially complex patterns of red across the wings. Finally, we use phylogenetic analyses to demonstrate that mimetic red patterns evolved only once within each species but likely evolved independently in the co-mimics H. erato and H. melpomene. These two distantly related mimetic species appear to generate their convergent phenotypes through different changes in the same genetic architecture. This study refines the regulatory regions driving adaptive phenotypic variation in a classic mimetic radiation and provides an improved understanding of the repeatability of evolution at the genomic level.
Results
Annotation, synteny, and conservation of the H. erato red color pattern (D) interval
Using transcriptome alignments, protein homology, and ab initio predictions, we annotated 30 protein coding genes across 1 Mb of the H. erato D interval (Supplemental Table S4; GenBank accession KC469894). The genes across the D interval are in perfect synteny with the corresponding red pattern region of the co-mimic H. melpomene (Supplemental Fig. S1). The annotated region contains a 200-kb gene desert that includes a single gene, optix. This gene desert contains the 65-kb peak that shows strong signatures of selection and association (described below). Sequence alignments of the H. erato D interval and the orthologous scaffolds from the H. melpomene genome identified 182 highly conserved regions (>90% sequence similarity in a 500-bp window), covering a total of 63 kb of sequence, of which 25 kb is located in noncoding regions. The 65-kb peak we have identified contains 20% of the conserved noncoding regions, but transcriptome alignments show no transcriptional activity. Analysis of potential transcription factor binding sites in the 65-kb region revealed binding sites for numerous transcription factors across the region, but no clear candidates for optix regulation (see Supplemental Material section S1; Supplemental Fig. S2).
Summary of resequencing data and genotyping
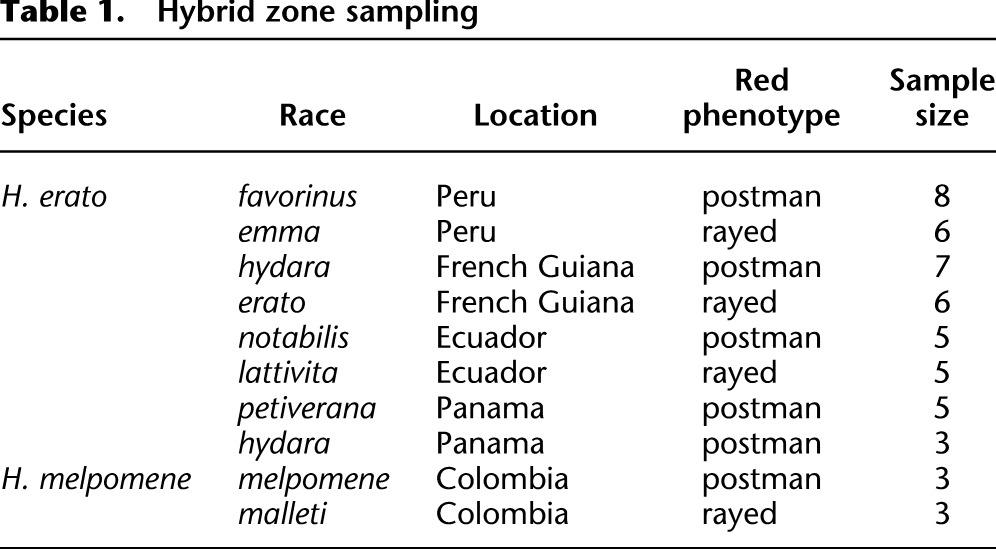
We examined whole genome data from multiple individuals of divergent color pattern races across four geographically distinct H. erato hybrid zones and one H. melpomene hybrid zone (Fig. 1; Table 1). We resequenced 45 H. erato individuals to a median per base coverage between 15× and 35× per individual and aligned sequence reads across 2.2 Mb of the H. erato partial reference genome sequence (1 Mb of the D interval and 1.2 Mb from other regions of the genome). For each individual, we identified, on average, one SNP for every 22 bases genotyped (Supplemental Table S6). Additionally, we resequenced six H. melpomene individuals to a median per base coverage between 19× and 34× per individual and aligned sequences to the whole H. melpomene reference genome (The Heliconius Genome Consortium 2012). For each individual, we identified, on average, one SNP for every 48 bases genotyped across the B/D interval (Supplemental Table S7).
Table 1.
Hybrid zone sampling
Genomic divergence and genotype by phenotype association
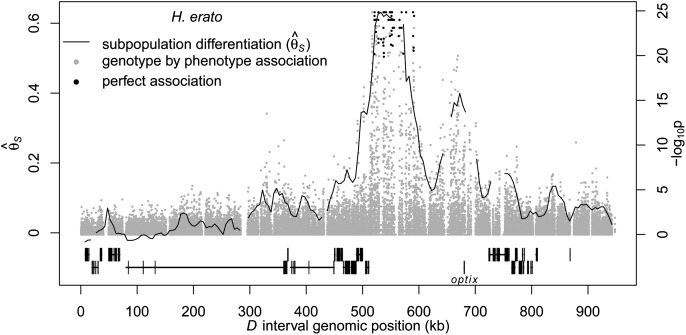
The sliding window divergence analysis between subpopulations of the two common H. erato phenotypes—the postman and the rayed—showed peaks of genetic divergence at two distinctive regions within a 200-kb stretch of the D interval (Fig. 2). The first region (the second peak in Fig. 2 spanned ∼40 kb [650–690 kb]) had moderate levels of differentiation (0.3 <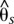 <0.4) (Fig. 2) and reduced nucleotide diversity (Fig. 3A). This region was centered immediately 3′ of the transcription factor optix, which is the only gene located within the peaks of divergence. The second region spanned ∼65 kb of noncoding sequence more distally 3′ of optix (515–580 kb). This 65-kb region had very high levels of differentiation (0.6 <
<0.4) (Fig. 2) and reduced nucleotide diversity (Fig. 3A). This region was centered immediately 3′ of the transcription factor optix, which is the only gene located within the peaks of divergence. The second region spanned ∼65 kb of noncoding sequence more distally 3′ of optix (515–580 kb). This 65-kb region had very high levels of differentiation (0.6 <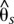 <0.7) between hybridizing races, low levels of nucleotide diversity within races (Fig. 3A), and elevated linkage disequilibrium relative to other regions of the genome (Supplemental Fig. S3), which are all indicative of recent positive selection driving the fixation of haplotypes. Of these two regions that showed signatures of selection in H. erato, only the 65-kb peak, which is furthest from optix, contained SNPs that were perfectly associated with color pattern phenotype across our entire sampling of individuals (Fig. 2).
<0.7) between hybridizing races, low levels of nucleotide diversity within races (Fig. 3A), and elevated linkage disequilibrium relative to other regions of the genome (Supplemental Fig. S3), which are all indicative of recent positive selection driving the fixation of haplotypes. Of these two regions that showed signatures of selection in H. erato, only the 65-kb peak, which is furthest from optix, contained SNPs that were perfectly associated with color pattern phenotype across our entire sampling of individuals (Fig. 2).
Figure 2.
Divergence and association between divergent H. erato color pattern races across the D interval. The solid line indicates sliding window (15-kb window size, 5-kb step size) subpopulation differentiation (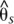 ) between the H. erato postman and rayed phenotypes for individuals from three hybrid zones (Peru, Ecuador, French Guiana; npostman = 20; nrayed = 17), requiring a minimum of 75% of individuals genotyped for each phenotype at each position and data for at least 20% of positions in each window. A baseline subpopulation differentiation of
) between the H. erato postman and rayed phenotypes for individuals from three hybrid zones (Peru, Ecuador, French Guiana; npostman = 20; nrayed = 17), requiring a minimum of 75% of individuals genotyped for each phenotype at each position and data for at least 20% of positions in each window. A baseline subpopulation differentiation of 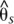 = −0.07 was calculated from genomic intervals unlinked to color pattern. The dots indicate genotype by phenotype association calculated for biallelic SNPs using a Fisher's exact test for all four hybrid zones (npostman = 28; nrayed = 17), requiring a minimum of 75% of individuals genotyped for each phenotype at each SNP. The black dots indicate association for the 76 SNPs perfectly associated with phenotype. The gene annotations are shown below for the plus strand (top) and minus strand (bottom), with the single exon gene optix denoted. Wider boxes represent coding exons and the narrower boxes represent introns.
= −0.07 was calculated from genomic intervals unlinked to color pattern. The dots indicate genotype by phenotype association calculated for biallelic SNPs using a Fisher's exact test for all four hybrid zones (npostman = 28; nrayed = 17), requiring a minimum of 75% of individuals genotyped for each phenotype at each SNP. The black dots indicate association for the 76 SNPs perfectly associated with phenotype. The gene annotations are shown below for the plus strand (top) and minus strand (bottom), with the single exon gene optix denoted. Wider boxes represent coding exons and the narrower boxes represent introns.
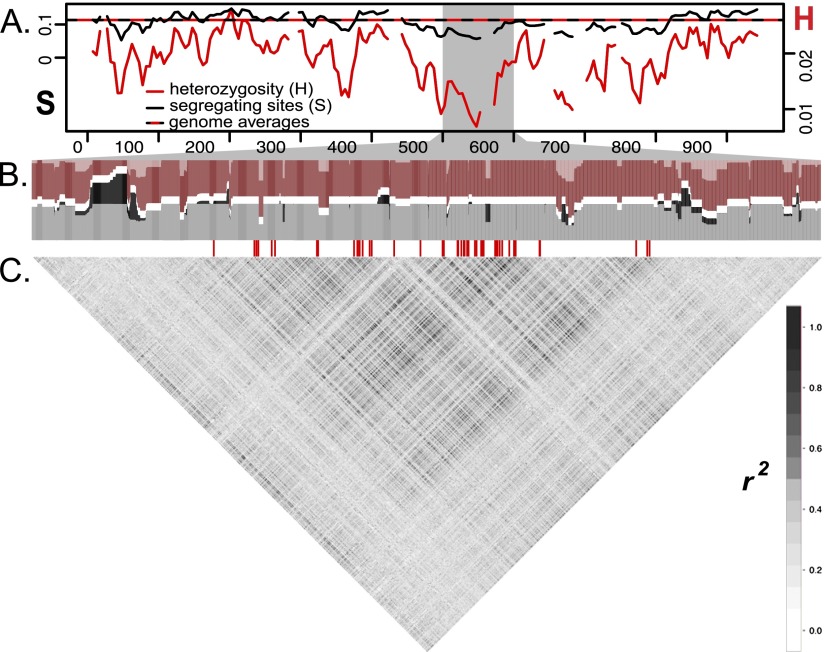
Figure 3.
Signatures of selection and recombination across the D interval in the H. erato Peruvian hybrid zone (n = 14). (A) Solid lines show sliding window (15-kb window size, 5-kb step size) values for the number of segregating sites (S, black) and heterozygosity (H, red). Horizontal dashed line represents averages of S and H from genomic regions unlinked to color, with axes scaled to match averages. (B) Haplotypes clustered from 3227 SNPs from the 500–600 kb region, which includes the peak of divergence. Haplotypes from rayed (shaded dark) and postman (shaded light) races were clustered into two groups (K = 2) (colored red and gray). A horizontal white row separates the two clusters. Blocks that have different haplotypes fixed between the two color pattern races are represented by regions with sets of neighboring SNPs in which all rayed individuals are assigned exclusively to the red cluster (red columns shaded entirely dark) and all postman individuals are assigned exclusively to the gray cluster (gray columns shaded entirely light). Blocks where the two color pattern races share haplotypes are represented by regions where individuals of a single color pattern race are assigned to more than one cluster (red columns with light and dark shading; gray columns with light and dark shading). SNPs with fixed allelic differences between color patterns are denoted by red hash marks below the haplotype clustering (data from Fig. 2). (C) Correlation plot of linkage disequilibrium (r2) among 3187 biallelic SNPs across the 500–600-kb window on the D interval.
Overall, the patterns of divergence and association in the gene desert near optix were consistent across the geographically distinct H. erato hybrid zones. All hybrid zones between rayed and postman phenotypes (Ecuador, French Guiana, and Peru) showed strong genetic differentiation ( > 0.6) and numerous fixed differences between phenotypes in the region 3′ of optix (Supplemental Fig. S4). In the 40-kb peak, which includes optix, only the hybrid zones in Ecuador and Peru showed strong differentiation and fixed differences between races. Individuals from French Guiana showed only moderate genetic differences and no perfectly associated SNPs in this region. The Panama hybrid zone is between two different postman phenotypes and, as expected, contained no indication of genetic divergence between phenotypes across the D interval and no regions of consistent association with phenotype (Supplemental Fig. S4D).
> 0.6) and numerous fixed differences between phenotypes in the region 3′ of optix (Supplemental Fig. S4). In the 40-kb peak, which includes optix, only the hybrid zones in Ecuador and Peru showed strong differentiation and fixed differences between races. Individuals from French Guiana showed only moderate genetic differences and no perfectly associated SNPs in this region. The Panama hybrid zone is between two different postman phenotypes and, as expected, contained no indication of genetic divergence between phenotypes across the D interval and no regions of consistent association with phenotype (Supplemental Fig. S4D).
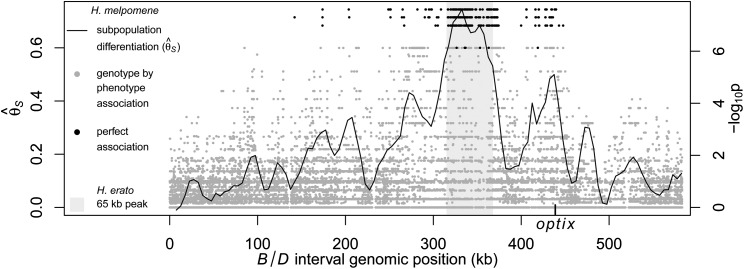
There were striking similarities in patterns of differentiation and association between H. erato and H. melpomene, two species that show identical shifts in their mimetic wing patterns across their geographic ranges (Turner 1975). The same two peaks of population differentiation and phenotypic association were seen when examining H. erato and H. melpomene in the Peruvian hybrid zone (Supplemental Fig. S1). The major peak of divergence and association identified in the H. erato hybrid zones (Fig. 2) perfectly coincides with the region of peak divergence and association identified in H. melpomene hybrid zones (Fig. 4; Supplemental Fig. S1).
Figure 4.
Divergence and association between divergent H. melpomene color pattern races across the B/D interval. The gray shaded area indicates the region of peak divergence and association identified in the co-mimic H. erato (Fig. 2). The solid line indicates sliding window (15-kb window size, 5-kb step size) subpopulation differentiation (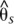 ) between the H. melpomene postman and rayed phenotypes for individuals from two hybrid zones (Peru, Colombia; npostman = 7; nrayed = 7), requiring a minimum of 75% of individuals genotyped for each phenotype at each position and data for at least 20% of positions in the window. A baseline subpopulation differentiation of
) between the H. melpomene postman and rayed phenotypes for individuals from two hybrid zones (Peru, Colombia; npostman = 7; nrayed = 7), requiring a minimum of 75% of individuals genotyped for each phenotype at each position and data for at least 20% of positions in the window. A baseline subpopulation differentiation of 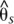 = −0.03 was calculated from genomic scaffolds unlinked to color pattern. The dots indicate genotype by phenotype association calculated for biallelic SNPs using a Fisher's exact test for both hybrid zones (npostman = 7; nrayed = 7), requiring a minimum of 75% of individuals genotyped for each phenotype at each SNP. The black dots indicate association for the 430 SNPs perfectly associated with phenotype.
= −0.03 was calculated from genomic scaffolds unlinked to color pattern. The dots indicate genotype by phenotype association calculated for biallelic SNPs using a Fisher's exact test for both hybrid zones (npostman = 7; nrayed = 7), requiring a minimum of 75% of individuals genotyped for each phenotype at each SNP. The black dots indicate association for the 430 SNPs perfectly associated with phenotype.
Haplotype structure and recombination under the divergence peak
To investigate what might be driving the high divergence in the 65-kb peak, we looked for evidence of chromosomal rearrangements, which could reduce recombination between color pattern races, resulting in high divergence. Using BreakDancer (Chen et al. 2009) to look for paired-end alignments indicative of inversions, we found no evidence of a chromosomal inversion between color pattern races in the H. erato and H. melpomene genomic data across this region. These analyses have limited power due to the necessity of having good coverage across inversion breakpoints from read pairs with one pair in the inverted region and the other outside. Despite these limitations, these results, along with the perfect gene synteny between species, suggest that divergent color pattern races are collinear and can recombine across the D locus.
We more closely examined haplotypes and linkage disequilibrium (LD) across the 65-kb peak of divergence for patterns of recombination. In genomic regions unlinked to color, LD decayed very rapidly, falling to background levels within a few thousand bases (Supplemental Fig. S3). Across the three hybrid zones between rayed and postman races, LD was consistently higher across the 65-kb peak of divergence than across other regions of the D interval (Supplemental Fig. S3).
Although recombination in the 65-kb peak region appears to be substantially reduced, haplotype reconstruction suggests that recombination is present. In the Peruvian hybrid zone, clustering of reconstructed haplotypes into two groups does not support a single large haplotype block distinguishing the two color pattern races. Rather, the clustering revealed several smaller blocks with haplotypes fixed between the color pattern races that were intervened by narrow regions where both races shared the same haplotypes. Similar to the LD analyses, haplotypes clustered more strongly by color pattern in the 65-kb peak than in the regions flanking the peak (Fig. 3B). Further examination of the distribution of SNPs across races revealed that nearly half of the SNPs that differ between hybridizing races were polymorphic in only a single hybrid zone. This large proportion of polymorphic sites private to a specific hybrid zone indicates that alleles and haplotypes shared between hybridizing races across the 65-kb peak likely result from recent gene flow at each hybrid zone, rather than incomplete sorting of ancestral variation among the different races.
The evolutionary history of red wing patterns
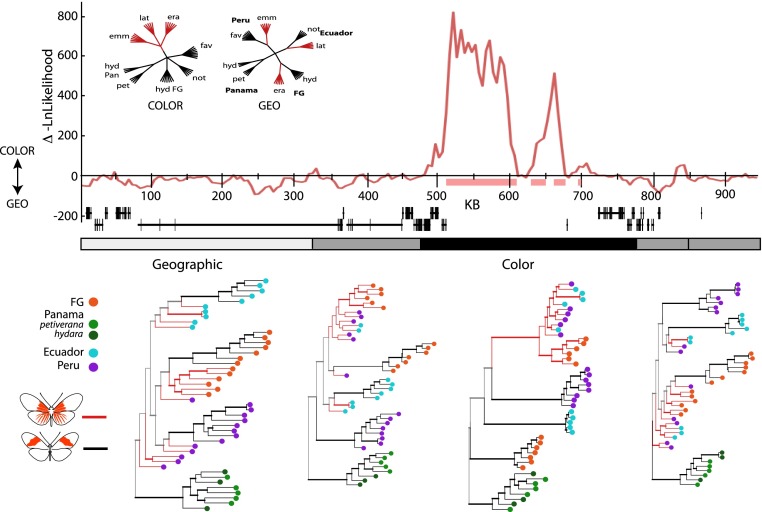
We examined phylogenetic relationships across the D interval using two methods—a sliding window likelihood tree preference and optimally partitioned phylogenetic topologies (Fig. 5). We identified a major shift in evolutionary history across the 200-kb region supported by the divergence and association analyses. Likelihood scores strongly support a tree that clusters samples by color pattern phenotype, rather than the tree that clusters samples by geographic population (Fig. 5). Additionally, the optimally partitioned phylogenetic topologies showed a partition encompassing the 200-kb region where the tree perfectly clusters by color. Moving away from this 200-kb region, phylogenetic topology shifted to a transitional topology between color-based and geography-based trees before transitioning to a geography-based tree on the far edge of the interval (Fig. 5). The tree in the color partition supports a single origin of the rayed color pattern, clustering rayed phenotypes separate from nonrayed phenotypes, as does a tree inferred from the 65-kb region of highest divergence (Supplemental Fig. S5). Branch lengths across topologies show a signature of reduced gene flow, whereby color pattern alleles have reduced gene flow among races and less variation among individuals relative to markers further from the color pattern locus (Fig. 5; Supplemental Fig. S5).
Figure 5.
Phylogenetic trees across the D interval. The top panel is the likelihood preference for color-based versus geography-based tree models (inset). Pink bars beneath the plot represent the region where neighbor-joining trees in this sliding window have a monophyletic lineage for the rayed phenotype. These peaks are shown relative to the annotated genes in the interval. The gray bar in the middle represents the five most optimal topological splits used for phylogenetic tree reconstruction, colored by the general history inferred, with black representing a tree clustering by color pattern, light gray a tree clustering by geographic region, and medium gray for transitional topologies. Tree topologies for these divisions are shown below, excluding the tree for the fourth interval, which does not differ substantially from the fifth tree. Nodes supported by a posterior probability >0.95 are represented with bold branches preceding them. Phenotypes are represented by branch color and geographic regions by terminal node color. Internal branches are colored only in cases of clade monophyly of that pattern, otherwise branches are represented in gray. All unrooted tree topologies were arbitrarily rooted at the Panamanian lineage for presentation.
We compared the evolutionary history of the red color pattern interval between the two co-mimics, H. erato and H. melpomene, which showed concordant peaks of divergence, indicating that they use the same genetic architecture to generate their convergent phenotypes. We aligned 71 regions showing high conservation between the two reference sequences within a 300-kb window that included both peaks of divergence. Phylogenetic analyses of 69 of the 71 fragments support complete monophyly of each species, rather than the clustering of samples by phenotype that would be expected if the phenotypes shared a common origin. Nonmonophyly in the two remaining fragments was driven by poor alignments of a few samples with extensive missing data. We more closely examined histories of SNPs within the 65-kb peak of divergence. After manual removal of regions with poor alignment and SNPs with missing data for more than 25% of samples, we obtained 1164 SNPs. Heliconius erato and H. melpomene share allelic variation across 73 of these 1164 SNPs, none of which show patterns of association with phenotype across both species. Phylogenetic analysis of the 1164 SNPs resolved H. erato and H. melpomene as separate lineages with high support, while clustering races by phenotype within each species (Supplemental Fig. S6). Results from these data support an independent origin of red patterns within each species.
Discussion
The high definition genomic data sets provided by next-generation sequencing techniques can give important insights into how different ecological pressures, complex genetic architectures, and evolutionary histories shape patterns of genomic variation. In this study, we used whole genome resequencing to successfully narrow the region modulating adaptive red color pattern variation in H. erato to a 65-kb interval of peak divergence that showed strong signatures of selection and included almost all of the SNPs perfectly associated with wing pattern phenotype (Fig. 2). There were no predicted genes nor any evidence of transcriptional activity across this region, which strongly supports the assertion that red pattern variation is caused by cis-regulatory changes controlling gene expression during wing development (Reed et al. 2011). Although this genetic architecture appears to be highly conserved across the Heliconius genus, it most likely arose independently in the co-mimics H. erato and H. melpomene.
Signatures of strong selection and recombination across the 65-kb region
The patterns of genomic variation across the H. erato D interval reflect a history of high rates of gene flow coupled with strong selection. Across most of the 2.2 Mb of the genome we analyzed, gene flow has homogenized genomic variation between hybridizing color pattern races of H. erato, resulting in levels of differentiation near zero. Within narrow sections of the D interval, however, there are sharply defined regions of divergence (Fig. 2), consistent with strong natural selection operating on mimetic color patterns (Benson 1972; Mallet and Barton 1989; Mallet et al. 1990; Kapan 2001). We observed a 65-kb region that showed hallmark signatures of strong selection—high divergence between populations, low nucleotide diversity, and strong LD. This region also contained almost all of the fixed allelic differences between divergent color pattern races.
The 65-kb region showing high divergence is much wider than expected given the rapid decay of LD to background levels in regions unlinked to color pattern, often within just a few thousand bases in H. erato (Supplemental Fig. S3; Counterman et al. 2010). Broad and sharp peaks of divergence can be driven by chromosomal rearrangements (e.g., inversion), which are known to suppress recombination (Hartl and Jones 2005), causing elevated patterns of divergence (McGaugh and Noor 2012). Recently, chromosomal inversions were shown to be important in maintaining wing color pattern polymorphisms among sympatric forms of Heliconius numata (Joron et al. 2011). However, we found no evidence for large chromosomal inversions between different color pattern races in our analyses of both gene synteny and the orientation of paired-end alignments.
Additional support against chromosomal inversions comes from finer analysis of haplotype structure, which suggests that recombination, while reduced, is occurring in the 65-kb peak of divergence. Across this peak we identified several regions where individuals from both color pattern races in the H. erato Peruvian hybrid zone shared similar haplotypes (Fig. 3). These blocks of shared haplotypes likely reflect gene flow between divergent races across these genomic regions, which suggests regions of this peak can recombine and introgress between rayed and postman individuals. The presence of recombination in this region indicates that genotyping of more individuals will provide a finer scale picture of how strong selection and recombination between divergent wing pattern races shape patterns of introgression across this 65-kb region of the genome.
In addition to the 65-kb peak of high divergence, we detected a second peak of moderate divergence located at the gene optix. This peak occurred in both H. erato and H. melpomene; however, the height and exact location of the peak varied between replicate H. erato hybrid zones. The peak showed high divergence in all of the western hybrid zones, whereas French Guiana, the only eastern hybrid zone, showed a low peak of divergence. Although the signatures of selection indicate that this region is functionally important, the different patterns of divergence between hybrid zones could be a result of either the unique histories of each hybrid zone or a complex genetic architecture regulating red color pattern variation.
The regulatory architecture of optix
We predict that the peaks of divergence identified in our analyses harbor the key regulatory regions of the transcription factor optix. Previous work has shown that expression patterns of optix are consistent with a critical role in red color pattern variation and the amino acid sequence is conserved across multiple Heliconius species, implicating a regulatory mechanism modulating red variation (Reed et al. 2011). We see strong signatures of selection across a 200-kb gene desert that contains only the gene optix. We believe that the major 65-kb peak of divergence, which is located in the gene desert ∼130 kb 3′ of optix, contains multiple cis-regulatory elements of optix.
The D locus segregates with a simple Mendelian inheritance pattern (Mallet 1989; Kapan et al. 2006); however, phenotypic patterns from H. erato hybrid zones suggest that different red color pattern elements are genetically distinct. Phenotypic hybrid forms having a red forewing dennis patch but no red hindwing rays should not exist if the same genetic variant at the D locus controls both those red pattern elements. Yet such individuals have been found on very rare occasions in hybrid zones (Mallet 1989), and they exist as naturally occurring color pattern races in both H. erato (amalfreda) and H. melpomene (meriana) (Sheppard et al. 1985).
We hypothesize that the 65-kb divergence and association peak actually contains a cluster of tightly linked cis-regulatory elements of optix, each controlling one of the three distinct red pattern elements. There is increasing evidence in other systems that some loci, originally thought to be single large effect loci, are actually clusters of tightly linked genetic changes (Rebeiz et al. 2009; Frankel et al. 2011; Studer and Doebley 2011; Loehlin and Werren 2012; Linnen et al. 2013). Our haplotype analysis supports this hypothesis because the 65-kb peak is rather broad and contains a number of haplotype blocks that are perfectly associated with phenotype, with haplotype blocks of low association interspersed between them. Although this could be a consequence of the history of recombination alone, it could also be indicative of several conserved subregions that control different aspects of phenotypic variation (Fig. 3).
In Heliconius, these clustered regulatory elements may bind different upstream regulatory genes that control the spatially specific expression patterns of optix and ultimately determine scale coloration across the wings. The region could thus act as an evolutionary hotspot, with the flexibility to generate diverse phenotypes, but where strong selection and physical linkage ensure that mimetic patterns are inherited as a whole.
Independent origins of a shared genetic architecture between co-mimics
The genetic architecture driving color pattern convergence and divergence appears to be highly conserved across the Heliconius genus. The D interval showed strikingly consistent differentiation across replicate H. erato hybrid zones between divergent color pattern races (Supplemental Fig. S4). Moreover, the 65-kb divergence and association peak identified in our H. erato analysis is nearly identical to the peak of divergence in the co-mimic, H. melpomene (Fig. 4; Supplemental Fig. S1). These observations raise some interesting questions about the broader evolutionary history of this region. Did the alleles responsible for mimetic patterns in H. erato and H. melpomene evolve independently or do they share a common origin, through either adaptive introgression or shared ancestral variation?
The history of color pattern evolution within H. erato indicates that races with similar red wing patterns likely share a common origin. Consistent with a previous gene-based study (Hines et al. 2011), sliding window phylogenetic analyses and inferred phylogenetic trees for the 65-kb region strongly support a common origin of the rayed pattern in H. erato (Fig. 5; Supplemental Fig. S5).
Adaptive introgression of red color pattern alleles between species could be argued to explain a shared origin of mimetic phenotypes across Heliconius, as it does within the H. melpomene clade (The Heliconius Genome Consortium 2012; Pardo-Diaz et al. 2012). Similarly, ancestral variation, shared between the co-mimics, could also be elicited to explain the shared mimetic phenotypes. Yet, in our comparative analysis across the 65-kb peak that we have identified as responsible for red patterning, we found no evidence for a common origin of shared color pattern variation in these co-mimics. No SNPs were perfectly associated with color pattern in both species, and all genealogies from this region were reciprocally monophyletic for both species. These results are consistent with the deep genetic divergence (Pohl et al. 2009) and lack of hybridization between H. erato and H. melpomene.
Although color patterns likely have a single origin within each species, patterns of variation suggest similar red patterns evolved independently in the co-mimetic species. Analyses between color pattern races within a species show numerous shared SNPs and strong phylogenetic clustering—indicating a shared origin or the rayed phenotype. Analyses between co-mimetic species show no shared SNPs and no phylogenetic clustering—indicating that it is most likely that the two parallel radiations independently evolved mimetic red patterns through changes in the same regulatory regions.
Although we found no evidence for a shared history of red color patterns between the co-mimics H. erato and H. melpomene, it is important to point out several caveats. Foremost, we are only beginning to understand the evolutionary dynamics of this region. Patterns of variation, including SNPs and insertions/deletions, across this noncoding genomic interval are complex, and our strategy of mapping short sequencing reads onto a static reference sequence may miss variation that is functionally important. In addition, mapping of these data onto different species-specific references may introduce some bias. The improvement of algorithms for de novo assembly from short read sequence data (Miller et al. 2010; Schatz et al. 2010), coupled with emerging technologies for generating long sequencing reads from single molecules (Schadt et al. 2010), will allow more detailed dissection of the evolutionary dynamics of this region in the future. Resolution of the question of the origin of mimicry in Heliconius awaits final discovery of the precise functional changes regulating phenotypic variation in both species.
In conclusion, using genomic sequencing of a small number of individuals from multiple admixed hybrid zones, we were able to confidently localize the noncoding genetic switch modulating an adaptive phenotype. We showed that both distantly related co-mimics used this same genetic architecture, but although regulatory alleles appear to have a single origin within species, mutations were most likely independently acquired in the co-mimetic species to produce the shared phenotypes. This study highlights how next-generation sequencing techniques can be leveraged to identify functional variation and to understand the evolutionary history of adaptive radiations.
Methods
Annotation, synteny, and conservation
We annotated 2.2 Mb of Heliconius erato petiverana BAC sequences from the ∼400-Mb H. erato genome (see Supplemental Material section S1 for further details). Approximately 1 Mb of this reference sequence, called the D interval, was previously identified as being involved in red wing color pattern elements (Counterman et al. 2010). We masked repetitive elements in this partial reference genome using RepeatMasker v3-2-9 (Smit et al. 2010) and a Heliconius repetitive elements database (The Heliconius Genome Consortium 2012).
We generated a partial reference transcriptome for H. erato wing tissue using reference-based assembly of Illumina RNA-seq short-read data. We obtained whole transcriptome short-read sequences from hindwing cDNA for 18 individuals, including two divergent H. erato color pattern races and three developmental stages. Transcripts were generated from these pooled samples using the Bowtie/TopHat/Cufflinks pipeline. We aligned each sample to the masked reference sequence using TopHat v1.2.0 (Trapnell et al. 2009) and Bowtie v0.12.7.0 (Langmead et al. 2009), with stringent mapping parameters to minimize false alignments. We generated transcripts by analyzing alignments with Cufflinks v1.0.1 (Trapnell et al. 2010).
We produced automated gene annotations across the H. erato partial genomic reference sequence using the MAKER pipeline v2.09 (Holt and Yandell 2011). This analysis involved masking repetitive elements in the reference sequence, aligning peptide sequences from the UniRef90 (Suzek et al. 2007) and Bombyx mori (Duan et al. 2010) protein databases, and aligning transcripts from the RNA-seq data and previously published EST sequences from wing tissues of H. erato and H. himera (Papanicolaou et al. 2009). MAKER generated ab initio gene models for both the masked and unmasked reference using AUGUSTUS v2.5.5 (Stanke et al. 2006) trained for H. melpomene (The Heliconius Genome Consortium 2012) and SNAP v2010-07-28 (Korf 2004) trained for Bombyx mori. MAKER generated gene predictions by promoting ab initio models with enough supporting evidence from protein homology or transcriptome alignments. We manually curated the predicted genes within the D interval based on the supporting evidence and alignments to annotated Heliconius proteins and curated insect proteins. We assigned gene descriptions and putative functions based on homology with known proteins and protein domains. In addition to protein coding genes, we predicted transcription factor binding sites using the Transcription Element Search System (TESS) (Schug and Overton 1997), searching against known Drosophila binding sites from the TRANSFAC and JASPAR databases.
We examined gene synteny across the D interval between the curated H. erato peptide sequences and H. melpomene v1.0 peptide sequences (The Heliconius Genome Consortium 2012) (see Supplemental Material section S2 for further details). We used Inparanoid v4.0 (Ostlund et al. 2010) to identify one-to-one orthologs and examined genes for consistent order and orientation using OrthoCluster release 2 (Vergara and Chen 2009).
We examined the level of sequence conservation between H. erato and H. melpomene across the D interval (see Supplemental Material section S2 for further details). We used mVista LAGAN (Brudno et al. 2003) to globally align the H. erato D interval sequence and the H. melpomene scaffolds containing the orthologous genes identified by the Inparanoid analysis. We examined sequence conservation in 500-bp windows across the interval, identifying regions of >90% similarity.
Sequencing and genotyping
To determine where in the D interval different red phenotypes diverge genetically, and therefore where the genetic control of the red phenotype is most likely located, we examined genomic sequence data for divergent red color pattern races of H. erato (see Supplemental Material section S3 for further details). We collected 45 individual H. erato butterflies from hybrid zones in Peru (n = 14), French Guiana (n = 13), Ecuador (n = 10), and Panama (n = 8) (Fig. 1; Supplemental Table S6). We collected phenotypically pure individuals of each color pattern race from admixed populations where the ranges of two color pattern races overlap. For dissecting red color pattern variation, the hybrid zones in Peru, French Guiana, and Ecuador are considered replicate hybrid zones since each involves hybridization between rayed and postman races. The Panamanian hybrid zone serves as a control in that both races are postman phenotypes, showing variation only in the yellow phenotypic elements, which are under independent genetic control from the red elements (Mallet 1986). Additionally, we collected six H. melpomene individuals near a hybrid zone in eastern Colombia, three samples representing each of the two major red phenotypes—the postman and the rayed (Supplemental Table S7). We assessed variation across a second H. melpomene hybrid zone in Peru, which is also represented by postman and rayed phenotypes, using published targeted resequencing data (Nadeau et al. 2012).
We sequenced the whole genome of each sample on the Illumina platform, producing 100-bp paired-end reads. We aligned the H. erato sequencing reads to our unmasked H. erato partial reference genome and H. melpomene reads to the H. melpomene genome v1.1 (The Heliconius Genome Consortium 2012) using BWA v0.5.9-r16 (Li and Durbin 2009) with relaxed mapping parameters. We called multisample genotypes across samples for each race using GATK's (DePristo et al. 2011; McKenna et al. 2010) UnifiedGenotyper with heterozygosity set to 0.025 and filtered genotype calls for quality using GATK's VariantFiltration, applying both site and individual sample filters to remove low quality genotypes, low coverage regions, and hypercoverage regions indicative of repetitive elements. We used BreakDancer v1.2.6 (Chen et al. 2009) to identify regions of the reference sequence that showed paired end alignments with incorrect orientations and unexpected distances between pairs, indicating possible structural rearrangement.
Divergence and association analyses
We examined signatures of selection and genotype by phenotype association between divergent color pattern races (see Supplemental Material section S4 for further details). We calculated sliding window genomic divergence between pairs of H. erato color pattern races at each hybrid zone independently and across all hybrid zones combined, with samples classified as either postman or rayed phenotypes. We used a model for diploid data with populations as random effects ( ) (Weir 1996) and no simplifying assumptions regarding sample sizes or number of populations (Weir and Cockerham 1984). For analyzing all hybrid zones combined, we incorporated the geographic structure of the populations by using a three-level hierarchy method (
) (Weir 1996) and no simplifying assumptions regarding sample sizes or number of populations (Weir and Cockerham 1984). For analyzing all hybrid zones combined, we incorporated the geographic structure of the populations by using a three-level hierarchy method (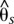 ) (Weir 1996). For all comparisons, we calculated divergence at a position only if at least 75% of the individuals were genotyped for each phenotype. We evaluated 15-kb sliding windows at 5-kb steps across the genomic intervals and required a window to have divergence calculated for at least 20% of the positions in the window. We calculated a baseline level of divergence for each comparison as the level of divergence observed across intervals unlinked to color pattern (H. erato: three unlinked BACs; H. melpomene: 38 unlinked scaffolds).
) (Weir 1996). For all comparisons, we calculated divergence at a position only if at least 75% of the individuals were genotyped for each phenotype. We evaluated 15-kb sliding windows at 5-kb steps across the genomic intervals and required a window to have divergence calculated for at least 20% of the positions in the window. We calculated a baseline level of divergence for each comparison as the level of divergence observed across intervals unlinked to color pattern (H. erato: three unlinked BACs; H. melpomene: 38 unlinked scaffolds).
We estimated genotype by phenotype association at each H. erato hybrid zone independently and across all four H. erato hybrid zones combined. We examined each biallelic SNP using a two-tailed Fisher's exact test based on allele counts. Positions were excluded if <75% of individuals were genotyped for each phenotype.
To look for signatures of selection, we calculated sliding window values for the proportion of segregating sites and heterozygosity for the H. erato Peruvian hybrid zone. We calculated estimates of these parameters for a genomic position only if at least 75% of individuals were genotyped and then looked at 15-kb windows with a 5-kb step size, for windows with at least 20% of positions with parameter estimates. To explore linkage disequilibrium (LD), we examined all biallelic SNPs with at least 75% of individuals genotyped. We calculated correlations (r2) between all pairwise SNPs using PLINK (Purcell et al. 2007), which for unphased data is based on genotype allele counts.
We assessed divergence and association in the H. melpomene hybrid zones as described above. Additionally, for each H. erato fixed SNP, we attempted to identify an orthologous SNP in H. melpomene and determined if the SNP was associated with phenotype in both species.
Linkage disequilibrium and haplotype clustering
We explored haplotype structure in the Peruvian hybrid zone by estimating haplotypes across a 100-kb window of the D interval (500–600 kb) containing the 65-kb peak of divergence and flanking regions using fastPHASE v1.2 (Scheet and Stephens 2006) (see Supplemental Material section S6 for further details). We filtered biallelic SNPs across this 100-kb region to remove sites that had genotypes from <75% of the individuals of each race, resulting in 3227 SNPs. Haplotypes were clustered during phase estimation to two clusters (K = 2) and the proportion of rayed and postman individuals assigned to each cluster at each SNP was determined. We used Haploscope (San Lucas et al. 2012) to visualize regions where the two races had fixed haplotype block differences and where individuals from both races shared the same haplotypes. Using the haplotype estimations from fastPHASE, for each SNP, Haploscope visualizes the portion of individuals from a race (light vs. dark) assigned to each cluster (red vs. gray) across the 100-kb region (San Lucas et al. 2012).
Phylogenetic analyses
We constructed phylogenetic trees across sliding windows in the D interval, sampling 15 kb of sequence every 5 kb (see Supplemental Material section S5 for further details). For each window, we tested the log likelihood of the data with two alternative trees: The geographic tree assumes samples cluster by geographic hybrid zone and the color based tree groups races with a similar color pattern (rayed or postman) (Fig. 5). Likelihood values were calculated for each interval and tree topology using scripts in PAUP* 4b10 (Swofford 2002), using a GTR + G model inferred for the interval as a whole using Modeltest v3.7 (Posada and Crandall 1998). Neighbor-joining trees across these sliding windows constructed in PAUP* were used to infer regions of monophyly by color phenotype.
To summarize variation in phylogenetic topology across the interval, we constrained division of the interval into the five most distinct topologies using the MDL method, raising the likelihood score penalty until five clusters of SNP blocks were reached (Ané 2011). Tree topologies for each of these five regions of the interval were constructed using MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). Analyses involved three runs for 3 million generations each, sampling every 500 generations and removing 33% burn-in and runs that did not converge (as assessed in MrBayes and Tracer v1.5 [Rambaut and Drummond 2007]). Models were assigned using MrModeltest v2.3 (Nylander 2004) and included the GTR model for the second and third regions of the interval and GTR + G for the remaining regions. In addition to these phylogenies, phylogenetic and network-based trees were constructed for both the 65-kb peak region of population differentiation and unlinked genetic regions.
To test whether shared color patterns between the mimics could result from a common origin, we also performed phylogenetic analyses combining H. erato and H. melpomene sequences along this interval. We focused on regions of high conservation between H. erato and H. melpomene (>80% conservation in a 500-bp window) from our mVista alignment within the 515–580 kb region of peak divergence. We used ClustalW2 (Larkin et al. 2007) and manual edits to align sequences from all 45 H. erato individuals (Supplemental Table S6) and 14 H. melpomene individuals (Supplemental Table S7). After filtering, 1134 SNPs were concatenated and used for a Bayesian analysis using the same parameters above and a GTR model.
Data access
Annotated gene models are available on the Heliconius Genome Project website (http://butterflygenome.org/) through the Genome Browser (Data Source: Hera_D_Jan2012) and as a downloadable gff file. Reference sequences are available on Genbank (http://www.ncbi.nlm.nih.gov/genbank) under accession numbers KC469892–KC469895 and AC208805–AC208806. Aligned sequencing reads are available at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRA059512 (resequencing) and SRA060220 (RNA-seq). SNP data, scripts, and tree files for phylogenetic analyses are available at Dryad (datadryad.org) under doi: 10.5061/dryad.rr65n.
Acknowledgments
We wish to thank Claudia Rosales for the tremendous amount of work she did in the laboratory; Jamie Walters and Chris Jiggins for contributing sequencing data; and Chris Smith for computational support. We thank Arnaud Martin, the editor, and three reviewers for their insightful comments on previous versions of the manuscript. We also thank the following permitting agencies for permission to collect butterflies: Peruvian Ministerio de Agricultura and Instituto Nacional de Recursos Naturales (004-2008-INRENA-IFFS-DCB and 011756-AG-INRENA); Ecuadorian Ministerio del Ambiente Ecuadorian (013-09 IC-FAU-DNB/MA); French Guiana Ministere de L'Ecologie, de L'Energie, du Developpemet Durable et de la Mar (BIODAD-2010-0433); Panamanian Autoridad Nacional del Ambiente (SC/A-7-11); and Colombian Ministerio de Ambiente, Vivienda y Desarrollo Territorial (RGE0027-LAM3483). This work was funded by the following awards: CNRS Nouraugues (B.A.C.); NIH F32 GM889942 (H.M.H.) and T32 HD060555 (J.J.L.); NSF DEB-1257839 (B.A.C.), DEB-0844244 (W.O.M.), DEB-0715096 (W.O.M.), and IOS-1305686 (J.J.L.); and the Smithsonian Institution.
Author contributions: Experimental design is credited to W.O.M., B.A.C., H.M.H., D.M.N., and M.A.S.; data collection was carried out by H.M.H., B.A.C., M.A.S., W.O.M., and C.S.; data analysis was done by M.A.S., H.M.H., B.A.C., K.K.D., J.J.L., and W.O.M. with assistance from D.A.R. and C.L.; and the manuscript was prepared by M.A.S., B.A.C., W.O.M., and H.M.H.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.150615.112.
References
- Ané C 2011. Detecting phylogenetic breakpoints and discordance from genome-wide alignments for species tree reconstruction. Genome Biol Evol 3: 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SW, Nadeau NJ, Maroja LS, Wilkinson P, Counterman BA, Dawson A, Beltran M, Perez-Espona S, Chamberlain N, Ferguson L, et al. 2010. Genomic hotspots for adaptation: The population genetics of Müllerian mimicry in the Heliconius melpomene clade. PLoS Genet 6: e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson WW 1972. Natural selection for Müllerian mimicry in Heliconius erato in Costa Rica. Science 176: 936–939 [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Program NCS, Green ED, Sidow A, Batzoglou S 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai P 1986. Field observations and feeding experiments on the responses of rufous-tailed jacamars (Galbula ruficauda) to free-flying butterflies in a tropical rainforest. Biol J Linn Soc Lond 29: 161–189 [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6: 677–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counterman BA, Araujo-Perez F, Hines HM, Baxter SW, Morrison CM, Lindstrom DP, Papa R, Ferguson L, Joron M, ffrench-Constant RH, et al. 2010. Genomic hotspots for adaptation: The population genetics of Müllerian mimicry in Heliconius erato. PLoS Genet 6: e1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Li R, Cheng D, Fan W, Zha X, Cheng T, Wu Y, Wang J, Mita K, Xiang Z, et al. 2010. SilkDB v2.0: A platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res 38: D453–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL 2011. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Jones EW 2005. Genetics: Analysis of genes and genomes, 6th ed. Jones & Bartlett, Sudbury, MA. [Google Scholar]
- The Heliconius Genome Consortium 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines HM, Counterman BA, Papa R, Albuquerque de Moura P, Cardoso MZ, Linares M, Mallet J, Reed RD, Jiggins CD, Kronforst MR, et al. 2011. Wing patterning gene redefines the mimetic history of Heliconius butterflies. Proc Natl Acad Sci 108: 19666–19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M 2011. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, Whibley A, Becuwe M, Baxter SW, Ferguson L, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapan DD 2001. Three-butterfly system provides a field test of Müllerian mimicry. Nature 409: 338–340 [DOI] [PubMed] [Google Scholar]
- Kapan DD, Flanagan NS, Tobler A, Papa R, Reed RD, Acevedo Gonzalez J, Ramirez Restrepo M, Martinez L, Maldonado K, Ritschoff C, et al. 2006. Localization of Müllerian mimicry genes on a dense linkage map of Heliconius erato. Genetics 173: 735–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen CR, Poh YP, Peterson BK, Barrett RD, Larson JG, Jensen JD, Hoekstra HE 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339: 1312–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin DW, Werren JH 2012. Evolution of shape by multiple regulatory changes to a growth gene. Science 335: 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J 1986. Hybrid zones of Heliconius butterflies in Panama and the stability and movement of warning colour clines. Heredity 56: 191–202 [Google Scholar]
- Mallet J 1989. The genetics of warning colour in Peruvian hybrid zones of Heliconius erato and H. melpomene. Proc R Soc Lond B Biol Sci 236: 163–185 [Google Scholar]
- Mallet J, Barton NH 1989. Strong natural selection in a warning-color hybrid zone. Evolution 43: 421–431 [DOI] [PubMed] [Google Scholar]
- Mallet J, Barton N, Lamas G, Santisteban J, Muedas M, Eeley H 1990. Estimates of selection and gene flow from measures of cline width and linkage disequilibrium in Heliconius hybrid zones. Genetics 124: 921–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Orgogozo V 2013. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250 [DOI] [PubMed] [Google Scholar]
- McGaugh SE, Noor MAF 2012. Genomic impacts of chromosomal inversions in parapatric Drosophila species. Philos Trans R Soc Lond B Biol Sci 367: 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G 2010. Assembly algorithms for next-generation sequencing data. Genomics 95: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW, Quail MA, Joron M, ffrench-Constant RH, Blaxter ML, et al. 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos Trans R Soc Lond B Biol Sci 367: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA 2004. MrModeltest v2. (Program distributed by the author.) Evolutionary Biology Centre, Uppsala University, Sweden.
- Ostlund G, Schmitt T, Forslund K, Köstler T, Messina DN, Roopra S, Frings O, Sonnhammer ELL 2010. Inparanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38: D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R, Martin A, Reed RD 2008. Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr Opin Genet Dev 18: 559–564 [DOI] [PubMed] [Google Scholar]
- Papanicolaou A, Stierli R, ffrench-Constant R, Heckel D 2009. Next generation transcriptomes for next generation genomes using est2assembly. BMC Bioinformatics 10: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl N, Sison-Mangus M, Yee E, Liswi S, Briscoe A 2009. Impact of duplicate gene copies on phylogenetic analysis and divergence time estimates in butterflies. BMC Evol Biol 9: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA 1998. MODELTEST: Testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ 2007. Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. 2011. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333: 1137–1141 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rosser N, Phillimore AB, Huertas B, Willmott KR, Mallet J 2012. Testing historical explanations for gradients in species richness in heliconiine butterflies of tropical America. Biol J Linn Soc Lond 105: 479–497 [Google Scholar]
- San Lucas FA, Rosenberg NA, Scheet P 2012. Haploscope: A tool for the graphical display of haplotype structure in populations. Genet Epidemiol 36: 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Turner S, Kasarskis A 2010. A window into third-generation sequencing. Hum Mol Genet 19: R227–R240 [DOI] [PubMed] [Google Scholar]
- Schatz MC, Delcher AL, Salzberg SL 2010. Assembly of large genomes using second-generation sequencing. Genome Res 20: 1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M 2006. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Overton GC 1997. TESS: Transcription element search software on the www. In Technical Report CBIL-TR-1997-1001-v0.0 Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania. http://www.cbil.upenn.edu/node/30
- Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC 1985. Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Philos Trans R Soc Lond B Biol Sci 308: 433–610 [Google Scholar]
- Smit AFA, Hubley R, Green P 2010. RepeatMasker Open-3.0. http://www.repeatmasker.org
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B 2006. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res 34: W435–W439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V 2009. Is genetic evolution predictable? Science 323: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer AJ, Doebley JF 2011. Do large effect QTL fractionate? A case study at the maize domestication QTL teosinte branched1. Genetics 188: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH 2007. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23: 1282–1288 [DOI] [PubMed] [Google Scholar]
- Swofford DL 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JRG 1975. A tale of two butterflies. Nat Hist 84: 28–37 [Google Scholar]
- Vergara IA, Chen N 2009. Using OrthoCluster for the detection of synteny blocks among multiple genomes. Curr Protoc Bioinformatics 27: 6.10.1–6.10.18 [DOI] [PubMed] [Google Scholar]
- Weir BS 1996. Genetic data analysis II Sinauer Associates, Sunderland, MA. [Google Scholar]
- Weir BS, Cockerham CC 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]