Abstract
The epigenetic mark of the centromere is thought to be a unique centromeric nucleosome that contains the histone H3 variant, centromere protein-A (CENP-A). The deposition of new centromeric nucleosomes requires the CENP-A-specific chromatin assembly factor HJURP (Holliday junction recognition protein). Crystallographic and biochemical data demonstrate that the Scm3-like domain of HJURP binds a single CENP-A–histone H4 heterodimer. However, several lines of evidence suggest that HJURP forms an octameric CENP-A nucleosome. How an octameric CENP-A nucleosome forms from individual CENP-A/histone H4 heterodimers is unknown. Here, we show that HJURP forms a homodimer through its C-terminal domain that includes the second HJURP_C domain. HJURP exists as a dimer in the soluble preassembly complex and at chromatin when new CENP-A is deposited. Dimerization of HJURP is essential for the deposition of new CENP-A nucleosomes. The recruitment of HJURP to centromeres occurs independent of dimerization and CENP-A binding. These data provide a mechanism whereby the CENP-A pre-nucleosomal complex achieves assembly of the octameric CENP-A nucleosome through the dimerization of the CENP-A chaperone HJURP.
Keywords: CENP-A, centromere, chromatin, mitosis, nucleosome
Introduction
The equal distribution of chromosomes into daughter cells during mitosis depends on the proper assembly of a centromere on each chromosome. Centromere assembly occurs independently of DNA sequence, with the exception of budding yeast point centromeres (Cleveland et al, 2003; Allshire and Karpen, 2008; Stellfox et al, 2012). All eukaryotes use a conserved, CenH3 containing, centromere-specific nucleosome to determine (or mark) the site of the centromere. Human centromere-specific nucleosomes contain centromere protein-A (CENP-A) in place of histone H3. CENP-A containing nucleosomes are found interspersed with canonical histone H3 nucleosomes within human centromeres (Blower et al, 2002). CENP-A nucleosomes direct the recruitment of a constitutive centromere-associated network (CCAN) and the kinetochore proteins that together orchestrate the attachment of chromosomes to the mitotic spindle and regulate cycle progression through the mitotic checkpoint.
Existing CENP-A is quantitatively retained at centromeres following DNA replication and redistributed to sister centromeres (Jansen et al, 2007). Thus, continuous inheritance of centromere position requires new CENP-A deposition that occurs in every cell cycle in order to maintain a sufficient number of CENP-A nucleosomes to specify the centromeric locus. The assembly of new centromeric nucleosomes depends on the CENP-A-specific chromatin assembly factor, HJURP (Holliday junction recognition protein) (Dunleavy et al, 2009; Foltz et al, 2009; Bernad et al, 2011). CENP-A interacts with HJURP as a soluble pre-nucleosomal complex. The deposition of centromeric nucleosomes in yeast requires the HJURP homologue, Scm3 (Camahort et al, 2007; Mizuguchi et al, 2007; Stoler et al, 2007; Sanchez-Pulido et al, 2009; Williams et al, 2009; Dechassa et al, 2011). HJURP and Scm3 share close to 69% homology within a small 52 amino-acid region in the amino terminus of HJURP, which is required for CENP-A binding (Sanchez-Pulido et al, 2009; Shuaib et al, 2010). The Scm3 domain of HJURP is sufficient to facilitate the formation of CENP-A nucleosomes in vitro and in vivo (Barnhart et al, 2011). The recruitment of HJURP and the deposition of CENP-A occur during early G1 (Jansen et al, 2007; Schuh et al, 2007; Dunleavy et al, 2009; Foltz et al, 2009). HJURP recruitment to centromeres depends on the activity of the Mis18 complex (Barnhart et al, 2011; Moree et al, 2011), which influences the histone modification and DNA methylation status of centromeres (Fujita et al, 2007; Kim et al, 2012). However, the mechanism by which Mis18 directs HJURP to centromeres remains unclear.
The crystal structures of the CENP-A–histone H4 heterotetramer, containing two copies each of CENP-A and H4, as well as the CENP-A octameric nucleosome have been solved (Sekulic et al, 2010; Tachiwana et al, 2011). Additional evidence suggests that the CENP-A nucleosome may transition from an octameric nucleosome to hemisome, containing a single copy of CENP-A and H4, as a cell progresses through the cell cycle (Bui et al, 2012; Shivaraju et al, 2012). Similarly to the H3–H3 interface in the canonical nucleosome, dimerization of CENP-A is required for stable CENP-A deposition. Mutants of human CENP-A or the Drosophila homologue, CID, in which the CENP-A–CENP-A dimerization interface is disrupted, are unable to form stable nucleosomes in vivo (Bassett et al, 2012; Zhang et al, 2012), suggesting that formation of a CENP-A octamer is required for stable nucleosome formation. Human HJURP and yeast Scm3 mediate the formation of octameric nucleosomes in vitro (Barnhart et al, 2011; Dechassa et al, 2011; Kingston et al, 2011; Shivaraju et al, 2011). Interestingly, several recent biochemical studies of HJURP/Scm3 in complex with CENP-A have demonstrated that CENP-A interacts with HJURP as a heterodimer containing a single copy of CENP-A and histone H4 (Cho and Harrison, 2011; Feng et al, 2011; Zhou et al, 2011; Bassett et al, 2012). These observations raise the question of how an octameric CENP-A nucleosome may be assembled from a heterodimeric intermediate.
Vertebrate HJURP proteins are significantly larger than their yeast orthologues and contain additional conserved domains (CDs) (Sanchez-Pulido et al, 2009). Human HJURP contains two HJURP_C-terminal domains (HCTD) within the carboxyl terminal half of the protein. HJURP_C domains are also found in the myocyte enhancer factor 2 (MEF2) transcription factors (Potthoff and Olson, 2007), but a functional role for this domain in MEF2 has not been determined. Only the Scm3 domain has been implicated in CENP-A deposition.
Here, we demonstrate that HJURP, in the pre-nucleosomal complex, self-associates through its carboxyl terminus. The C-terminal portion of HJURP required for self-association in vivo forms a dimer when expressed and purified in vitro. We show that the targeting of HJURP to centromeres occurs independently of HJURP dimerization and requires a region of HJURP between the CD and the second HCTD (HCTD2) (Sanchez-Pulido et al, 2009). Importantly, we also find that self-association of HJURP is essential for the assembly of CENP-A nucleosomes at centromeres. These data identify the region of HJURP sufficient for centromere targeting and provide a potential mechanism by which octameric CENP-A nucleosomes are assembled from a heterodimeric intermediate.
Results
Centromeric localization of HJURP through the carboxyl terminus
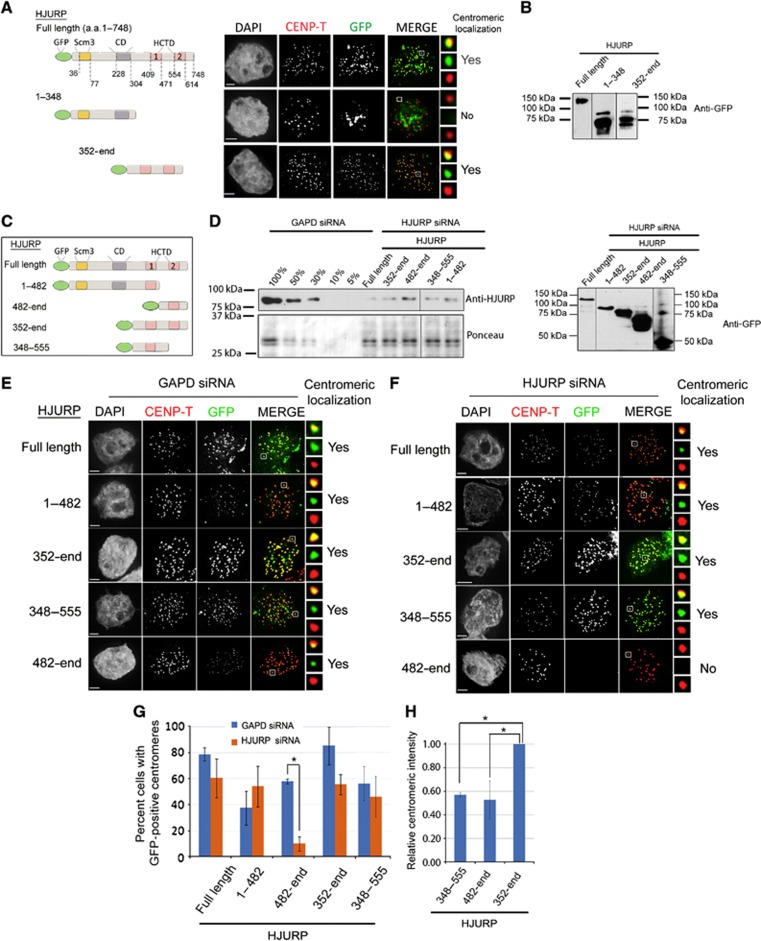
A set of non-overlapping HJURP truncation proteins were examined to determine which domains of HJURP mediate its specific recruitment to centromeres. Centromeric localization was assessed in early G1 cells (midbody positive) at 24 h post transfection. Full-length HJURP localized to centromeres in ∼82% (±5) of G1 cells (n>60 cells, two independent experiments) (Figure 1A and B). The Scm3 domain alone was not recruited to centromeres, as shown previously (Barnhart et al, 2011). The HJURP CD is a distinguishing feature of vertebrate HJURP orthologues (Sanchez-Pulido et al, 2009), but is absent from HJURP/Scm3 orthologues in fungi. The HJURP1–348 deletion mutant, which contains the Scm3 and CD domains of HJURP, did not localize to centromeres; however, the complementary deletion mutant, containing the remaining carboxyl half (HJURP352-end), was recruited to centromeres (Figure 1A and B). The HJURP352-end mutant was as efficient at centromere recruitment in G1 cells as the full-length protein (91±1%). When the recruitment was compared between all cells (asynchronously dividing) and just those in G1 (based on the presence of a mid-body), HJURP352-end was enriched at centromeres three-fold during G1 similar to full-length HJURP. These data demonstrate that centromere targeting is controlled by the carboxyl half of HJURP and that this protein truncation is under the same cell-cycle control as endogenous HJURP. The carboxyl half of HJURP contains the HCTD and based on these results also contains the specific centromere-targeting domain. We conclude that centromere targeting does not require the CD or CENP-A binding through the Scm3 domain.
Figure 1.
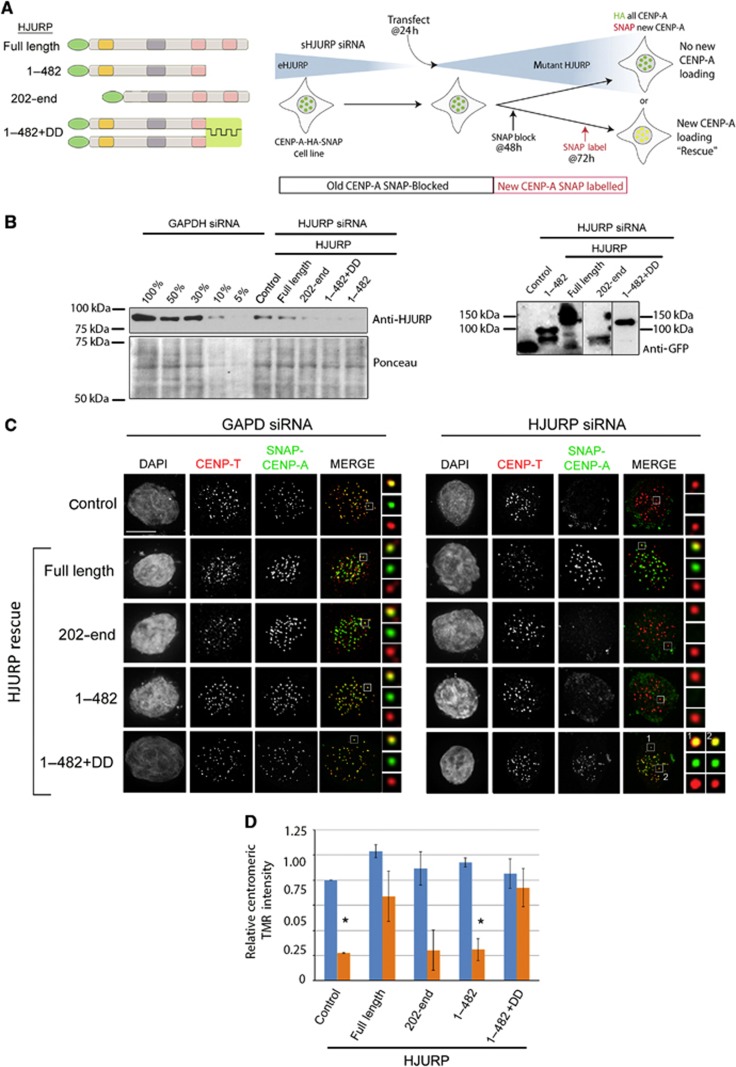
Identification of CIS-acting elements within HJURP required for centromere recruitment. (A) Schematic representations of transfected HJURP fragments (left panel) and corresponding representative images of cells transfected with GFP-tagged HJURP fragments (right panel). DNA was visualized by DAPI staining; anti-CENP-T is shown in red, the GFP-tagged fragments are in green. Merge includes CENP-T and GFP signals. Scale bar is 2 μm in all panels. Boxed regions are magnified to the right of merged images. (B) Anti-GFP western blot shows the expression of GFP-HJURP fusion proteins expressed in (A). (C) Schematic representations of transfected GFP-HJURP fragments used in (D–F). (D) Western blot showing the efficiency of HJURP depletion (left blot) and expression of transfected constructs (right blot) used in (E) and (F). Efficiency of HJURP siRNA treatment was assessed by anti-HJURP antibody, the expression efficiency was assessed by anti-GFP antibody, Ponceau staining serves as a loading control. (E, F) Representative images of cells expressing GFP-HJURP fragments and treated with either GAPD (E) or HJURP (F) siRNA. DNA was visualized by DAPI; CENP-T is shown in red, the GFP-tagged fragments are in green. (G) Quantification of the percentage of G1 cells in which GFP-HJURP was recruited to centromeres. Data are from at least two independent experiments, >60 cells per condition. Error bars represent the standard deviation. *Indicates P<0.01. (H) Relative centromeric intensity of HJURP fragments. n>180 centromeres per condition. *Indicates P<0.05.
Recruitment of HJURP to centromeres through direct recruitment and self-association
To further refine the centromere-targeting domain, a pair of HJURP truncation mutants (HJURP1–482 and HJURP482-end) was expressed, which divided HJURP into the carboxyl terminus (Figure 1C and D). By separating the two C-terminal repeats (HCTD’s) into two fragments, we predicted only one of these two truncation mutants would localize to centromeres. However, we observed that both HJURP1–482 and HJURP482-end could localize to centromeres (Figure 1E).
We hypothesized that the recruitment of both non-overlapping HJURP carboxyl-terminal fragments to centromeres may occur directly through the centromere-targeting domain and indirectly through dimerization with endogenous HJURP. The recruitment of an HJURP fragment via dimerization should be dependent on endogenous HJURP for centromere localization. Conversely, the recruitment of a fragment containing the direct centromere-targeting domain should localize to centromeres independently of endogenous HJURP. Therefore, we tested the recruitment of a series of HJURP truncation mutants in cells where endogenous HJURP was depleted by siRNA.
Endogenous HJURP was depleted for 24 h using an siRNA directed against the 3′UTR of HJURP and followed by expression of HJURP truncation mutants (Figure 1D–F). Endogenous HJURP expression was decreased by siRNA treatment to ∼20% of GAPD-treated levels (Figure 1D). Centromeric localization of the exogenously expressed HJURP fragments was analysed in early G1-phase cells (Figure 1E and F). Exogenous full-length HJURP efficiently associated with the centromeric chromatin in control (GAPD) and HJURP siRNA-treated cells. HJURP352-end was recruited to centromeres in control and HJURP siRNA-treated cells (Figure 1E and F). The number of cells that recruited HJURP352-end under control and HJURP siRNA conditions recapitulated the same degree of recruitment as the exogenously expressed full-length HJURP under similar conditions (Figure 1G).
Centromeric recruitment of the complementary pair of HJURP deletion mutants, HJURP1–482 and the reciprocal fragment, HJURP482-end, was tested following siRNA treatment. HJURP1–482 was recruited to centromeres with similar efficiency in control and HJURP siRNA-treated cells. It was also recruited to centromeres in a similar fraction of cells as exogenous full-length HJURP in the HJURP depletion condition (Figure 1G). In contrast, HJURP depletion abolished centromeric recruitment of HJURP482-end despite its recruitment to centromeres in the control GAPD depletion (Figure 1E–G). The dependence of HJURP482-end on endogenous HJURP for its recruitment is consistent with dimerization of HJURP at centromeres. Together, these data suggest that dimerization and recognition of the centromere are mediated by distinct domains within HJURP.
We compared the centromeric intensities between HJURP352-end (HCTD1 and HCTD2) with HJURP348-555 (HCTD1) and HJURP482-end (HCTD2), each of which contains only a single HCTD. HJURP352-end contains both repeats and should therefore be recruited by both direct targeting to the centromere and through dimerization. Consistent with this, HJURP352-end GFP signal was 1.5 × more intense at centromeres than either HJURP348-555 or HJURP482-end, which contain the individual HCTD1 and HCTD2 regions, respectively (Figure 1E and H). Therefore, the HCTD2 domain mediates a multimerization of HJURP, and amino acids 348–482 directly recruit HJURP to centromeres.
Dimerization of HJURP through the carboxyl terminus
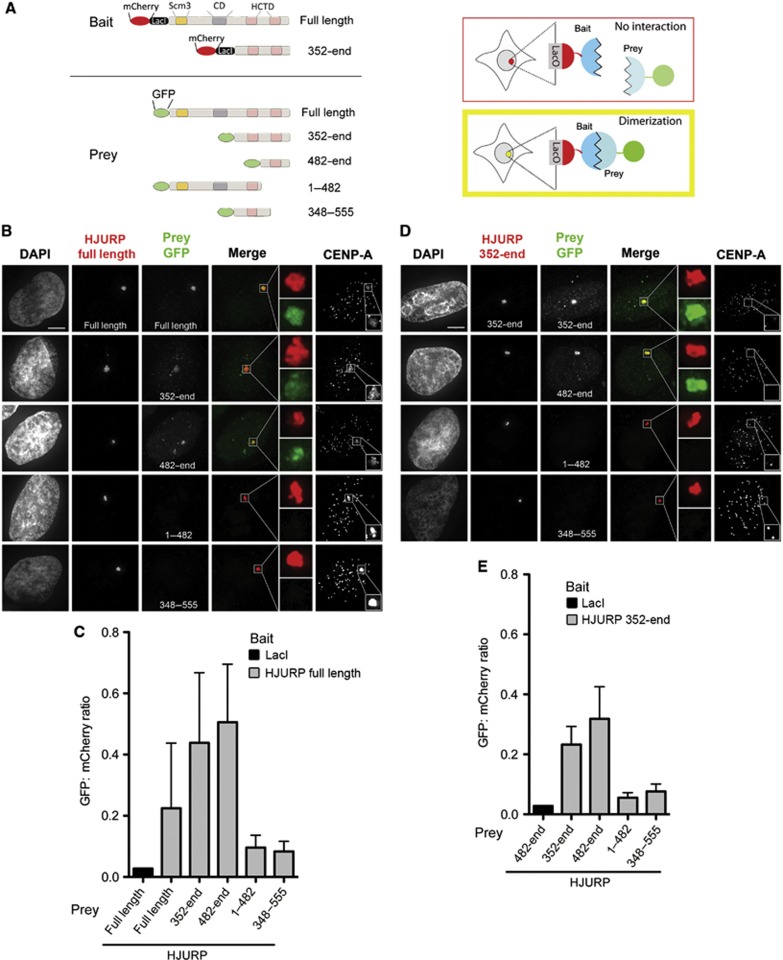
The ability of HJURP to self-associate was examined using a LacI/LacO-based in vivo interaction assay to directly assess if HJURP multimerizes in vivo (Figure 2A). Full length or HJURP352-end was fused to the lac repressor (LacI) and expressed as bait in cells that have a stably integrated LacO array (Janicki et al, 2004; Barnhart et al, 2011). The interaction between HJURP proteins was tested by expressing GFP-HJURP fragments as prey. Tethering HJURP to the LacO array resulted in GFP-HJURP recruitment (Figure 2B and C). Furthermore, LacI-HJURP was able to recruit carboxyl-terminal fragments of HJURP (Figure 2B and C). This interaction only required the HJURP carboxyl terminus because tethering the LacI-HJURP352-end fragment to the array was sufficient to recruit GFP-carboxyl fragments, containing amino acids 352-end and 482-end (Figure 2D and E). Full length and HJURP352-end showed minimal recruitment of an HJURP fragment containing amino acids 1–482 to the array, showing that HCTD2 in the carboxyl terminus is the primary site of HJURP self-association. The HCTD1 domain present in HJURP348–555 was unable to be efficiently recruited by either the full-length or HJURP352-end bait protein. We conclude that the C-terminal region of HJURP containing the second HCTD2 domain of HJURP is sufficient to mediate self-interaction in vivo.
Figure 2.
In vivo recruitment of HJURP through the carboxyl terminus. (A) Schematic of LacO-LacI interaction assay and the bait and prey constructs used in the study. (B) U2OS-LacO cells were co-transfected with mCherry-LacI-HJURPFullLength and indicated GFP-tagged prey fragments. DNA is stained with DAPI. Centromere staining and endogenous CENP-A recruitment to the arrays are shown using anti-CENP-A antibody. Scale bar represents 5 μM. All images are scaled equally. Boxed regions are magnified to the right of merged images. (C) Quantitation of prey protein recruitment to the array when HJURPFullLength (grey) or control mCherry-LacI alone (black) is targeted. Recruitment is expressed as the ratio of GFP to mCherry integrated intensity at the array. (D) Cells co-transfected with mCherry-LacI-HJURP352-end or mCherry-LacI alone as bait with the indicated GFP-tagged prey fragments. Cells were stained with DAPI to visualize DNA and anti-CENP-A as in (B). Scale bar represents 5 μM. (E) Prey protein recruitment to the array in response to mCherry-LacI-HJURP352-end or control mCherry-LacI targeting is quantified as in (C). The GFP:mCherry ratios for LacI-HJURP are plotted as the mean of n=3 experiments at ≥30 arrays per condition. Error bars represent standard deviation.
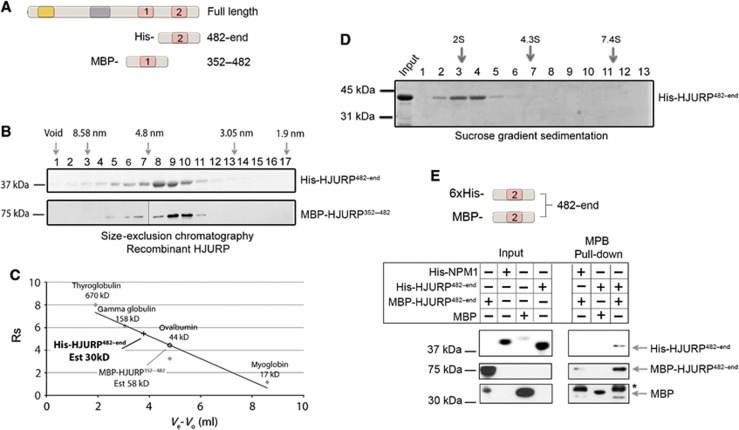
In order to elucidate whether HJURP self-association through its carboxyl terminus is a direct interaction that occurs without any additional factors, the putative dimerization domain (HJURP482-end) was expressed and purified from bacteria (Figure 3A; Supplementary Figure S1A). His-tagged HJURP482-end protein migrates on a denaturing SDS–PAGE gel as a 36-kDa protein, which is consistent with its calculated molecular weight of 30 kDa. Based on size-exclusion chromatography, the Stokes radius of His-HJURP482-end was calculated as 5.46 nm (Figure 3B and C; Supplementary Figure S1B), which is twice as large as the expected 2.48 nm Stokes radius of a 30-kDa globular protein. This larger than expected Stokes radius may indicate either that HJURP482-end is an elongated protein or that it exists as a multimer. For comparison, MBP-tagged HJURP352–482, which should not dimerize, and is a larger protein than HJURP482-end (58 kDa versus 30 kDa), elutes from the size-exclusion column with a smaller Stokes radius of 4.46 nm (Figure 3B and C; Supplementary Figure S1B). The sedimentation coefficient for HJURP482-end was determined by sucrose gradient (Figure 3D; Supplementary Figure S1C and D). Based on these analyses, the molecular weight of HJURP482-end was calculated as 59 kDa, consistent with the formation of a dimer (Siegel and Monty, 1966) (Supplementary Figure S1D). An MBP pull-down assay using two differently tagged recombinant proteins, MBP-HJURP482-end and His-HJURP482-end, demonstrates that the carboxyl terminus of HJURP self associates (Figure 3E). The in vivo data lead us to conclude that HJURP forms a dimer through a direct self-interaction mediated by a region that includes the HCTD2 domain. In vitro data are consistent with the same region of HJURP forming a multimer in vivo.
Figure 3.
Dimerization of HJURP in vitro. (A) Schematic of recombinant carboxyl terminal HJURP fragments expressed in bacteria. (B) Recombinant MBP-HJURP352–482 and His-HJURP482-end were analysed by size-exclusion chromatography. The arrows indicate the elution of protein standards. His-HJURP482-end has a predicted molecular weight of 30 kDa and was detected by Coomassie stain. MBP-tagged HJURP352–482 has a predicted molecular weight of 58 kDa and was detected by immunoblot using anti-MBP antibody. (C) The elution of the standards and HJURP fragments are plotted relative to their Stokes radius (Rs). (D) Coomassie stained SDS–PAGE gel of fractions collected after sucrose gradient ultracentrifugation of His-HJURP482-end fragment. (E) Schematic of recombinant carboxyl terminal HJURP fragments expressed in bacteria (top panel). MBP pull-down experiment demonstrating direct interaction between differently tagged HJURP 482-end fragments (bottom panel). His and MBP-tagged proteins were visualized by antibody staining. Asterisk indicates MBP-HJURP breakdown product.
Dimerization of HJURP forms a high molecular weight pre-nucleosomal complex
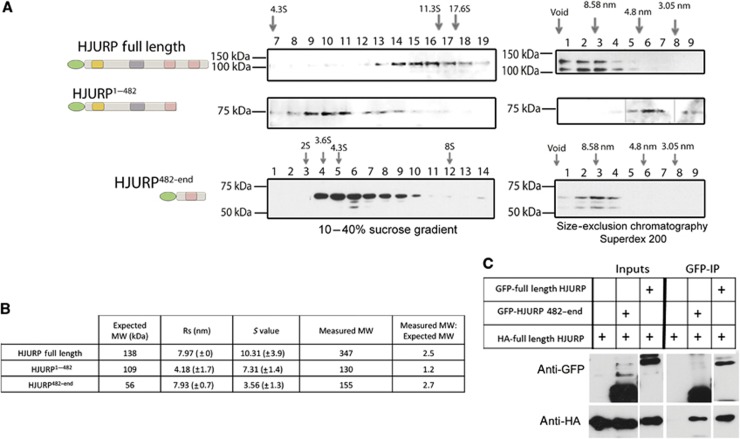
To determine whether HJURP is present as a self-associate in the prenucleosomal CENP-A complex, we determined the native molecular weight of the HJURP complex from cells transiently expressing GFP-tagged full-length and truncated HJURP. The prenucleosomal complex containing full-length GFP-tagged HJURP protein migrates on a sucrose gradient as a large complex with a sedimentation coefficient of 10.3S. This is slightly higher than the 10S reported previously for the endogenous CENP-A prenucleosomal complex, possibly due to the addition of the 30-kDa GFP tag (Foltz et al, 2009). The calculated molecular weight of the pre-nucleosomal complex based on sedimentation and size-exclusion chromatography was ∼347 kDa (Figure 4A and B), 2.5 times larger than the expected size of a heterotrimer containing a single copy of CENP-A, histone H4, and GFP-HJURP (∼138 kDa).
Figure 4.
In vivo dimerization of HJURP in the pre-nucleosomal complex. (A) Anti-GFP immunoblots of fractions collected after sucrose gradient ultracentrifugation (left) or size-exclusion chromatography (SEC) (right) of chromatin-free extracts from GFP-HJURP full-length, GFP-HJURP1–482 and GFP-HJURP482-end expressing HEK293 cells. The arrows indicate the migration of protein standards. (B) Table showing the measured Rs, sedimentation coefficient (S), and expected molecular weights of the HJURP proteins analysed by SEC and sucrose gradient sedimentation. Expected molecular weights include CENP-A and histone H4 (28 kDa) for proteins that contain the CENP-A binding domain. (C) Cell extracts co-expressing full-length GFP-HJURP or GFP-HJURP482-end with HA-tagged full-length HJURP were subjected to anti-GFP immunoprecipitation followed by immunoblot using antibodies against HA and GFP.
Amino acids 482–748 were sufficient to interact with full-length HJURP in the LacO/LacI interaction assay and formed a dimer in vitro, making this region a prime candidate to mediate multimerization of the complex in vivo. To determine if this domain is responsible for forming the high molecular weight HJURP pre-nucleosomal complex, we examined the size of the soluble HJURP complex formed by HJURP1–482, which lacks the putative dimerization domain. As we expected, HJURP1–482 formed a significantly smaller complex with a molecular weight of ∼130 kDa (Figure 4A and B; Supplementary Figure S2A and B). This value is consistent with a complex that contains only a single HJURP, along with the CENP-A, histone H4 heterodimer, calculated to be 109 kDa. We found that HJURP482-end alone was sufficient to form a multimer in vivo. We calculated the native molecular weight of HJURP482-end as 155 kDa (Figure 4A and B). The theoretical molecular mass for this fragment is expected to be 56 kDa.
To determine if the HJURP pre-nucleosomal complex contains more than one HJURP molecule, we co-transfected cells with constructs expressing GFP-tagged full-length HJURP and HA-tagged full-length HJURP and immunoprecipitated using anti-GFP antibodies. Under these conditions, GFP-HJURP associated with HA-HJURP (Figure 4C). The immunoprecipitations did not contain histone H2B and were therefore not chromatin-associated complexes but represent soluble HJURP multimers (Supplementary Figure S2C). We observed an interaction between full-length HJURP and the HJURP482-end fragment in co-immunoprecipitations from cells transiently transfected with GFP-HJURP482-end and HA-tagged HJURP protein, but not in control immunoprecipitations (Figure 4C; Supplementary Figure S2D). Together, these data demonstrate the HJURP carboxyl terminal tail (amino acids 482–748) is required for the formation of the multimeric HJURP pre-nucleosomal complex.
Dimerization of HJURP is required for CENP-A deposition
Since HJURP self-association occurs on centromeric chromatin, we hypothesized that new CENP-A deposition requires HJURP dimerization. A CENP-A SNAP tag assay was used to determine if new CENP-A was recruited to centromeres when endogenous HJURP was depleted by siRNA and replaced with exogenous HJURP that lacked the proposed dimerization domain (HJURP1–482). The SNAP tag assay specifically follows the incorporation of new SNAP-tagged CENP-A nucleosomes by blocking detection of existing CENP-A with a non-fluorescent SNAP substrate and labelling new CENP-A with an SNAP substrate that is fluorescent (Jansen et al, 2007; Foltz et al, 2009). SNAP-tagged CENP-A cells were treated with HJURP 3’UTR siRNA for 24 h to deplete endogenous HJURP followed by expression of either full-length or truncated HJURP replacement fragments (Figure 5A). Cells were given 24 h to express the HJURP replacement fragments, and then new CENP-A assembly was assayed over the following 24 h during which time CENP-A deposition was dependent on the replacement construct. As expected, HJURP siRNA treatment of CENP-A SNAP cells significantly decreased the percentage of cells with new SNAP labelled CENP-A at centromeres and reduced the amount of new SNAP-labelled CENP-A at centromeres (Figure 5B–D; Supplementary Figure S3A). The expression of full-length HJURP restored new CENP-A assembly in HJURP siRNA-treated cells (Figure 5C and D). As a negative control, we expressed the HJURP202-end fragment, which lacks the CENP-A binding domain and therefore should not rescue CENP-A deposition. New CENP-A recruitment to centromeres in cells transfected with the HJURP202-end was significantly impaired relative to the GAPD siRNA control (Figure 5C and D; Supplementary Figure S3A).
Figure 5.
CENP-A assembly requires HJURP dimerization. (A) Schematic showing the GFP-HJURP constructs used and the design of the SNAP-tag experiment testing new CENP-A recruitment. The 1–482+DD (dimerization domain) mutant contains the Lac repressor fused the carboxyl terminus of HJURP1–482. (B) Western blot showing the efficiency of HJURP depletion and expression of transfected constructs used in (C) and (D). HJURP siRNA treatment efficiency was assessed using an anti-HJURP antibody (left). Ponceau staining is a loading control. Expression efficiency of HJURP truncation mutants was assessed by anti-GFP antibody (right). (C) Representative images of new CENP-A loading in the SNAP-tagging experiment. Cells were treated with either GAPD (left panel) or HJURP siRNA (right panel). New CENP-A (TMR-star labelled SNAP-CENP-A, green in merge) is recruited to centromeres in control and HJURP rescue conditions. Immunostaining for CENP-T (red) identifies centromeres. Scale bar is 5 μm in all panels. Boxed regions are magnified to the right of merged images. Two boxed regions (1, 2) are shown for HJURP siRNA-treated HJURP1–482+DD. (D) Fluorescence intensity of centromeric TMR-star labelled SNAP-CENP-A was measured relative to GAPD siRNA-treated control. n>180 centromeres per condition from two independent experiments. *Indicates P<0.05.
If dimerization is required for deposition, then we expect expression of HJURP1–482, lacking the dimerization domain, to reduce new CENP-A deposition, similarly to the CENP-A binding domain mutant (HJURP202-end). New CENP-A deposition was not affected in GAPD siRNA-treated cells co-transfected with HJURP1–482; however, new CENP-A deposition was significantly decreased when HJURP was depleted (Figure 5C and D; Supplementary Figure S3A). The reduction in new CENP-A assembly was similar to that observed in the control and the CENP-A binding mutant HJURP202-end. These results demonstrate that the dimerization domain of HJURP is required for CENP-A deposition.
Using an independent assay, a similar dependence of CENP-A deposition on the carboxyl terminus of HJURP was observed. New CENP-A deposition was assessed in HeLa cells treated with siRNA against endogenous HJURP for 24 h and subsequently co-transfected mCherry-CENP-A together with either full-length or the HJURP1–482 dimerization deficient fragment. Under these conditions, only full-length HJURP rescues mCherry-CENP-A incorporation, while removing the amino acids 482–748(end) almost completely eliminates the localization of mCherry-CENP-A to centromeres (Supplementary Figure S4A–E).
In order to determine if HJURP dimerization is the primary function of the HJURP carboxyl terminus, amino acids 483–748 were replaced with an exogenous dimerization domain (HJURP1–482+DD). We used the Lac repressor, which has been previously engineered to form a dimer (Chen and Matthews, 1992). The HJURP1–482+DD fusion protein was recruited to centromeres as expected (Supplementary Figure S3B). We determined the native molecular weight of the HJURP1–482+DD protein expressed in cells based on hydrodynamic analysis to be 402 kDa, more than twice its predicted size (148 kDa when complexed with CENP-A and histone H4) and consistent with formation of a multimer (Supplementary Figure S3C and D).
We tested if the addition of the dimerization domain was sufficient to rescue the CENP-A deposition defect of HJURP1–482. HJURP1–482+DD rescued new CENP-A deposition in HJURP siRNA-treated cells when compared to HJURP that lacked amino acids 482–748(end) (Figure 5C and D; Supplementary Figure S3A). The percentage of cells with new CENP-A deposited at centromeres in HJURP siRNA-treated cells was similar to GAPD siRNA controls when HJURP1–482+DD was expressed (Figure 5C and D; Supplementary Figure S3A). Importantly, HJURP1–482+DD was able to rescue the degree of new CENP-A deposition per centromere to the same level as full-length HJURP (Figure 5D). These data demonstrate that the primary function of the carboxyl terminus of HJURP is to form an HJURP dimer and that dimerization is required for the stable assembly of new CENP-A nucleosomes.
Discussion
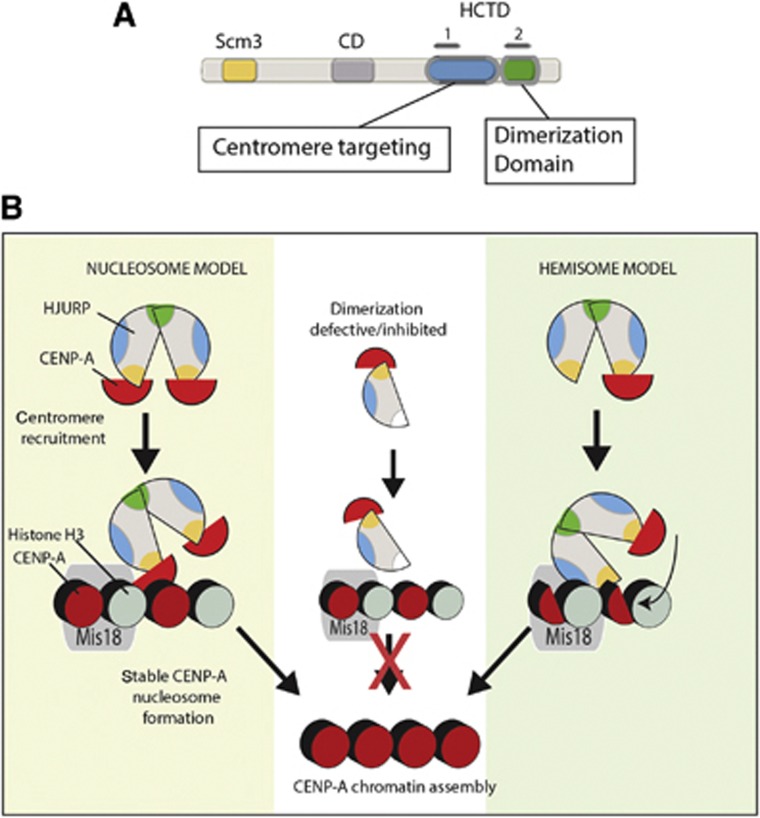
Crystal structures of the CENP-A/HJURP pre-nucleosomal complex demonstrate that HJURP precludes the formation of a pre-nucleosomal CENP-A/histone H4 heterotetramer by blocking the CENP-A self-dimerization domain (Hu et al, 2011). Yet, the HJURP-mediated deposition of stable centromeric nucleosomes requires an intact dimerization surface within CENP-A (Bassett et al, 2012; Zhang et al, 2012). Additionally, in vitro chromatin assembly assays show that human HJURP and yeast Scm3 mediate the assembly of octameric nucleosomes onto DNA templates (Barnhart et al, 2011; Dechassa et al, 2011; Shivaraju et al, 2011). Together, these observations suggest that the in vivo deposition of CENP-A nucleosomes by HJURP results in a CENP-A nucleosome that contains two copies of CENP-A, which is consistent with an octameric nucleosome. Therefore, the formation of a CENP-A nucleosome requires two HJURP proteins to be recruited to each site of new CENP-A deposition. Here, we show that HJURP forms a self-associate through its c-terminus, which contains the second HCTD repeat (Figure 6A). The same HCTD2 containing domain forms a dimer in vitro. HJURP self associates in the pre-nucleosomal and chromatin-associated complexes. HJURP dimerization is required for new CENP-A deposition, providing a mechanism by which an octameric nucleosome is assembled at the centromere from two new CENP-A–histone H4 heterodimers (Figure 6B, nucleosome model).
Figure 6.
Model of HJURP dimerization in CENP-A deposition. (A) Distinct regions within the carboxyl terminus of HJURP mediate centromere targeting and dimerization. Amino acids 482-end of HJURP include the HCTD2 and are sufficient to mediate HJURP dimerization. The amino acids that contribute to direct centromere targeting of HJURP are between residues 352 and 452 and include HCTD1. (B) Centromeric chromatin contains both CENP-A and histone H3 nucleosomes. During G1, new CENP-A nucleosomes are assembled and may displace existing H3 nucleosomes. HJURP recruitment depends on the Mis18 complex through an unknown process (grey ellipse). Dimerization of HJURP facilitates the assembly of CENP-A nucleosomes. HJURP dimerization may be required to bring two new CENP-A–H4 heterodimers to centromeres in order to form an octameric nucleosome de novo (left side). Alternatively, dimerization of HJURP may be required for formation of CENP-A nucleosomes from a pre-existing hemisome and new a CENP-A–histone H4 heterodimer (right side).
Recent work in both human and budding yeast suggests that CENP-A nucleosomes occupy two distinct states during the cell cycle, an octameric and hemisome (tetrameric) form (Bui et al, 2012; Shivaraju et al, 2012). The hemisome form contains a single copy of each histone: CENP-A, H4, H2A and H2B. Based on these observations, an alternative model exists whereby HJURP dimerization links the existing CENP-A hemisome to the incoming new CENP-A–histone H4 heterodimer (Figure 6B, hemisome model). In this model, the Scm3 domain of one HJURP dimer subunit interacts with the existing centromeric hemisome. The Scm3 domain of the second HJURP dimer subunit binds a new CENP-A/H4 heterodimer. In this way, octameric CENP-A nucleosome formation can be coupled to the pre-existing CENP-A hemisome. Cell-cycle analysis suggests that the CENP-A hemisome may be present at centromeres at the time when HJURP is recruited (Bui et al, 2012). However, CENP-A deposition does not absolutely require an existing hemisome as a substrate for new CENP-A deposition as CENP-A nucleosomes can be deposited at initially non-centromeric loci (Barnhart et al, 2011). These two models are not mutually exclusive, and it is possible that both modes of CENP-A deposition occur at centromeres.
Human HJURP contains two HCTD repeat domains within its carboxyl terminus. Duplication of the repeat domain is an evolutionarily recent event, which is restricted to mammals (excluding the egg laying monotremes) (Sanchez-Pulido et al, 2009). The two HCTD repeats are more similar between species than between the two repeats present within a species (Supplementary Figure S5A). HCTD are also found in the MEF2 transcription factor family (Potthoff and Olson, 2007). HCTD2 of HJURP is more similar to the HCTD of the MEF2 transcription factors (Supplementary Figure S5). Our data suggest that a domain of HJURP that includes HCTD2 is sufficient to mediate self-association and without this domain HJURP does not form a multimeric pre-nucleosomal complex or efficiently deposit new CENP-A.
Previous experiments demonstrated that the Scm3 domain was sufficient to deposit CENP-A nucleosomes in vivo at non-centromeric loci (Barnhart et al, 2011). These in vivo experiments were conducted using a LacI-tagged HJURP-Scm3 domain, and we show here that exogenous dimerization driven by LacI is sufficient for the deposition of CENP-A at endogenous centromeres in the absence of the endogenous HJURP dimerization domain. The number of HJURP binding sites within the endogenous centromeres is unknown. Binding domains for exogenously expressed HJURP may be very dense in the LacO array compared to endogenous centromeres and the high concentration of HJURP may circumvent the need for HJURP dimerization. Alternatively, dimerization of the LacI tag may functionally substitute for the HJURP dimerization domain to facilitate CENP-A deposition at the LacO array, as we demonstrated for CENP-A deposition at endogenous centromeres using HJURP1–482+DD. Previously reported in vitro experiments used purified proteins that did not dimerize, and we hypothesize that in vitro assembly may occur without the need for dimerization because these assays were conducted with a very high histone/HJURP protein-to-DNA ratios, which would favour the frequent interaction of HJURP–CENP-A heterotrimers and DNA.
Restricting CENP-A deposition to the centromere depends on the specific recruitment of HJURP. The Mis18 complex is required for HJURP recruitment to centromeres (Barnhart et al, 2011; Moree et al, 2011), although we know very little about how this process occurs. Our data suggest that the direct recognition of the CENP-A nucleosome or hemisome through the HJURP Scm3 domain is not the mechanism by which HJURP is recruited to centromeres, since the presence of the Scm3 domain is not sufficient to recruit HJURP to centromeres (Figure 1A and B). Instead, we have demonstrated targeting of HJURP to centromeres depends on amino acids 348–482 of HJURP, a region that includes the HCTD1 domain (Figure 1E and F). The direct recruitment of HJURP to centromeres, presumably through Mis18, is therefore independent of the HJURP dimerization.
Histone chaperone dimerization may also be involved in the assembly of general chromatin. Histone chaperones involved in canonical histone H3 delivery and deposition also interact with a histone H3–H4 heterodimer at a one-to-one stoichiometric ratio similar to HJURP. These include ASF1, Vps75, NAP1 and NASP (English et al, 2006; Natsume et al, 2007; Campos et al, 2010; Su et al, 2011). Several of these chaperones bring two histone H3 heterodimers into a single complex through dimerization of the chaperone. For example, Vps75 forms an α-β earmuff structure, which contains a dimerization domain within the amino terminus (Tang et al, 2008). Moreover, Vsp75 and ASF1 form a complex with the Rtt109 histone acetyltransferase and facilitate the acetylation of pre-nucleosomal histone H3 on Lysine 56 (Schneider et al, 2006; Driscoll et al, 2007; Han et al, 2007; Su et al, 2011).
The ability of centromeric nucleosome assembly factors to dimerize may be a conserved mechanism from yeast. HJURP and its yeast orthologue Scm3 share homology within the CENP-A binding domain (Sanchez-Pulido et al, 2009). Human HJURP is significantly larger than Scm3 and the dimerization domain of HJURP, which we identified here, is not found in Scm3 proteins. Despite the lack of conservation, the dimerization of yeast Scm3 proteins may also occur. Mizuguchi et al (2007) observed that in high-salt conditions the Scm3–Cse4–histone H4 complex formed a hexamer with a 1:1:1 stoichiometry. A subsequent study by Cho and Harrison (2011) using physiologically salt concentrations observed a stable trimer. Since the heterotetramerization of Cse4 and histone H4 is precluded by Scm3 binding, the hexamer formation observed under high salt conditions may be formed by an interaction between Scm3 proteins. Dimerization of human HJURP requires the HCTD2 domain, which is absent from yeast Scm3. Therefore, yeast Scm3 proteins may also dimerize similar to HJURP, albeit through a distinct mechanism. Our study demonstrates that dimerization of human HJURP is required for the stable deposition of CENP-A nucleosomes at centromeres and provides a mechanism by which octameric CENP-A nucleosomes may be formed at the centromere from heterdimeric subunits, a mechanism that may also apply to canonical nucleosome formation.
Materials and methods
DNA constructs
All constructs were generated by PCR amplification using Vent polymerase, digested by restriction enzyme and ligated into the indicated plasmid. Primers used for PCR amplifications, restriction enzymes used for cloning, and parent vectors are listed in Supplementary Table 1. GFP-tagged plasmids were constructed using the pIC113 plasmid (Cheeseman and Desai, 2005). LacI-mCherry fusions were constructed as previously published (Barnhart et al, 2011). GFP-HJURP1–482-LacI construct was created by a two-step cloning approach. The HJURP 1–482 amino acid fragment lacking a stop codon was PCR amplified (see Supplementary Table 1 for primers) and cloned into pIC113 vector (Cheeseman and Desai, 2005) using Not1 and XhoI restriction sites. LacI was amplified from a vector provided by T Misteli and was cloned into the Xho1 and KpnI sites of pIC113 containing HJURP 1–482.
Transfection
Cells were cultured under standard conditions. DNA and siRNA transfection cells were seeded onto six-well plates at a density of 2.25 × 105 (HeLa and HEK293) or 4.5 × 105 cells/well 24 h prior to transfection. DNA transfection was conducted using Effectene (Qiagen) with 0.4 μg of plasmid DNA per well. siRNA rescue experiments were conducted by treating cells with siRNA 24 h after plating using RNAiMAX (Invitrogen). For each condition cells were treated with either 10 nM of HJURP 3′UTR siRNA (5′-GAGAUAACCUCGAGUUCUUUU-3′) (Dharmacon) or GAPD control siRNA (Invitrogen). Following 24 h of siRNA treatment, cells were transfected using Effectene (Qiagen) as indicated. Immunoblots were conducted using previously established protocols. Antibodies used: anti-HJURP 1:5000 (Bethyl Inc.), anti-GFP 1:1000 (Covance), HA 1.1–1:1000, Anti-tubulin (AA2) 1:100, anti-MBP-1:1000, H2B 1:2000 (Millipore).
Indirect immunofluorescence and SNAP labelling
Cells were plated onto poly-lysine-coated glass coverslips prior to transfection. Following transfection, cells were pre-extracted with 0.1% Triton-X in PBS, fixed with 4% formaldehyde and quenched with 100 mM Tris, pH 7.5. Cells were blocked in 0.3% Triton-X in PBS, 2% BSA, 2% FBS for 1.5 h at room temperature and incubated with primary antibody for 1.5 h. Anti-CENP-T and anti-CENP-A (Barnhart et al, 2011) was used in 1:3000 and 1:1000 dilution, respectively and detected using fluorescently conjugated secondary antibodies (Cy3 or Cy5, Jackson Immuno Inc.). DNA was stained with 0.2 g/ml DAPI in PBS and coverslips were mounted in Prolong Gold (Invitrogen).
A stable cell line expressing SNAP-tagged CENP-A (Jansen et al, 2007) was treated with siRNA for 24 h prior to transfection of the HJURP rescue constructs. After an additional 24 h, the pre-assembled CENP-A was blocked with 10 μM O6-BG (BG-block; Covalys) for 30 min at 37°C followed by a PBS wash and three washes with DMEM over 30 min. Cells were incubated in opti-MEM and siRNA for 24 h and labelled with 2 μM TMR-Star (Covalys) in complete growth medium for 60 min at 37°C. Labelling was followed by one wash each wtih PBS, and DMEM and incubated for 30 min, and washed with PBS prior to fixation.
Images were collected using a × 100 oil-immersion Olympus objective lens on a DeltaVision microscope (Applied Precision Inc.) using a Photometrics CoolSNAP HQ2 camera and Softwrox acquisition software. Images were deconvolved and presented as maximum stacked images. Within siRNA experiments, GFP and TMR-star images for presentation and analysis were collected with identical exposure times and were scaled equally. Integrated intensities were measured from raw images using ImageJ. Intensities of GFP and TMR-star at centromeres were measured using a consistent set area for each experiment. Centromeres were identified based on the presence of the centromere marker (CENP-T). All quantitation of centromere recruitment was restricted to transfected cells by selecting only GFP-expressing cells. GFP intensities were averaged and background corrected using local background correction (Howell et al, 2000). TMR-star intensities were background corrected using an average background calculated from three non-centromeric sites within the nucleus. G1-phase cells were identified by the presence of a mid-body, apparent by DIC optics.
Immunoprecipitation
Cells were lysed 24 h post transfection in RIPA buffer (150 mM NaCl, 1% NP-40, 0.3% deoxycholate, 0.15% SDS, 50 mM Tris HCl pH 7.5, 1 mM EDTA, 10% glycerol, Protease Inhibitors (Roche), 200 μM NaV, 0.5 mM PMSF, 5 mM NaF, 50 mM β-mercaptoethanol, 5 μM microcystin) on ice for 15 min with occasional vortexing. Extracts were DNAseI (1:200, NEB Biolabs) treated and sonicated where indicated. Lysates were centrifuged at 18 000 g for 10 min at 4°C and pre-cleared with Protein A agarose (Biorad) for 2 h at 4°C. Pre-cleared lysates were incubated with anti-GFP antibody (1:1000, Cell Signaling) at 4°C overnight. Antibody-bound complexes were recovered on Protein A Dynabeads (Invitrogen) at room temperature for 45 min, washed with RIPA buffer followed three times in PBS including 0.1–0.5% Tween-20. Complexes were eluted by boiling in SDS sample buffer.
Purification of recombinant proteins
HIS-HJURP482–748 and MBP-HJURP352–482 were expressed in Rosetta BL21 (pLsyS). Cultures were grown in LB at 37°C to OD600nm of 0.6 and induced with 0.5 mM IPTG for 3 h. Bacteria were lysed by French press and sonication in 25 mM Tris-Cl, pH 7.2, 200 mM NaCl, 20 mM MgCl2, 10% Glycerol, 5 mM β-mercaptoethanol, 10 mM β-glycerophosphate, 0.2 mM PMSF, 1 mM benzamidine, Protease Inhibitors (Roche). Lysis buffer for HIS-HJURP482–748 purification was suplemented with 10 mM imidazole. Lysates were centrifuged at 27 000 r.p.m. for 15 at 4°C and the protiens were purified on Ni-NTA agarose (Qiagen) or Amylose resin (BioLabs). Proteins were eluted with lysis buffer plus 250 mM imidzole (Ni-NTA) or 10 mM maltose (amylose).
Sucrose gradient and size-exclusion chromatography
Chromatin-free extracts were prepared from transfected HEK293 as described previously (Foltz et al, 2009) except chromatin isolation buffer (CIB) contained 300 mM NaCl. Chromatin-free extracts were applied to a 14 ml 10–40% sucrose gradient in CIB except digitonin was replaced with 0.05% NP-40. Sucrose gradients were centrifuged at 4°C for 20 h at 40 000 r.p.m. in SW41 Ti swinging bucket rotor and the gradient was separated into 0.5 ml fractions using a BioComp Gradient Station. Sedimentation standards included Catalase (11.3S), Alcohol Dehydrogenase (7.45), BSA (4.3S), and RNaseA (2S). For size-exclusion chromatography, chromatin-free extracts were separated on an AKTA-Micro using a Superdex200 PC 3.2/30 column (GE Healthcare) in 3.75 mM Tris, pH 7.5, 300 mM KCl, 0.5 mM EDTA. Recombinant proteins were analysed on a Superdex200 10/300 GL column (GE Healthcare) in 25 mM Tris-Cl, pH 7.2, 200 mM NaCl, 20 mM MgCl2, 5 mM β-mercaptoethanol. Peak fractions of HIS-HJURP482–748 were applied to a 14 ml 5–20% sucrose gradient and analysed as described above. Apparent molecular weights were calculated using the Siegel and Monty equation (Siegel and Monty, 1966).
Supplementary Material
Acknowledgments
We thank Dan Burke, Todd Stukenberg and members of the Foltz laboratory for their comments on this manuscript, and Madison Stellfox for editing the manuscript. We thank Lars Jansen and Susan Janicki for cell lines. This work was supported by a Research Scholar Grant from the American Cancer Society (DRF) and a Basil O’Connor Award from the March of Dimes (DRF).
Author contributions: EZ, MCBD and PHLK performed experiments. DRF, EZ, PHLK and MCBD designed the experiments and analysed the data. DRF and EZ wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE (2012) HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell 22: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A (2011) Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol 192: 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y (2012) Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150: 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL (2007) Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26: 853–865 [DOI] [PubMed] [Google Scholar]
- Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D (2010) The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol 17: 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005: pl1. [DOI] [PubMed] [Google Scholar]
- Chen J, Matthews KS (1992) Deletion of lactose repressor carboxyl-terminal domain affects tetramer formation. J Biol Chem 267: 13843–13850 [PubMed] [Google Scholar]
- Cho US, Harrison SC (2011) Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci USA 108: 9367–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K (2011) Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun 2: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhou Z, Zhou BR, Bai Y (2011) Structure of the budding yeast Saccharomyces cerevisiae centromeric histones Cse4-H4 complexed with the chaperone Scm3. Proc Natl Acad Sci USA 108: E596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol 150: 1233–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu RM (2011) Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev 25: 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL (2004) From silencing to gene expression: real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, Kim KI (2012) Roles of Mis18alpha in Epigenetic Regulation of Centromeric Chromatin and CENP-A Loading. Mol Cell 46: 260–273 [DOI] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, Singleton MR (2011) Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem 286: 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C (2007) Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF (2011) CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194: 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T (2007) Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446: 338–341 [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN (2007) MEF2: a central regulator of diverse developmental programs. Development 134: 4131–4140 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC (2009) Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137: 1173–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A (2006) Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem 281: 37270–37274 [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S (2007) Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17: 237–243 [DOI] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE (2010) The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 467: 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Camahort R, Mattingly M, Gerton JL (2011) Scm3 is a centromeric nucleosome assembly factor. J Biol Chem 286: 12016–12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL (2012) Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 150: 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A (2010) HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA 107: 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ (1966) Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta 112: 346–362 [DOI] [PubMed] [Google Scholar]
- Stellfox ME, Bailey AO, Foltz DR (2012) Putting CENP-A in its place. Cell Mol Life Sci 70: 387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE (2007) Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA 104: 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Hu Q, Zhou H, Thompson JR, Xu RM, Zhang Z, Mer G (2011) Structure and histone binding properties of the Vps75-Rtt109 chaperone-lysine acetyltransferase complex. J Biol Chem 286: 15625–15629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H (2011) Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476: 232–235 [DOI] [PubMed] [Google Scholar]
- Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R (2008) Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci USA 105: 12206–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P (2009) Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Colmenares SU, Karpen GH (2012) Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell 45: 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y (2011) Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472: 234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.