Abstract
Quorum sensing is a chemical communication process that bacteria use to control collective behaviours including bioluminescence, biofilm formation, and virulence factor production. In Vibrio harveyi, five homologous small RNAs (sRNAs) called Qrr1–5, control quorum-sensing transitions. Here, we identify 16 new targets of the Qrr sRNAs. Mutagenesis reveals that particular sequence differences among the Qrr sRNAs determine their target specificities. Modelling coupled with biochemical and genetic analyses show that all five of the Qrr sRNAs possess four stem-loops: the first stem-loop is crucial for base pairing with a subset of targets. This stem-loop also protects the Qrr sRNAs from RNase E-mediated degradation. The second stem-loop contains conserved sequences required for base pairing with the majority of the target mRNAs. The third stem-loop plays an accessory role in base pairing and stability. The fourth stem-loop functions as a rho-independent terminator. In the quorum-sensing regulon, Qrr sRNAs-controlled genes are the most rapid to respond to quorum-sensing autoinducers. The Qrr sRNAs are conserved throughout vibrios, thus insights from this work could apply generally to Vibrio quorum sensing.
Keywords: quorum sensing, regulation, sRNAs
Introduction
Quorum sensing is a cell-to-cell communication process that bacteria use to monitor changes in cell-population density. By producing, releasing, and detecting extracellular signal molecules called autoinducers, bacteria transition between individual and group behaviours. Quorum sensing ensures that bacteria execute collective behaviours such as bioluminescence, biofilm formation, and virulence factor production only at appropriate cell densities (Waters and Bassler, 2005; Ng and Bassler, 2009; Rutherford and Bassler, 2012). In the model bacterium Vibrio harveyi, three quorum-sensing pathways function in parallel (Henke and Bassler, 2004). At low cell density (LCD), the concentrations of the three autoinducers AI-1, AI-2, and CAI-1 are low. Under this condition, the cognate receptors LuxN, LuxPQ, and CqsS act as kinases, and they phosphorylate the phosphotransfer protein LuxU (Freeman and Bassler, 1999b; Neiditch et al, 2006; Swem et al, 2008; Wei et al, 2012). LuxU∼P passes its phosphate to the response regulator LuxO (Freeman and Bassler, 1999a, 1999b). LuxO∼P, together with σ54, activates the transcription of genes encoding five non-coding quorum-regulated small RNAs (sRNAs) called Qrr1–5 (Lilley and Bassler, 2000; Lenz et al, 2004; Tu and Bassler, 2007). The Qrr sRNAs activate the translation of the LCD master regulator AphA and repress the translation of the high cell density (HCD) master regulator LuxR (Tu and Bassler, 2007; Rutherford et al, 2011; Shao and Bassler, 2012). At HCD, the concentrations of the three autoinducers are high. Under this condition, the three receptors act as phosphatases, and they initiate a reversal of phospho flow through the circuit. LuxO, when unphosphorylated, is unable to activate the transcription of qrr1–5 (Tu and Bassler, 2007). Therefore, aphA translation is not activated and luxR translation is not repressed (Tu and Bassler, 2007; Rutherford et al, 2011; Shao and Bassler, 2012). This regulatory architecture ensures that maximum AphA is produced at LCD, while maximum LuxR exists at HCD (Rutherford et al, 2011; van Kessel et al, 2012). AphA and LuxR, in turn, direct the proper LCD to HCD quorum-sensing gene expression patterns, respectively (Rutherford et al, 2011; van Kessel et al, 2012). In addition to the two quorum-sensing master regulators AphA and LuxR, at LCD, the Qrr sRNAs also repress luxO and the genes encoding the AI-1 pathway synthase/receptor luxMN (Tu et al, 2010; Teng et al, 2011). The former is crucial for controlling Qrr sRNA levels, and the latter is important for adjusting the sensitivity to different autoinducers at different cell densities (Tu et al, 2010; Teng et al, 2011).
The Qrr sRNAs belong to a large group of trans-encoded regulatory sRNAs in bacteria (Waters and Storz, 2009). Typically, sRNA-mediated activation of targets occurs through base pairing with and alteration of secondary structures in the 5′ UTRs of target mRNAs. Generally, pairing reveals the ribosome-binding sites and promotes translation (Fröhlich and Vogel, 2009). Alternative activation mechanisms include generating accessible ribosome-binding sites via endonucleolytic cleavage and protection from endonucleolytic destruction (Obana et al, 2010; Ramirez-Peña et al, 2010; Papenfort et al, 2013). The canonical sRNA repression mechanism is through base pairing with the mRNA region encoding the ribosome-binding site to occlude ribosome access. This mechanism leads to degradation or sequestration of the target mRNAs; in both cases, no translation of the mRNA targets occurs (Waters and Storz, 2009). Alternative repression mechanisms include base pairing within target mRNA coding regions or within intergenic regions of polycistronic transcripts, which leads to endonucleolytic cleavage (Desnoyers et al, 2009; Pfeiffer et al, 2009). Interactions between sRNAs and their mRNA targets are often mediated by the RNA chaperone Hfq. Hfq stabilizes the sRNAs, brings together sRNAs and target mRNAs, and interacts with RNase E (Vogel and Luisi, 2011; Mackie, 2012). Hfq can also be recruited, at least by the Spot42 sRNA, to act as a direct repressor of translation (Desnoyers and Massé, 2012).
In the present study, we identify 16 new mRNA targets of the Qrr sRNAs. Particular sequence differences among the Qrr sRNAs determine whether each Qrr sRNA regulates all 16 of the new targets or only a subset of them. Using the newly identified target genes coupled with mutagenesis, we pinpoint the role of each portion of the Qrr sRNAs in target regulation. The first two stem-loops are involved in base pairing with the mRNA targets. The most 5′ stem-loop also protects the Qrr sRNAs from RNase E-mediated degradation. The third stem-loop plays an accessory role in base pairing and stability. The fourth stem-loop functions as the terminator. Analyses of regulation of the newly identified targets show that, of all of the genes in the quorum-sensing regulon, those that are directly controlled by the Qrr sRNAs are the most rapid to respond when bacteria transit from HCD to LCD. We find that the Qrr sRNAs can independently regulate particular target genes, and they can also act in conjunction with AphA or LuxR to control target gene expression.
Results
Identification of targets of the Qrr sRNAs in V. harveyi
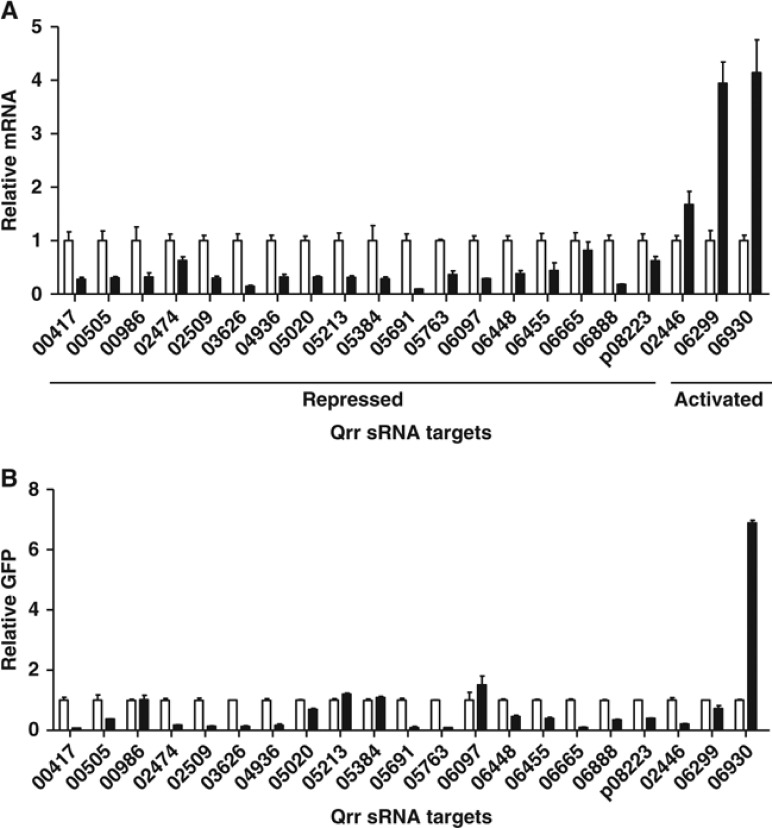
The V. harveyi Qrr sRNAs regulate luxR, luxO, luxMN, and aphA (Tu and Bassler, 2007; Tu et al, 2010; Rutherford et al, 2011; Teng et al, 2011; Shao and Bassler, 2012). All of these targets are members of the quorum-sensing regulatory circuit. Thus, to date, the only known role of the Qrr sRNAs is to regulate quorum-sensing regulators. We wondered whether the Qrr sRNAs control targets in addition to those in the quorum-sensing cascade. To explore this possibility, we constructed a plasmid containing qrr4 under an arabinose-inducible promoter and mobilized the plasmid into a V. harveyi Δqrr1–5 strain. We chose Qrr4 for this analysis because it is the most highly expressed Qrr at LCD and thus the most likely to be capable of controlling additional targets (Tu and Bassler, 2007). Qrr4 production was induced for 15 min, and global mRNA changes were measured by microarray and compared to the same strain in the absence of arabinose (see Materials and methods, Supplementary Figure S1). The microarray revealed 30 genes that changed expression more than two-fold (Supplementary Table S1). This set of genes includes the expected quorum-sensing regulators luxR, luxMN, and aphA. The level of luxO transcript did not change following Qrr4 induction, likely because Qrr control of luxO occurs via sequestration rather than degradation (Tu et al, 2010). Thus, 26 new genes were identified. In order to eliminate genes that are induced by arabinose, we performed qRT–PCR analysis on the putative targets following arabinose induction of the empty vector in V. harveyi. Of the 26 new genes, four in the gal operon are induced by arabinose but do not require Qrr4. One other target, vibhar_03460, is located directly downstream of luxR and has only a short transcript. This gene is likely co-transcribed with luxR, so we did not investigate it further. The remaining 21 genes are located in 18 operons: 16 are repressed and two are activated by Qrr4. To confirm the microarray results, we measured mRNA changes for all 18 operons by qRT–PCR. Figure 1A shows that, indeed, all of these genes are regulated following qrr4 induction.
Figure 1.
Regulation of target genes by Qrr4. (A) Regulation of genes identified from the microarray following a pulse of production of Qrr4 was confirmed by qRT–PCR. Target mRNA expression levels were compared at mid-logarithmic phase in a V. harveyi Δqrr1–5 strain (KT282) harbouring a plasmid with an arabinose-inducible qrr4 (pLF575) without (white bars) or with (black bars) addition of 0.2% arabinose for 15 min. Mean and s.e.m. values of triplicate cultures are shown. (B) Fluorescence from E. coli carrying IPTG-inducible translational GFP fusions to potential Qrr targets was measured in the presence of an empty vector (pLF253, white bars) or a plasmid carrying tetracycline-inducible qrr4 (pLF127, black bars). GFP levels were normalized to the vector control for each target. Mean and s.e.m. values of triplicate samples are shown.
Qrr4 regulates target mRNAs through direct base pairing
The above experiment, using a pulse of expression of qrr4, was designed to reveal direct Qrr4 targets. However, it is possible that, within the 15 min of induction, Qrr4 could regulate a factor that, in turn, controls some or all of the newly identified targets. To define which of the 18 target mRNAs are directly controlled by Qrr4, we measured their regulation by Qrr4 in E. coli in the absence of other V. harveyi components. To do this, we constructed translational GFP fusions to each of the 18 targets on plasmids and measured GFP fluorescence in the presence and absence of qrr4 expression (Supplementary Figure S2). Fourteen of the 18 targets exhibited altered production when Qrr4 was induced (Figure 1B). Four targets (vibhar_00986, vibhar_05213, vibhar_05384, and vibhar_06097) showed no regulation in E. coli, suggesting that these targets are not directly controlled by Qrr4 or the regions responsible for regulation are not included in the reporter constructs (vibhar_00986: −44 to +171, vibhar_05213: −78 to +60, vibhar_05384: −156 to +42, vibhar_06097: −56 to +51). Interestingly, one activated target vibhar_02446 did not follow the expected pattern. The vibhar_02446 mRNA increased following overexpression of Qrr4 in V. harveyi (Figure 1A). However, the VIBHAR_02446-GFP translational fusion was repressed by Qrr4 in E. coli (Figure 1B). One possibility is that base pairing between Qrr4 and vibhar_02446 mRNA prevents the mRNA from being degraded in V. harveyi, but in E. coli, blocking the ribosome-binding site prevents the protein from being translated.
The 14 direct targets include three metabolic enzymes (vibhar_00417/prephenate dehydratase, vibhar_03626/deacetylase DA1, and vibhar_04936/glutathione-dependent formaldehyde-activating-like protein), two potential transcription factors (vibhar_00504 within operon vibhar_00506-vibhar_00504 and vibhar_05763), one hemagglutinin/protease (vibhar_02509), one RTX toxin transporter operon (vibhar_06455-vibhar_06452), one methyl-accepting chemotaxis protein (vibhar_05691), and one operon potentially involved in polysaccharide export (vibhar_06665–vibhar_06667) (Table I).
Table 1. Novel Qrr sRNA target genes.
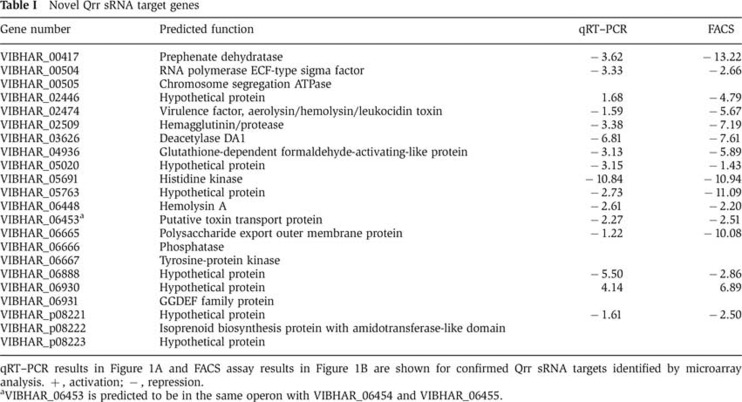
To demonstrate that the response of the 14 new targets was due to base pairing with Qrr4, we engineered mutations disrupting the putative pairing regions in Qrr4. We also constructed compensatory mutations in the targets to restore pairing. We show our analysis for two representative targets vibhar_05691 and vibhar_06930 that are repressed and activated, respectively (Figure 1, Figures 2A, B, and Table I). Computational prediction of the interaction between the 5′ UTR of the repressed vibhar_05691 mRNA and Qrr4 suggests pairing between −10 to −2 and −32 to −26 relative to the vibhar_05691 translation start site. Mutating AGCC to UCGG at nucleotides 13–16 of Qrr4 (Qrr4mut1) substantially reduced the ∼50-fold repression exhibited by wild-type Qrr4. By contrast, mutating CAACU to GUUGA between nucleotides 31–35 of Qrr4 (Qrr4mut2) only modestly affected repression. Consistent with these findings, altering GGCU to CCGA at −9 to −6 in vibhar_05691 (vibhar_05691-MutI) abolished regulation by wild-type Qrr4. However, this mutation restored regulation by Qrr4mut1 (Figure 2A). We suspect that the low basal expression of vibhar_05691MutI is due to a weakened ribosome-binding site (Figure 2A). Mutating AGUUG to UCAAC at −31 to −27 in vibhar_05691 (vibhar_05691MutII) somewhat impaired repression by wild-type Qrr4. Qrr4mut2 containing the compensatory changes fully restored regulation, whereas the Qrr4mut1 changes did not (Figure 2A). Together, these data suggest that pairing of the Qrr4 nucleotides 9–17 with the vibhar_05691 translation initiation region is most critical for regulation, however, the second pairing site (nucleotides −32 to −26 in vibhar_05691 and 30–36 in Qrr4) is required for full target control.
Figure 2.
Qrr4 regulates target genes through direct base pairing. (A) Qrr4 represses vibhar_05691 through base pairing. Fluorescence from E. coli carrying a plasmid with an IPTG-inducible translational GFP fusion to wild-type vibhar_05691 (pLF767), vibhar_05691MutI (pYS256), or vibhar_05691MutII (pYS257) was measured in the presence of an empty vector (pLF253), a vector with wild-type Qrr4 (pLF127), a vector with Qrr4mut1 (pYS258), or a vector with Qrr4mut2 (pYS259). The mutations are highlighted in the sequences by over or underlines. Mean and s.e.m. values of triplicate samples are shown. (B) Qrr4 activates vibhar_06930 through base pairing. Fluorescence from E. coli carrying a plasmid with an IPTG-inducible translational GFP fusion to wild-type vibhar_06930 (pLF1285), vibhar_06930 truncation (Δ−129 to −79) (pLF1730), or vibhar_6930MutI (pLF840) was measured in the presence of an empty vector (pLF253), a vector with wild-type Qrr4 (pLF127), or a vector with Qrr4mut3 (pLF770). The mutations in the sequences are highlighted by over or underlines. Mean and s.e.m. values of triplicate samples are shown. Base pairings between the mRNA targets (vibhar_05691 and vibhar_06930) and Qrr4 were predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Nucleotides involved in base pairing are shown in red. Translational start sites are denoted as +1. The structure of the 5′ UTR of vibhar_06930 was predicted by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Base pairing between other mRNA targets and Qrr4 are shown in Supplementary Figure S3.
In the case of the activated target vibhar_06930, the 125 nucleotide 5′ UTR is predicted to form a secondary structure that conceals the ribosome-binding site (Figure 2B). Deletion of the first 47 nucleotides (Δ−125 to −79) increased production of the VIBHAR_06930-GFP fusion by ∼12.5-fold, indicating that this region is crucial for intrinsic translation inhibition (Figure 2B). Qrr4 is predicted to base pair with this self-inhibitory loop to relieve repression (Figure 2B). Indeed, wild-type Qrr4 activated production of VIBHAR_06930-GFP by ∼7.5-fold and did not activate the truncated VIBHAR_06930-GFP fusion. Mutating GGC to CCG at −95 to −97 nucleotides in the 5′ UTR of vibhar_06930 (vibhar_06930MutI) eliminated activation by wild-type Qrr4. Mutating GCC to CGG at positions 29–31 in Qrr4 (Qrr4mut3) impaired regulation of wild-type vibhar_06930, but restored regulation to vibhar_06930MutI (Figure 2B). Basal GFP production from vibhar_06930MutI is higher than that of wild type but lower than that from the truncated construct. Likely, the self-inhibitory loop is only partially disrupted in vibhar_06930MutI whereas it is completely eliminated in the truncation mutant.
Qrr2–5 regulate an identical set of target mRNAs
The above experiments examined the function of Qrr4 in target mRNA regulation. Given that the other Qrr sRNAs possess similar sequences to Qrr4, we wondered whether they likewise regulate the same or other additional targets. Using an identical strategy, we performed microarray experiments following a pulse of expression of each Qrr sRNA (Supplementary Figure S1). In all cases, the genes that exhibited two-fold or higher changes in levels corresponded well to those identified in the Qrr4 experiment (Supplementary Table S1). Four additional targets (vibhar_02474, vibhar_06299, vibhar_06448, and vibhar_06895) were identified as controlled by Qrr2, Qrr3, or Qrr5. However, qRT–PCR showed that vibhar_06895 did not respond to induction of any Qrr sRNA. We conclude that vibhar_06895 was a false positive. We tested vibhar_02474, vibhar_06299, and vibhar_06448 for control by Qrr4, and these genes did in fact exhibit expression changes upon qrr4 induction (Figure 1A). We suspect that these three genes were not identified in the original Qrr4 pulse microarray experiment because they fell below the two-fold cut-off. Qrr4 also controlled GFP translational fusions to vibhar_02474 and vibhar_06448, but not vibhar_06299 in E. coli (Figure 1B). VIBHAR_06299-GFP was also not regulated by Qrr2, Qrr3, or Qrr5 in E. coli, so we did not investigate it further. The two newly identified targets are potential virulence factors: vibhar_02474 contains an aerolysin toxin motif and vibhar_06448 encodes a hemolysin A protein. This brings the total to 16 new Qrr targets (Table I).
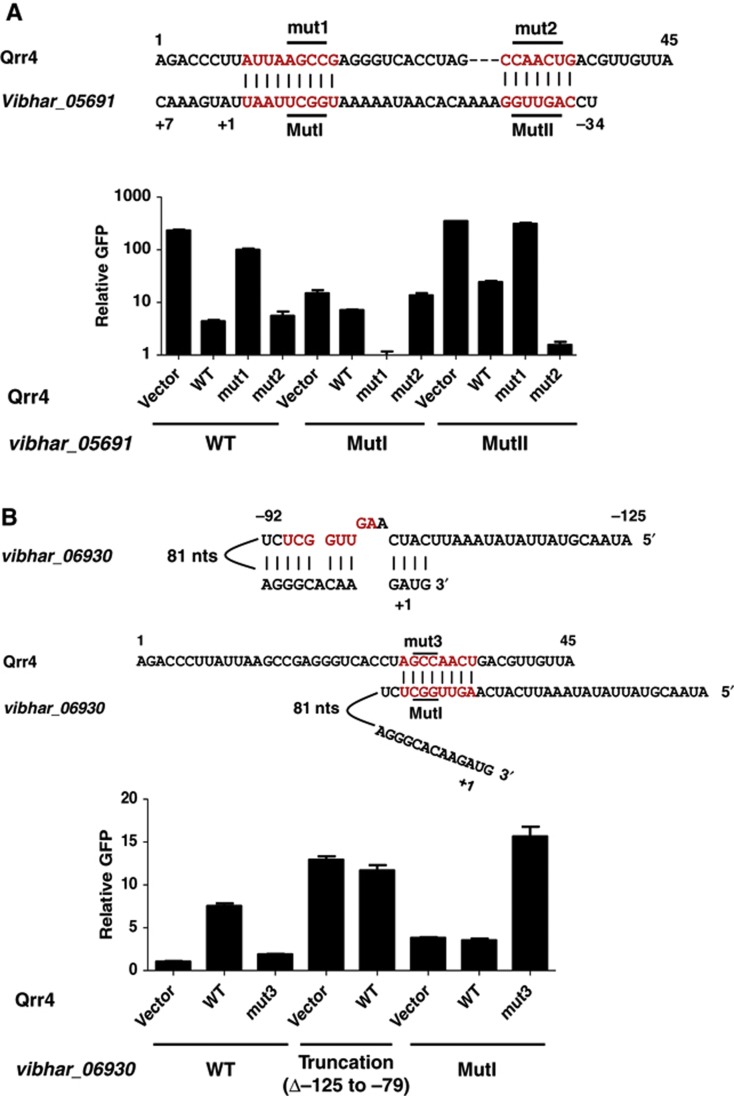
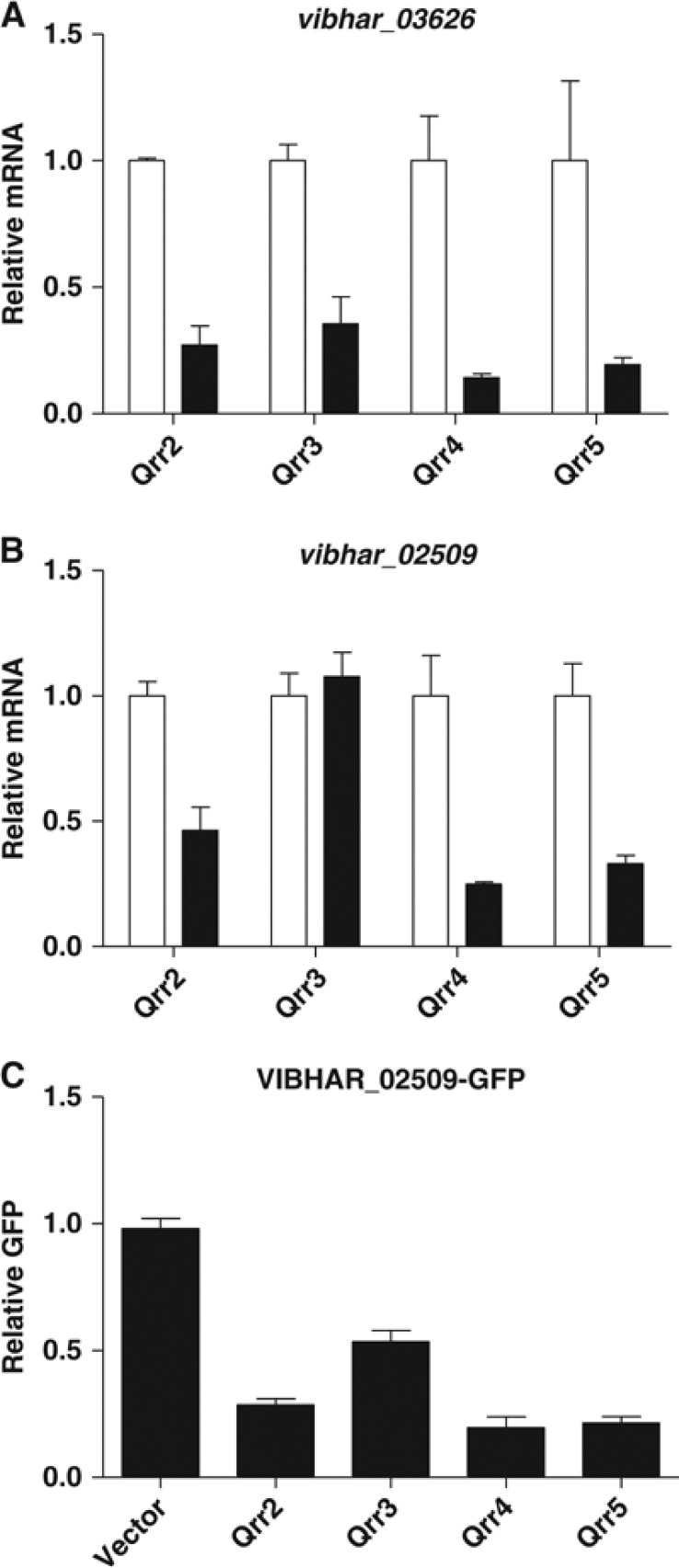
qRT–PCR following pulse induction of each Qrr sRNA was used to verify the microarray results. Thirteen of the 16 targets are regulated by Qrr2–5 (Supplementary Figure S4). As an example, we use vibhar_03626 and show that Qrr2, Qrr3, Qrr4, and Qrr5 control its expression (Figure 3A). Three targets, vibhar_02474, vibhar_02509, and vibhar_06665, are regulated by Qrr2, Qrr4, and Qrr5, but not by Qrr3 in V. harveyi. The data for vibhar_02509 are shown in Figure 3B and results for vibhar_02474 and vibhar_06665 are provided in Supplementary Figure S4. Of these three targets, vibhar_02509 showed the strongest defect. This is borne out in recombinant E. coli; Qrr2, Qrr4, and Qrr5 repress VIBHAR_02509-GFP while Qrr3 is somewhat defective (Figure 3C). The difference between the V. harveyi and E. coli results could come from the fact that Qrr targets in addition to the one we are measuring exist in V. harveyi and compete for regulation by the Qrr sRNAs. Thus, in vivo differences in the roles of the Qrr sRNAs can be revealed. Because no competition for the Qrr occurs in E. coli, even when a Qrr is defective in V. harveyi, residual regulatory capability can occur in E. coli.
Figure 3.
Regulation of targets by Qrr2–5. qRT–PCR of vibhar_03626 (A) and vibhar_02509 (B) without (white bars) and with (black bars) arabinose induction of the specific Qrr. Mean and s.e.m. values of replicates are shown. (C) Fluorescence from E. coli carrying a plasmid with an IPTG-inducible VIBHAR_02509-GFP fusion (pYS214) was measured in the presence of an empty vector (pLF253), a vector with Qrr2 (pLF186), Qrr3 (pLF126), Qrr4 (pLF127), or Qrr5 (pLF187). Mean and s.e.m. values of triplicate samples are shown.
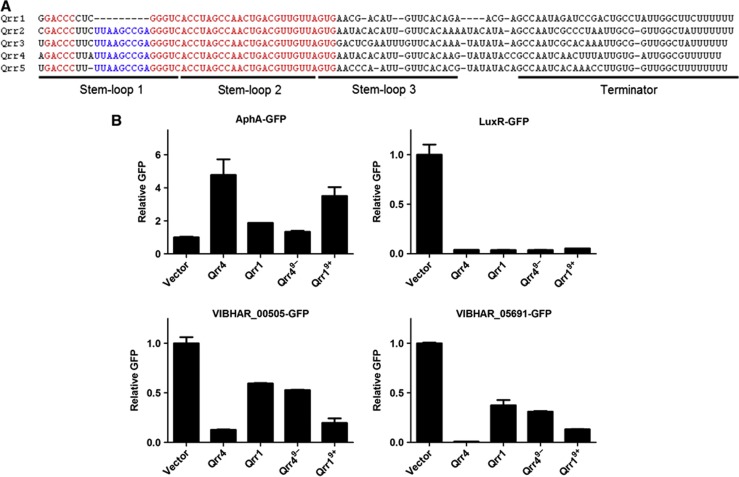
Sequence differences at the 5′ terminus dictate Qrr1 target selectivity
Qrr1 regulates all of the target genes that are controlled by Qrr2–5 except vibhar_00505, vibhar_05691, and aphA (Figure 4B; Supplementary Figure S3 and S4) (Shao and Bassler, 2012). Qrr1 lacks nine nucleotides that are conserved in Qrr2–5 near the 5′ terminus (Figure 4A) (Tu and Bassler, 2007). This gap makes Qrr1 unable to activate aphA translation but has no effect on Qrr1 repression of luxR (Figure 4B, top graphs) (Shao and Bassler, 2012). Reintroducing the missing 9 nucleotides into Qrr1 (denoted Qrr19+) restores regulation of aphA, and does not alter luxR regulation (Figure 4B, top graphs). We deleted the corresponding 9 nucleotides from Qrr4 to ‘convert’ it to Qrr1. We call this construct Qrr49−. Like Qrr1, Qrr49− is impaired in activation of aphA, but it is wild type for repression of luxR (Figure 4B, top graphs). We predict that these 9 conserved nucleotides could also be important for regulating vibhar_00505 and vibhar_05691. If so, Qrr19+ should be functional at these two targets, and indeed Figure 4B shows this is the case (bottom graphs). Likewise, Qrr49−, which lacks these 9 nucleotides, cannot control vibhar_00505 and vibhar_05691 (Figure 4B, bottom graphs). Thus, we conclude that the 9 nucleotides that Qrr1 lacks are necessary for regulating a subset of the Qrr targets including aphA, vibhar_00505, and vibhar_05691. These results are consistent with the base pairing patterns we mapped through mutagenesis analysis (Figure 2A; Supplementary Figure S5) (Shao and Bassler, 2012).
Figure 4.
5′ sequence differences confer distinct regulatory capabilities to Qrr1. (A) RNA sequence alignment of V. harveyi Qrr1–5. The conserved 9 nucleotides that are absent in Qrr1 are shown in blue and other highly conserved sequences in the 5′ region are shown in red. Sequences corresponding to predicted stem-loops and to the terminator are indicated with underlines. (B) Fluorescence from plasmid-encoded V. harveyi AphA-GFP (pLF255), LuxR-GFP (pLF128), VIBHAR_00505-GFP (pLF804), and VIBHAR_05691-GFP (pLF767) translational fusions were measured in E. coli carrying an empty vector (pLF253), a vector expressing a tetracycline-inducible qrr1 (pLF396), qrr1 with 9 nucleotides reintroduced (Qrr19+, pYS241), qrr4 (pLF127), or qrr4 with 9 nucleotides deleted (Qrr49−, pYS239). GFP from three independent cultures was measured for each strain and the mean and s.e.m. values are shown. All measurements were normalized to the means of the vector controls.
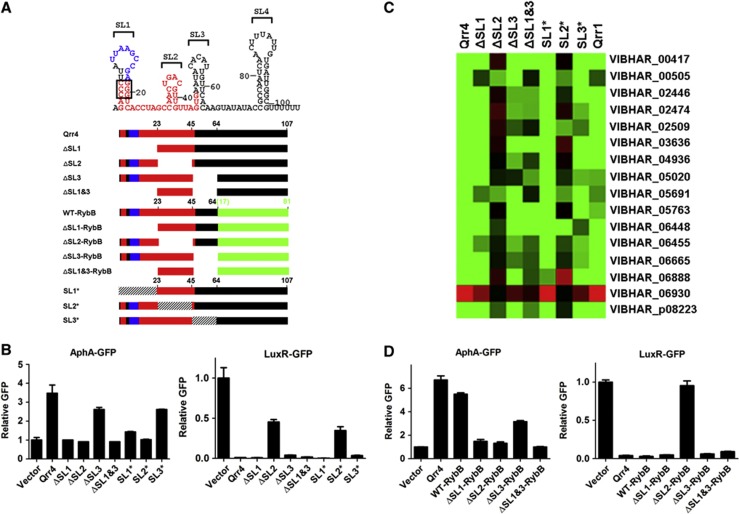
Contribution of each stem-loop to base pairing between Qrr sRNAs and target mRNAs
Based on secondary structure predictions, there exist four stem-loops in the Qrr sRNAs (Tu and Bassler, 2007). We name them, from 5′ to 3′: SL1, SL2, SL3, and SL4 (Figures 4A and 5A). Each of the Qrr sRNAs has all four stem-loops but, as mentioned, Qrr1 is the most different from the other Qrr sRNAs, because it lacks 9 nucleotides in SL1. SL4 contains the rho-independent terminator. Having a large set of Qrr sRNA targets in hand allows us to investigate the individual and combined roles of each of the stem-loops in Qrr function. We constructed a series of stem-loop deletions in Qrr4 and measured the effects on target regulation (Figure 5A, ΔSL1, ΔSL2, ΔSL3, and ΔSL1 and SL3 with deleted sequences shown as blanks). To examine the effects of these changes, we started with two well-studied targets, aphA and luxR. Deletion of SL1 eliminated aphA activation but did not affect luxR repression (Figure 5B). Deletion of SL2 eliminated regulation of both aphA and luxR. Deletion of SL3 had only a modest effect on each target (Figure 5B). Deletion of both SL1 and SL3 gave results identical to the SL1 deletion alone (Figure 5B). We used this exact strategy to test the role of each stem-loop in regulation of each of the 16 newly identified Qrr sRNA targets. The results are summarized in Figure 5C. All of the data are shown in Supplementary Figure S6. In brief, deletion of SL1 primarily affects regulation of only two targets in addition to aphA; vibhar_00505 and vibhar_05691. Deletion of SL2 abolishes regulation of all of the targets with the exception of vibhar_00505 and vibhar_05691. Deletion of SL3 affects regulation of several targets to different extents, especially vibhar_02509 and vibhar_05020. Additive effects occur in most cases when both SL1 and SL3 are deleted (Figure 5C; Supplementary Figure S6). Thus, SL2 contains conserved sequences required for base pairing with the majority of the target mRNAs, and SL1 and SL3 are crucial for base pairing with a subset of targets. In cases in which SL2 is not crucial, SL1 has increased importance (for example, see vibhar_00505 and vibhar_05691). These results are consistent with the above findings that vibhar_00505 and vibhar_05691 are regulated by Qrr4 but not by Qrr1 (Figure 4).
Figure 5.
Stem-loop 2 functions as the core base pairing region of the Qrr sRNAs. (A) Predicted secondary structure of Qrr4 and schematics of WT Qrr4, Qrr4 deletion mutants, chimeric Qrr4 sRNAs, and Qrr4 inversion mutants are shown. Colour codes are the same as in Figure 4, with the conserved 9 nucleotides missing in Qrr1 in blue, other 5′ highly conserved sequences in red, deleted sequences left blank, inverted sequences hatched, and RybB sequences in green. Nucleotides mutated to construct the SL1 disruption and restoration mutants are highlighted in the box. Data for those mutants are in Figure 6. (B) Fluorescence from plasmid-encoded V. harveyi AphA-GFP (pLF255) and LuxR-GFP (pLF128) translational fusions was measured in E. coli carrying an empty vector (pLF253), a vector expressing a tetracycline-inducible qrr4 (pLF127), qrr4 stem-loop 1 deletion (ΔSL1, pYS225), qrr4 stem-loop 2 deletion (ΔSL2, pYS226), qrr4 stem-loop 3 deletion (ΔSL3, pYS227), qrr4 stem-loop 1 and stem-loop 3 double deletion (ΔSL1&3, pYS229), qrr4 stem-loop 1 inversion (SL1*, pYS230), qrr4 stem-loop 2 inversion (SL2*, pYS231), or qrr4 stem-loop 3 inversion (SL3*, pYS232). GFP from three independent cultures was measured for each strain and the mean and s.e.m. values are shown, with all measurements normalized to the mean of the vector controls. (C) Fluorescence from 16 plasmid-encoded Qrr sRNA target-GFP translational fusions was measured in E. coli carrying an empty vector (pLF253), a vector expressing a tetracycline-inducible qrr4 (pLF127), qrr1 (pLF396), or individual qrr4 deletion/inversion mutants as in panel B. GFP from three independent cultures was measured for each strain. All fluorescence changes were normalized to the changes of WT Qrr4. Mean values are shown, with activation coloured in red and repression coloured in green, see Supplementary Figure S6 for specific data for each individual target gene. (D) Fluorescence from plasmid-encoded V. harveyi AphA-GFP (pLF255) and LuxR-GFP (pLF128) translational fusions was measured in E. coli carrying an empty vector (pLF253), a vector expressing a tetracycline-inducible qrr4 (pLF127), WT chimeric qrr4 (WT-RybB, pYS280), stem-loop 1 deletion chimeric qrr4 (ΔSL1-RybB, pYS277), stem-loop 2 deletion chimeric qrr4 (ΔSL2-RybB, pYS274), stem-loop 3 deletion chimeric qrr4 (ΔSL3-RybB, pYS271), or the double stem-loop 1 and stem-loop 3 deletion chimeric qrr4 (ΔSL1&3-RybB, pYS268). GFP from three independent cultures was measured for each strain and the mean and s.e.m. values are shown. All measurements were normalized to the mean of the vector controls.
To examine the function of each stem-loop in the context of the remainder of the Qrr sRNA sequence, we engineered chimeric sRNAs using a collection of previously well-studied sRNAs such as RybB. RybB uses the most 5′ 16 nucleotides to base-pair with target mRNAs (Papenfort et al, 2010). We fused the first three stem-loops of Qrr4 to RybB lacking its 16 critical base pairing nucleotides (Figure 5A, WT-RybB with RybB sequences coloured in green). The WT-RybB chimera regulates aphA and luxR exactly like wild-type Qrr4 (Figure 5D). We also tested target regulation by each of the Qrr4 stem-loop deletion constructs fused to the same portion of RybB (Figure 5A, ΔSL1-RybB, ΔSL2-RybB, ΔSL3-RybB, and ΔSL1 and 3-RybB with RybB sequences coloured in green). ΔSL1-RybB could not activate aphA, ΔSL2-RybB does not regulate either aphA or luxR, ΔSL3-RybB regulates both aphA and luxR, and ΔSL1&3-RybB has the phenotype of the ΔSL1-RybB construct (Figure 5D). These results are entirely consistent with the data from the above Qrr4 stem-loop deletion mutants. Similar results were obtained when the MicA and OmrB sRNAs were used to construct the chimeras rather than RybB (Supplementary Figure S7). Thus, the first three stem-loops of Qrr4 contain all of the sequences required for its base pairing functions.
Beyond affecting base pairing, deletion of sRNA stem-loops could affect the overall sRNA structure, stability, and/or interaction with the chaperone Hfq. To distinguish between these mechanisms, we constructed three more Qrr4 sRNA mutants harbouring stem-loop inversions. Our idea was to preserve the overall structure while eliminating base pairing (Figure 5A, SL1*, SL2*, and SL3* with inverted sequences hatched). Again, we relied on aphA and luxR as the representative targets to test regulation. SL1* could not activate aphA, SL2* could not regulate either aphA or luxR, and SL3* functions like WT Qrr4 (Figure 5B). We also tested the set of stem-loop inversion constructs for regulation of the 16 newly identified targets. SL2* abolishes regulation of the majority of targets, while SL1* and SL3* function like WT Qrr4 in most cases (Figure 5C and Supplementary Figure S6). These data suggest that the effects caused by deletion of stem-loops arise from defects in base pairing rather than from structural or stability issues. We propose that SL2 contains the core base-pairing region of the Qrr sRNAs, and SL1 and SL3 participate in base pairing with only a select few targets.
Contribution of each stem-loop to the overall structure of Qrr sRNAs
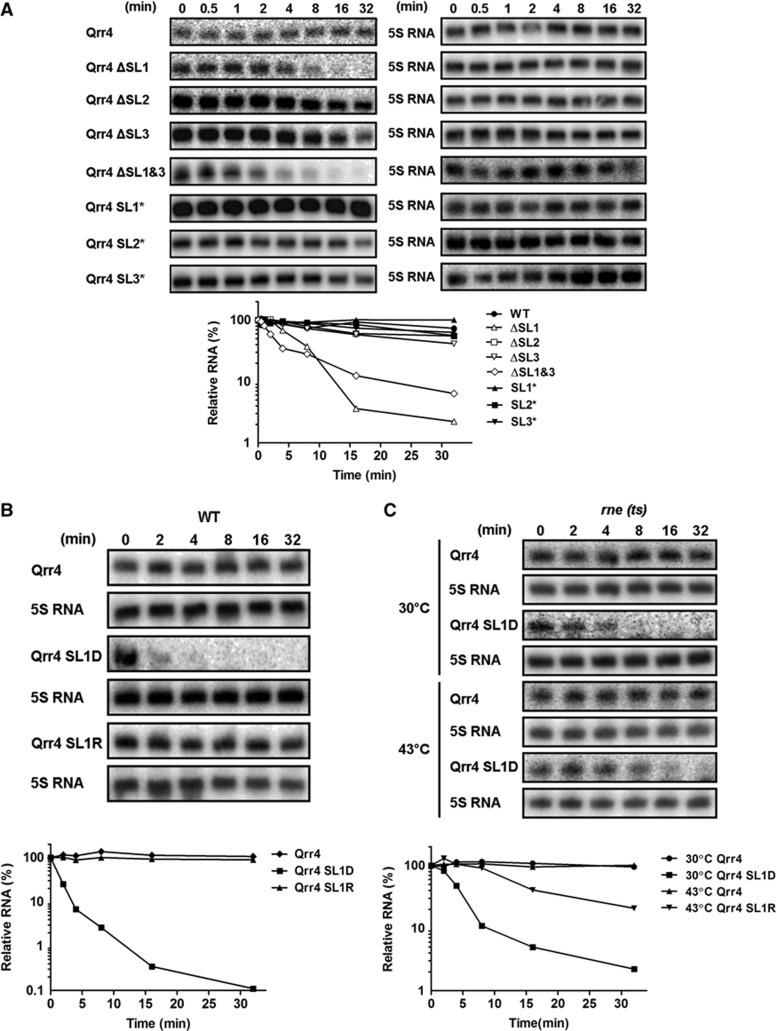
To pinpoint the function of each stem-loop beyond base pairing, we examined their contributions to stability and Hfq interaction. First, regarding stability, we measured the effects of each stem-loop on the stability of Qrr4. We used rifampicin to terminate transcription, collected cells over time, and measured the half-lives of wild-type Qrr4 and the Qrr4 stem-loop deletion mutants. Wild-type Qrr4 has a half-life of over 32 min, deletion of SL1 dramatically reduced the half-life to ∼5 min, deletion of SL2 and SL3 had little effect on the half-life, and deletion of both SL1 and SL3 also reduced the half-life to less than 5 min (Figure 6A). We propose that SL1 is the main stem-loop responsible for Qrr4 stability. SL3 does play somewhat of a role in Qrr stability when measured in V. harveyi, which could be due to the presence of the complete set of target mRNAs (unpublished data).
Figure 6.
Stem-loop 1 protects the Qrr sRNAs from RNase E-mediated degradation. (A) Half-lives of plasmid-encoded V. harveyi WT Qrr4 and Qrr4 mutants from Figures 4 and 5 were measured in E. coli by northern blot. (B) Half-lives of plasmid-encoded V. harveyi WT Qrr4 (pLF127), the SL1 disrupted Qrr4 mutant (Qrr4 SL1D, pYS287), and the corresponding SL1 restored Qrr4 mutant (Qrr4 SL1R, pYS296) were measured in E. coli by northern blot. (C) Half-lives of plasmid-encoded V. harveyi WT Qrr4 (pLF127) and the SL1 disrupted Qrr4 mutant (Qrr4 SL1D, pYS287) were measured in E. coli containing a temperature-sensitive RNase E allele rne-50 (LF1018) (Massé et al, 2003). Strains were grown to OD600 ∼1.0 and shifted to 43°C for 20 min prior to sample collection. In all panels, northern blots are shown with the data plotted below. 5S rRNA was used as the control.
5′ stem-loop structures are known to be important for protecting mRNAs from degradation (Belasco, 2010). As the Qrr4 SL1 deletion mutant exhibited a reduced half-life, we wondered whether it is the stem-loop structure that matters or if there are specific sequences that are required for stability. To explore this, we measured the half-lives of the stem-loop inversion mutants (Qrr4 SL1*, SL2*, and SL3*). They all have the same half-life as wild-type Qrr4 (Figure 6A). Thus, the contribution of SL1 to Qrr stability is indeed due to the stem-loop structure. To verify this result, we introduced a mutation (CCC to GGG, denoted Qrr4 SL1D) to disrupt the SL1 structure (see Figure 5A). The half-life of Qrr4 SL1D decreased dramatically, to less than 2 min. Introduction of a compensatory mutation (GGG to CCC, denoted Qrr4 SL1R) to restore the stem-loop structure also restored the half-life to that of WT Qrr4 (Figure 6B).
Secondary structure predictions indicate that the SL1 structure exists in Qrr1 even though Qrr1 lacks the 9 nucleotides that are critical for base pairing with three target mRNAs (Tu and Bassler, 2007). We hypothesized that the SL1 structure should therefore also be critical for Qrr1 stability. Indeed, disruption of SL1 in Qrr1 (CCC to GGG, denoted Qrr1 SL1D) decreased the half-life to ∼15 min (Supplementary Figure S8). Again, stability could be restored by introducing a compensatory mutation (GGG to CCC, denoted Qrr1 SL1R) (Supplementary Figure S8). Thus, the SL1 stabilization mechanism is maintained across all five Qrr sRNAs. To understand whether the instability of the SL1 deletion mutant is caused by RNase E-mediated degradation, we exploited a temperature-sensitive E. coli rne mutant (Massé et al, 2003). The Qrr4 SL1D mutant exhibited a half-life of ∼5 min at the permissive temperature (30°C) and an elongated half-life of ∼15 min at the non-permissive temperature (43°C) (Figure 6C). Thus, the instability of the Qrr4 SL1 disruption mutant is due to RNase E-mediated degradation. Finally, the contribution of SL1 to Qrr sRNA stability could also depend on its interaction with Hfq. To investigate this possibility, we performed Hfq–sRNA binding gel mobility shift assays. The SL1, SL2, and SL3 Qrr4 deletion mutants all exhibited a modestly reduced ability to bind Hfq compared to wild-type Qrr4. However, there was no difference between the Qrr4 SL deletion mutants in their ability to bind Hfq (unpublished data). Together, our results suggest that the instability of the Qrr4 SL1 deletion/disruption mutants is due to increased vulnerability to RNase E-mediated degradation rather than due to any defect in Hfq interaction. We suspect that, even though the SL1 deletion and disruption mutants exhibit decreased half-lives, there remains sufficient sRNA present to regulate most targets in our GFP reporter assay.
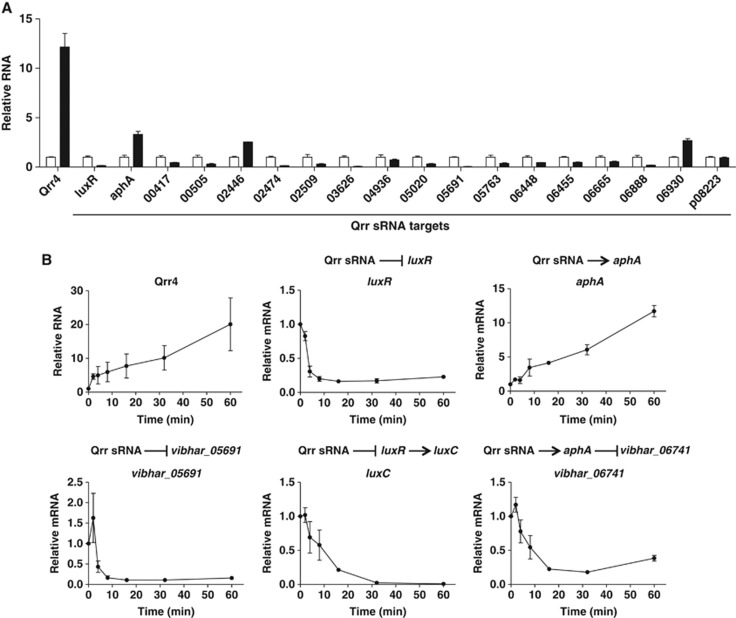
Qrr sRNA targets rapidly respond to quorum-sensing signals in vivo
In signalling networks, one advantage of sRNA regulation over protein transcription factor regulation is the presumed rapid response to external stimuli (Waters and Storz, 2009). We wondered whether the 16 targets identified here respond to quorum-sensing molecules under in vivo conditions and if so, how rapidly relative to other quorum-sensing targets that are indirectly controlled by the Qrr sRNAs (i.e., via AphA or LuxR). To examine this, we used a V. harveyi ΔluxM ΔluxPQ ΔcqsS strain, which responds only to exogenously added AI-1. We grew this strain in the presence of saturating AI-1 (to mimic the HCD state and eliminate Qrr production). We added the AI-1 antagonist 3-oxo-C12-HSL for 15 min (to mimic the HCD to LCD transition and induce Qrr production), and performed qRT–PCR to assess changes in expression of the 16 new targets. As a control, we performed the same experiment with a V. harveyi ΔluxM ΔluxPQ ΔcqsS Δqrr1–5 strain. This control allows us to eliminate non-Qrr effects. Addition of the antagonist caused a 10-fold increase in Qrr sRNA production. The luxR mRNA was repressed five-fold, and aphA was activated three-fold. These results show that the antagonist does elicit the HCD to LCD transition (Figure 7A). By contrast, in the control experiment, expression of both luxR and aphA did not change (Supplementary Figure S9). Fifteen of the 16 new targets showed Qrr-dependent changes in expression in response to the antagonist; only the target vibhar_p08223 did not change (Figure 7A and Supplementary Figure S9). Figure 7B shows a time course of RNA changes for some representative targets following addition of the antagonist. Targets that are directly repressed by Qrr4, such as luxR and vibhar_05691 decreased to their minimal levels within 8min. By contrast, vibhar_06741, which is an indirect target that is repressed by AphA, reached its minimal level only after 16 min following the addition of antagonist. Likewise, luxC, which is indirectly controlled by the Qrr sRNAs via LuxR, did not reach its minimal level until 30 min after the addition of the antagonist. Importantly, luxC is the most highly responsive LuxR-controlled gene identified to date (van Kessel et al, 2012). In contrast to luxR and vibhar_05691 that exhibit rapid decreases in mRNA levels following an alteration in Qrr levels, aphA undergoes a gradual increase in mRNA level. This phenomenon might arise from the different mechanisms governing target repression versus target activation by sRNAs. Positive regulation depends on the intrinsic half-life of the stabilized mRNA, whereas negative regulation often involves active degradation of the target mRNA (Beisel and Storz, 2010). Together, these results indicate that targets under direct control of the Qrr sRNAs are the most rapid to respond to cell density changes, followed by those under the control of the protein regulators AphA and LuxR, which are themselves controlled by the Qrr sRNAs. Together, the Qrr sRNAs, AphA, and LuxR establish a precisely timed quorum-sensing gene expression programme.
Figure 7.
Direct Qrr targets rapidly respond to quorum-sensing autoinducers. V. harveyi ΔluxM ΔluxPQ ΔcqsS strain (TL25) was grown in the presence of 1 μM AI-1 to mid-logarithmic phase. DMSO (white bars) or 100 μM 3-oxo-C12-HSL (black bars) was added to the culture (Long et al, 2009). (A) Samples were collected 15 min after the addition of 3-oxo-C12-HSL and the mRNA levels of the target genes were measured by qRT–PCR. Mean and s.e.m. values of triplicate samples are shown. (B) Samples were collected at different time points. Relative mRNA levels of Qrr4, aphA, luxR, vibhar_05691, luxC, and vibhar_06741 were measured by qRT–PCR. Mean and s.e.m. values of triplicate samples are shown.
Discussion
Five homologous Qrr sRNAs function at the centre of the V. harveyi quorum-sensing circuit (Tu and Bassler, 2007). At LCD, they activate translation of aphA encoding the LCD master regulator and repress translation of luxR encoding the HCD master regulator (Tu and Bassler, 2007; Rutherford et al, 2011; Shao and Bassler, 2012). Because AphA and LuxR control hundreds of downstream target genes, the precise levels of the Qrr sRNAs dictate the precise timing and level of expression of genes in the quorum-sensing programme (Rutherford et al, 2011; van Kessel et al, 2012). Qrr sRNA-mediated feedback loops also play roles in V. harveyi quorum-sensing regulatory dynamics. Qrr sRNAs repress translation of the quorum-sensing response regulator LuxO, which in turn controls Qrr sRNA levels. This feedback loop further influences the timing of transitions between LCD and HCD (Tu et al, 2010). Qrr sRNAs also repress translation of the AI-1 pathway components luxMN, which adjusts the input–output response range to different autoinducer signals (Teng et al, 2011). All of these previously discovered Qrr sRNA targets are members of the quorum-sensing circuit. Thus, until the work presented here, the Qrr sRNAs were only known to function to control quorum-sensing regulatory components, and thus they acted only as indirect regulators of quorum-sensing targets. Here, we identify 16 additional Qrr sRNA targets and all of them reside outside of the central quorum-sensing regulatory pathway (Figure 8, Table I).
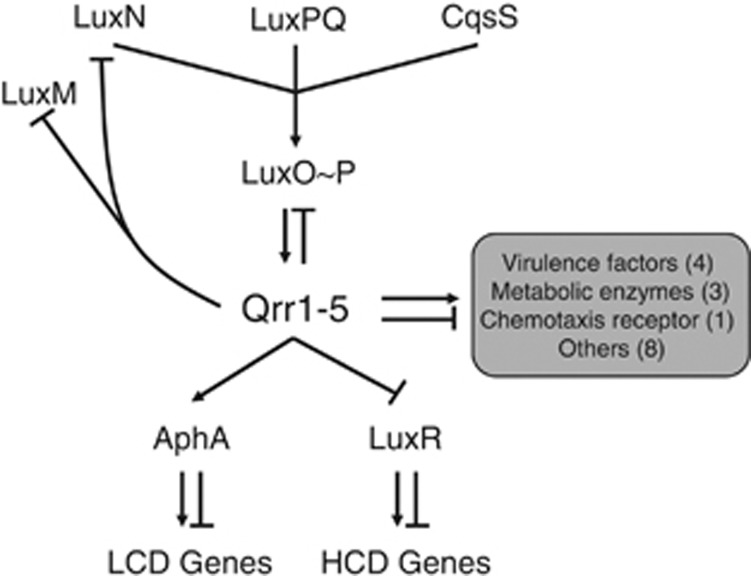
Figure 8.
Model for Qrr control of quorum-sensing targets. The V. harveyi quorum-sensing circuit is shown. At low cell density (LCD), in the absence of autoinducers, the three autoinducer receptors (LuxN, LuxPQ, and CqsS) act as kinases. They transfer phosphate to the response regulator LuxO. Phosphorylated LuxO activates the production of Qrr1–5. The Qrr sRNAs repress the translation of the high cell density (HCD) master transcriptional regulator LuxR and they activate production of the LCD master transcriptional regulator AphA. The Qrr sRNAs also repress production of the autoinducer synthase LuxM, the receptor LuxN, and the response regulator LuxO. The Qrr sRNAs control 16 newly identified target genes or operons that encode functions that act outside of the central quorum-sensing cascade. These targets include four virulence factors, three metabolic enzymes, and one chemotaxis receptor.
Genome-wide studies identifying targets of sRNAs in bacteria, particularly in E. coli and Salmonella, show that individual sRNAs commonly regulate multiple targets (Storz et al, 2011). Our study expands the size of the Qrr sRNA regulon from four to at least 20 targets. Additional genes directly regulated by the Qrr sRNAs could remain to be discovered. If they are regulated, for example, by sequestration rather than degradation or stabilization, no significant changes at the transcript level would occur, and we would not have identified such targets by our microarray studies.
The 16 targets identified here expand the roles of the Qrr sRNAs from indirect (via AphA and LuxR) to direct controllers of quorum-sensing targets. We propose that genes directly controlled by the Qrr sRNAs form the immediate response to changes in autoinducer concentration, while those controlled by AphA and/or LuxR form the secondary response. Within the set of homologous Qrr sRNAs, there are distinct preferences for particular Qrr sRNAs and particular targets. Using the newly identified targets as probes for Qrr function, we defined the specificity, structural, and functional domains of the Qrr sRNAs: SL1 and SL2 are involved in direct base pairing with the mRNA targets, SL1 also protects Qrr sRNAs from RNase E-mediated degradation, SL3 plays an accessory role in base pairing and stability, and SL4 harbours the terminator.
Quorum sensing is crucial for bacteria to monitor cell-population density changes and for synchronizing population-wide gene expression. One advantage of using sRNA regulators in the quorum-sensing circuit is the presumed rapid response to cell density changes they enable (Figure 7). At the transition from HCD to LCD, qrr expression is activated within seconds. In turn, the Qrr sRNAs activate the LCD master regulator AphA and repress the HCD master regulator LuxR. However, it takes time for AphA protein to accumulate to its functional level and for LuxR protein to be diluted to below its functional level. Our discovery of 16 direct Qrr targets signifies the identification of the ‘first response’ genes in quorum sensing. Many of the target genes encode proteins of unknown functions. However, we do know that direct Qrr targets include virulence factors, a chemotaxis receptor, and metabolic enzymes (Figure 8, Table I). Quorum sensing regulates virulence factors in numerous bacterial species. To our knowledge, prior to the present work, all known quorum-sensing-controlled virulence factors were regulated by protein transcription factors. Virulence factors are considered energetically costly, thus we speculate that placing them under direct Qrr sRNA control provides a mechanism to rapidly repress their production and thereby conserve energy at LCD when such factors are ineffective. Likewise, repression of a chemotaxis receptor (vibhar_05691) by the Qrr sRNAs may help cells rapidly respond to the disappearance of a specific attractant, again, providing an environmental advantage. Perhaps the targets that are directly activated and repressed by the Qrr sRNAs encode functions required to prepare V. harveyi for the HCD to LCD transition. Subsequently, the ∼200 AphA-controlled genes and the ∼600 LuxR-controlled genes could encode the functions required for commitment to the LCD or HCD lifestyle (Figure 8) (van Kessel et al, 2012). Interestingly, some of the targets that are directly controlled by the Qrr sRNAs are also regulated by AphA or LuxR. For example, vibhar_02509 is repressed by the Qrr sRNAs and AphA, and activated by LuxR; vibhar_03626 and vibhar_05020 are repressed by the Qrr sRNAs, and activated by LuxR (van Kessel et al, 2012). Combining all of these regulators at a particular target likely reinforces regulatory patterns at specific cell densities, further ensuring the commitment to the LCD or HCD programme.
Other sets of homologous sRNAs exist in bacteria, such as OmrA/OmrB, Prr1/Prr2, 6S homologues, CsrB homologues, GlmY/GlmZ, and several toxin/anti-toxin modules (Waters and Storz, 2009). Having multiple homologous sRNAs could allow the sRNAs within each group to diversify their functions and to adopt distinct preferences for particular targets. For example, unlike Qrr2–5, Qrr1 is unable to activate aphA translation due to the lack of 9 nucleotides at its 5′ end. This result suggests that Qrr1 prefers other targets such as luxR and luxO (Shao and Bassler, 2012). In the present study, by systematically comparing each Qrr sRNA’s regulon, we could define the regulatory functions that are shared and that are exclusive to each Qrr sRNA. Qrr1–5 are all capable of regulating 13 of the 16 newly identified targets, suggesting that the major function of the Qrr sRNAs are conserved across Qrr1–5. Qrr1 is unable to regulate aphA, vibhar_00505, and vibhar_05691, and Qrr3 is unable to regulate vibhar_02509. These findings suggest that the functions of Qrr1 and Qrr3 have diverged or may continue to diverge. The five Qrr sRNAs are controlled by five distinct promoters and hallmarks suggestive of different transcription factor binding sites exist in each promoter (Tu and Bassler, 2007). Thus, specific regulation of a particular Qrr sRNA could be possible under environmental conditions. If so, more elaborate differential regulation of quorum-sensing targets than we have shown here could occur in response to environmental changes.
Trans-encoded Hfq-dependent sRNAs function through direct base pairing with target mRNAs. They can act as both activators and repressors by revealing or occluding ribosome-binding site sequences, respectively. Unlike eukaryotic ∼22 nt unstructured microRNAs, which always use 5′ seed regions for base pairing, bacterial trans-encoded sRNAs have a variety of lengths and secondary structures and they use complicated non-contiguous base pairing strategies (Bartel, 2009). Several base pairing schemes have been reported: first, a single, short conserved region of an sRNA, such as in RybB, can base pair with multiple targets (Papenfort et al, 2010). Second, two separate regions of an sRNA, such as in Spot42, can base pair with two different regions of the same mRNA target (Beisel and Storz, 2011). Third, multiple regions of an sRNA, such as in GcvB, can base pair with the same target redundantly and each region is sufficient for regulation (Sharma et al, 2011). The first two schemes are used by the Qrr sRNAs. 5′ terminal conserved sequences of the Qrr sRNAs are responsible for base pairing, and this conserved region can be further divided into two sub-regions, SL1 and SL2. SL2 is critical for base pairing with most targets, while SL1 regulates only a small subset of targets.
Most of the targets identified here are likely regulated through the classical sRNA regulatory mechanisms. Base pairing over the ribosome-binding site of the first gene in an operon typically regulates the entire operon and leads to mRNA degradation. However, there are a few special cases. For example vibhar_00505, which is in an operon with vibhar_00506 and vibhar_00504. vibhar_00506 is the first gene in the operon followed by vibhar_00505 and vibhar_00504. The Qrr sRNAs base pair over the ribosome-binding site of vibhar_00505 and reduce the mRNA levels of vibhar_00505 and vibhar_00504, but not vibhar_00506 (unpublished data). Presumably, by base pairing in the vicinity of the second gene in the operon, the Qrr sRNAs can uncouple regulation of vibhar_00505 and vibhar_00504 from regulation of vibhar_00506. Uncoordinated regulation of polycistronic transcripts has been described for other sRNAs, such as Spot42, GlmY, and RyhB (Møller et al, 2002; Urban et al, 2007; Desnoyers et al, 2009). We speculate that this mechanism of sRNA regulation facilitates gene expression patterns for operons that would be difficult to achieve using protein transcription factors. Another special case is vibhar_02446. The mRNA of vibhar_02446 is stabilized by the Qrr sRNAs. However, VIBHAR_02446-GFP production is repressed by the Qrr sRNAs in E. coli. This finding indicates that vibhar_02446 mRNA could be sequestered by the Qrr sRNAs until the quorum-sensing LCD to HCD transition occurs, and then the vibhar_02446 mRNA can be immediately translated to protein. Alternatively, sequestration could be a mechanism for V. harveyi to hedge against the transient loss of autoinducer. If an mRNA such as vibhar_02446 is sequestered rather than destroyed, if autoinducer reappears, the mRNA is present and the cell is primed to rapidly respond.
In addition to base pairing roles, our study also pinpoints the structural roles of each portion of the Qrr sRNAs. SL1 is critical for Qrr sRNA stability. Consistent with this, 5′ stem-loops commonly protect mRNAs from RppH- and RNase E-mediated degradation (Belasco, 2010). Here, we show that the instability caused by disruption of SL1 is RNase E-dependent, which indicates that the same protective mechanism used for mRNAs is used to protect the Qrr sRNAs.
The Qrr sRNAs and other components of the quorum-sensing circuit are highly conserved among all vibrio species. Thus, the specificity, structural, and functional insights gained in this study of the V. harveyi Qrr sRNAs likely apply to other vibrios. Studying whether the Qrr sRNAs regulate genes encoding similar or different functions in other vibrio species could reveal how quorum sensing promotes different diseases, for example, in pathogenic vibrios, or could reveal the basis for the unique environmental niches different vibrios occupy. As the Qrr sRNAs rapidly respond to cell density changes, different vibrio species may use the Qrr sRNAs to repress their most costly HCD-specific genes and activate their most critical LCD-specific genes. Thus, defining by bioinformatics or by experimentation direct Qrr targets among vibrios could yield clues as to the most evolutionarily costly products or those most critical for fitness.
Our work reveals how bacteria integrate sRNA and protein regulators into a rapid alternation between social and asocial lifestyles. When such transitions occur, for example, when bacteria exit a biofilm or a host, the Qrr sRNAs are used to rapidly transition between gene expression programs. This switch is followed up by regulation via the transcription factors AphA and LuxR that commit the cells to specific gene expression patterns at specific cell densities. Combining sRNA and protein regulators provides a rapid but staged mechanism to alternate between two dramatically different genetic programs.
Materials and methods
Bacterial strains and growth conditions
V. harveyi strain BB120 (BAA-1116) (Bassler et al, 1997) and derivatives were grown aerobically in Luria-Murine (LM) medium at 30°C. E. coli strains S17-1λpir (De Lorenzo and Timmis, 1994), BW-RI (Levine et al, 2007) and derivatives were grown aerobically in LB medium or M9 medium (0.5% glucose) at 37°C. Strains used in this study are described in Supplementary Table S2. Antibiotics (Sigma-Aldrich) were used at the following concentrations: 50 U ml−1 polymyxin B (Pb), 100 μg ml−1 ampicillin (Amp), 100 μg ml−1 kanamycin (Kan), 10 μg ml−1 chloramphenicol (Cm), and 60 μg ml−1 spectinomycin (Spec). Plasmids were introduced into electrocompetent E. coli S17-1λpir, BW-RI and derivatives using 0.1 cm gap cuvettes (USA Scientific) and a Bio-Rad MicroPulser.
DNA manipulations and mutant constructions
E. coli S17-1λpir was used for cloning. DNA manipulations were performed as in Sambrook et al (1989). iProof DNA polymerase (Bio-Rad) was used for regular PCR reactions, and PfuUltra DNA polymerase (Agilent) was used for constructing point mutations, deletions, and insertions. Restriction enzymes, T4 DNA ligase, T4 polynucleotide kinase, and Antarctic phosphatase were purchased from New England Biolabs. Plasmids were constructed as described in Supplementary Table S3 using primers listed in Supplementary Table S4 from Integrated DNA Technologies (IDT). All plasmids were confirmed by sequencing at Genewiz. The anhydrotetracycline-inducible qrr genes as well as qrr mutants and chimeras were blunt cloned under the PLtet-O1 promoter of the pZA31-lucNB plasmid, replacing the luc gene (Levine et al, 2007). Target-GFP translational fusions were constructed under the PLlac-O1 promoter of the pZE12G plasmid (Levine et al, 2007). Arabinose-inducible qrr genes were blunt cloned under the araC-pBAD promoter of the pBAD/myc-His A plasmid (Invitrogen) and then moved onto pEVS143 (Dunn et al, 2006). V. harveyi mutants were constructed as described previously using λ red recombineering in E. coli, followed by homologous recombination in V. harveyi (Datsenko and Wanner, 2000; Rutherford et al, 2011). To construct the rne temperature-sensitive mutant strain LF1018, the rne-50 zce-726::Tn10 allele from EM1371 (Massé et al, 2003) was introduced into BW-RI by P1 transduction followed by selection for tetracycline resistant colonies at 30 °C.
Microarray analysis and qRT–PCR
The V. harveyi Δqrr1–5 mutant strain KT282 (Rutherford et al, 2011) with plasmids containing arabinose-inducible qrr genes was grown in LM overnight with 0.2% arabinose. Cultures were diluted into LM to OD600∼0.001 in the absence of arabinose, and 0.2% arabinose was added at OD600∼0.5 or OD600∼1.0 to induce qrr expression for 15 min followed by harvesting cells by centrifugation. RNA preparation, cDNA synthesis, microarray hybridization conditions, and data acquisition were carried out as described (Rutherford et al, 2011). In every case, four arrays were performed comparing three independent cultures as well as a dye-swap comparison. Data analysis was performed using the Princeton University Microarray Database (PUMAdb) (http://puma.princeton.edu/). These data are publicly available at PUMAdb (http://puma.princeton.edu/cgi-bin/publication/viewPublication.pl?pub_no=549). All qRT–PCR analyses were carried out as described (Rutherford et al, 2011). 5S rRNA was used as the control.
5′ RACE
The transcription start sites of vibhar_00417, vibhar_02446, vibhar_02509, vibhar_04936, vibhar_05213, vibhar_05763, vibhar_05691, and vibhar_06930 were mapped using FirstChoice® RLM-RACE Kit (Invitrogen), following the manufactures’ instructions.
GFP reporter assay
E. coli strains were grown overnight aerobically at 37°C in LB medium. Cultures were diluted 1:1000 in triplicate into M9 medium (0.5% glucose). Upon dilution, 100 ng ml−1 anhydrotetracycline (Clontech) was added to induce qrr expression and target-GFP translational fusions were induced with 0.5 mM IPTG. GFP fluorescence was measured after 8–10 h of growth using FACS (BD Biosciences FACSAria cell sorter).
Northern blot analysis
E. coli BW-RI and derivatives containing plasmids encoding qrr genes were grown in LB to OD600 ∼1.0 in the presence of 100 ng ml−1 anhydrotetracycline. 250 μg ml−1 rifampicin was added to stop transcription followed by collection of cells at different time points. Total RNA was isolated using phenol/chloroform extraction. Northern blot was carried out as previously described (Urban and Vogel, 2007). 5 μg of total RNA was resolved on 6% polyacrylamide gels (7M urea) followed by transfer to Hybond-XL membranes (GE Healthcare). For Qrr sRNA detection, membranes were hybridized with the Qrr Riboprobe at 68°C in Rapid-hyb buffer (GE Healthcare) and washed in three steps with SSC wash buffers (2X, 1X, 0.5X, respectively). For 5S RNA detection, membranes were hybridized with 5′ end-labelled DNA probe at 42°C in Rapid-hyb buffer (GE Healthcare) and washed in three steps with SSC wash buffers (5X, 1X, 0.5X, respectively). Wash buffers were supplemented with 0.1% SDS. The blots were exposed to a PhosphorImager screen (GE Healthcare), scanned with Typhoon 9410 (GE Healthcare), and band intensities were quantified with Image J (http://imagej.nih.gov/ij/). The Riboprobe was synthesized by T7-mediated in vitro transcription of 200 ng template DNA in the presence of 32P-α-UTP with the MAXIscript kit (Ambion). The 5′ end-labelled DNA probe was synthesized using T4 polynucleotide kinase in the presence of 32P-γ-ATP.
Supplementary Material
Acknowledgments
We thank Susan Gottesman for generously providing us the RNase E temperature-sensitive mutant strain. We are grateful to Terence Hwa for giving us the BW-RI strain and the pZA31-lucNB and pZE12G plasmids. We thank Donna Storton and Jessica Buckles for assistance with microarray experiments and analyses. We thank Igor Zhulin, Ned Wingreen, and other members of the Bassler and Wingreen laboratories for insightful discussions and suggestions. PUMAdb is funded in part by NIH grant P50 GM071508 and is a project within the Lewis-Sigler Institute for Integrative Genomics at Princeton University. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) Grant 5R01GM065859 and National Science Foundation (NSF) Grant MCB-0343821 to BLB. KP is supported by a post-doctoral fellowship from the Human frontiers in Science program (HFSP). STR is supported by a NIH fellowship F32AI085922.
Author contributions: YS, LF, STR, KP, and BLB designed the experiments; YS, LF, and STR performed the experiments; YS, LF, and BLB analysed the data; and YS, LF, and BLB wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM (1997) Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. JBacteriol 179: 4043–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G (2010) Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiology Rev 34: 866–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G (2011) The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 41: 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco JG (2010) All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nature reviews. Mol Cell Biol 11: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci UStatesA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo V, Timmis KN (1994) Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235: 386–405 [DOI] [PubMed] [Google Scholar]
- Desnoyers G, Massé E (2012) Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 26: 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prévost K, Massé E (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 28: 1551–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Jeffrey L, Stabb E V, Bose JL. (2006) New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Micriobiol 72: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL (1999a) A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31: 665–677 [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL (1999b) Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol 181: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J (2009) Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682 [DOI] [PubMed] [Google Scholar]
- Henke JM, Bassler BL (2004) Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol 186: 6902–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni R V, Wingreen NS, Bassler BL (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82 [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T (2007) Quantitative characteristics of gene regulation by small RNA. PLoS Biol 5: e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Bassler BL (2000) Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol 36: 940–954 [DOI] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS (2009) Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol 7: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GA (2012) RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11: 45–57 [DOI] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P (2002) Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16: 1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM (2006) Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL (2009) Bacterial quorum-sensing network architectures. Ann Rev Genet 43: 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana N, Shirahama Y, Abe K, Nakamura K (2010) Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol Microbiol 77: 1416–1428 [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J (2010) Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA 107: 20435–20440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J (2013) Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16: 840–846 [DOI] [PubMed] [Google Scholar]
- Ramirez-Peña E, Treviño J, Liu Z, Perez N, Sumby P (2010) The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 78: 1332–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, van Kessel JC, Shao Y, Bassler BL (2011) AphA and LuxR/HapR reciprocally control quorum sensing in Vibrios. Genes Dev 25: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning A Laboratory Manual Cold Spring Harbor Laboratory Press: NY, Vol. 1, 2, 3: [Google Scholar]
- Shao Y, Bassler BL (2012) Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol 83: 599–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf H-J, Hinton JCD, Vogel J (2011) Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81: 1144–1165 [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, Wingreen NS, Bassler BL (2008) Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell 134: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S-W, Schaffer JN, Tu KC, Mehta P, Lu W, Ong NP, Bassler BL, Wingreen NS (2011) Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol Sys Biol 7: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL (2007) Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL (2010) Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell 37: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Papenfort K, Thomsen J, Schmitz RA, Vogel J (2007) A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J Mol Biol 373: 521–528 [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35: 1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL (2012) Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195: 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Ann Rev Cell Dev Biol 21: 319–346 [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Ng W-L, Cong J, Bassler BL (2012) Ligand and antagonist driven regulation of the Vibrio cholerae quorum-sensing receptor CqsS. Mol Microbiol 83: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.