Abstract
Next-generation sequencing has led to a revolution in the study of hematological malignancies with a substantial number of publications and discoveries in the last few years. Significant discoveries associated with disease diagnosis, risk stratification, clonal evolution and therapeutic intervention have been generated by this powerful technology. As part of the post-genomic era, sequencing analysis will likely become part of routine clinical testing and the challenge will ultimately be successfully transitioning from gene discovery to preventive and therapeutic intervention as part of individualized medicine strategies. In this report, we review recent advances in the understanding of hematological malignancies derived through genome-wide sequence analysis.
Keywords: hematological malignancy, genome sequencing, RNA sequencing, clonal architecture
Introduction
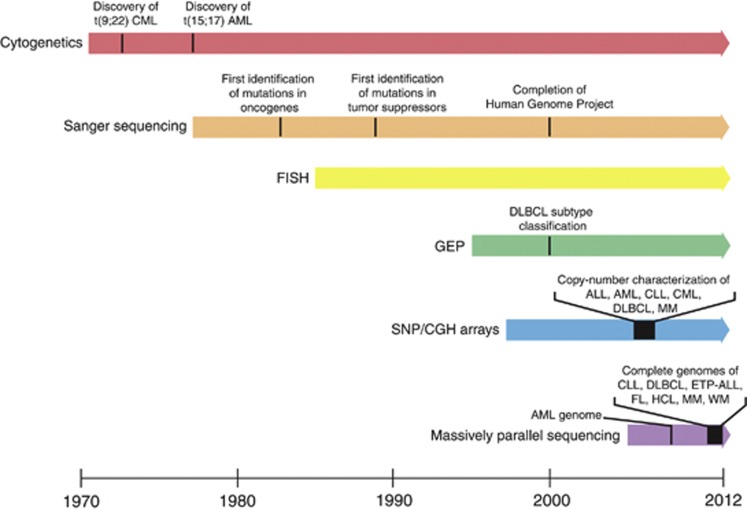
Over the past few years, a remarkable effort has been underway to identify the genetic basis of hematological malignancies catalyzed by increasing availability and more refined sequencing technologies (Figure 1). The growth in high-throughput sequencing, which has facilitated this effort, has been exponential with a dramatic increase in efficiency and correlating drop in price per base pair. Modern platforms can now perform whole-genome sequencing (WGS) of an individual for less than $5000 and a few days of work; notable progress compared with the resources and time that were used just a few years ago by an international consortium when completing the first human genome.1 Strikingly, this technological revolution occurred in <10 years.
Figure 1.
Evolution of genetic detection methods and discoveries. Landmark findings from each method are indicated.
A representative example of how increasingly more powerful technologies continuously improve our knowledge of the genetic basis of hematological malignancies comes from the study of the genome of an acute myeloid leukemia (AML) patient with normal cytogenetics, which was studied twice in a period of 2 years. The initial WGS analysis study only revealed small insertions and deletions affecting two genes, and nonsynonymous somatic mutations, in another eight genes.2 Two years later, the same genome was resequenced utilizing more advanced sequencing technology and analytical methods resulting in the detection of a previously unidentified frameshift deletion in DNMT3A.3 After the initial discovery, DNMT3A mutations were screened in large cohorts and it is now known that 22–30% of AML patients have mutations in this gene, being currently one of the most relevant and potentially targetable mutations found in AML.
Next-generation sequencing (NGS) encompasses several different methodologies that allow the investigation of genomics, transcriptomics and epigenomics. A summary of the different sequencing approaches is briefly described below and summarized in Table 1. For more in-depth information, we direct the reader to a number of excellent reviews.4, 5, 6, 7, 8, 9, 10
Table 1. Summary of high-throughput sequencing methods.
| Method | Minimum input quantitya | Strengths | Weaknesses |
|---|---|---|---|
| Whole genome | 10 ng–1 μg genomic DNA | Small input DNA requirement | Lower DNA input may reduce library complexity and representation |
| Variant detection in all regions of the genome | PCR duplicates can impact accuracy of variant detection software | ||
| Computationally intensive analysis | |||
| Mate pair | 5–10 μg genomic DNA | Identification of large structural rearrangements | Large input DNA requirement |
| High false discovery rate | |||
| Whole exome | 1 μg genomic DNA | Deep coverage of exome enabling precise interrogation of coding regions | Non-coding regions excluded |
| Multiple samples can be pooled and run together reducing time and cost per sample | Standard capture kits do not capture all exons | ||
| mRNASeq | 100–400 ng total RNAb | Dynamic range of expression detection can be much broader than using microarrays | RNA fragmentation methods can bias the resulting library |
| Detection of rare and hybrid transcripts | Artifacts from amplified cDNA libraries8 | ||
| Precise quantitation of highly expressed transcripts and multiple isoforms | Appropriate normal controls may be difficult to obtain for tumor/normal comparison | ||
| Investigation of 3'UTR and promoters | |||
| ChIPSeq | 10 ng ChIP enriched DNA | Detection of DNA–protein interactions | Quality of sequencing results dependent on the quality of ChIP assay |
| Discovery of new interactions in regions not represented on microarray chips | Library preparation can introduce GC-rich region bias | ||
| Avoids hybridization problems associated with array-based ChIP assays | |||
| Single molecule | 1 μg genomic DNA | No amplification step resulting in no PCR duplicates | High error rate |
| Long-read length (>1 kb) | Throughput not comparable to current platforms |
Abbreviations: ChIPSeq, chromatin immunoprecipitation sequencing; UTR, untranslated region.
Cost per sample is highly variable depending on the platform and on the amount of multiplexing utilized.
May vary by platform and approach.
May require polyA RNA- or rRNA-depleted total RNA.
Whole-genome sequencing
Two major approaches are utilized in the preparation of DNA libraries for WGS. The first is called paired-end sequencing, where ∼100 bp are sequenced from each end of ∼400-bp DNA fragments. By this method, single nucleotide variants (SNV), insertions and deletions and copy-number changes can be identified. Paired-end WGS needs low-input quantities of DNA (<1 μg) for generating the libraries, which is a critical advantage in the study of hematological malignancies, where the amount of tumor tissue is usually scarce.
The second approach used in the DNA library preparation is named mate-pair sequencing. Mate-pair is based on the generation of much larger DNA fragments than paired-end sequencing with fragments ranging in length from 1 to 10 kb. Longer distance between the read pairs enables improved detection of structural rearrangements because the read pairs can span repeat and duplicate regions, thereby capturing regions not adequately captured with smaller insert sizes utilized with paired-end sequencing. Very low coverage of the genome is enough for studies focused on the detection of structural abnormalities, thus reducing costs and complexity of the analysis. On the other hand, if the genome is covered in enough depth (>30X mean coverage), mate-pair sequencing can be used for the simultaneous detection of mutations, copy-number changes and structural abnormalities. A disadvantage of the mate-pair approach is that quite a large quantity of DNA is required for the library preparation, thus limiting its use in a significant number of tumors.
Whole-exome sequencing
Whole-exome sequencing (WES) is useful for those interested in studying only what lies within the exome (coding genome) and untranslated regions. This method is based on an initial enrichment step of exonic regions followed by targeted sequencing. As the exome represents only 1.4% of the genome, multiple samples can be pooled and sequenced together in a single instrument run. The major weakness of WES is the inability of the available enrichment kits to capture the totality of the exome.
Ideally a non-tumoral, reference DNA sample from the individual patient is simultaneously analyzed with each tumor. The amount of normal variation between individuals is in the order of thousands of variants, and performing a paired analysis enables subtraction of the nontumor-specific from the tumor-specific variants. In the event that normal tissue is not available for comparison, an increased number of publicly available databases, such as dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), HapMap (http://hapmap.ncbi.nlm.nih.gov/) and 1000 genomes (http://www.1000genomes.org/), can be utilized to identify and clean previously reported variants in the general population that are normal genetic variation rather than somatically acquired mutations.
Messenger RNA sequencing
Besides the detection of mutations, messenger RNA sequencing is also a powerful tool for gene expression analysis. The dynamic range of expression obtained by messenger RNA sequencing can be much broader than that obtained by gene expression microarrays, allowing the detection of rare transcripts and more precise quantitation of expressed transcripts.8, 11 In addition, messenger RNA sequencing can be utilized for the identification of hybrid transcripts and quantitation of multiple isoforms resulting from alternative splicing. Major weaknesses include the biases in library preparation caused by the RNA fragmentation methods utilized, introducing artifacts into the resulting reads.
New approaches and frontiers
It is still the subject of debate which approach is the most appropriate for studying the cancer genome. WGS is the most inclusive approach, but major limitations remain related to the high cost and the difficulty associated with managing the data storage and intensive computational analysis. The study of the exome reduces these limitations. WES is a well-established strategy for analyzing coding regions at low cost, making this approach the most popular in the analysis of the tumor genome nowadays. However, eliminating 98% of the genome from the analysis brings the obvious risks associated with omitting crucial information. This concern is supported by recent findings performed by the Encyclopedia of DNA Elements Project, where integrated analysis demonstrated that >80% of the genome is biochemically active.12 This new paradigm will require reconsideration of the best strategic approach to optimize the cost/benefit ratio in the analysis of the cancer genome.
Single-molecule, long-read, sequencing approaches are now available and allow the simultaneous search for single-allele mutations and methylation profiles. Furthermore, several new platforms are currently under development that promise to sequence the whole genome in few hours for less than $1000. These technological advances open a new world of opportunities and soon will put the use of WGS within reach of the clinical labs.
Challenges in data analysis
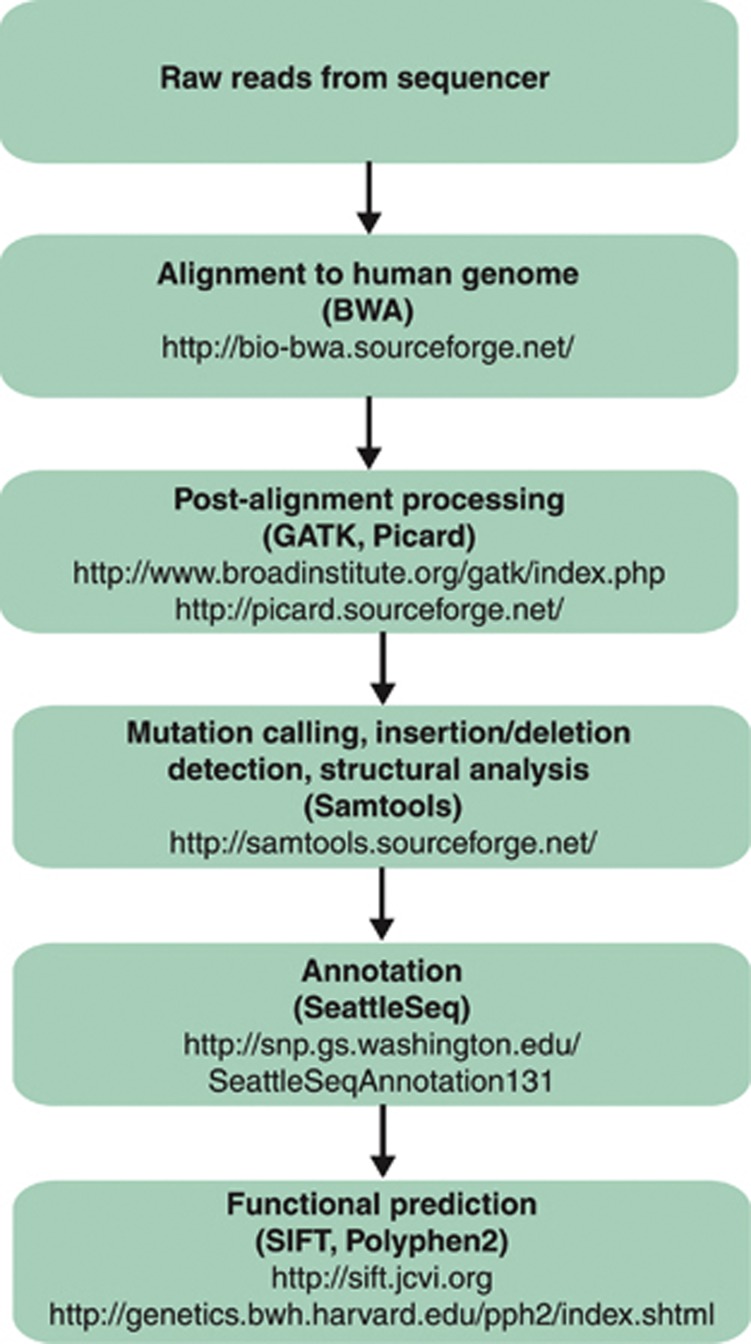
Data generation is just a small facet of the much bigger challenge associated with data analysis. The goal of data analysis is to utilize bioinformatics tools in a data analysis ‘pipeline' (Figure 2) to transform the raw data into results that can ultimately be seen in a user-friendly visualization tool. Detailed information about the most commonly utilized alignment and functional prediction tools are beyond the scope of this manuscript, and thus we direct the readers' attention to several informative manuscripts.13, 14, 15, 16
Figure 2.
Schematic of a bioinformatics pipeline. Examples of the most commonly used publicly available software programs utilized at a particular step are in parentheses. The programs listed were the most commonly used in 2012 hematological malignancy sequencing analyses. These are only examples and are not intended to be an exhaustive list. The number of publicly available tools is rapidly expanding and review of these tools is beyond the scope of this report.
The ability to quickly generate large quantities of data at relatively low cost is limited by these constantly evolving data analysis pipelines, failure to report analytical methods with the level of detail expected from traditional experimental data and the lack of consensus regarding which tools to use when transforming the data into a useable form. Nekrutenko and Taylor17 reviewed 50 papers that used the Burrows–Wheeler Aligner for mapping sequencing reads, and they found that most of the papers neither provide access to the raw data nor specify the parameters utilized or identify the precise version of the genomic reference sequence. From the remaining analyses, only four provide settings, eight list the version used and seven list all necessary details. Furthermore, of 19 sequencing articles that cited and had a similar experimental design to that of the 1000 Genomes project, only 4 used the workflow recommended. Different analysis pipelines can yield similar, yet different results suggesting that more than one pipeline may be currently necessary to successfully analyze the data and ultimately introduce this technology into the clinic.
Sequencing in hematological malignancies
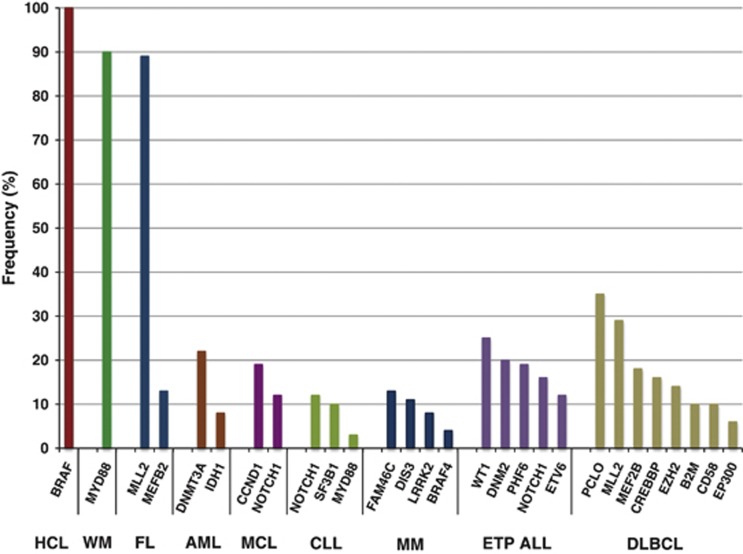
At this point, where medium-sized cohorts of most of the hematological malignancies have been sequenced, it is time to wonder what have these data revealed thus far in this group of malignancies and what are the opportunities ahead? As we expected, genes such as TP53, ATM and RAS among others were confirmed as mutated in a wide variety of malignancies. However, promising and exciting findings came from the discovery of a completely novel group of genes and pathways impaired in hematological malignancies. A few malignancies seem to be driven by mutations in only one or a few genes, suggesting a unique pathway, pathognomonic to the disease. However, most of the malignancies show considerable genetic heterogeneity, with multiple genes and pathways affected. In this review, we aim to summarize the current knowledge of the genetic background on different hematological malignancies and how this knowledge could facilitate targeting of dysregulated signaling pathways by therapeutic targets. The most recurrent novel somatic genetic mutations per malignancy are summarized in Figure 3 and Table 2.
Figure 3.
Most frequent somatic genetic mutations per hematological malignancy. Only original data from massively parallel sequencing were included, excluding confirmation data in previously mutated genes (for example, ATM and TP53 in CLL, FLT3 in AML, RAS and TP53 in MM, MYD88 in DLBCL, mutations in the nuclear factor-kB pathways in DLBCL and MM). In cases where data were obtained from multiple studies, the data originating from the largest cohort were included.
Table 2. Summary of high-throughput sequencing studies performed so far in hematological malignancies.
| Disease | Discovery cohort (N) | Validation cohort (N) | Method | Platform | Mean coverage depth (X) | Highlights | Year | Reference |
|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 24 | mRNASeq | GAII | NR | DPEP1 (4%), longitudinal detection of PLXNB2and CXorf21 | 2012 | 80 |
| ALL(Phlike) | 15 | 231 | WGS/mRNASeq | GAIIx/HiSeq | NR | NUP214-ABL1 fusion (2%), IK2F1 (67%) | 2012 | 81 |
| ALL (ETP) | 12 | 94 | WGS | GAII | 33 | RAS pathway (67%), hematopoiesis and lymphoid development (58%), histone modification (42%) | 2012 | 40 |
| ALL (T) | 11 | — | WES/mRNASeq | HiSeq | 55/15 | DNMT3A (17%), JARID2 (8%), IDH2 (8%),EZH2(17%) | 2012 | 82 |
| AML | 1 | — | WES | HiSeq2000 | NR | Leukemic transformation from SCN to AML | 2012 | 83 |
| AML | 8 | — | WGS/deep sequencing | GAIIx | 25/590 | Clonal evolution | 2012 | 73 |
| AML | 5 | 160 | WES | GAIIx | NR | GATA2 (39%) with biallelic CEBPA mutation | 2012 | 84 |
| AML | 2 | 3 | ChIPSeq | GAII/HiSeq | NR | Differential H3K4me3/H3K27me3 gene enrichment of stem and progenitor cells | 2013 | 85 |
| AML-CN | 1 | 95 | mRNASeq | GAIIx | NR | TLE4 (2%), SHKBP1 (2%) | 2011 | 86 |
| AML-CN | 1 | 262 | WES | GAIIx | 69 | BCOR (4%), DNMT3A (13%) | 2011 | 87 |
| AML-CN | 7 | 230 | mRNASeq | HiScanSQ | 36 | CBFA2T3-GLIS2 fusion (8%) | 2013 | 88 |
| AML-M1 | 1 | — | WGS | GA | 33 | First genome sequenced | 2008 | 2 |
| AML-M1 | 1 | 187 | WGS | GAII | 23 | IDH1 (8%) | 2009 | 50 |
| AML-M1 | 1 | 281 | WGS | GAII | 39 | DNMT3A (22%) | 2010 | 3 |
| AML-M5 | 14 | 98 | WES | GAIIx | 97 | DNMT3A (21%) | 2011 | 89 |
| sAML | 7 | 200 | WGS/WES | GAIIx/HiSeq | 34 | UMODL1 (29%), SMC3 (14%), CDH23, ZSWIM4 (14%) | 2012 | 90 |
| BL | 28 | 78 | mRNASeq | HiSeq2000 | NR | ID3 (59%), TCF3 (29%), CCND3 (15%) | 2012 | 91 |
| BL | 4 | 97 | WGS/WES/mRNASeq/MethylSeq | GAII/HiSeq | 32/121 | ID3 (42%) | 2012 | 92 |
| BL | 14 | 45 | WES | GAIIx/HiSeq | 47 | ID3 (34%) | 2012 | 62 |
| CLL | 4 | 363 | WGS/WES | GAIIx | 40/119 | NOTCH1 (12%), MYD88 (3%) | 2011 | 59 |
| CLL | 5 | 226 | WES | Genome Sequencer FLX | 10 | NOTCH1 (17%) | 2011 | 63 |
| CLL | 3/88 | 101 | WGS/WES | GAII | 38/132 | SF3B1 (15%), MYD88 (10%) | 2011 | 36 |
| CLL | 105 | 279 | WES | GAIIx | 62 | SF3B1 (10%), NOTCH1 (10%) | 2012 | 37 |
| CLL | 7 | 103 | RNASeq/WGS | GAII | NR/12 | YPEL5-PPP1CB fusion (95%) | 2013 | 32 |
| CLL | 160 | — | WES | GAIIx/HiSeq | 112 | Patterns of clonal evolution | 2013 | 76 |
| DLBCL | 6 | 105 | WES | Genome Sequencer FLX | 10 | MLL2 (24%), regulation of immune response (63% ABC, 31% GCB) | 2011 | 29 |
| DLBCL | 13/83 | 37 | WGS/mRNASeq | GAIIx/HiSeq | 32/41 | MLL2 (32%) MEF2B (11%), histone modification (13%), lymphocyte activationa, differentiationa and apoptosisa | 2011 | 28 |
| DLBCL | 49 | — | WES | HiSeq | 150 | Histone H1 proteins (69%), ACTB (10%),P2RY8 (12%), PCLO (35%) | 2012 | 38 |
| DLBCL | 34 | 39 | WGS/WES | GAII/HiSeq | 29/47 | Signal transductiona, chromatin modificationa | 2013 | 39 |
| FL | 1/12 | 35 | WGS/mRNASeq | GAIIx/HiSeq | 9/28 | MLL2 (89%), MEF2B (13%), histone modification (15%), lymphocyte activationa, differentiationa and apoptosisa | 2011 | 28 |
| HCL | 1 | 47 | WES | GAIIx | 71 | BRAF V600E (100%) | 2011 | 18 |
| MCL | 18 | 108 | mRNASeq | GAII | NR | NOTCH1 (12%), CCND1 (19%) | 2012 | 60 |
| MM | 23/16 | 161 | WGS/WES | GAII | 33/104 | Protein translation (42%), HOX9 pathway (29%) | 2011 | 19 |
| MM | 22 | 127 | WES | GAIIx | 61 | Distinct mutation patterns between t(4;14)and t(11;14) | 2012 | 75 |
| MM | 1 | — | WGS | SOLiD/HiSeq | 30 | Genomic evolution and clonal tides over course of disease | 2012 | 74 |
| MDS | 9 | 354 | WES | GAIIx | NR | SF3B1 (67%) | 2011 | 34 |
| MDS | 29 | 582 | WES | GAIIx/HiSeq | 134 | RNA splicing (55%) | 2011 | 33 |
| MDS | 1 | 150 | WGS | GAIIx/HiSeq | 39 | U2AF1 (9%) | 2012 | 43 |
| MDS/MPN | 15 | 310 | WESmRNASeq | HiSeq | NR | RNA splicing, SRSF2 (24%), clinical outcomes associated with mutations | 2012 | 35 |
| MPN | 40 | — | WES | HiSeq | NR | SUZ12 (3%) | 2011 | 93 |
| NHL(B-cell) | 2 | 263 | mRNASeq | GAII | NR | CIITA translocations (16%) | 2011 | 94 |
| PCNSL | 4 | 25 | WES | GAIIx | NR | MYD88 L265P (38%), TBL1XR1 (14%) | 2012 | 95 |
| SMZL | 6 | 93 | WGS | Complete genomics | 80 | MLL2 (50%), NOTCH2 (25%) | 2012 | 69 |
| SMZL | 8 | 109 | WES | HiSeq | 111 | NOTCH2 (21%), NOTCH1 (5%), SPEN (5%),DTX1 (2%) | 2012 | 68 |
| WM | 30 | 54 | WGS | Complete genomics | 66 | MYD88 L265P (91%) | 2012 | 26 |
Abbreviations: ALL, acute lymphocytic leukemia; AML, acute myelogenous leukemia; BL, Burkitt's lymphoma; ChIPSeq, chromatin immunoprecipitation sequencing; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; ETP ALL, early T-cell precursor acute lymphoblastic leukemia; FL, follicular lymphoma; HCL, hairy cell leukemia; MCL, mantle cell lymphoma; MDS, myelodysplastic syndrome; MethylSeq, methylation sequencing; MM, multiple myeloma; MPN, myeloproliferative neoplasms; mRNASeq, messenger RNA sequencing; NHL, non-hodgkins lymphoma; NR not reported; PCNSL, primary central nervous system lymphoma; SMZL, splenic marginal zone lymphoma; WM, Waldenströms macroglobulinemia.
Platforms: GAII, GAIIx, HiScanSQ and HiSeq2000 are from Illumina, Genome Sequencer FLX from 454 Sequencing Roche and SOLiD from Life Technologies.
Indicates significant pathway enrichment.
Single causative mutations: hairy cell leukemia and Waldenström's macroglobulinemia as paradigms
Probably, the most representative example of a single hit identified by sequencing are the hairy cell leukemias (HCL). Initially, WES was performed on a single HCL tumor/normal pair with somatic mutations identified in five genes: BRAF, CSMD3, SLC5A1, CNTN6 and OR8J1.18 Another 47 HCL cases were screened for BRAF mutations and strikingly, the BRAF V600E substitution was found in all 47 patients evaluated. Conversely, BRAF mutations were absent in related peripheral B-cell lymphomas and chronic lymphocytic leukemia (CLL), and were only found in a small subset of multiple myeloma (MM) patients (4%).19 The same activating mutation and its damaging effect has been previously reported in solid tumors such as in melanoma20 and papillary thyroid cancer.21 The presence of a common mutation across HCL provides a central novel therapeutic avenue in HCL based on V600E BRAF inhibitors22, 23 alone or in combination with MEK or ERK inhibitors. The success of vemurafenib, a BRAF inhibitor, in the treatment of V600E BRAF-mutated melanoma patients led two groups to investigate the effectiveness of this small molecule inhibitor in one HCL case study each. In both cases, including one with a biallelic V600E BRAF mutation, treatment with vemurafenib resulted in successful disease treatment,24, 25 thus providing evidence for clinical trials to evaluate the use of BRAF inhibitors in HCL.
A similar situation was found in Waldenström's macroglobulinemia (WM). Remarkably, a MYD88 L265P-activating mutation was recently found in 90% of WM cases.26 MYD88 encodes for an adapter protein that affects the interleukin-1 and toll-like receptor pathway, with the L265P mutation leading to the dysregulation of the nuclear factor-kB and the JAK-signaling pathways.27 The same mutation has been found, but to a lesser extent, in additional B-cell lymphomas, such as diffuse large B-cell lymphomas (DLBCL) of the ABC type (∼40%), MALT lymphomas and CLL (<10%), supporting the key role of MYD88 in the pathogenesis of these neoplasias.27, 28, 29 Interestingly, a recent study evaluating the association between MYD88 L265P and clinical characteristics of WM patients reported more involvement of the bone marrow disease, higher serum IgM, and lower IgA and IgG levels.30 Another group conducting a case–control study evaluating the association between MYD88 L265P and IgM MGUS patients progressing to WM or other lymphoproliferative disorders reported a trend toward progression in patients with the presence of the mutation when compared with patients with wild-type MYD88.31 These findings highlight the potential value of MYD88 as a potential biomarker of disease progression in WM.
In CLL, Velsusamy et al.32 recently identified the presence of a YPEL5-PPP1CB RNA fusion in 95% of CLL patients screened. Interestingly, WGS in the two index cases possessing the chimera did not reveal the presence of a gene fusion at the DNA level.32 These findings emphasize the importance of concurrently utilizing multiple methodologies such as WGS and RNASeq when studying tumors to better screen for genetic abnormalities.
One of the recurrent findings of sequencing research efforts has been the epistatic nature of discoveries. This notion reinforces the thought of classifying disease more along the line of functional aberrant pathways, rather than on specific genetic changes. In fact, excluding HCL and WM, the majority of the malignancies show a considerable genetic heterogeneity, affecting multiple genes and pathways. Presented here are some of the most remarkable recent discoveries.
Mutations affecting the splicing machinery
Recent sequencing studies identified recurrent mutations affecting genes of the splicing machinery in myelodysplastic syndrome (MDS).33, 34, 35 Interestingly, six of these genes (SF3A1, SF3B1, SRSF2, U2AF35, ZRSR2 and PRPF40B) affect the initial steps of RNA splicing; thus, mutations leading to the impaired recognition of the 39 splice site result in the production of abnormal mRNA splicing. Mutations of the spliceosome are highly prevalent in MDS and other myeloproliferative disorders, ranging from 44% of cases without increased sideroblasts to 85% of cases with increased sideroblasts.33 The mutations were mutually exclusive in disease subtypes,33, 34, 35 suggesting a key role of the spliceosome mutations in the pathogenesis of myeloproliferative disorders. On the other hand, mutations affecting the spliceosome were significantly lower in de novo AML and myeloproliferative neoplasms.33, 34
SF3B1 was the most commonly mutated of these genes, and it was significantly enriched in the group of MDS with increased ring sideroblasts, refractory anemia with ring sideroblasts, and refractory cytopenia with multilineage dysplasia and ring sideroblasts (P<0.001).34 Clinically, SF3B1 mutations were associated with fewer cytopenias and longer event-free survival.34, 35 The high prevalence of SF3B1 mutations in diseases with ring sideroblasts and the confirmation that the mutation can be identified in peripheral blood suggest that SF3B1 could potentially be used as a biomarker.
SF3B1 mutation was also one of the most significant discoveries in CLL, found in 10–15% of cases.36, 37 Mutations in SF3B1 were associated with deletion 11q (P=0.004).36 Moreover, SF3B1 mutations and/or deletion 11q were predictive markers of an earlier need for treatment (P<0.0001).36 Altogether, these results indicate that mutations of the spliceosome are involved in hematological malignancies and offer a novel therapeutic avenue for MDS and CLL.
Mutations modulating transcription and translation
One of the most interesting themes arising from the study of hematological malignancies is that alterations of genes modulating transcription and expression are a recurrent finding. Sequencing studies in DLBCL and follicular lymphomas reported genes involved in the histone modification process.28, 29, 38, 39 MLL2, a histone methyltransferase specific to the H3K4 residue, was the most commonly mutated gene in follicular lymphoma, affecting almost 90% of cases.28 The vast majority of mutations had an inactivating effect and included missense and frameshift mutations affecting or truncating the C-terminal domains, including the SET domain.28, 29 These findings place MLL2 collectively with the t(14;18)(q32;q21), as the two most common abnormalities in follicular lymphoma.
In addition, genes involved in histone modification were collectively identified in ∼20–40% of DLBCL29, 39 and early T-cell precursor acute lymphoblastic leukemia (ETP ALL).40 Furthermore, EZH2, which is involved in histone methylation, is mutated in DLBCL and follicular lymphoma,28, 29 with the mutations occurring in a critical SET domain.41 Preclinical studies in DLBCL have found the inhibition of EZH2 an effective therapeutic approach for tumors containing activating mutations, thus presenting a novel therapeutic target for the treatment of DLBCL.41
Chromatin modifiers are also recurrently affected in MM. MMSET, a histone methyltransferase transcriptional repressor, is overexpressed in ∼15% of MM as a consequence of the t(4;14)(p16;q32).42 Sequencing studies show that other chromatin modifiers are mutated in a significant subset of MM, including KDM6A and HOXA9.19 In addition, in the analysis of MM there was an enrichment of mutations within genes involved in protein translation. Thus, 42% of MM cases had mutations in this pathway, mainly affecting FAM46C (13%), DIS3 (11%) and LRRK2 (8%).19
A major finding in MDS and AML was the identification of mutations in a set of genes associated with DNA methylation. DNMT3A, a methyltransferase, is the most commonly mutated gene in AML found in around 20–30% of AML cases. Interestingly, no mutations were found in the related genes DNMT1, DNMT3B or DNMT3L.3 DNMT3A mutations were associated with poor survival (P<0.001). In addition, mutations have been identified in U2AF1 in MDS patients, and those harboring U2AF1 mutations were more likely to progress to secondary AML.43
Other breakthrough discoveries by NGS: IDH1 and IDH2 mutations in AML
Another major discovery in AML was the identification of mutations in IDH1, which encodes isocitrate dehydrogenase 1, and the related IDH2 gene. IDH1 mutations have been observed in DLBCL,39 cartilaginous tumors,44 astrocytoma45, 46 and glioblastoma,47 whereas IDH2 mutations have been reported in astrocytoma.45 Interestingly, patients with grade II astrocytoma who have IDH1 mutations show significantly shorter progression-free survival than tumors with wild-type IDH1.46 Studies in AML have reported mutations in 10–15% of cases, preferentially found in the intermediate-risk cytogenetic group, and their association with worse prognosis in a subset of AML patients that have been confirmed.48, 49 Mutations in IDH1 and IDH2 are mutually exclusive and primarily affect IDH1 at codon R132 and IDH2 at codons R140 or R172.50 Mutations in IDH1 were enriched in cases possessing DNMT3A mutations.3 Conversely, IDH2 mutations are rarely found together with other known recurrent mutations.48, 49 In addition, mutations in IDH1 and IDH2 seem to be mutually exclusive with TET2 mutations.51, 52 A recent study reported that mutations in IDH1 or IDH2 disrupted TET2 function and led to a hypermethylation phenotype with impaired hematopoietic differentiation.53
It becomes clear then that the morphological and clinicopathological classification of AML is now challenged by these new genetic findings. How many subcategories of AML exist? How does this heterogeneity exist, or not, in the better defined entities at the chromosome level (for example, M3)? In short, the various new perspectives to classify AML may ultimately lead more toward a molecular and pathway approach, but in some cases they might still have very significant resemblance to older cytogenetic classification.
Other breakthrough discoveries by NGS: NOTCH mutations
Aberrant NOTCH1 signaling has been identified in both solid and hematological tumors, and is a therapeutic target of interest currently in preclinical and clinical trials.54, 55 NOTCH1 encodes a transcription factor that transduces extracellular signals into expression changes in targets genes, including MYC56 and PI3K–AKT signaling pathways.57 NOTCH receptors are involved in cell fate determination, having a critical role in T-cell development. In fact, impaired NOTCH1 results in a block at the earliest stages of T-cell lymphopoiesis.58 Mutations in NOTCH1 lead to an active protein isoform lacking the C-terminal domain, and have been identified in over 50% of T-cell ALL and, to a lesser extent, in CLL, MCL and Burkitt's lymphoma.59, 60, 61, 62 These mutations mainly target the PEST domain, which is required for NOTCH1 interaction with FBW7, and subsequent NOTCH1 targeting for proteosomal degradation.
Data suggest that NOTCH1 mutations are a progressive event in CLL, increasing in prevalence from newly diagnosed CLL to chemorefractory CLL to CLL patients with Ritcher syndrome that underwent transformation to DLBCL.63 NOTCH1 mutations were associated with trisomy 12 (P=0.009) and with IGHV-unmutated status.36, 59, 64 In addition to the association with more advanced stages of the disease and with transformation to DLBCL, NOTCH1 mutations were associated with adverse biological course and worse overall survival in CLL (P=0.03)59, 63 and MCL (P=0.003).60 In T-cell ALL, NOTCH1 mutations were associated with improved response to glucocorticoid therapy; however, the association of NOTCH activation and clinical outcome seems to be therapy dependent.65, 66, 67
On the other hand, recurrent NOTCH2-activating mutations were identified in 21–25% splenic marginal zone lymhomas, but only rarely in nonsplenic MZLs and other low-grade B-cell lymphomas and leukemias.68, 69 Although these studies evaluated the association of NOTCH2 mutations and clinical outcomes, the findings are conflicting and additional work is necessary to clarify the potential clinical impact of mutations in NOTCH2.68, 69 Small molecule pan-NOTCH inhibitors have not shown significant effects as single agents targeting T-cell ALL, but there is an improved antileukemia effect when used in combination with inhibitors of PI3K–AKT–mTOR pathway or CDK inhibitors.70, 71
NGS as a tool for discrimination of related diseases
Besides the importance of identifying pathognomonic mutations and pathways, sequencing is also a powerful tool to differentiate related entities. Overall, 67% of ETP ALL had mutations in the RAS signaling pathway (BRAF, JAK1, JAK3, KRAS, NRAS) or cytokine receptors (IL7R), which was significantly higher than that in non-ETP ALL (19% P<0.0001).40 Furthermore, genes involved in hematopoiesis and lymphoid development (RUNX1, IKZF1, ETV6, GATA3 and EP300) were also more frequently mutated in ETP ALL (58%) than that in non-ETP ALL (17% P<0.0001). Altogether, 81% of ETP ALL cases have mutations in either of these pathways compared with 31% of non-ETP ALL cases (P<0.0001). A similar enrichment was identified in genes involved in histone modification (EED, EZH2 and SUZ12), which were more commonly mutated in ETP ALL (42%) compared with non-ETP ALL (12% P=0.0001).
Mutations in genes affecting the RAS pathway, cytokine receptor and epigenetic modification are common in AML, but are rare in B- and T-cell neoplasias.40, 72 These findings together with previous data demonstrate that ETP ALL has a gene expression signature closer to leukemic stem cells and granulocyte precursors, suggesting that ETP ALL is a distinct entity from non-ETP ALL with a less mature phenotype that retains the potential to become a myeloid cell.
Genomic sequencing in the analysis of clonal architecture and clonal evolution
Genomic sequencing performed in high-coverage depth is a useful tool for characterizing the clonal architecture and analyzing the clonal evolution in disease progression and in response to therapy. Ding et al.73 have provided a good example of the power of genomic sequencing in sequential analysis. WGS was performed in eight AML cases utilizing normal skin biopsies paired with tumor samples collected at diagnosis and after relapse. Candidate somatic events were analyzed by deep sequencing with a median of 590X coverage. In five out of eight cases, the primary sample was characterized by up to four mutation clusters, thus indicating the existence of multiple (sub)clones. Two major patterns of clonal evolution were identified when comparing primary versus relapse samples. Either the original clone in the primary tumor sample acquired additional mutations and evolved into the relapse clone, or most of the (sub)clones were eradicated by therapy leaving one clone. This clone is usually observed at a low frequency in the primary sample, it then survives the initial therapy, gains additional mutations and expands, becoming the predominant clone at relapse. Another interesting finding was obtained by comparing the transition with transversion mutation rate between primary and relapse samples. The data obtained strongly suggest that the chemotherapy regimen used (cytarabine and anthracycline for induction and additional cytotoxic chemotherapy for consolidation) have had a significant effect in the origin of novel mutations in the AML relapse sample.
In our sequencing analysis performed in MM, we have observed the presence of single nucleotide variants waxing and waning over the course of several longitudinally collected samples from a single patient.74 This shift in the presence of single nucleotide variants suggests the presence of multiple clones rising and falling in dominance over time. In the work by Walker et al.,75 a single MM patient, from whom they had WES data, identified three populations containing mutations in four genes: ATM, FSIP2, CLTC and GLMN. When they then evaluated which mutations were shared in a single cell, they identified one population with only an ATM mutation, a second with ATM and FSIP2 and a third with ATM, CLTC and GLMN mutations.75 The observed presence of these different clones suggests that if these patients were to be followed longitudinally, clonal dominance would likely shift as the tumor evolves with time and treatment.
The study of clonal complexity and clonal evolution is an old field that has been reinvigorated since the introduction of NGS. We believe NGS will help us to elucidate several unanswered questions such as what are the driver-initiating mutations in the different hematological malignancies? What are the specific mutations associated with disease progression in the different hematological malignancies? What mutations are the primary contributors to chemoresistance? Does clonal heterogeneity need to be considered in the context of determining therapeutic options? Do all the clones need to be targeted?
Some of these questions have been at least partially answered in a recent study.76 The authors analyzed 149 CLL cases, including 18 that were analyzed at two time points, using WES and SNP arrays. Data obtained from this study confirm previous findings showing the existence of linear and multibranching clonal evolution in CLL.77, 78 Furthermore, the authors were able to infer the order of genetic changes occurring in CLL pathogenesis. Thus, it was suggested that the clonal driver mutations, which are proposed to be initiating events, mainly affect genes that selectively affect B cells, such as MYD88 and del13q, whereas subclonal driver mutations associated with disease progression affect genes more ubiquitously involved in carcinogenesis such as TP53 and ATM. The number of subclonal mutations increases in treated compared with untreated cases; thus, the therapy would be a trigger for natural selection leading to the emergence of more aggressive subclones. Furthermore, the study shows the importance of subclonal driver mutations as an independent risk factor for rapid disease progression and poor outcome. Thus, this study suggests that dissecting the clonal architecture of CLL is crucial not only for developing novel risk-stratification algorithms but also for designing novel therapeutic approaches, considering the presence of driver mutations as well as the genomic landscape.
We expect to see similar efforts in several other hematological malignancies, which will ultimately help to elucidate the clonal complexity and its importance in each particular disease.
How are new discoveries translating into novel therapeutic approaches?
Several novel somatic mutations, such as SF3B1, IDH1, IDH2, DNMT3A, MYD88 and MLL2 have been identified as a consequence of NGS efforts, leading to the discovery of previously unrecognized genes and molecular processes/pathways with pathogenic effects. The genomic profiling of each individual cancer will potentially have a key role clinically assisting in early disease diagnosis, risk stratification, longitudinal analyses of tumor evolution and selection of the most favorable and personalized therapeutic intervention.
One of the most emblematic examples is AML. The unprecedented characterization of the AML cancer genome may substantially affect the clinical management and the therapeutic decisions. The prior characterization of mutations in FLT3, NPM1, RUNX1 and CEBPA together with the recent identification of mutations in IDH1, IDH2, DNMT3A and TET2 encourage the incorporation of genomic studies as part of routine clinical tests and may enable optimization of therapeutic plans based on this patient-specific genomic background.
However, the genetic characterization of AML will not improve patient survival per se, unless it is synchronized with the development of alternative therapeutic approaches. One of the major limitations in the treatment of AML is the intrinsic drug resistance of the tumor cells. Standard induction chemotherapy regimens, consisting of cytarabine and anthracycline combinations, have remained largely unchanged in the treatment of AML over decades. Thus, the major challenge is to provide the AML patients with alternative drug combinations targeting novel genes/pathways discovered in chemoresistant cases.
The discovery of novel genes/pathways not only increases our understanding of the pathogenesis of the disease but also opens new therapeutic avenues. The existence of potential ‘Achilles' heels to be exploited for generating a unifying targeted therapy for all patients is very provocative and opens an exciting era for translational research. Exploiting this knowledge is critical in hematological malignancies when considering that most of them are still incurable and more effective therapies are urgently needed.
So far, we have discovered a different range of genetic heterogeneity across tumor types. We have learnt that some malignancies have a mutated gene or pathway that affect most or all cases. An excellent example is provided by the BRAF V600E, common to all HCL patients,18 or MYD88 L265P, found in most WM.26, 28, 79 For example, V600E BRAF can be targeted with BRAF inhibitors22, 23 alone or in combination with MEK or ERK inhibitors.
Conversely, the majority of hematological malignancies are characterized by considerable tumor heterogeneity, making the search for therapeutic targets more difficult. One of the biggest challenges is to reduce the complexity of the generated data by first, distinguishing the driver over the passenger mutations and clones, and second, generating systematic and more sophisticated approaches for data analysis integration, thus unifying the vast genomic heterogeneity of these cancers into more homogeneous groupings based on cellular pathways rather than on single genes. As in the case of single gene mutations, the disruption of specific pathways may be exploited therapeutically.
With the dawn of the $1000 genome drawing close, we anticipate an ever-increasing role of genomic sequencing in the diagnosis and treatment of patients. As this technology moves ever closer to widespread clinical application, there are several challenges that must be addressed. First, the management of the data obtained from sequencing must be addressed. Not only does the physical storage of the data present a challenge but how the information obtained is reported to the patient, retained over time and/or destroyed, are important issues that also must be discussed. Second, incidental findings of mutations in genes unrelated to the medical reason, a patient is seeking genome sequencing can result in legal and ethical dilemmas for the care providers. Furthermore, knowledge about genomics and disease is rapidly expanding, thus consideration of whether a patient's genome should be re-evaluated at a later time must be considered.
As our focus shifts from large population-based studies with large cohorts to the ‘N of one' with individualized genomic medicine, it is imperative that the recommendations made to patients be based on evidence from well-designed functional studies. Translating this genetic data into the clinic is challenging and a significant amount of functional work is still required to better understand the biological significance of these hits using both in vitro and in vivo models. In the near future, we anticipate a standard of care for personalized medicine that involves sending samples for sequencing at the time of biopsy. A variant report will be generated for the physician who will then base treatment decisions on the findings from sequencing in addition to pathology and clinical diagnostics.
The ultimate goal in the post-genomic era will be to extend to other hematological malignancies the successful transition from gene discovery to therapeutic intervention observed in the paradigmatic BCR-ABL CML cases treated with imatinib.
Acknowledgments
This work was supported by NIH Grant CA133115-01 and Mayo Clinic Cancer Center. Rafael Fonseca is a Clinical Investigator of the Damon Runyon Cancer Research Fund. His work is supported by Grants SPORE CA90297052, P01 CA62242, R01 CA83724, ECOG CA 21115T, Predolin Foundation, Mayo Clinic Cancer Center and the Mayo Foundation.
RF has received a patent for the prognostication of MM based on genetic categorization of the disease. He has received consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS, Lilly, Millenium and AMGEN. He also has sponsored research from Cylene and Onyx. EB is a recipient of the Marriott Specialized Workforce Development awards in Individualized Medicine, The Henry Predolin Foundation Career Development award and the George Haub Family Career Development award fund in Cancer Research. JBE is a recipient of the Multiple Myeloma Research Foundation Research Fellow award. The remaining authors declare no conflict of interest.
Footnotes
Author Contributions
EB, JBE, RF, AKS wrote the manuscript. All authors reviewed and gave final approval of the manuscript.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- Cullum R, Alder O, Hoodless PA. The next generation: using new sequencing technologies to analyse gene regulation. Respirology. 2011;16:210–222. doi: 10.1111/j.1440-1843.2010.01899.x. [DOI] [PubMed] [Google Scholar]
- Tucker T, Marra M, Friedman JM. Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet. 2009;85:142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol. 2011;136:527–539. doi: 10.1309/AJCPR1SVT1VHUGXW. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Grimmond SM. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008;9:234. doi: 10.1186/gb-2008-9-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- Hao Da C, Feng Y, Xiao R, Xiao PG. Non-neutral nonsynonymous single nucleotide polymorphisms in human ABC transporters: the first comparison of six prediction methods. Pharmacol Rep. 2011;63:924–934. doi: 10.1016/s1734-1140(11)70608-9. [DOI] [PubMed] [Google Scholar]
- Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, Taylor J. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat Rev Genet. 2012;13:667–672. doi: 10.1038/nrg3305. [DOI] [PubMed] [Google Scholar]
- Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Hurlstone A. BRAF as therapeutic target in melanoma. Biochem Pharmacol. 2010;80:561–567. doi: 10.1016/j.bcp.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Li Y, Nakamura M, Kakudo K. Targeting of the BRAF gene in papillary thyroid carcinoma (review) Oncol Rep. 2009;22:671–681. doi: 10.3892/or_00000487. [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follows GA, Sims H, Bloxham DM, Zenz T, Hopper MA, Liu H, et al. Rapid response of biallelic BRAF V600E mutated hairy cell leukaemia to low dose vemurafenib. Br J Haematol. 2013;161:150–153. doi: 10.1111/bjh.12201. [DOI] [PubMed] [Google Scholar]
- Dietrich S, Glimm H, Andrulis M, von Kalle C, Ho AD, Zenz T. BRAF inhibition in refractory hairy-cell leukemia. N Engl J Med. 2012;366:2038–2040. doi: 10.1056/NEJMc1202124. [DOI] [PubMed] [Google Scholar]
- Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao Y, et al. MYD88 L265P somatic mutation in Waldenstrom's macroglobulinemia. N Engl J Med. 2012;367:826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Hunter ZR, Yang G, Zhou Y, Cao Y, Liu X, et al. MYD88 L265P in Waldenstrom macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood. 2013;121:2051–2058. doi: 10.1182/blood-2012-09-454355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varettoni M, Arcaini L, Zibellini S, Boveri E, Rattotti S, Riboni R, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom's macroglobulinemia and related lymphoid neoplasms. Blood. 2013;121:2522–2528. doi: 10.1182/blood-2012-09-457101. [DOI] [PubMed] [Google Scholar]
- Velusamy T, Palanisamy N, Kalyana-Sundaram S, Sahasrabuddhe AA, Maher CA, Robinson DR, et al. Recurrent reciprocal RNA chimera involving YPEL5 and PPP1CB in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2013;110:3035–3040. doi: 10.1073/pnas.1214326110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, et al. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- Stockhammer F, Misch M, Helms HJ, Lengler U, Prall F, von Deimling A, et al. IDH1/2 mutations in WHO grade II astrocytomas associated with localization and seizure as the initial symptom. Seizure. 2012;21:194–197. doi: 10.1016/j.seizure.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Thon N, Eigenbrod S, Kreth S, Lutz J, Tonn JC, Kretzschmar H, et al. IDH1 mutations in grade II astrocytomas are associated with unfavorable progression-free survival and prolonged postrecurrence survival. Cancer. 2012;118:452–460. doi: 10.1002/cncr.26298. [DOI] [PubMed] [Google Scholar]
- Yan W, Zhang W, You G, Bao Z, Wang Y, Liu Y, et al. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One. 2012;7:e30339. doi: 10.1371/journal.pone.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Heuser M, Morgan M, Wagner K, Gorlich K, Grosshennig A, et al. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. 2011;117:4561–4568. doi: 10.1182/blood-2010-08-303479. [DOI] [PubMed] [Google Scholar]
- Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. 2010;116:5486–5496. doi: 10.1182/blood-2010-02-267955. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JC, Lorenzi MV. Drug discovery approaches to target Wnt signaling in cancer stem cells. Oncotarget. 2010;1:563–577. doi: 10.18632/oncotarget.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, JPt Morris, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2011;119:329–331. doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuurbier L, Homminga I, Calvert V, te Winkel ML, Buijs-Gladdines JG, Kooi C, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24:2014–2022. doi: 10.1038/leu.2010.204. [DOI] [PubMed] [Google Scholar]
- Clappier E, Collette S, Grardel N, Girard S, Suarez L, Brunie G, et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24:2023–2031. doi: 10.1038/leu.2010.205. [DOI] [PubMed] [Google Scholar]
- Kox C, Zimmermann M, Stanulla M, Leible S, Schrappe M, Ludwig WD, et al. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010;24:2005–2013. doi: 10.1038/leu.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209:1553–1565. doi: 10.1084/jem.20120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–6181. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, O'Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69:3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39 (4 Suppl 3:6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120:1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–1086. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braggio E, Kay NE, Vanwier S, Tschumper RC, Smoley S, Eckel-Passow JE, et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia. 2012;26:1698–1701. doi: 10.1038/leu.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Yau C, Clifford R, Timbs AT, Sadighi Akha E, Dreau HM, et al. Quantification of subclonal distributions of recurrent genomic aberrations in paired pre-treatment and relapse samples from patients with B-cell chronic lymphocytic leukemia. Leukemia. 2012;26:1564–1575. doi: 10.1038/leu.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Sohani AR, Arcaini L, Hunter Z, Yang G, Zhou Y, et al. A somatic variant in MYD88 (L265P) revealed by whole genome sequencing differentiates lymphoplasmacytic lymphoma from marginal zone lymphomas Blood (ASH Ann Meeting Abstr) 2011118Abstract 261. [Google Scholar]
- Iacobucci I, Ferrarini A, Sazzini M, Giacomelli E, Lonetti A, Xumerle L, et al. Application of the whole-transcriptome shotgun sequencing approach to the study of Philadelphia-positive acute lymphoblastic leukemia. Blood Cancer J. 2012;2:e61. doi: 10.1038/bcj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman R, Valkhof MG, Sanders MA, van Strien PM, Haanstra JR, Broeders L, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119:5071–5077. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- Greif PA, Dufour A, Konstandin NP, Ksienzyk B, Zellmeier E, Tizazu B, et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood. 2012;120:395–403. doi: 10.1182/blood-2012-01-403220. [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Estecio MR, Lu Y, Long H, Malouf GG, Graber D, et al. The epigenome of AML stem and progenitor cells. Epigenetics. 2013;8:92–104. doi: 10.4161/epi.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greif PA, Eck SH, Konstandin NP, Benet-Pages A, Ksienzyk B, Dufour A, et al. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia. 2011;25:821–827. doi: 10.1038/leu.2011.19. [DOI] [PubMed] [Google Scholar]
- Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2012;118:6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- Masetti R, Pigazzi M, Togni M, Astolfi A, Indio V, Manara E, et al. CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood. 2013;121:3469–3472. doi: 10.1182/blood-2012-11-469825. [DOI] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44:1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- Puda A, Milosevic JD, Berg T, Klampfl T, Harutyunyan AS, Gisslinger B, et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am J Hematol. 2011;87:245–250. doi: 10.1002/ajh.22257. [DOI] [PubMed] [Google Scholar]
- Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilar A, Idbaih A, Boisselier B, Habbita N, Rossetto M, Laurenge A, et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res. 2012;18:5203–5211. doi: 10.1158/1078-0432.CCR-12-0845. [DOI] [PubMed] [Google Scholar]