Abstract
Inflammatory bowel disease (IBD) is a debilitating and widespread immune-mediated illness characterized by excessive inflammatory and effector mucosal responses leading to tissue destruction at the gastrointestinal tract. Interactions among the immune system, the commensal microbiota and the host genotype are thought to underlie the pathogenesis of IBD. However, the precise etiology of IBD remains unknown. Diet-induced changes in the composition of the gut microbiome can modulate the induction of regulatory versus effector immune responses at the gut mucosa and improve health outcomes. Therefore, manipulation of gut microbiota composition and the local production of microbial-derived metabolites by using prebiotics, probiotics and dietary fibers is being explored as a promising avenue of prophylactic and therapeutic intervention against gut inflammation. Prebiotics and fiber carbohydrates are fermented by resident microflora into short chain fatty acids (SCFAs) in the colon. SCFAs then activate peroxisome proliferator-activated receptor (PPAR)γ, a nuclear transcription factor with widely demonstrated anti-inflammatory efficacy in experimental IBD. The activation of PPARγ by naturally ocurring compounds such as conjugated linoleic acid, pomegranate seed oil-derived punicic acid, eleostearic acid and abscisic acid has been explored as nutritional interventions that suppress colitis by directly modulating the host immune response. The aim of this review is to summarize the status of innovative nutritional interventions against gastrointestinal inflammation, their proposed mechanisms of action, preclinical and clinical efficacy as well as bioinformatics and computational modeling approaches that accelerate discovery in nutritional and mucosal immunology research.
Keywords: Inflammatory bowel disease, Prebiotics, Fibers, Probiotics, Conjugated linoleic acid, Punicic acid, Abscisic acid, Animal models, Computational modeling
1. Introduction
Inflammatory bowel disease (IBD) is a chronic immune-mediated illness of unknown etiology associated with a dysregulated mucosal immune response to intestinal microorganisms in a genetically susceptible host [1]. IBD is characterized by the destruction of gut tissue, which is initiated from alterations of the intestinal epithelium barrier function involving increased tight junction permeability and maintained by a defective down-regulation of mucosal immunity toward the intestinal microflora [2]. The two main clinical manifestations of IBD are Crohn’s disease (CD) and ulcerative colitis (UC). UC is characterized by the presence of localized inflammation and superficial lesions in the colon, whereas CD is associated with discontinuous and transmural lesions of the gut wall which can affect the whole intestine, although a colonic presentation is most typical [1]. Both innate immunity and adaptive immunity play a role in IBD pathogenesis, but dysregulation of T cell responses contributes to the chronicity of IBD [2]. The immunopathology of CD is associated with dysregulated T helper (Th) 1 responses characterized by over-production of interferon-γ (IFN-γ) and tumor necrosis factor α (TNF-α) [1]. More recent studies demonstrate an important role of Th17 cells as key contributors to the immunopathology of IBD [3]. In contrast to the predominance of Th1 and Th17 responses in CD, UC is mediated by a predominant Th2 response [4].
IBD affects up to 0.5% of the human population in developed countries, and numbers are increasing in the developing countries [5]. The total number of IBD cases in the United States is estimated to be around 1 to 1.5 million [6], which results in annual direct health costs of $6.3 billion, with pharmaceutical claims accounting for ~30% of those expenses [7]. Current treatments for IBD include corticosteroids (i.e., 6-methylprednisolone and budesonide), aminosalicylates and immunomodulators (i.e., azathioprine, 6-mercaptopurine, cyclosporine and methotrexate) and the Food and Drug Administration (FDA)-approved, anti-TNF-α humanized antibodies [8]. These therapies ameliorate IBD by inducing and maintaining clinical remission, but cannot be considered for the long-term management of the disease due to their significant adverse side effects, which include immune suppression, enhanced susceptibility to malignancies and suppressed resistance against infectious diseases [9]. In addition, none of these approaches has been approved as a prophylactic. Dissatisfaction with current traditional therapies has resulted in increased use of complementary and alternative medicine approaches such as prebiotics and probiotics, with an estimated incidence of 49.5% among IBD patients [10]. Thus, exploring novel therapeutic and preventive approaches for IBD and their mechanisms of anti-inflammatory activity is both novel and important. This review will summarize the status of innovative nutritional interventions against gastrointestinal inflammation, their proposed mechanisms of action, and their preclinical and clinical efficacy, and present novel computational modeling approaches that can be applied to accelerate knowledge discovery in nutritional and mucosal immunology research.
2. Mucosal and nutritional immunology of relevance to gut inflammation
The principal challenge of the intestinal immune system is balancing the host response to pathogens while not responding to stimuli derived from commensal microbiota and food antigens [7]. Commensal bacteria residing in the intestinal lumen are thought to contribute to immune tolerance, although translocation of the same commensal microfloral antigens to the lamina propria (LP) results in effector and inflammatory responses. Also, mice are better protected from colitis in a germ-free environment [11], suggesting a role of the gut microbiota in the pathogenesis of IBD. Mucosal immune homeostasis can be disrupted in IBD due to alterations in (a) barrier function of the epithelium, (b) the innate immune cells [e.g., macrophages and dendritic cells (DC)] which provide the initial innate response to invading bacteria and (c) lymphocyte function in both the LP and the mesenteric lymph nodes (MLN), including the T cell population of the normal gut mucosa [12].
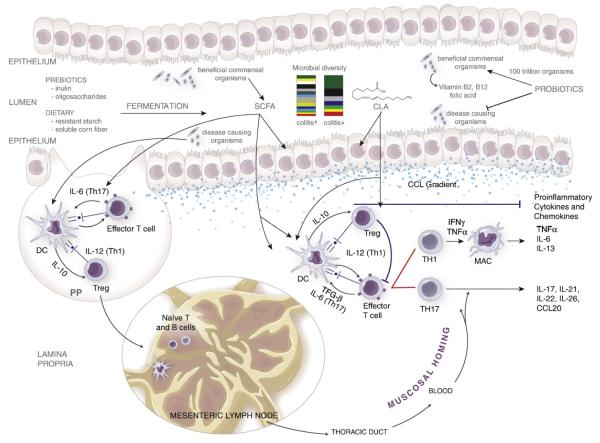
Absorptive and secretory cells, as well as different epithelial cell subsets such as microfold cells (M cells), goblet cells and Paneth cells, form a single cell layer that constitutes the epithelial barrier. Goblet cells form a protective mucus layer, whereas Paneth cells secrete potent antimicrobial peptides known as defensins in the base of small intestinal crypts. M cells are found in the follicle-associated epithelium of the Peyer’s patches sampling intestinal mucosal contents and delivering them via transcytosis to DC and lymphocytes. The disruption of the epithelial barrier or the infiltration of pathogenic bacteria into the LP activates DC and macrophages, which process the antigen and present it on their surfaces through the MHC class II complex. These antigen-presenting cells (APC) are transported to the MLN, where they promote the differentiation of naïve T cells into effector and regulatory T cells (Treg). The cytokine environment secreted in part by the APC skews the differentiation of naïve CD4+ T cells into T helper (Th1, Th2, Th17, Th9, T follicular helper, Th22) or induced/adaptive Treg cell subsets. These now differentiated T cells migrate back to the LP, where they start the adaptive immune response towards the antigen. A predominance of dysregulated Th1 and Th17 responses in the colonic LP has been associated with CD [12]. Fig. 1 illustrates some key aspects of mucosal immunity relevant to the pathogenesis of gut inflammation in the context of nutritional protective mechanisms.
Fig. 1.
Mucosal immune mechanisms of nutritional protection against colitis. The disruption of the epithelial barrier or the infiltration of pathogenic bacteria into the LP or Peyer’s patches (PP) activates DC and macrophages (MAC), which process the antigen and present it on their surfaces through the MHC class II complex. These APC either stay in the PP or are transported to the MLN, where they promote the differentiation of naïve T cells into effector and regulatory T cells. The cytokine environment secreted in part by the APC skews the differentiation of naïve CD4+ T cells into T helper (Th1 and Th17) or Treg subsets. Th1 cells differentiate in the presence of IL-12; IL-6 and TGF-β induce a Th17 phenotype, whereas IL-10 induces a Treg cell differentiation. These mature T cells migrate back to the LP following a chemokine gradient (CCL), where they start the adaptive immune response towards the antigen, resulting in the production of effector and proinflammatory cytokines. The administration of prebiotics (inulin, oligofructose), dietary fibers (resistant starch, soluble corn fiber) and probiotics decreases effector responses and proinflammatory cytokine expression by the production of SCFAs. Dietary compounds such as CLA, PUA, ESA and ABA, as well as probiotic-derived SCFA, can activate PPARγ to suppress inflammation.
3. Animal models of gut inflammation
The development of animal models of colitis as a means of studying the pathogenesis of human IBD and testing novel therapeutics started almost half a century ago [13]. The recent introduction of multiple models of mucosal inflammation indicates that a number of immune imbalances can result in loss of tolerance to mucosal antigens and therefore induction of gastrointestinal immunoinflammatory responses. In this review, we will summarize two models of acute colitis, dextran sodium sulfate (DSS)-induced and trinitrobenzene sulfonic acid (TNBS)-induced colitis, and two models of chronic colitis, adoptive transfer studies using severe combined immunodeficient mice (SCID), lacking mature T or B cells due to a loss-of-function mutation of the protein kinase, DNA-activated, catalytic polypeptide (Prkdc) gene, or recombination activating gene 2-deficient (Rag2−/−) mice, which fail to produce B or T lymphocytes, and the interleukin-10 (IL-10) deficient mice.
The models of mucosal inflammation can be classified into two broad categories: “type 1 models,” including TNBS- and DSS-induced colitis, in which the defect lies with the effector mechanism of the mucosal response, and “type 2 models,” including SCID-transfer colitis and IL-10 deficiency, wherein the effector response is normal but the regulatory cell response is impaired [2].
3.1. TNBS-induced colitis
The rectal administration of TNBS, a hapten, results in a transmural infiltrative disease limited to the colon and characterized by an IL-12-driven, Th1-mediated response based on CD40–CD40L interactions [14]. Some studies have shown the importance of TNF-α in both the initiation and the persistence of the Th1 response since TNBS could not induce colitis in TNF-α-deficient mice [15]. On the other hand, transforming growth factor (TGF)-β seems to be the main regulatory cytokine, although IL-10 is necessary for the effectiveness of the TGF-β response [16]. Hapten-induced colitis is a relevant model of gut inflammation since it allows the study of early events in the development of mucosal inflammation, as well as the analysis of the relation of the response to a specific antigen (hapten) to the overall mucosal immune response leading to colonic inflammation [2]. However, this is a severe model of acute colitis, and it might not be ideal for testing nutritional protective mechanisms.
3.2. DSS-induced colitis
This is an old acute model that is frequently used to study the efficacy of potential therapeutic compounds since a consistent level of colitis is easily induced by the administration of DSS in the drinking water [17]. Early changes in the epithelial barrier result in the permeabilization of the intestinal wall, setting the stage for macrophage activation and the release of proinflammatory cytokines [17,18]. This model has been widely utilized for testing both efficacy and mechanism of action of both drugs and nutritional compounds. The severity of disease can be calibrated by the amount of DSS added and the length of administration, although the most typical regimen consists of 2.5% DSS in the drinking water for 7 days, which we have used to test the anti-inflammatory mechanisms of conjugated linoleic acid (CLA) [9], abscisic acid (ABA) [19,20] and pomegranate seed oil (PSO) [21].
3.3. The SCID-transfer model of colitis
This chronic model of mucosal inflammation is produced by the transfer of SCID or Rag2−/− mice with either CD45RBhi T cells (naïve T cells), which leads to severe colitis in 3–5 weeks, or CD45RBhi T cells and CD45RBlo T cells (mature T cells), in which no inflammation occurs. Therefore, this model allows the identification and study of two cell populations: the effector cells in CD45RBhi T cell populations and regulatory cells in CD45RBlo T cell populations [2]. In this case, the inflammation is limited to the colon, and it is also characterized by a Th1 response driven by IL-12 and mediated by IFN-γ [22], although there is recent evidence showing that Th17 cells also play an important role in mediating colonic immunopathology in this model. Several studies have also suggested TGF-β as the major suppressor cytokine and the requirement of IL-10 to facilitate TGF-β secretion and activity [9,23,24]. Moreover, cell activation in SCID-transferred mice is an antigen-driven event occurring as a result of a dysregulated response to antigens in the mucosal microbiota. Therefore, this model demonstrates that abnormal reactivity to antigen in the mucosal microflora can occur in the absence of genetic abnormality and does not require disruption of the epithelial barrier [2]. This model was used to investigate the mechanisms of anti-inflammatory action of CLA [9] and probiotic bacteria [25].
3.4. IL-10 deficiency colitis
IL-10-deficient mice develop colitis characterized by epithelial cell hyperplasia and transmural inflammation. The early stage of the disease is characterized by a Th1 response that can be ameliorated by anti-IL-12 antibody treatment. However, Th2 mediates the response later on due to the absence of IL-10, and the lesions are no longer treatable with anti-IL-12 [26]. Interestingly, IL-10-deficient mice do not develop colitis under germ-free conditions [27]. Therefore, antigens in the mucosal microflora contribute to the disease pathogenesis in this model. IL-10-deficient mice show increased intestinal permeability even before they develop colitis. This might increase the stimulation by antigens in the mucosal microflora, thus facilitating the development of inflammation [28]. We have used this model to test the anti-inflammatory efficacy of dietary fibers [29], punicic acid (PUA) [21,29] and probiotics [25].
3.5. Pig models of colitis
There are three distinctive advantages of using pig and gnotobiotic (Gn) pig models to study the immunomodulating mechanism of nutritional interventions: (a) neonatal Gn pigs avoid the interference of maternal antibodies/immunoregulators and confounding factors from commensal organisms and therefore provide an immunologically naïve background that allows clear identification of the immune-modulating effects of probiotics in hosts colonized with the uniform and clearly defined microflora; (b) the gastrointestinal, nutritional, metabolic and immunologic similarities between pigs and humans [30]; and (c) relatively large numbers of immune cells can be harvested from each tissue of the pig, allowing comprehensive studies of many immunological parameters to identify the mechanism underlying the immunomodulating effect of nutritional interventions. This makes pig models ideal for translational gut inflammation research.
We have used pig models successfully to investigate the anti-inflammatory mechanisms of fish oil and CLA [31]. A pilot study to titrate the optimal dose of DSS was performed, showing that the administration of 4% DSS in 100 ml of water by gastric intubation once daily for 7 days is optimal to induce colitis in pigs [31]. We have also developed a bacterial-induced model of colitis in pigs, in which pigs are challenged with two doses of 1010 colony forming units of Brachyspira hyodysenteriae given in two consecutive days to study the anti-inflammatory effects of CLA. This model results in dysentery, a severe mucohemorrhagic diarrheal disease and colitis characterized by mucosal enlargement as a result of crypt elongation and epithelial necrosis [32,33]. We have used the Gn pig model of human rotavirus infection to characterize the dose effects of probiotic Lactobacillus rhamnosus GG and L. acidophilus NCFM strains on APC, natural and induced Treg cell, IFN-γ-producing T cell, and B cell responses in the intestinal and systemic lymphoid tissues [34]. More specifically, we demonstrated that, in Gn pigs receiving an oral attenuated rotavirus vaccine, high-dose L. acidophilus induced strong Treg cell responses and promoted IL-10 and TGF-β production by tissue-residing Treg cells, whereas low-dose L. acidophilus significantly enhanced IFN-γ-producing T cell and decreased Treg cell responses, but it did not enhance virus-specific B cell and antibody responses. The low- and intermediate-dose L. rhamnosus GG and intermediate-dose L. acidophilus significantly increased rotavirus-specific effector/memory T cell, B cell and antibody responses and down-regulated the Treg cell responses, leading to significantly increased protection rate against virulent rotavirus challenge [34,35]. Together, these animal models provide an ideal window to explore the immunological mechanisms controlling inflammation at the gut mucosa and how they can be modulated nutritionally.
4. The role of prebiotics and dietary fibers in gut inflammation
Recent evidence suggests that the intestinal microflora contributes to modulating immune responses and protecting from gut inflammatory diseases [36]. Accordingly, the use of specific prebiotics to stimulate growth and activity of intestinal microbiota has been successful in animal models of colitis [37], although knowledge regarding their mechanism of action is limited. Prebiotics are nondigestible oligosaccharides, defined as “selectively fermented ingredients that allow specific changes, both in the composition and/or activity of the gastrointestinal microflora that confers benefits upon host wellbeing and health” [38]. Nowadays, only two dietary nondigestible oligosaccharides fulfill all the criteria for prebiotic classification: inulin and oligofructose, which are natural food ingredients or dietary fibers present in certain plants as storage carbohydrates [39]. Many other food components, including other dietary fibers, have been claimed to have prebiotic activity [38] even though they do not meet the required criteria: (a) resistance to gastric acidity, hydrolysis by mammalian enzymes and absorption in the upper part of the gastrointestinal tract; (b) fermentation by beneficial bacteria in the intestine; and (c) selective stimulation of growth and/or activity of colonic microflora toward a healthier composition [40].
Prebiotics and fiber carbohydrates are not digested in the upper gastrointestinal tract, and they are thought to be selectively fermented by residential bacteria into short chain fatty acids (SCFAs) and lactate once they reach the colon. Besides increasing the production of SCFAs such as acetate, propionate and butyrate, other protective mechanisms of prebiotic activity have been proposed including changes in the intestinal microbiota, improvement of the intestinal barrier and regulation of the mucosal and systemic immune response [41]. However, the mechanisms underlying these effects remain unknown.
Dietary prebiotics can be applied to the prevention of human enteric inflammatory disorders while maintaining optimal levels of immune surveillance. In this regard, dietary fibers can act as effective prebiotics by altering the intestinal microbial composition and promoting the growth of beneficial bacterial communities within the large intestine, resulting in colitis reduction [42,43]. Recent studies showed how prebiotics increased the number of beneficial bacteria such as lactobacilli and bifidobacteria, while decreasing the disease-causing bacteria in both animal models and human clinical studies [42,44]. Moreover, prebiotics can also provide resistance to colonization by pathogenic bacteria by inhibiting the adherence of pathogens to the gut epithelium. For instance, inulin inhibits the in vitro intestinal colonization of Clostridium difficile [45].
C. difficile is typically a harmless anaerobic bacterium, but recently, it has reemerged as a pathogen that can cause nosocomial diarrhea, colitis and even death [46], particularly after antibiotic treatment. It grows in the intestine of individuals with altered commensal microflora [47] due to treatment with antimicrobials, immunosuppressants, cytostatic agents or proton pump inhibitors [48]. An increase in both incidence and severity of C. difficile-associated disease has been reported over the last years [49], probably due to the emergence of new hypervirulent strains such as NAP1/BI/027, which has resulted in increased morbidity and mortality in the United States, Canada and Europe [50].
There are several explanations for the selective stimulation of protective microflora in detriment of disease-causing bacteria. Fermentation of nondigestible carbohydrates in the colon decreases the pH, which inhibits the growth of certain organisms such as Bacteroides spp. [50,51]. Some protective bacteria like Bifidobacterium infantis have specific enzymes that hydrolyze saccharides, resulting in their own proliferation [52]. Other organisms can induce the proliferation of beneficial bacteria by a cross-feeding mechanism. For instance, B. longum releases free fructose during oligofructose degradation, which results in the proliferation of other organisms, which are not able to ferment oligofructose themselves [53]. Although several bacteria can ferment prebiotics, lactobacilli and bifidobacteria are their most capable fermenters [54,55].
Results of a recent study demonstrated the efficacy of resistant starch (RS), soluble corn fiber (SCF) and inulin to ameliorate clinical disease and prevent inflammatory lesions in an IL-10−/− mouse model of IBD [29]. The protective effect of RS was proposed to be associated with an increase in SCFA production, butyrate in particular [56]. A decrease in “healthy” microbiota and of SCFAs is characteristic of patients with IBD [36]. Therefore, the increase of colonic SCFA production by the fermentation of RS-75 and SCF was considered a likely mechanism of action [57,58]. The changes occurring in the gastrointestinal lumen due to administration of dietary fiber of RS are well characterized, but little is known about their immunomodulatory effects in the mucosal immune system. We found that the preventive effect of RS-75 is also associated with increased percentages of Treg in the spleen and Peyer’s patches as well as a reduced production of IFN-γ, suggesting a suppression of Th1 cells in the gut. Dietary RS-75 and inulin supplementation modulates the Treg compartment, causing changes in the gut’s microbial ecology and resulting in increased butyrate levels that subsequently activate peroxisome proliferator-activated receptor (PPAR)γ [59]. PPARγ is a nuclear receptor and transcription factor involved in lipid metabolism and glucose homeostasis [60]. Interestingly, PPARγ was required for the protective anti-inflammatory actions of Treg against effector T cell-induced colitis in mouse adoptive co-transfer studies [61]. Moreover, PPARγ antagonizes several proinflammatory pathways such as STAT, AP-1 and NF-κB. Therefore, activation of PPARγ represents a conserved anti-inflammatory mechanism involved in the prevention of inflammatory and immune-mediated diseases [9,32,61,31]. PPARγ was first shown to be efficacious in suppressing intestinal inflammation based on results of DSS challenge studies using synthetic agonists known as thiazolidinediones [62]. To date, mounting evidence supports the prophylactic and therapeutic effects of PPARγ activation in multiple strains of mice, rats and pigs with acute colitis induced by chemical compounds [62,63] or gut pathogens [32], as well as chronic colitis occurring after the transfer of naive CD4+ T cells in SCID mice [9] or spontaneous panenteritis in IL-10-deficient mice [64]. Therefore, targeting PPARγ is a promising avenue for developing novel prophylactic and therapeutic interventions for gut inflammation. A recent study in UC patients demonstrated that rosiglitazone (Avandia), a thiazolidinedione agonist of PPARγ and a US FDA-approved drug for treating type II diabetes, is also efficacious in the treatment of mild to moderately active UC [65]. However, rosiglitazone is unlikely to be adopted for treating IBD due to its significant side effects [66,67]. Hence, the discovery of novel naturally ocurring agonists of PPARγ that exert therapeutic and prophylactic actions against IBD with no adverse side effects is timely and necessary.
5. Activation of PPARγ by naturally occurring compounds
Many polyunsaturated fatty acids (PUFA) and their metabolites are natural ligands for PPARγ [68] and have been proposed as a promising avenue for developing safer nutritional interventions against gut inflammation without adverse side effects. In an effort to expedite the discovery of anti-inflammatory natural products, virtual screening (VS) can complement traditional experimental methods for identification of novel PPARγ agonists. VS allows the screening of thousands of compounds within a collective of large compound libraries in a cost-effective manner [69]. Our group has successfully established a protocol for screening fatty acid compounds against PPARγ as a means to identify novel agonists, which can be later verified through in vitro assays [70,71]. We performed a small-scale screen focusing on conjugated trienes due in part to their structural similarity to CLA, which is a well-known PPARγ agonist (Fig. 2). Moreover, conjugated trienes are effective at ameliorating chronic inflammation [72,73]. Hence, the integration of molecular modeling with wet nutritional immunology experimental validation represents a cost- and time-efficient approach for the discovery of novel anti-inflammatories.
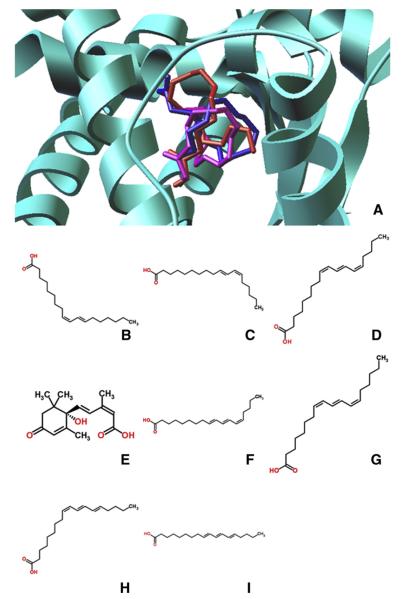
Fig. 2.
Chemical structure of naturally occuring agonists of PPARγ. (A) Representative binding mode of the most stable docked orientation of ligands with PPARγ illustrated in ribbon mode. Ligand poses are generated by AutoDock Vina. Cis-9, trans-11 CLA is in red; trans-10, cis-12 CLA is in magenta; PUA is in blue. (B) c-9,t-11 CLA. (C) t-10,c-12 CLA. (D) 2-D PUA. (E) ABA. (F) 2-CAA. (G) JAA. (H) α-ESA. (I) β-ESA. (J) Butyric acid, a type of SCFA.
5.1. Conjugated linoleic acid
CLA refers to a mixture of positional and geometric isomers of linoleic acid (LA), of which cis-9,trans-11 (c9,t11) and trans-10,cis-12 (t10,c12) CLAs predominate. CLA is naturally present in milk, cheese and ruminant products and can be produced industrially by partial hydrogenation of LA [74,75]. CLA has been considered for the prevention and treatment of gut inflammation since 2002 [76]. By using a bacterial-induced colitis pig model, we found that dietary CLA supplementation suppresses colonic inflammation and up-regulates colonic PPARγ expression. Specifically, CLA ameliorated tissue inflammation and weight loss associated with B. hyodysenteriae-induced colitis [32]. In 2004, similar findings were observed in mouse models of DSS- and CD4-induced colitis [9]. CLA repressed TNF-α expression and NF-kappaB activation while inducing the immuno-regulatory cytokine TGF-β1. This seminal publication provided molecular evidence in vivo demonstrating that the loss of the PPARγ gene in the colon abrogated the beneficial effects of CLA in DSS colitis, suggesting that CLA ameliorates colitis through a PPARγ-dependent mechanism. PPARγ is mainly expressed in the colon by epithelial cells and LP mononuclear cells such as macrophages and T and B cells. Although some studies show the importance of PPARγ in each cell type, additional investigations in animals with cell type-specific expression of PPARγ are required to determine the main cellular source responsible for the therapeutic effect of PPARγ [77].
More recently, we investigated the immunoregulatory efficacy of CLA in patients with mild to moderate CD [78]. Overall, oral CLA administration was well tolerated and suppressed the ability of peripheral blood T cells to produce proinflammatory cytokines such as IFN-γ, TNF-α and IL-17, decreased disease activity and increased the quality of life of patients with CD. Thus, the results of this clinical trial validated the findings of previous pig and mouse CLA-feeding studies. In contrast to the promising clinical findings with CLA in CD patients, clinical data on n-3 PUFA in IBD remain generally unimpressive. The gut anti-inflammatory activity of CLA versus n-3 PUFA was reviewed previously [79].
5.2. PSO-derived PUA
PUA, also known as trichosanic acid, is a conjugated triene fatty acid naturally found at high concentrations in the seed of Punica granatum (Punicaceae, pomegranate) and Trichosanthes kirilowii [73]. PUA constitutes 64% to 83 % of the PSO, but it also contains minor amounts of α-eleostearic acid (ESA), catalpic acid (CAA) and jacaric acid (JAA) [80]. All these acids are collectively known as conjugated linolenic acids (CLnAs) and are stereo- or regioisomers of each other (Fig. 2). PUA is a conjugated octadecatrienoic acid containing c9, t11, c13 double bonds that resembles the cis-9, trans-11 CLA isomer [76,81]. Due to the similarities between the chemical structure of PUA and the c9, t11 CLA isomer, PUA was suggested as a potential PPARγ agonist. PUA was shown to robustly bind and activate PPARγ, therefore increasing PPARγ-responsive gene expression and ameliorating diabetes and gut inflammation. The loss of PPARγ in immune cells impaired its ability to modulate glucose homeostasis and obesity-related inflammation [73]. PUA was later shown to ameliorate DSS-induced colitis and spontaneous panenteritis in IL-10−/− mice [70]. Specifically, PUA intake down-regulated RORγt expression in the colon and suppressed colonic and M1 macrophage-derived TNF-α. Dietary PUA also increased the levels of IL-17 and IFN-γ in CD8+ T cells in the MLN. In line with previous studies, the loss of PPARγ in immune cells impaired the ability of PUA to ameliorate experimental colitis, strongly suggesting that PUA modulates mucosal immune responses and ameliorates gut inflammation through a PPARγ-dependent mechanism. Recent studies by our group show that the immune modulatory actions of ESA may be both PPARγ dependent and PPARγ independent to ameliorate disease activity and intestinal lesions in mice with DSS colitis.
A recently published study showed that supplementation of PSO into milk formula reduces the incidence and severity of necrotizing enterocolitis (NEC) in rats [82]. NEC is a devastating disease associated with severe and excessive intestinal inflammation, and it is the major cause of morbidity and mortality in premature infants. Although its etiology is unknown, it is thought that the combination of intestinal immaturity, high immunoreactivity of the intestinal mucosa and genetic predisposition lead to the development of NEC [83]. The administration of PSO protects against NEC in a neonatal rat model. PSO’s beneficial effect is associated with improved enterocyte proliferation, protection of intestinal architecture and reduced expression of proinflammatory cytokines.
In addition to CLnAs, PSO also contains a diverse array of phytochemicals, including polyphenolic constituents, hydrolyzable tannins and condensed tannins [84]. Phytosterols (i.e., β-sitosterol, campesterol and stigmasterol) are also found in quite high concentrations in the PSO (4089–6205 mg/kg of PSO) [85]. Some of these compounds seem to have potent antioxidant and anti-inflammatory properties implicated in the prevention and intervention of several inflammation-related diseases, including cancer [84,86]. It has been demonstrated that these pomegranate phytoconstituents utilize Nrf2-mediated antioxidant mechanisms to abrogate the oxidative stress generated during diethylnitrosamine (DENA) hepatocarcinogenesis [87]. Nrf2 may also act as a key modulator of inflammatory pathways through its interaction with NF-κB, a master regulator of the proinflammatory response [88]. A study of the same group showed how pomegranate constituents are able to suppress DENA-mediated inflammatory cascade by down-regulating the expression of inflammatory mediators, including iNOS, 3-NT, HSP70, HSP90, COX-2 and NF-κB [89].
5.3. Activation of PPARγ by ABA
ABA is an isoprenoid phytohormone discovered in the early 1960s that has received some recent attention due to its medicinal applications [90]. Specifically, oral ABA administration has shown prophylactic and therapeutic efficacy in mouse models of IBD [19,20,91–93]. However, little is known about the role of ABA in the modulation of immune and inflammatory responses and the cellular and molecular mechanisms underlying its health effects.
ABA has been shown to ameliorate colonic inflammation by suppressing immune cell infiltration. These changes are associated with significant down-regulation of cellular adhesion molecule expression and an increase in Treg cells systemically [19]. Interestingly, the loss of PPARγ in T cells abrogated the anti-inflammatory efficacy of ABA against experimental IBD. Dietary ABA worsened colonic inflammation and enhanced cellular adhesion molecule expression in T cell-specific PPARγ null mice. Therefore, ABA-mediated activation of anti-inflammatory pathways might predominate when PPARγ is expressed in T cells, whereas proinflammatory pathways, such as NF-κB, become ABA’s targets when PPARγ is deleted from T cells [94]. Although ABA has been shown to activate PPARγ [92], it does not bind to its ligand-binding domain, and such activation can be blocked by inhibiting intracellular cyclic adenosine monophosphate production or protein kinase A activity [91], suggesting that ABA triggers an alternative mechanism of PPARγ activation. A recent study suggested that ABA can indirectly activate PPARγ through LANCL2/cAMP-initiated signaling and LANCL catalytic functions [95]. Further research is necessary to understand the cell-specific response of ABA-induced PPARγ activation and how it resembles and differs from the mechanism of PPARγ activation by other compounds.
6. Manipulation of the gut microbiota by probiotics
The alimentary tract is a sterile organ at very early stages of development (i.e., embryonic and fetal phases). However, after birth, the gastrointestinal mucosa evolves to become densely colonized by bacteria. Specifically, from birth to weaning, successive waves of microorganisms will colonize the mucosa with a final result of 500–1000 species, which amount to 100 trillion microorganisms, residing in the large intestine of adult humans [96]. The number of gut microorganisms is 10 times greater than the total number of somatic and germ cells [97]. At the same time that intestinal host tolerance is established towards foods and the commensal microflora, additional mechanisms are in place to recognize and eliminate pathogens and their toxins. A unique feature of the gastrointestinal mucosa is that, in healthy individuals, it maintains a delicate equilibrium between induction of effector immune responses and tolerance. The latter involves a controlled down-regulation of mucosal immunity [98]. The mutualism that exists between the host and the commensal microflora is illustrated by the actions of Bacteroides thetaiotaomicron. Hooper and colleagues demonstrated that this commensal bacterium modulated the expression of genes involved in nutrient absorption, intestinal maturation, angiogenesis and strengthening of the mucosal barrier [99].
Recent estimates indicate that the human gastrointestinal tract contains about 1014 bacteria, with about 103/ml in the stomach with a predominance of Helicobacter pylori, rising with descent of the tract to 1011–1012/ml in the colon [100]. Among other beneficial effects, the intestinal microflora serve important roles in the healthy intestine, from the digestion, absorption and storage of nutrients to the protection against pathogen colonization by the production of antimicrobial substances and competition for nutrients. Moreover, they are important for the development and optimal function of the immune system [101].
The trigger of chronic intestinal inflammation is thought to depend on the microflora since several studies have failed to induce inflammation in germ-free animals. Also, intestinal inflammation will not occur in IL-10-deficient mice or chemically treated rats without the presence of the intestinal microflora [102]. The hypothesis that the intestinal bacterial flora contribute to the pathogenesis of IBD is supported by several experimental and clinical observations. Recent studies revealed that IBD patients show a loss in microflora biodiversity as well as an increase in the proportion of fungi [4]. A decrease in bifidobacteria but not lactobacilli has also been reported [103]. Moreover, UC and CD patients have lower levels of Bifidobacterium in samples collected from inflamed tissue compared with healthy tissue [104].
Due to the evidence correlating the intestinal bacterial flora with IBD pathogenesis, various attempts have been made to modify the microflora by administering probiotics. Probiotics are live microbial supplements which beneficially impact host health. These products rely on introducing particular exogenous strains into the gut microflora [105]. Several probiotics have been shown to be efficacious in the treatment of IBD, specially the commercially available mixture VSL#3, the Escherichia coli strain Nissle 1917 and several Lactobacillus species [106]. The E. coli strain Nissle 1917 has been demonstrated to improve intestinal homeostasis and minimize the bacteria-induced reduction of the intestinal barrier, thus decreasing the invasion of intestinal epithelial cells by several pathogens [106].
Orally or rectally administered lactobacilli improve experimental colitis in mice and rats. L. plantarum 299v prevented and reduced established colitis in an IL-10-deficient mouse model [107] and in chemically treated rats [108]. However, it failed to ameliorate established colitis in TNBS-induced colitis in rats [109]. L. reuteri (R2LC) and L. salivarius UCC118 decreased mucosal inflammatory activity in IL-10-deficient mice [110,111]. Some of these probiotics have been proposed for the prevention and treatment of antibiotic-associated diarrhea and C. difficile-associated disease. These include several bacterial strains such as Bifidobacterium, Lactobacillus GG, L. rhamnosus, L. casei, L. plantarum 299v and Enterococcus faecium [SF68], which are commonly available as lyophilized capsules or in the form of a fermented drink [112]. The disturbance of normal flora, particularly after antibiotic treatment, is believed to facilitate the colonization by C. difficile. The delivery of bacteria to the gastrointestinal tract by probiotic administration is believed to restore equilibrium in the altered microflora, thus protecting against C. difficile colonization [113].
VSL#3 is a mixture composed of four strains of lactobacilli (L. casei, L. plantarum, L. bulgaricus and L. acidophilus), three strains of bifidobacteria (B. longum, B. breve and B. infantis) and Streptococcus thermophilus. Of these, L. casei has been identified as the beneficial strain. VSL#3 has demonstrated efficacy in patients with UC [114] and in animal models of colitis [115]. Interestingly, some of the strains present in VSL#3 are able to produce CLA and CLnA isomers in vitro from LA or α-LA [116]. In this line, a recently published study provides novel in vivo evidence that VSL#3 administration changes microbial diversity and local CLA production, which results in PPARγ-dependent anti-inflammatory effects in different models of experimental colitis in mice [106].
7. Computational modeling of mucosal immune responses in nutritional immunology research
We constructed a computational model of mucosal immune responses during IBD [117]. Overall, our modeling approach involved the following: (a) creation of a structural network using Cell Designer, a software package that enables users to describe molecular interactions using a well-defined and consistent graphical notation [118]; (b) parameter estimation based on published or newly generated experimental data [119]; and (c) in silico experimentation. Our model synthesized knowledge related to immunological mechanisms involving T cells, macrophages, DC and epithelial cells as they move and interact with bacteria in the lumen, LP and MLN (Fig. 3). In silico experiments revealed a positive inflammatory feedback loop formed by inflammatory M1 macrophage activation of T cells as a driving force underlying the immunopathology of IBD, thus suggesting macrophage plasticity as key interventions points in preventive and therapeutic nutritional interventions against gut inflammation. Such computational prediction was later validated in vivo [120]. Specifically, results of DSS challenge studies in mice showed that macrophage PPARγ deficiency worsens experimental IBD by modulating the expression of inflammatory/effector and metabolic genes, impairing the peripheral Treg compartment and increasing the recruitment and activation of LP macrophages and T cells at the colonic mucosa during DSS-induced colitis in mice. Moreover, the induction of M2 macrophages by activation of PPARγ by pioglitazone favored tissue reconstitution in mice with colitis [120].
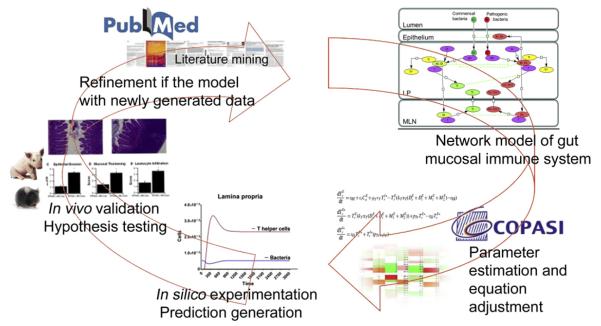
Fig. 3.
Integration of animal and computational modeling approaches in nutritional immunology research. The modeling approaches include fully integrated computational strategies and experimental validation studies. After literature search and generation of calibration database, a comprehensive network is created using a Systems Biology Markup Language (SBML) compliant format. A fully calibrated model that synthesizes existing knowledge is created using Complex Pathway Simulator (COPASI). Computer simulations are conducted with the model to generate new hypotheses. These hypotheses will then be tested in vivo using mouse models of colitis. Finally, the new data generated will be used to recalibrate the model through an iterative process.
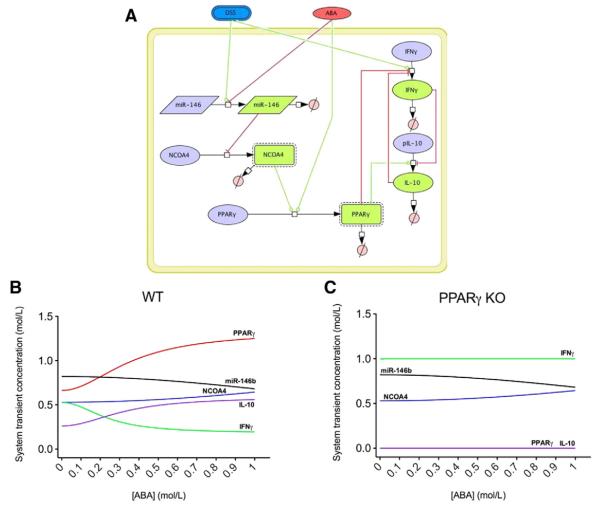
In line with the previous modeling effort, we have also created a small model representing a novel potential pathway by which ABA might elicit its anti-inflammatory effects during DSS-induced colitis in mice. Experimental findings showed that dietary ABA down-regulates the expression of miRNA-146b during DSS-induced colitis. miRNAs are small (~22–24-nucleotide), noncoding, single-stranded RNA molecules that lead to the degradation and/or inhibition of translation of specific mRNAs depending on the type of base-pairing between the miRNA and its mRNA target [121]. Reverse transcriptase polymerase chain reaction results demonstrate that the expression of nuclear receptor coactivator 4 (NCOA4), a potential target of miR-146b and a PPARγ coactivator, was up-regulated during ABA treatment. Although changes in miRNA and mRNA expression in IBD patients have been identified [122], many of the mechanistic links between miRNA alterations and gene targeting remain to be determined. Therefore, these experimental data were used to create and calibrate a computational model with the aim to determine how ABA impacts the mRNA expression of several cytokines via PPARγ and miRNA-146b. In silico simulations illustrate a down-regulation of miR-146b when ABA is administered to the system, resulting in increased concentrations of NCOA4, which, in turn, activates PPARγ and therefore favors IL-10 production and decreases IFN-γ levels. However, when PPARγ is deleted from the system, IL-10 production is drastically decreased. As a result, IFN-γ predominates and NCOA4 is unable to up-regulate PPARγ (Fig. 4).
Fig. 4.
In silico effect of ABA administration on miRNA-146b expression. The model represents the interaction between DSS, ABA, miRNA-146, NCOA4, PPARγ, IL-10 and IFN-γ in SBML format (A). A COPASI scan task shows a down-regulation of miR-146b when ABA is administered to the system, resulting in increased concentrations of NCOA4, which, in turn, activates PPARγ and therefore favors IL-10 production and decreases IFN-γ levels (B). However, when PPARγ is deleted from the system, IL-10 production is drastically decreased. As a result, IFN-γ predominates due to DSS activation, and NCOA4 is unable to up-regulate PPARγ (C).
In conclusion, the use of computational approaches not only can help us discover new nutritional interventions against gut inflammation but also can facilitate discovering the mechanisms of action by which such nutritional compounds elicit their anti-inflammatory effects.
8. Conclusions
Dissatisfaction with current therapies for IBD has spurred interest in the discovery of novel interventions without adverse side effects. In this regard, the nutritional manipulation of gut microbiome and the mucosal immune system represent promising prophylactic interventions against gut inflammation. Dietary activation of PPARγ by fiber, probiotics as well as other naturally ocurring compounds such as CLA, CLnAs and ABA represents an efficacious and safe approach for the prevention and amelioration of IBD. The combination of computational and animal modeling provides novel avenues for synthesizing and rationalizing existing knowledge in a systematic way that accelerates mechanistic nutritional immunology research and discovery to improve gastrointestinal health outcomes.
Acknowledgments
Supported by award number 5R01AT004308 of the National Center for Complementary and Alternative Medicine at the National Institutes of Health awarded to J.B.-R., funds from the Nutritional Immunology and Molecular Medicine Laboratory and National Institute of Allergy and Infectious Diseases Contract No. HHSN272201000056C to JB-R and funds of the Nutritional Immunology and Molecular Medicine Laboratory. We would also like to thank Rachel Robinson for kind support in graphic design.
Footnotes
Disclosures: none.
References
- [1].Neuman MG. Immune dysfunction in inflammatory bowel disease. Transl Res. 2007;149:173–86. doi: 10.1016/j.trsl.2006.11.009. [DOI] [PubMed] [Google Scholar]
- [2].Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- [3].Pene J, Chevalier S, Preisser L, Venereau E, Guilleux MH, et al. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180:7423–30. doi: 10.4049/jimmunol.180.11.7423. [DOI] [PubMed] [Google Scholar]
- [4].Reiff C, Kelly D. Inflammatory bowel disease, gut bacteria and probiotic therapy. Int J Med Microbiol. 2010;300:25–33. doi: 10.1016/j.ijmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- [5].Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–17. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- [6].Kappelman MD, Rifas-Shiman SL, Kleinman K, Ollendorf D, Bousvaros A, et al. The prevalence and geographic distribution of Crohn’s disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol. 2007;5:1424–9. doi: 10.1016/j.cgh.2007.07.012. [DOI] [PubMed] [Google Scholar]
- [7].Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- [8].Camilleri M. GI clinical research 2002–2003: the year in review. Clin Gastroenterol Hepatol. 2003;1:415–20. doi: 10.1016/s1542-3565(03)00220-9. [DOI] [PubMed] [Google Scholar]
- [9].Bassaganya-Riera J, Reynolds K, Martino-Catt S, Cui Y, Hennighausen L, et al. Activation of PPAR gamma and delta by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology. 2004;127:777–91. doi: 10.1053/j.gastro.2004.06.049. [DOI] [PubMed] [Google Scholar]
- [10].Clarke JO, Mullin GE. A review of complementary and alternative approaches to immunomodulation. Nutr Clin Pract. 2008;23:49–62. doi: 10.1177/011542650802300149. [DOI] [PubMed] [Google Scholar]
- [11].Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–76. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- [12].Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- [13].Strober W. Animal models of inflammatory bowel disease–an overview. Dig Dis Sci. 1985;30:3S–10S. doi: 10.1007/BF01296964. [DOI] [PubMed] [Google Scholar]
- [14].Stuber E, Strober W, Neurath M. Blocking the CD40L-CD40 interaction in vivo specifically prevents the priming of T helper 1 cells through the inhibition of interleukin 12 secretion. J Exp Med. 1996;183:693–8. doi: 10.1084/jem.183.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Neurath MF, Fuss I, Pasparakis M, Alexopoulou L, Haralambous S, et al. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur J Immunol. 1997;27:1743–50. doi: 10.1002/eji.1830270722. [DOI] [PubMed] [Google Scholar]
- [16].Fuss IJ, Boirivant M, Lacy B, Strober W. The interrelated roles of TGF-beta and IL-10 in the regulation of experimental colitis. J Immunol. 2002;168:900–8. doi: 10.4049/jimmunol.168.2.900. [DOI] [PubMed] [Google Scholar]
- [17].Kitajima S, Takuma S, Morimoto M. Changes in colonic mucosal permeability in mouse colitis induced with dextran sulfate sodium. Exp Anim. 1999;48:137–43. doi: 10.1538/expanim.48.137. [DOI] [PubMed] [Google Scholar]
- [18].Egger B, Bajaj-Elliott M, MacDonald TT, Inglin R, Eysselein VE, et al. Characterisation of acute murine dextran sodium sulphate colitis: cytokine profile and dose dependency. Digestion. 2000;62:240–8. doi: 10.1159/000007822. [DOI] [PubMed] [Google Scholar]
- [19].Guri AJ, Hontecillas R, Bassaganya-Riera J. Abscisic acid ameliorates experimental IBD by downregulating cellular adhesion molecule expression and suppressing immune cell infiltration. Clin Nutr. 2010;29:824–31. doi: 10.1016/j.clnu.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guri AJ, Hontecillas R, Ferrer G, Casagran O, Wankhade U, et al. Loss of PPAR gamma in immune cells impairs the ability of abscisic acid to improve insulin sensitivity by suppressing monocyte chemoattractant protein-1 expression and macrophage infiltration into white adipose tissue. J Nutr Biochem. 2008;19:216–28. doi: 10.1016/j.jnutbio.2007.02.010. [DOI] [PubMed] [Google Scholar]
- [21].Bassaganya-Riera J, DiGuardo M, Climent M, Vives C, Carbo A, et al. Activation of PPARgamma and delta by dietary punicic acid ameliorates intestinal inflammation in mice. Br J Nutr. 2011;106:878–86. doi: 10.1017/S0007114511001188. [DOI] [PubMed] [Google Scholar]
- [22].Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, et al. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–62. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- [23].Powrie F, Carlino J, Leach MW, Mauze S, Coffman RL. A critical role for transforming growth factor-beta but not interleukin 4 in the suppression of T helper type 1-mediated colitis by CD45RB(low) CD4+ T cells. J Exp Med. 1996;183:2669–74. doi: 10.1084/jem.183.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Carbo A, et al. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR gamma to suppress colitis. PLoS One. 2012;7:e31238. doi: 10.1371/journal.pone.0031238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- [27].Dieleman LA, Arends A, Tonkonogy SL, Goerres MS, Craft DW, et al. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect Immun. 2000;68:5107–13. doi: 10.1128/iai.68.9.5107-5113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, et al. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis. 1999;5:262–70. doi: 10.1097/00054725-199911000-00004. [DOI] [PubMed] [Google Scholar]
- [29].Bassaganya-Riera J, DiGuardo M, Viladomiu M, de Horna A, Sanchez S, et al. Soluble fibers and resistant starch ameliorate disease activity in interleukin-10-deficient mice with inflammatory bowel disease. J Nutr. 2011;141:1318–25. doi: 10.3945/jn.111.139022. [DOI] [PubMed] [Google Scholar]
- [30].Bassaganya-Riera J, King J, Hontecillas R. Health benefits of conjugated linoleic acid: lessons from pig models in biomedical research. Eur J Lipid Sci Tech. 2004;106:856–61. [Google Scholar]
- [31].Bassaganya-Riera J, Hontecillas R. CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Clin Nutr. 2006;25:454–65. doi: 10.1016/j.clnu.2005.12.008. [DOI] [PubMed] [Google Scholar]
- [32].Hontecillas R, Wannemeulher MJ, Zimmerman DR, Hutto DL, Wilson JH, et al. Nutritional regulation of porcine bacterial-induced colitis by conjugated linoleic acid. J Nutr. 2002;132:2019–27. doi: 10.1093/jn/132.7.2019. [DOI] [PubMed] [Google Scholar]
- [33].Hontecillas R, Bassaganya-Riera J, Wilson J, Hutto DL, Wannemuehler MJ. CD4+ T-cell responses and distribution at the colonic mucosa during Brachyspira hyodysenteriae-induced colitis in pigs. Immunology. 2005;115:127–35. doi: 10.1111/j.1365-2567.2005.02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wen K, Li G, Bui T, Liu F, Li Y, et al. High dose and low dose Lactobacillus acidophilus exerted differential immune modulating effects on T cell immune responses induced by an oral human rotavirus vaccine in gnotobiotic pigs. Vaccine. 2012;30:1198–207. doi: 10.1016/j.vaccine.2011.11.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yuan L, Wen K, Liu F, Li G. Kongo JM, editor. Dose Effects of LAB on modulation of rotavirus vaccine induced immune responses, lactic acid bacteria, R & D for food, health and livestock purposes. ISBN: 978-953-51-0955-6, InTech, http://dx.doi.org/10.5772/50379.
- [36].Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leenen CH, Dieleman LA. Inulin and oligofructose in chronic inflammatory bowel disease. J Nutr. 2007;137:2572S–5S. doi: 10.1093/jn/137.11.2572S. [DOI] [PubMed] [Google Scholar]
- [38].Roberfroid M. Prebiotics: the concept revisited. J Nutr. 2007;137:830S–7S. doi: 10.1093/jn/137.3.830S. [DOI] [PubMed] [Google Scholar]
- [39].Guarner F. Inulin and oligofructose: impact on intestinal diseases and disorders. Br J Nutr. 2005;93(Suppl 1):S61–5. doi: 10.1079/bjn20041345. [DOI] [PubMed] [Google Scholar]
- [40].Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev. 2004;17:259–75. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- [41].Looijer-van Langen MA, Dieleman LA. Prebiotics in chronic intestinal inflammation. Inflamm Bowel Dis. 2009;15:454–62. doi: 10.1002/ibd.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- [43].Young GP, Hu Y, Le Leu RK, Nyskohus L. Dietary fibre and colorectal cancer: a model for environment–gene interactions. Mol Nutr Food Res. 2005;49:571–84. doi: 10.1002/mnfr.200500026. [DOI] [PubMed] [Google Scholar]
- [44].Langlands SJ, Hopkins MJ, Coleman N, Cummings JH. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut. 2004;53:1610–6. doi: 10.1136/gut.2003.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hopkins MJ, Macfarlane GT. Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl Environ Microbiol. 2003;69:1920–7. doi: 10.1128/AEM.69.4.1920-1927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hookman P, Barkin JS. Clostridium difficile associated infection, diarrhea and colitis. World J Gastroenterol. 2009;15:1554–80. doi: 10.3748/wjg.15.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].George RH, Symonds JM, Dimock F, Brown JD, Arabi Y, et al. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br Med J. 1978;1:695. doi: 10.1136/bmj.1.6114.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim JW, Lee KL, Jeong JB, Kim BG, Shin S, et al. Proton pump inhibitors as a risk factor for recurrence of Clostridium-difficile-associated diarrhea. World J Gastroenterol. 2010;16:3573–7. doi: 10.3748/wjg.v16.i28.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lim PL, Barkham TM, Ling LM, Dimatatac F, Alfred T, et al. Increasing incidence of Clostridium difficile-associated disease, Singapore. Emerg Infect Dis. 2008;14:1487–9. doi: 10.3201/eid1409.070043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Barbut F, Jones G, Eckert C. Epidemiology and control of Clostridium difficile infections in healthcare settings: an update. Curr Opin Infect Dis. 2011;24:370–6. doi: 10.1097/QCO.0b013e32834748e5. [DOI] [PubMed] [Google Scholar]
- [51].Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005;71:3692–700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Perrin S, Warchol M, Grill JP, Schneider F. Fermentations of fructooligosaccharides and their components by Bifidobacterium infantis ATCC 15697 on batch culture in semi-synthetic medium. J Appl Microbiol. 2001;90:859–65. doi: 10.1046/j.1365-2672.2001.01317.x. [DOI] [PubMed] [Google Scholar]
- [53].Falony G, Vlachou A, Verbrugghe K, De Vuyst L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol. 2006;72:7835–41. doi: 10.1128/AEM.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hartemink R, Van Laere KM, Rombouts FM. Growth of enterobacteria on fructooligosaccharides. J Appl Microbiol. 1997;83:367–74. doi: 10.1046/j.1365-2672.1997.00239.x. [DOI] [PubMed] [Google Scholar]
- [55].Mitsuoka T, Hidaka H, Eida T. Effect of fructo-oligosaccharides on intestinal microflora. Nahrung. 1987;31:427–36. doi: 10.1002/food.19870310528. [DOI] [PubMed] [Google Scholar]
- [56].Simpson EJ, Chapman MA, Dawson J, Berry D, Macdonald IA, et al. In vivo measurement of colonic butyrate metabolism in patients with quiescent ulcerative colitis. Gut. 2000;46:73–7. doi: 10.1136/gut.46.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Maathuis A, Hoffman A, Evans A, Sanders L, Venema K. The effect of the undigested fraction of maize products on the activity and composition of the microbiota determined in a dynamic in vitro model of the human proximal large intestine. J Am Coll Nutr. 2009;28:657–66. doi: 10.1080/07315724.2009.10719798. [DOI] [PubMed] [Google Scholar]
- [58].Weaver CM, Martin BR, Story JA, Hutchinson I, Sanders L. Novel fibers increase bone calcium content and strength beyond efficiency of large intestine fermentation. J Agric Food Chem. 2010;58:8952–7. doi: 10.1021/jf904086d. [DOI] [PubMed] [Google Scholar]
- [59].Schwab M, Reynders V, Ulrich S, Zahn N, Stein J, et al. PPARgamma is a key target of butyrate-induced caspase-3 activation in the colorectal cancer cell line Caco-2. Apoptosis. 2006;11:1801–11. doi: 10.1007/s10495-006-9788-2. [DOI] [PubMed] [Google Scholar]
- [60].Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, et al. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci U S A. 2005;102:6207–12. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hontecillas R, Bassaganya-Riera J. Peroxisome proliferator-activated receptor gamma is required for regulatory CD4+ T cell-mediated protection against colitis. J Immunol. 2007;178:2940–9. doi: 10.4049/jimmunol.178.5.2940. [DOI] [PubMed] [Google Scholar]
- [62].Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, et al. A novel therapy for colitis utilizing PPAR-gamma ligands to inhibit the epithelial inflammatory response. J Clin Invest. 1999;104:383–9. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Desreumaux P, Dubuquoy L, Nutten S, Peuchmaur M, Englaro W, et al. Attenuation of colon inflammation through activators of the retinoid X receptor (RXR)/peroxisome proliferator-activated receptor gamma (PPAR-gamma) heterodimer. A basis for new therapeutic strategies. J Exp Med. 2001;193:827–38. doi: 10.1084/jem.193.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lytle C, Tod TJ, Vo KT, Lee JW, Atkinson RD, et al. The peroxisome proliferator-activated receptor gamma ligand rosiglitazone delays the onset of inflammatory bowel disease in mice with interleukin 10 deficiency. Inflamm Bowel Dis. 2005;11:231–43. doi: 10.1097/01.mib.0000160805.46235.eb. [DOI] [PubMed] [Google Scholar]
- [65].Lewis JD, Lichtenstein GR, Deren JJ, Sands BE, Hanauer SB, et al. Rosiglitazone for active ulcerative colitis: a randomized placebo-controlled trial. Gastroenterology. 2008;134:688–95. doi: 10.1053/j.gastro.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nesto RW, Bell D, Bonow RO, Fonseca V, Grundy SM, et al. Thiazolidinedione use, fluid retention, and congestive heart failure: a consensus statement from the American Heart Association and American Diabetes Association. October 7, 2003. Circulation. 2003;108:2941–8. doi: 10.1161/01.CIR.0000103683.99399.7E. [DOI] [PubMed] [Google Scholar]
- [67].Marcy TR, Britton ML, Blevins SM. Second-generation thiazolidinediones and hepatotoxicity. Ann Pharmacother. 2004;38:1419–23. doi: 10.1345/aph.1E072. [DOI] [PubMed] [Google Scholar]
- [68].Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- [69].Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today. 2006;11:580–94. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lewis SN, Brannan L, Guri AJ, Lu P, Hontecillas R, et al. Dietary alpha-eleostearic acid ameliorates experimental inflammatory bowel disease in mice by activating peroxisome proliferator-activated receptor-gamma. PLoS One. 2011;6:e24031. doi: 10.1371/journal.pone.0024031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lewis SN, Bassaganya-Riera J, Bevan DR. Virtual screening as a technique for PPAR modulator discovery. PPAR Res. 2010;2010:861238. doi: 10.1155/2010/861238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hontecillas R, Diguardo M, Duran E, Orpi M, Bassaganya-Riera J. Catalpic acid decreases abdominal fat deposition, improves glucose homeostasis and upregulates PPAR alpha expression in adipose tissue. Clin Nutr. 2008;27:764–72. doi: 10.1016/j.clnu.2008.07.007. [DOI] [PubMed] [Google Scholar]
- [73].Hontecillas R, O’Shea M, Einerhand A, Diguardo M, Bassaganya-Riera J. Activation of PPAR gamma and alpha by punicic acid ameliorates glucose tolerance and suppresses obesity-related inflammation. J Am Coll Nutr. 2009;28:184–95. doi: 10.1080/07315724.2009.10719770. [DOI] [PubMed] [Google Scholar]
- [74].Chin SF, Storkson JM, Liu W, Albright KJ, Pariza MW. Conjugated linoleic acid (9,11- and 10,12-octadecadienoic acid) is produced in conventional but not germ-free rats fed linoleic acid. J Nutr. 1994;124:694–701. doi: 10.1093/jn/124.5.694. [DOI] [PubMed] [Google Scholar]
- [75].Parodi PW. Conjugated linoleic acid and other anticarcinogenic agents of bovine milk fat. J Dairy Sci. 1999;82:1339–49. doi: 10.3168/jds.S0022-0302(99)75358-0. [DOI] [PubMed] [Google Scholar]
- [76].Bassaganya-Riera J, Hontecillas R, Beitz DC. Colonic anti-inflammatory mechanisms of conjugated linoleic acid. Clin Nutr. 2002;21:451–9. doi: 10.1054/clnu.2002.0594. [DOI] [PubMed] [Google Scholar]
- [77].Dubuquoy L, Rousseaux C, Thuru X, Peyrin-Biroulet L, Romano O, et al. PPARgamma as a new therapeutic target in inflammatory bowel diseases. Gut. 2006;55:1341–9. doi: 10.1136/gut.2006.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bassaganya-Riera J, Hontecillas R, Horne WT, Sandridge M, Herfarth HH, et al. Conjugated linoleic acid modulates immune responses in patients with mild to moderately active Crohn’s disease. Clin Nutr. 2012;31:721–7. doi: 10.1016/j.clnu.2012.03.002. [DOI] [PubMed] [Google Scholar]
- [79].Bassaganya-Riera J, Hontecillas R. Dietary conjugated linoleic acid and n-3 polyunsaturated fatty acids in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care. 2010;13:569–73. doi: 10.1097/MCO.0b013e32833b648e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sassano GP, Sanderson J, Franx P, Groot J, van Straalen J, et al. Analysis of pomegranate seed oil for the presence of jacaric acid. J Food Sci Agric. 2009;6:1046–52. [Google Scholar]
- [81].O’Shea M, Bassaganya-Riera J, Mohede IC. Immunomodulatory properties of conjugated linoleic acid. Am J Clin Nutr. 2004;79:1199S–206S. doi: 10.1093/ajcn/79.6.1199S. [DOI] [PubMed] [Google Scholar]
- [82].Coursodon-Boyiddle CF, Snarrenberg CL, Adkins-Rieck CK, Bassaganya-Riera J, Hontecillas R, et al. Pomegranate seed oil reduces intestinal damage in a rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G744–51. doi: 10.1152/ajpgi.00248.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Carlisle EM, Poroyko V, Caplan MS, Alverdy JA, Liu D. Gram negative bacteria are associated with the early stages of necrotizing enterocolitis. PLoS One. 2011;6:e18084. doi: 10.1371/journal.pone.0018084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lansky EP, Newman RA. Punica granatum (pomegranate) and its potential for prevention and treatment of inflammation and cancer. J Ethnopharmacol. 2007;109:177–206. doi: 10.1016/j.jep.2006.09.006. [DOI] [PubMed] [Google Scholar]
- [85].Kaufman M, Wiesman Z. Pomegranate oil analysis with emphasis on MALDI-TOF/MS triacylglycerol fingerprinting. J Agric Food Chem. 2007;55:10405–13. doi: 10.1021/jf072741q. [DOI] [PubMed] [Google Scholar]
- [86].Faria A, Calhau C. The bioactivity of pomegranate: impact on health and disease. Crit Rev Food Sci Nutr. 2011;51:626–34. doi: 10.1080/10408391003748100. [DOI] [PubMed] [Google Scholar]
- [87].Bishayee A, Bhatia D, Thoppil RJ, Darvesh AS, Nevo E, et al. Pomegranate-mediated chemoprevention of experimental hepatocarcinogenesis involves Nrf2-regulated antioxidant mechanisms. Carcinogenesis. 2011;32:888–96. doi: 10.1093/carcin/bgr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Nair S, Doh ST, Chan JY, Kong AN, Cai L. Regulatory potential for concerted modulation of Nrf2- and Nfkb1-mediated gene expression in inflammation and carcinogenesis. Br J Cancer. 2008;99:2070–82. doi: 10.1038/sj.bjc.6604703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bishayee A, Thoppil RJ, Darvesh AS, Ohanyan V, Meszaros JG, et al. Pomegranate phytoconstituents blunt the inflammatory cascade in a chemically induced rodent model of hepatocellular carcinogenesis. J Nutr Biochem. 2012;24:178–87. doi: 10.1016/j.jnutbio.2012.04.009. [DOI] [PubMed] [Google Scholar]
- [90].Bassaganya-Riera J, Skoneczka J, Kingston DG, Krishnan A, Misyak SA, et al. Mechanisms of action and medicinal applications of abscisic Acid. Curr Med Chem. 2010;17:467–78. doi: 10.2174/092986710790226110. [DOI] [PubMed] [Google Scholar]
- [91].Guri AJ, Hontecillas R, Bassaganya-Riera J. Abscisic acid synergizes with rosiglitazone to improve glucose tolerance and down-modulate macrophage accumulation in adipose tissue: possible action of the cAMP/PKA/PPAR gamma axis. Clin Nutr. 2010;29:646–53. doi: 10.1016/j.clnu.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Guri AJ, Hontecillas R, Si H, Liu D, Bassaganya-Riera J. Dietary abscisic acid ameliorates glucose tolerance and obesity-related inflammation in db/db mice fed high-fat diets. Clin Nutr. 2007;26:107–16. doi: 10.1016/j.clnu.2006.07.008. [DOI] [PubMed] [Google Scholar]
- [93].Guri AJ, Misyak SA, Hontecillas R, Hasty A, Liu D, et al. Abscisic acid ameliorates atherosclerosis by suppressing macrophage and CD4+ T cell recruitment into the aortic wall. J Nutr Biochem. 2010;21:1178–85. doi: 10.1016/j.jnutbio.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Guri AJ, Evans NP, Hontecillas R, Bassaganya-Riera J. T cell PPARgamma is required for the anti-inflammatory efficacy of abscisic acid against experimental IBD. J Nutr Biochem. 2011;22:812–9. doi: 10.1016/j.jnutbio.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bassaganya-Riera J, Guri AJ, Lu P, Climent M, Carbo A, et al. Abscisic acid regulates inflammation via ligand-binding domain-independent activation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2011;286:2504–16. doi: 10.1074/jbc.M110.160077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Greicius G, Arulampalam V, Pettersson S. A CLA’s act: feeding away inflammation. Gastroenterology. 2004;127:994–6. doi: 10.1053/j.gastro.2004.07.038. [DOI] [PubMed] [Google Scholar]
- [97].Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- [98].Hayday A, Theodoridis E, Ramsburg E, Shires J. Intraepithelial lymphocytes: exploring the Third Way in immunology. Nat Immunol. 2001;2:997–1003. doi: 10.1038/ni1101-997. [DOI] [PubMed] [Google Scholar]
- [99].Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–4. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- [100].Linskens RK, Huijsdens XW, Savelkoul PH, Vandenbroucke-Grauls CM, Meuwissen SG. The bacterial flora in inflammatory bowel disease: current insights in pathogenesis and the influence of antibiotics and probiotics. Scand J Gastroenterol Suppl. 2001:29–40. doi: 10.1080/003655201753265082. [DOI] [PubMed] [Google Scholar]
- [101].Neuman MG, Nanau RM. Inflammatory bowel disease: role of diet, microbiota, life style. Transl Res. 2012;160:29–44. doi: 10.1016/j.trsl.2011.09.001. [DOI] [PubMed] [Google Scholar]
- [102].Jonkers D, Stockbrugger R. Probiotics and inflammatory bowel disease. J R Soc Med. 2003;96:167–71. [PMC free article] [PubMed] [Google Scholar]
- [103].Favier C, Neut C, Mizon C, Cortot A, Colombel JF, et al. Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig Dis Sci. 1997;42:817–22. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- [104].Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol. 2009;15:5287–94. doi: 10.3748/wjg.15.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Shanahan F. Physiological basis for novel drug therapies used to treat the inflammatory bowel diseases I. Pathophysiological basis and prospects for probiotic therapy in inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2005;288:G417–21. doi: 10.1152/ajpgi.00421.2004. [DOI] [PubMed] [Google Scholar]
- [106].Hormannsperger G, Haller D. Molecular crosstalk of probiotic bacteria with the intestinal immune system: clinical relevance in the context of inflammatory bowel disease. Int J Med Microbiol. 2010;300:63–73. doi: 10.1016/j.ijmm.2009.08.006. [DOI] [PubMed] [Google Scholar]
- [107].Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, et al. Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71–80. doi: 10.1097/00054725-200203000-00001. [DOI] [PubMed] [Google Scholar]
- [108].Mao Y, Nobaek S, Kasravi B, Adawi D, Stenram U, et al. The effects of Lactobacillus strains and oat fiber on methotrexate-induced enterocolitis in rats. Gastroenterology. 1996;111:334–44. doi: 10.1053/gast.1996.v111.pm8690198. [DOI] [PubMed] [Google Scholar]
- [109].Kennedy RJ, Hoper M, Deodhar K, Kirk SJ, Gardiner KR. Probiotic therapy fails to improve gut permeability in a hapten model of colitis. Scand J Gastroenterol. 2000;35:1266–71. doi: 10.1080/003655200453601. [DOI] [PubMed] [Google Scholar]
- [110].Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107–14. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- [111].O’Mahony L, Feeney M, O’Halloran S, Murphy L, Kiely B, et al. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther. 2001;15:1219–25. doi: 10.1046/j.1365-2036.2001.01027.x. [DOI] [PubMed] [Google Scholar]
- [112].Dendukuri N, Costa V, McGregor M, Brophy JM. Probiotic therapy for the prevention and treatment of Clostridium difficile-associated diarrhea: a systematic review. CMAJ. 2005;173:167–70. doi: 10.1503/cmaj.050350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sullivan A, Nord CE. Probiotics in human infections. J Antimicrob Chemother. 2002;50:625–7. doi: 10.1093/jac/dkf194. [DOI] [PubMed] [Google Scholar]
- [114].Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, et al. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100:1539–46. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- [115].Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–91. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- [116].Gorissen L, Raes K, Weckx S, Dannenberger D, Leroy F, et al. Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl Microbiol Biotechnol. 2010;87:2257–66. doi: 10.1007/s00253-010-2713-1. [DOI] [PubMed] [Google Scholar]
- [117].Wendelsdorf K, Bassaganya-Riera J, Hontecillas R, Eubank S. Model of colonic inflammation: immune modulatory mechanisms in inflammatory bowel disease. J Theor Biol. 2010;264:1225–39. doi: 10.1016/j.jtbi.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Funahashi A, Jouraku A, Matsuoka Y, Kitano H. Integration of Cell Designer and SABIO-RK. In Silico Biol. 2007;7:S81–90. [PubMed] [Google Scholar]
- [119].Hoops S, Sahle S, Gauges R, Lee C, Pahle J, et al. COPASI–a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–74. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- [120].Hontecillas R, Horne WT, Climent M, Guri AJ, Evans C, et al. Immunoregulatory mechanisms of macrophage PPAR-gamma in mice with experimental inflammatory bowel disease. Mucosal Immunol. 2011;4:304–13. doi: 10.1038/mi.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [122].Dalal SR, Kwon JH. The role of MicroRNA in inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2010;6:714–22. [PMC free article] [PubMed] [Google Scholar]