Summary
Plasmodium falciparum expresses on the host erythrocyte surface clonally variant antigens and ligands that mediate adherence to endothelial receptors. Both are central to pathogenesis, since they allow chronicity of infection and lead to concentration of infected erythrocytes in cerebral vessels. Here we show that expression of variant antigenic determinants is correlated with expression of individual members of a large, multigene family named var. Each var gene contains copies of a motif that has been previously shown to bind diverse host receptors; expression of a specific var gene correlated with binding to ICAM-1. Thus, our findings are consistent with the involvement of var genes in antigenic variation and binding to endothelium.
Introduction
Of the four species of malaria parasite that naturally infect humans, P. falciparum is responsible for a high proportion of the morbidity and nearly all the mortality (Warrell et al., 1990). The virulence of P. falciparum is thought to be due to modifications of the surface of infected erythrocytes (Berendt et al., 1994). During the latter half of the intraery-throcytic growth cycle, parasite-derived molecules are expressed on the host cell surface that mediate adherence of infected cells to endothelial cells lining postcapillary venules. Thus, only the younger developmental forms are detected in the peripheral circulation, since the mature forms are sequestered. It has been argued that by not circulating, the parasite avoids spleen-dependent killing mechanisms (Langreth and Peterson, 1985; Miller et al., 1994). All P. falciparum field isolates undergo this process of sequestration. In some cases, infected cells adhere to venules in the brain, leading to the syndrome of cerebral malaria, which is associated with a high mortality (Warrell, 1993). A number of endothelial receptors for infected erythrocytes have been identified, including CD38, intercellular adhesion molecule 1 (ICAM-l), thrombospondin, vascular cell adhesion molecule (VCAM), and E-selectin (Roberts et al., 1985; Barnwell et al., 1989; Berendt et al., 1989; Ockenhouse et al., 1992). Most isolates can adhere to CD36 and thrombospondin, whereas only a subset show the capacity to bind to ICAM-1 (Ockenhouse et al., 1991; Newbold et al., unpublished data). The up-regulation of ICAM-1 on cerebral vessels during acute disease may be an important contributory factor in the development of cerebral malaria (Berendt et al., 1994; Turner et al., 1994).
The process of malarial sequestration in humans is unique to P. falciparum; however, all malaria species studied appear to modify their host cell surface by insertion of clonally variant proteins into the erythrocyte membrane. These antigens, which were described originally in P. knowlesi, can be detected by antibody-mediated agglutination (Brown and Brown, 1965). During P. knowlesi infection, the parasitemia oscillates, and each new peak in parasitemia is associated with changes in parasite proteins expressed at the surface of the infected erythrocyte (Brown et al., 1970; Howard et al., 1983). Antibodies present at any point in time recognize infected erythrocytes from preceding peaks but not those present at the time sera is drawn or those appearing later. This parallels, at the phenotypic level, the well-studied phenomenon of anti-genic variation in African trypanosomes (Borst, 1991). In both P. knowlesi and in rodent malarial infection, the process of antigenic variation appears to be necessary for the establishment of chronic infection (Brown, 1971; McLean et al., 1982; Gilks et al., 1990). Moreover, antibody responses to the variant antigen are strongly implicated in the host protective immune response (Marsh et al., 1989; Gilks et al., 1990). At the biochemical level, sera have been used to immunoprecipitate a family of high molecular mass proteins (200–350 kDa) present on the surface of infected cells in a variant-specific fashion. These molecules were originally termed SICA (for schizont-infected cell agglutination) antigens in P. knowlesi (Howard et al., 1983), and the homolog in P. falciparum has been named PfEMPl (P. falciparum erythrocyte membrane protein 1) (Leech et al., 1984). They are defined operationally by their antigenic diversity, presence on the erythrocyte surface, high and variable molecular mass, and insolubility in nonionic detergents. Attempts to purify and sequence the proteins or to clone the relevant genes have been unsuccessful owing, in part, to the low abundance of these proteins and the difficulty of maintaining their expression in vitro. Thus, the question of whether the genes encoding PfEMP1 are members of a single gene family remains unresolved.
In P. falciparum and in some animal models where sequestration occurs, the balance of evidence suggests that binding to endothelium and antigenic variation are functions of the same molecule. Initially, it was shown that immune sera inhibited the binding of infected erythrocytes to vascular endothelium in vitro in a strain-specific manner (Udeinya et al., 1983). Similar sera immunoprecipitated a protein with the characteristics of PfEMP1 (Leech et al., 1984; van Schravendijk et al., 1993). More recently, it has been observed that selection of parasites for particular cytoadherence changes is accompanied by changes in PfEMP1 expression (Magowan et al., 1988; Biggs et al., 1992). In parallel studies, we developed an extended family of P. falciparum subclones in which we could study both the process of antigenic switching and its relationship to cytoadherence. We demonstrated that antigenic variation in vitro was extremely rapid, with a mean rate of 2.4% per generation. In this clone family, switches in antigenic type were always accompanied by a change in PfEMP1 molecular mass and a loss of ability to bind to ICAM-1 (Roberts et al., 1992). Thus, while there exists a very close link between adhesive and antigenic phenotypes, it has not been formally proven that the variant antigen genes encode the parasite ligands for cytoadherence. The absence of gene sequences for molecules central to both pathogenesis and the development of protective immunity is thus a fundamental barrier both to understanding the system at the molecular level and to the development of novel intervention strategies.
In the course of mapping and sequencing a segment of chromosome 7 containing the chloroquine resistance locus of P. falciparum, Su et al. (1995 [this issue of Cell]) identified several gene sequences that possess multiple domains similar to the binding region, region II, of erythrocyte binding proteins from P. vivax, P. knowlesi, and P. falciparum (Adams et al., 1992; Chitnis and Miller, 1994; Sim et al., 1994). Although the P. vivax and P. falciparum erythrocyte binding proteins bind different erythrocyte receptors (the Duffy antigen and glycophorin A, respectively), regions II of these parasite proteins share homology with each other and the genes described by Su et al. (1995).
The gene sequences from chromosome 7 hybridized with many bands in Southern blot analysis, suggesting that they belonged to a large family of genes, and have been named var genes (Su et al., 1995). var genes have many properties that might be expected of the variant antigen and cytoadherence ligands: they are polymorphic, contain large open reading frames that could encode proteins of over 200 kDa, and possess multiple DBL domains known to have diverse binding specificities in erythrocyte binding proteins. Taking advantage of homologies within the Duffy binding–like (DBL) domains, we have used degenerate oligonucleotide primers to study var message expression in antigenically variant clones. We show that the expression of distinct variant antigens at the surface of infected erythrocytes correlates with expression of distinct var genes. Thus, we conclude that var genes encode P. falciparum variant surface antigens.
Results
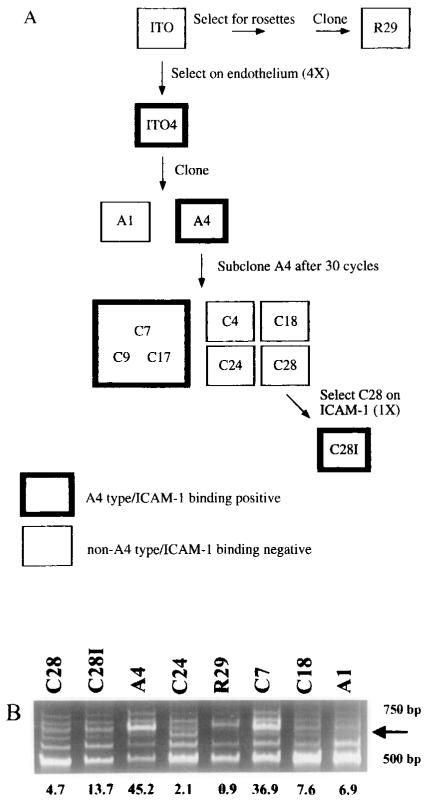
To test whether var genes encode the variant antigens, we have employed a P. falciparum clonal tree in which all members have been characterized as to their antigenic and cytoadherence phenotype (Figure 1A) (Roberts et al., 1992). Of the A4 subclones that were used in the present study, three were antigenically similar to A4 (C7, C9, and C17); four others (C4, C18, C24, and C28) were different from A4 and from each other. All parasitized erythrocytes that were antigenically similar to A4 bound ICAM-1, whereas antigenically variant subclones had greatly reduced binding to ICAM-1. C28, one of the non-A4 types, was selected on ICAM-1. The resulting line, C28l, had increased antigenic similarity to A4. Using these parasite clones, we expected certain results if the var genes encoded the variant antigens expressed on the erythrocyte surface. Specifically, var message should vary in a pattern that correlates with the antigenic type of the clone. Thus, all subclones that are antigenically similar to A4 should express a common var mRNA. Subclones that are antigenically distinct should express unique var genes. To investigate this hypothesis, we used reverse transcription–polymerase chain reaction (RT-PCR) to compare var gene expression in a series of antigenically defined clones.
Figure 1.
Degenerate Oligonucleotide Primers to DBL Domains Amplify a Message That Correlates with A4-Specific Agglutination of Members of the ITO Clonal Family
(A) P. falciparum clonal tree. Clones in this family tree were derived as described previously by Roberts et al. (1992). Bold boxes indicate that the clone is of the A4 antigenic type and binds ICAM-1. Plain boxes indicate that the parasite is of a different antigenic type from A4 and other clones and that it does not bind ICAM-1.
(B) RNA from antigenically similar or distinct clones was DNase treated, reverse transcribed, and amplified with the DBL primers UNIEBP5′ and UNIEBP3′. The arrow on the right side of the figure identifies a band whose intensity correlates with A4 antigenicity. Numbers shown below Figure 1B represent antigenic similarity to the A4 clone as defined by chance of heterologous cross-agglutination (see Experimental Procedures).
Identification of an Expressed var Gene in A4-Type Clones
To study var gene expression, we utilized primers to conserved sequences within the DBL domains of var genes (Peterson et al., in press). var genes have between two and four such domains (Su et al., 1995). Because DBL domains are sufficiently variable that no region of perfect amino acid conservation extends for as many as six amino acids, it was necessary to make the primers degenerate.
Initially, an experiment was performed to exclude the possibility of genomic DNA contamination of RNA samples. For all experiments, RNA samples were treated with DNase prior to reverse transcription. In the absence of reverse transcription, no bands were detected, confirming that the products of RT–PCR were derived from the mRNA of each clone (data not shown).
To identify DBL transcripts whose expression correlates with the variant-specific agglutination of infected erythrocytes, mRNAs from a blinded panel of clones were studied by using a range of different DBL primers based upon the sequences of Su et al. (1995; see Experimental Procedures). From these experiments, only the primer set developed by Peterson et al. (in press), UNIEBP5′ and UNIEBP3′, amplified a band whose presence correlated with A4 antigenicity (Figure 1 B). This 565 bp PCR product was cloned, and its sequence confirmed that it was a member of the DBL family. The DBL domains of var genes can be grouped into different types, each with a characteristic consensus amino acid sequence. When the predicted amino acid sequence of the A4 cDNA clone was compared with that of the var genes identified by Su et al. (1995) it aligned most closely with the consensus sequence for var DBL domain 3 (Figure 2A).
Figure 2.
Deduced Amino Acid Sequence of var Genes Isolated from Antigenically Variant Clones
(A) Alignment of the A4var sequence with the consensus sequence from the var genes cloned by Su et al. (1995). The deduced amino acid sequence encoded by A4var is compared with consensus sequence of the var DBL domains to which these genes are most similar (Su et al., 1995). Above the A4var sequence, a bold line indicates the regions of A4var homology with DBL domain 3 or DBL domain 2. At positions within the consensus sequence at which a number of similar amino acids can be found, the following symbols were used: U (I, L, V, M), at symbol (Y, F, W), ampersand (I, L, V, M, Y, F, W), percent (E, D), and dollar sign (S, T). The asterisk in the consensus sequence indicates gaps in the sequence where different numbers of amino acids are present. Boxed sequence identifies the original A4var sequence amplified with UNIEBP5′ and UNIEBP3′. A bold line, labeled hydrophobic, at the 3′ end of exon 1 indicates the putative transmembrane region. The position of a 950 bp intron is marked with an arrow, and the area of exon 2 homology is shown with consensus amino acid residues from other var sequences identified by Su et al. (1995).
(B) The predicted amino acid sequences of C18var, C24var, and R29var genes are homologous with var DBL domain 1. Predicted amino acid sequences from the C18, C24, and R29 var genes were aligned with the consensus amino acid sequence of var DBL domain 1 defined by Su et al. (1995). The same symbols were used as above. Amino acids in the var sequences are highlighted with reverse print if a consensus amino acid is always present at that position in the var sequences identified by Su et al. (1995) and are highlighted in stippled print if a consensus amino acid is the most common of more than one amino acid present at that position.
To establish that the A4 cDNA was a member of the var gene family, the sequence was extended in the 3′ direction by PCR using genomic DNA from A4 (Su et al., 1995). var genes are encoded in two exons, separated by an approximately 1 kb intron (Su et al., 1995). Exon 1 is 5–8 kb in length, highly polymorphic, contains two to four DBL domains, and has a hydrophobic sequence at the 3′ end that is consistent with a transmembrane region. Exon 2 is more conserved and encodes a putative cytoplasmic domain. The extended genomic DNA sequence contained one additional DBL domain, a putative transmembrane region at the 3′ end of exon 1, a putative 1 kb intron, and sequence from the highly conserved exon 2 (Figure 2A). Primers based on the genomic DNA sequence were used to clone a 2.5 kb cDNA from mRNA of parasite clone A4. This A4 var cDNA clone was sequenced at the 5′ and 3′ ends to confirm that the sequence was identical to the genomic sequence. In addition, it was found that the A4 var cDNA clone lacked the putative intron, establishing that the A4 var had the typical intron–exon structure found in var genes. The presence of multiple DBL domains in exon 1, the highly conserved exon 2, and the characteristic intron–exon organization proved that the A4 cDNA clone was a member of the var gene family described by Su et al. (1995). The gene from which the cDNA fragment was isolated is referred to as A4var.
The Expression of the A4var Gene in Different Clones Correlates with Their Antigenic Similarity to A4
To study the expression of A4var in different antigenic variants, specific primers were designed to a region of A4var that was highly variable among the var genes (Su et al., 1995). Initially, the A4var primers were tested on A4 cDNA at varying numbers of PCR cycles to determine cycling conditions in which the PCR reaction was within the exponential phase. These conditions were then used to analyze cDNA from each of the clones. RT–PCR products from different clones were separated on polyacrylamide gels and compared by Southern blot analysis with a labeled A4var probe.
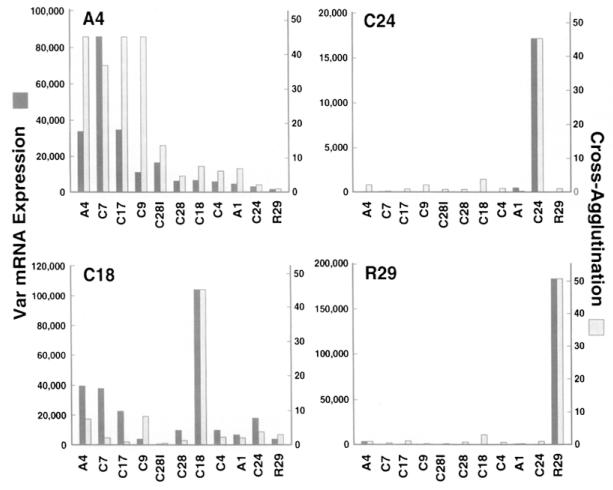
The clones included in this study possessed varying degrees of antigenic similarity to A4 as measured by cross-agglutination between parasitized erythrocytes of A4 and the other clones. By this criterion, they fell into three major groups: highly similar (C7, C17, and C9), moderately similar (C28l), and largely dissimilar (C4, C18, C28, C24, A1, and R29). With use of the A4var primers, a strong band of the appropriate size was present in A4 and the strongly cross-agglutinating C subclone lines C7 and C17 (Figure 3). This band was also present in the C4, C18, and C9 clones at a slightly reduced intensity, but the amount of this product was sharply reduced or absent in the R29, C24, A1, and C28 clones, which poorly cross-agglutinated with A4. Significantly, the selection of C28l from C28 by binding parasitized erythrocytes on ICAM-1 was accompanied by an increase in antigenic similarity to A4, and this increase was paralleled by an increase in the intensity of the A4var band. The geometric mean of three A4var RT–PCR experiments was compared with the serologically determined antigenic similarity of the clones. A highly significant correlation was observed between the amount of A4var RT–PCR product and the antigenic similarity of a clone with A4 (Figure 4; Spearman rank correlation coefficient of 0.933, p < 0.001).
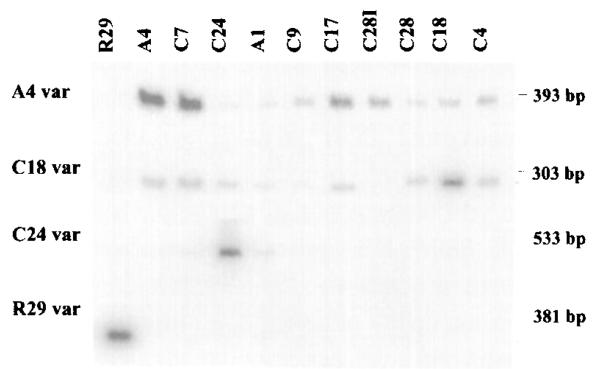
Figure 3.
Antigenically Distinct P. falciparum Clones Express Distinct var Messages
RT–PCR using specific primers to var genes isolated from the A4, C18, C24, or R29 clones (indicated at the left of the figure) was performed on RNA isolated from each of the 11 clones listed at the top of the figure. RT–PCR products were hybridized with the respective var probes. The results are representative of three experiments (A4var) or two experiments (other var genes) performed on the same preparation of cDNAs.
Figure 4.
Expression of Distinct var Transcripts Correlates with the Surface Antigenicty of P. falciparum–Infected Erythrocytes
The results of RT–PCR experiments with different var-specific primers on the panel of clones (Figure 3) were quantified by densitometry and compared with the cross-agglutination of each clone with either the A4, C24, C18, or R29 clone (indicated at the top left of each bar graph). The geometric means of RT–PCR products from three experiments using A4var primers and two experiments using C24var, C18var, or R29var primers are plotted next to cross-agglutination of each clone with A4, C18, C24, or R29 clones (see Experimental Procedures).
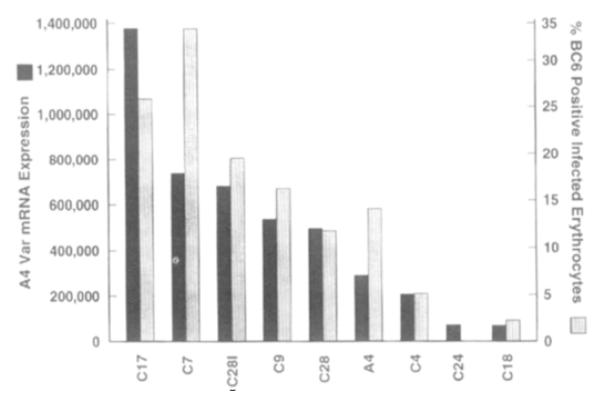
The relationship between A4 antigenicity and A4var expression was independently tested by using monoclonal antibody (MAb) BC6, which binds to infected erythrocytes in a pattern that correlates with the A4 antigenic type. Nine clones were freshly grown, and RNA was isolated at the same time that flow cytometry was performed with MAb BC6. As expected, clones that strongly cross-agglutinated with A4 contained a greater percentage of MAb BC6–reactive infected erythrocytes than subclones that had switched to other antigenic types. The unexpectedly low percentage of MAb BC6–reactive parasites in the A4 culture may be due to stochastic differences in switching during culture. Despite this, a strong correlation was found between A4var expression as measured by RT–PCR and the percentage of infected erythrocytes that expressed the MAb BC6 epitope as measured by flow cytometry (Figure 5; Spearman rank correlation of 0.97, p < 0.001). The fact that A4var expression and A4 antigenicity were so closely correlated strengthens the case for their causative association.
Figure 5.
A4var Expression Correlates with the Percentage of P. falciparum-Infected Erythrocytes Binding a Monoclonal Antibody (MAb BC6) That Is Specific for A4 Variants
The percentage of parasitized erythrocytes of the A4 type in different clones was determined by flow cytometry using MAb BC6. At the same time, RNA was extracted, and the level of A4var expression was determined by RT–PCR. The geometric mean of two RT–PCR experiments is plotted next to the percentage of A4-type variants in each clonal culture.
To confirm that the amplified product in different clones was the same and not a different var message that cross-hybridized with the A4var probe, we cloned this band from all of the clones that strongly cross-agglutinate with A4 (C7, C17, C9, and C28l). The sequence from each clone was identical to that of A4var. Thus, this portion of the expressed var gene in different clones of the A4 antigenic type is conserved.
The existing data on the protein known as PfEMP1, which carries the variant-specific epitopes, show it to be of high (200–350 kDa) and variable molecular mass (Howard, 1988). To determine the size of the A4var message, Northern blots of RNA from several variants were probed with the A4var fragment (data not shown). Clones of the A4 antigenic type, C7 and C281, shared an mRNA of approximately 8 kb that hybridized with this probe. This 8 kb band was absent in the antigenically distinct clone, C24, but a band of slightly higher mobility was evident. An additional weak band in the Northern blot of C7 at 9.4 kb may have been a cross-reaction with another var or unspliced A4 var mRNA. The fact that the A4var message is of a size that would be expected for the variant antigen, and that the expression of the A4var message correlates with A4 antigenicity as measured by mixed agglutination with immune sera (see Figure 4) or reactivity with MAb BC6 (Figure 5) leads us to conclude that A4var encodes a variant antigen at the surface of infected erythrocytes.
Antigenically Distinct P. falciparum Clones Express Distinct var Genes
To extend these observations of var gene expression to clones expressing different antigenic types, the degenerate DBL primers, 1 and UNIEBP3′, were used to clone and sequence three unique var cDNAs from the antigenically distinct clones C18, C24, and R29. The predicted amino acid sequence of each cDNA was different, but all had the consensus sequence of var DBL domain 1 (see Figure 2B; Su et al., 1995). The presence of sequences that are typical of var DBL domain 1 (see Figure 2B) indicated that these three cDNAs were members of the var gene family. The cloned var genes from C18, C24, and R29 will be referred to as C18var, C24var, and R29var, respectively.
Specific primers for C18var, C24var, and R29var were designed and used together with specific primers to A4var to compare the expression of these genes in the panel of clones (see Figure 3). The C18 and C24 subclones strongly expressed C18var and C24var, respectively, but weakly expressed A4var. By contrast, clones that were antigenically distinct from C18 and C24 had lower or no expression of C18var and C24var (see Figures 3 and 4). Thus, subclones of A4 that had switched antigenic type expressed new var genes. Data on the expression of R29var supported the fact that unique var mRNAs were expressed by clones that were antigenically distinct (see Figures 3 and 4). The DNA fingerprints of R29 and A4 were identical (Roberts et al., 1992) indicating that they were derived from the same parent clone, although they are not immediately related (see Figure 1A). R29var was expressed in R29 but was greatly reduced or absent in other clones. Thus, in all of the cases that we have examined, changes in antigenic phenotype were directly correlated with expression of different var genes.
Large-Scale Genomic Rearrangements Are Not Essential for Antigenic Switching
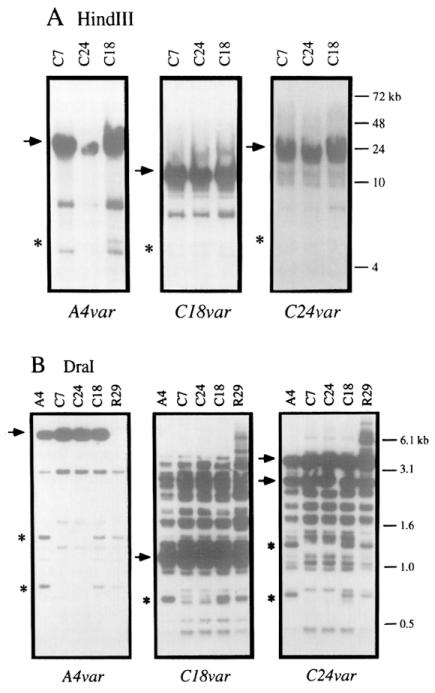
In addition to Plasmodium, clonal antigenic variation has been described for several protozoan species, including African trypanosomes, Giardia lamblia, and Paramecium (Borst, 1991; Nash, 1992; Preer, 1986). Of these, the mechanism of antigenic variation has been described in greatest molecular detail for African trypanosomes. Antigenic variation in trypanosomes is accomplished by the sequential expression of members of a large multigene family by mechanisms that frequently involve the duplicative transposition of silent sequences into expression sites. These rearrangements can be readily detected on Southern blots. To determine whether or not a similar mechanism was operative in the malaria parasite, cloned cDNA fragments from the expressed var genes of A4, C18, and C24 (A4var, C18var, and C24var) were used as probes in Southern blot analysis of genomic DNA isolated from members of the P. falciparum clonal tree.
To detect rearrangements of var genes in subclones of A4 that had switched antigenic type, probes for the A4var, C18var, and C24var genes were hybridized to HindIII-digested genomic DNA from the clones C7, C18, and C24. C7 is a subclone of A4 that is antigenically similar to A4. The A4 subclones, C18 and C24, are distinct from A4 and each other. HindIII cuts malarial DNA infrequently and produces large fragments, so the restricted DNA samples were resolved by pulsed-field gel (PFG) electrophoresis. Each probe hybridized to a single major band of HindIII-restricted DNA on each blot (Figure 6A). The major band identified by each probe was of a different size (Figure 6A), indicating that the three probes hybridized to different DNA fragments. However, there was no difference in the size of the major band in different clones hybridized with the same var probe, indicating that these probes did not detect DNA rearrangements associated with antigenic switching.
Figure 6.
Southern Blot Analysis of var Gene Organization in Clones That Have Switched Antigenic Type
(A) DNA from the C7, C24, and C18 clones was digested with HindIII and separated on a PFG.
(B) DNA from the A4, C7, C18, C24, and R29 clones was digested with Dral and separated by standard electrophoresis. The probes used were A4var, C24var, or C18var as indicated. Major bands of hybridization are indicated with an arrow. The R29 clone has deleted the major A4var band of hybridization. Minor bands that are deleted are indicated by an asterisk.
The restriction enzyme Dral (Figure 6B), which cuts malarial DNA frequently and produces small restriction fragments, was also used to identify deletions and rearrangements within the var gene family. Consistent with the absence of Dral sites in probes for A4var and C18var, these probes hybridized to a single major band of Dral-digested DNA. In the same analysis, the probe for C24var, which contains an internal Dral site, hybridized to two major bands of Dral-digested DNA. Except for the absence of the single major band identified by the A4var probe in DNA from R29, the probes did not identify any differences in the major bands of hybridization. Another frequently cutting restriction enzyme, Hinfl, gave similar results (data not shown). Thus, DNA rearrangements associated with antigenic switching were not detected.
Deletion of minor bands was detected in the Southern blot analysis with DNA restricted with HindIII as well as Dral. For instance, each var probe detected an additional minor band in HindIII-restricted DNA from C18 that was absent in the C7 and C24 clones (Figure 6A). Moreover, minor bands that were deleted in Dral-digested DNA from the C7 and C24 clones were present in A4, C18, and R29 clones. It is assumed that the minor bands represent var sequences that cross-react with the var probes, but this was not proven by cloning and sequence analysis. Deletion of minor bands reflected changes in the P. falciparum genome during culture, but none of the changes observed correlated with switches in the antigenic phenotype of the clones.
The A4var, C18var, C24var, and R29var genes were mapped to P. falciparum chromosomes. DNA from all the family members was separated by PFG electrophoresis under standard conditions and hybridized to probes for the cloned var genes. A4var, C24var, and R29var mapped to chromosome 12, and C18var mapped to chromosome 1. No differences in chromosomal localization were found between expressed and silent genes (data not shown).
Discussion
The absence of sequence information for the genes coding for the variant antigens and cytoadherence proteins of P. falciparum has been a major barrier to research on this organism, since these proteins are intimately involved in pathogenesis. We argue in this and the accompanying papers (Su et al., 1995; Baruch et al., 1995 [both in this issue of Cell]) that this barrier has now been overcome. Su et al. (1995) describe a large family of genes (var genes) that are polymorphic between genotypes, code for proteins of an appropriate molecular mass for the variant antigens in the erythrocyte surface, and contain two to four domains homologous with erythrocyte binding domains in P. vivax, P. knowlesi, and P. falciparum (Chitnis and Miller, 1994; Sim et al., 1994). The finding of potential host binding domains on the variant antigen is consistent with experimental evidence that places the phenotypes of cytoadherence and antigenic variation on a single molecule (Udeinya et al., 1983; Leech et al., 1984; Magowan et al., 1988; Roberts et al., 1992; van Schravendijk et al., 1993). We present evidence that expression of variant antigens on the erythrocyte surface and ICAM-1 binding correlate with expression of members of the var gene family.
We utilized the small regions of conservation in DBL domains to amplify sequences from RNAs by RT–PCR (Peterson et al., in press). We were encouraged to find that parasites of a similar antigenic type (in this case A4) shared a major RT–PCR band that was absent from non-cross-reactive clones. The RT–PCR band was cloned, and sequencing revealed that it contained sequences that were characteristic of the DBL domains found in var genes. RT–PCR using specific primers to the cloned A4var gene on RNA from an extensive panel of clones showed a correlation between the level of A4var expression and antigenic similarity to A4.
Interpretation of these data required that RNA was employed from clonally derived parasite populations of known variant antigen type. For antigenic typing, we were forced to rely primarily on the mixed agglutination assay (Newbold et al., 1992); which measures the frequency with which a polyspecific malarial immune serum can agglutinate infected erythrocytes from two different sources. Since we had already shown that the formation of mixed agglutinates occurred with the expected binomial distribution, we could use the results as a semiquantitative measure of the presence of common variant phenotypes within any of the parasites from the clone family (see Experimental Procedures).
The interpretation of the relationship between antigenic similarity as measured by agglutination and expression of the variant antigen is not straightforward. The actual percentage of parasites expressing the same antigenic type in two clonal cultures that are indistinguishable by cross-agglutination (>95% mixed agglutinates) cannot be determined directly by this method, since it can vary in different clonal cultures because of stochastic differences in antigenic variation postcloning. For instance, A4 and the clones C7, C9, and C17, which are indistinguishable from A4 by the mixed agglutination assay, contain differing proportions of parasites expressing the A4 variant. We were fortunate in the case of the A4 variant type to be able to circumvent this problem with the use of MAb BC6. This antibody showed that the A4-type parasites contained between 14% and 33% MAb BC6–positive infected erythrocytes. When we assessed A4var mRNA levels and BC6 positivity of different clones in the same cell cycle, we obtained a significant correlation.
The relation between antigenic variation and the expression of var genes is strengthened by the identification of distinct var cDNAs from three antigenically variant clones, C24, C18, and R29. For C24 and R29, specific var transcripts were virtually unique to the homologous clone. For C18, the specific var transcript was expressed at the highest level in the clone from which it was derived. Thus, our evidence indicates that distinct var transcripts are expressed by clones that display different antigenic types. This indicates that the var genes encode the variant surface antigens of P. falciparum and are the basis of antigenic variation.
The paper of Baruch et al. (1995) provides direct evidence that var genes encode parasite proteins expressed on the erythrocyte surface. These authors cloned a gene from the Malayan Camp strain of parasites by screening an expression library with antibody to the infected erythrocyte surface. They showed that this gene is a member of the var gene family and that antisera raised to the cloned product reacted with the surface of infected erythrocytes in a strain-specific pattern and immunoprecipitated a molecule with the characteristics of PfEMP1 Furthermore, the antisera reacted over the knobs, the points of contact with endothelium (Luse and Miller, 1971).
Are the DBL domains of var genes involved in binding to endothelium? Evidence from the role of DBL domains in other members of the family shows that they are capable of binding different receptors. The DBL domain in the erythrocyte binding protein of P. vivax binds the Duffy antigen on human erythrocytes (Chitnis and Miller, 1994) but the DBL domain from EBA-175 of P. falciparum binds to glycophorin A (Sim et al., 1994). Three further erythrocyte binding proteins have been found in P. knowlesi that contain DBL domains (Adams et al., 1990; Adams et al., 1992). One of these also binds the Duffy antigen, but the other two interact with undefined structures on rhesus monkey erythrocytes (Chitnis and Miller, 1994). It is therefore not unreasonable to expect that the DBL domains of the var gene products have the potential to mediate binding to different endothelial receptors: CD36, ICAM-1, thrombospondin, VCAM, and E-selectin. Because the var genes contain between two and four copies of the DBL motif (Su et al., 1995), a role for them in endothelial binding could explain some puzzling results in the literature. In some cases, selection of parasites for binding to one receptor decreases binding to others (Ockenhouse et al., 1991). In other cases, selection on one receptor results in an increased binding to both this and alternative receptors (Ockenhouse et al., 1992). The variable number of DBL domains in a given var would mean that both of these results are consistent with a single molecule mediating adherence. The inhibition of CD36 binding by anti-PfEMP1 sera (Baruch et al., 1995) and the association of the A4 antigenic type with ICAM-1 binding suggest that var genes encode cytoadherent ligands; however, formal proof will require direct measurement of the receptor binding function of these sequences.
How is the expression of this large repertoire of genes controlled? Data from Southern blots of variant types probed with A4var, C18var, and C24var would appear to rule out large-scale genomic rearrangement as a frequent or necessary event for antigenic switching. We also did not find evidence for duplicate copies of expressed genes, since the intensity of hybridization with var-specific probes was identical in all clones. Deletions in var genes were found, but these did not correlate with changes in antigenic phenotypes. We do provide evidence that multiple var expression sites exist, since the C24 and C18 subclones of A4 that have switched antigenic type express var transcripts from different chromosomes. Moreover, Southern blot analysis using a probe from the conserved var exon 2 suggested that var genes may be present on most or all P. falciparum chromosomes (Su et al., 1995). It is unclear at present whether all var genes can potentially be expressed in situ or whether only a subset is capable of being transcriptionally activated.
Despite the overall high frequency of switching in P. falciparum, a wide distribution of switch rates is necessary in order for the variant repertoire not to be rapidly exhausted during acute infection (Roberts et al., 1993). Indeed, wide variation of switch rates has been measured directly for P. chabaudi, African trypanosomes, and Candida (Slutsky et al., 1985; Turner and Barry, 1989; Brannan et al., 1994). The fact that the A4var and C18var transcripts are frequently present at low levels in clonel cultures that have switched antigenic type may reflect a higher rate of switching to these antigenic phenotypes. In contrast, C24var and R29var transcripts are rarely found in other clones and may reflect low frequency of switching to these antigenic phenotypes. It will be interesting to determine how the var genes are arranged and inherited within natural populations of interbreeding organisms. The fact that these genes are distributed throughout the genome would suggest that the repertoire would be continuously reshuffled by reassortment and recombination in the mosquito even at fairly low rates of outcrossing. If, however, the immune response to the variant antigen is an important determinant of parasite survival and, hence, gametocyte production in vivo, then there may be significant selective pressure to stratify the variant repertoire into groups of minimal overlap. Such a situation would be consistent with recent theoretical predictions (Gupta and Day, 1994).
With the identification of the genes encoding the variant antigens, a wide range of questions regarding the mechanism of antigenic switching, the relationship between antigenic and adhesive phenotypes, the role of the variant repertoire in evading the immune response as well as inducing protection, and the population genetics of the var locus can now be addressed.
Experimental Procedures
Growth of Parasites and Antigenic Phenotyping
Parasites were grown in vitro as previously described (Roberts et al., 1992). The antigenic phenotype of the cultures was determined by the mixed agglutination assay (Newbold et al., 1992). The chance of heterologous agglutination, q, of cells from two cultures can be estimated from the observed proportion of mixed-colored agglutinates, m, by the relationship q = 1 – (1 – m)1/(n − 1) for agglutinates of size n(Roberts et al., 1992). Since most agglutinating antibodies in immune serum are variant specific (Newbold et al., 1992), the chance of heterologous agglutination between two clones expressing different major variant types is related to the proportion of minor variants derived from switching in each of the populations. For example, if clone A expresses antigenic types a, b, c… in proportions p(a), p(b), p(c) where p(a) >> p(b) or p(c) (implied by the order in which antigenic switches have occurred), and similarly, clone B expresses antigenic types b, a, c in proportions p′(b), p′(a), p′(c) where p′(b)>>p′(a) or p′(c), then the chance of heterologous agglutination q is given by the following:
| (1) |
i.e., q ≈ k1(proportion of major variant a in clone A) (proportion of minor variant a in clone B) + k2(proportion of major variant b in clone B) (proportion of minor variant b in clone A).
Assuming the amount of RT–PCR is proportional to the level of variant antigen expression, the chance of heterologous agglutination q (as calculated from the results of the mixed agglutination reaction) for parasitized erythrocytes from two cultures will be proportional to the amount of RT–PCR product in each clone, provided that the second term in equation 1 is negligible. We would thus predict an approximately linear relationship between the chance of heterologous agglutination and the amount of any one RT–PCR product, but occasional combinations of clones would deviate significantly from linearity as a result of their minor variant composition.
Monoclonal Antibody Production and Flow Cytometry
Hybridomas were produced from BALB/c × CBAF1 hybrid mice immunized repeatedly with 20 μg of Triton X-100-insoluble extract from 109 parasitized erythrocytes infected with the A4 clone by standard techniques (Kohler and Milstein, 1975; Orlik and Altener, 1988). Screening was carried out by indirect immunoflorescence microscopy (Roberts et al., 1992) and flow cytometry (see below). The antibody BC6 was isolated from a single well, recloned, and isotyped as lgG1 by hemagglutination (Serotec, Bicester, England). It immunoprecipitated a high molecular mass protein with the characteristics of PfEMP1 (Leech et al., 1984) only from clones of the A4 antigenic type (Roberts et al., unpublished data).
Mature infected trophozoites were washed three times with PBS, 1% BSA (wash buffer) and incubated with tissue culture supernatant, MAb BC6, or a mouse lgG1 control antibody (0.5 μg per ml in wash buffer) at 10% hematocrit for 30 min at 37°C. Bound immunoglobulins were detected by incubating cells with FITC-conjugated rabbit anti-mouse immunoglobulins (DAKO, High Wycombe, England) and then with FITC-conjugated swine anti-rabbit immunoglobulins (DAKO, High Wycombe, England) with 200 μg/ml ethidium bromide. Cells were washed three times with buffer between each incubation and four times after the final incubation. The number of FITC- and ethidium-stained cells as a proportion of the ethidium-stained cells was analyzed by flow cytometry (EPICS Profile II. Coulter, Luton, England). Results are given as the mean of two measurements on two successive cycles counting 2000 infected cells on each occasion.
Preparation of RNA for Isolation of var Transcripts and for Analysis of var Expression In Different Clones
To control for possible stage-specific differences in var message expression, synchronized parasite cultures were enriched for trophozoite- and schizont-infected erythrocytes by plasmagel flotation (Pasvol et al., 1978). RNA was isolated by RNAzol (Tel-Test, Incorporated, Friendswood, TX). The RNAs from each of these clonal preparations were quantified by spectrophotometry, and the quality and quantity of RNA was confirmed in glyoxal gels.
Cloning and Sequencing of Unique var Genes Expressed by A4, C18, C24, and R29
To identify var messages whose expression correlated with variant-specific agglutination of infected erythrocytes, RNA from the panel of clones was studied by RT–PCR using a range of different degenerate oligonucleotide primers designed to conserved sequences in DBL domains. Of these primers, only the primers shown below amplified products that differed among clones, and only these primers were used to clone var sequences. The sequences of the other primers are available from the authors upon request. The sequences of the primers used in the present report were as follows, in which inosine is represented with the letter I. Sense strand: UNIEBP5′, 5′-CC(A/G)AG(A/G)AG (A/G)CAA(G/A)AA(C/T)TATG; primer 1,5′-GC(T/C/A)TG(T/C)GCICCIT (T/A)(C/T)(C/A)G. Antisense strand: UNIEBP3′, 5′-CCA(A/T)C(T/G) (T/G)A(A/G)(AlG)(A/G)AATTG(A/T)GG.
In all RT–PCR experiments, 10 μg of total RNA was pretreated with RQ1 DNase (Promega, Madison, WI). Half of the sample was reverse transcribed with random primers by use of Superscript (Bethesda Research Laboratories, Gaithersburg, MD), according to the protocol of Wynn et al. (1993). The remaining half was used in control experiments to confirm that the DNase treatment was complete. Reverse-transcribed material was resuspended in 100 μl and 1 μl used for PCR reactions. In ethidium bromide-stained gels, the amount of cDNA from different clones was similar, and no adjustments were made in the amount of cDNA added to PCR reactions. PCR conditions with each primer set were identical, except that 1.5 mM Mg2+ was used with the UNIEBP5′ and UNIEBP3′ combination, and 3.5 mM Mg2+ was used with other primer combinations. Taq polymerase (Boehringer, Mannheim, Federal Republic of Germany) was used for all PCR reactions. cDNA was denatured at 94°C for 1 min and then amplified for 30 cycles at 94°C for 25 s, 48°C for 25 s. and 72°C for 1 min and 30 s. PCR products were separated on a PCR Purity Plus polyacrylamide gel solution (AT Biochem, Malvern, PA). Bands that differed among clones were cloned into the TA cloning vector pCRll (Invitrogen, San Diego, CA) and sequenced with Sequenase version 2.0 (United States Biochemical Corporation, Cleveland, OH). A4var was cloned by using a similar protocol from C17, C9, C7, and C281 with A4var-specific primers (sense strand: A4REG5, 5′-AGTGAAAAAGCAACAGAAACTGAATTG; antisense strand: A4B, 5′-GGACGAAATTTTGGTGTATTTATTGTT). These primers amplify a 480 bp fragment of A4var.
Comparison of var Expression In Different Clones by RT–PCR
Specific primers were designed to the four cloned var sequences and used to study var expression in different clones by RT–PCR. The first primer in each set corresponds to the sense orientation and the second primer to the antisense orientation. A4var. A45B, 5′-AGAAAAGCTTTTATTGAATGTGC; A43C, 5′-TCCATTCTTAATATGATGAGATAAT. C24var. C24A, 5′-GAGGTAATAATGGTGGAGCCTC; C24B, 5′-AGTGCACGTATTTCCATTGGAG. C18var: C185C, 5′-TCCTTCAGACCATCACAA; C183C, 5′-ATATTGAGCAGTACCAGG. R29var. R295B, 5′-GTTAATAATCACCCAGATAAA; R293C, 5′-TTCACTATGCCCACAATATCC.
To determine conditions in which the PCR reaction was within the exponential phase, a PCR cycling optimization of 18–36 cycles was first performed on cDNA from which the var was cloned. On the basis of the intensity of ethidium bromide–stained agarose gels, PCR conditions were selected to compare var expression in all of the clones. The same preparation of cDNAs was used in all var gene expression studies. PCR conditions were similar to those with degenerate primers, except that different numbers of PCR cycles were used for each primer set. To quantitate the amount of var product in different samples, the PCR products were separated on PCR Purity polyacrylamide gels (AT Biochem, Malvern, PA) and transferred to Zeta-Probe GT blotting membranes (Bio-Rad, Hercules, CA). For hybridization, cloned var fragments were radioactively labeled by the random primer method (Boehringer, Mannheim, Federal Republic of Germany) with [α-32P]dCTP (Amersham, Arlington Heights, IL). Prehybridization and hybridization were performed in a solution of 0.25 M Na2HPO4, and 5% SDS. Probes were hybridized overnight at 50°C. The following day, membranes were washed two times for 1 hr at 50°C with 50 ml of washing solution (20 mM Na2HPO4, and 5% SDS). Washed membranes were exposed to Phosphorlmager screens (Molecular Dynamics, Sunnyvale, CA), and bands were quantified by densitometry using Image Quant (Molecular Dynamics).
Isolation of A4var cDNA
To clone the A4var cDNA, specific primers were designed based upon the sequence of A4var cloned from genomic DNA by Su et al. (1995). For this experiment, 5 μg of RNA was DNase-treated and reversed transcribed with XCR1 (antisense, 5′-GGTATATCATAA(A/T)CACTTTTGG) from an almost perfectly conserved region of exon 2 in different var sequences (Su et al., 1995). The first-strand cDNA was first amplified with A4REG5 primer and the XCR1 primer for 30 cycles (94°C for 25 s, 48°C for 25 s, and 72°C for 3 min). The PCR product from this reaction was diluted 50-fold and further amplified 30 cycles with the primers A45B (5′-AGAAAAGCTTTTATTGAATGTGC) and Gfh2 (5′-GGATATTAATAACACTGAAAAGGT) from regions adjacent to the original primers. The PCR product was cloned by using the TA vector (Invitrogen, San Diego, CA) and sequenced for several hundred base pairs in either direction to confirm its identity and the exon-intron splice boundary.
Southern Blot Analysis of Expressed var Sequences
DNA from the C7, A4, C24, C18, and R29 clones was extracted from plasmagel-enriched cultures as previously described (Robson and Jennings, 1991). DNA was digested with HindIII and separated on a PFG (1% agarose, 0.5× TBE, 180 V; switching time, 2–8 s; running time, 18 hr). Alternatively, DNA was digested with Hinfl or Dral and separated by standard electrophoresis. DNA was transferred to Hybond N+ (Amersham, Little Chalfont, England) according to the instructions of the manufacturer. Probes were labeled with (32P)dCTP (Amersham, Little Chalfont, England) by use of rediprime kits (Amersham, Little Chalfont, England). Hybridization was carried out at 60°C in 0.5 M sodium phosphate (pH 7.2). 7% SDS (Church and Gilbert, 1984) overnight followed by four washes in 40 mM sodium phosphate (pH 7.2) 1% SDS at 60°C.
Assignment of var Genes to P. falciparum Chromosomes by PFG Electrophoresis
DNA from all of the clones was separated by PFG electrophoresis in 1% rapid agarose (GIBCO), 0.5× TBE, 1–5 V; switching time, 200–400 s for 60 hr, and then 600–1000 s for 36 hr. The order of the chromosomes in the IT lineage (compared with the 3D7 reference clone) was determined by using a series of chromosome-specific probes. The probe for chromosome 12 was from the ABRA gene (Foote and Kemp, 1989) and that for chromosome 1 was from the RESA gene (Cappai et al., 1989).
Acknowledgments
Correspondence should be addressed to C. I. N. and L. H. M. We thank members of the laboratories of Thomas Wellems and Russell Howard for sharing results and for discussion of data prior to publication; Dr. David Alling and Ms. Claire Hallahan for statistical analysis of RT–PCR results and for graphing RT–PCR and agglutination results; and Mr. Peter Warn for growth of P. falciparum isolates and extraction of RNA. C. I. N., A. G. C., and D. J. R. gratefully acknowledge financial support from The Wellcome Trust.
Footnotes
References
- Adams JA, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, Miller LH. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- Adams JH, Sim BKL, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell JW, Asch AS, Nachman RL, Yamaya M, Aikawa M, Ingravallo P. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J. Clin. Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Xiahui B, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the Plasmodium falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82(this issue) doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Berendt AR, Simmons DL, Tansey J, Newbold CI, Marsh K. Intercellular adhesion molecule-1 is an endothelial adhesion receptor for Plasmodium falciparum. Nature. 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- Berendt AR, Ferguson DJ, Gardner J, Turner G, Rowe A, McCormick C. Molecular mechanisms of sequestration in malaria. Parasitology. 1994;108:S19–S28. doi: 10.1017/s0031182000075685. [DOI] [PubMed] [Google Scholar]
- Biggs B-A, Anders RF, Dillon HE, Davern KM, Martin M, Petersen C, Brown GV. Adherence of infected erythrocytes to venular endothelium selects for antigenic variants of Plasmodium falciparum. J. Immunol. 1992;149:2047–2054. [PubMed] [Google Scholar]
- Borst P. Molecular genetics of antigenic variation. Immunoparasitol. Today. 1991;12:A29–A33. doi: 10.1016/S0167-5699(05)80009-X. [DOI] [PubMed] [Google Scholar]
- Brannan LR, Turner CMR, Phillips RS. Malaria parasites undergo antigenic variation at high rates in vivo. Proc. R. Soc. Lond. (B) 1994;256:71–75. doi: 10.1098/rspb.1994.0051. [DOI] [PubMed] [Google Scholar]
- Brown K, Brown IN. Immunity to malaria: antigenic variation in chronic infections of Plasmodium knowlesi. Nature. 1965;208:1286–1288. doi: 10.1038/2081286a0. [DOI] [PubMed] [Google Scholar]
- Brown K, Brown IN, Trigg PI, Phillips RS, Hills LA. Immunity to malaria. II. Serological response of monkeys sensitised by drug-suppressed infected or parasitised cell in Freund’s complete adjuvant. Exp. Parasitol. 1970;28:318–338. doi: 10.1016/0014-4894(70)90102-5. [DOI] [PubMed] [Google Scholar]
- Brown KN. Protective immunity to malaria provides a model for the survival of cells in an immunologically hostile environment. Nature. 1971;230:163–167. [Google Scholar]
- Cappai R, van Schravendijk M, Anders RF, Peterson MG, Thomas LM, Cowan AF, Kemp DJ. Expression of the RESA gene in Plasmodium falciparum isolate FCR3 is prevented by a subtelomeric deletion. Mol. Cell. Biol. 1989;9:3584–3597. doi: 10.1128/mcb.9.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc. Natl. Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote SJ, Kemp DJ. Chromosomes of malaria parasites. Trends Genet. 1989;5:337–342. doi: 10.1016/0168-9525(89)90139-x. [DOI] [PubMed] [Google Scholar]
- Gilks CF, Walliker D, Newbold CI. Relationships between sequestration, antigenic variation and chronic parasitism in Plasmodium chabaudi chabaudi: a rodent malaria model. Parasite Immunol. 1990;12:45–64. doi: 10.1111/j.1365-3024.1990.tb00935.x. [DOI] [PubMed] [Google Scholar]
- Gupta S, Day K. A strain theory of malaria transmission. Parasitol. Today. 1994;12:476–481. doi: 10.1016/0169-4758(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Howard RJ, Barnwell JW, Kao V. Antigenic variation Of Plasmodium knowlesi malaria: identification of the variant antigen on infected erythrocytes. Proc. Natl. Acad. Sci. USA. 1983;80:4129–4133. doi: 10.1073/pnas.80.13.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard RJ. Malarial proteins at the membrane of Plasmodium falciparum-infected erythrocytes and their involvement in cytoadherence to endothelial cells. Prog. Allergy. 1988;41:98–147. doi: 10.1159/000415221. [DOI] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous culture of fused cells secreting antibodies of predetermined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Langreth SG, Peterson E. Pathogenicity, stability, and immunogenicity of a knobless clone of Plasmodium falciparum in Colombian monkeys. Infect. Immun. 1985;47:760–766. doi: 10.1128/iai.47.3.760-766.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech JH, Barnwell JW, Miller LH, Howard RJ. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J. Exp. Med. 1984;159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse SA, Miller LH. Plasmodium falciparum malaria ultrastructure of parasitized erythrocytes in cardiac vessels. Am. J. Trop. Med. Hyg. 1971;20:655–660. [PubMed] [Google Scholar]
- Magowan C, Wollish W, Anderson L, Leech J. Cytoadherence by Plasmodium falciparum-infected erythrocytes is correlated with the expression of a family of variable proteins on infected erythrocytes. J. Exp. Med. 1988;168:1307–1320. doi: 10.1084/jem.168.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh K, Otoo L, Hayes RJ, Carson DC, Greenwood BM. Antibodies to blood stage antigens of Plasmodium falciparum in rural Gambians and their relation to protection against infection. Trans. R. Soc. Trop. Med. Hyg. 1989;83:293–303. doi: 10.1016/0035-9203(89)90478-1. [DOI] [PubMed] [Google Scholar]
- McLean SA, Pearson CD, Phillips RS. Plasmodium chabaudi: antigenic variation during recrudescent parasitaemias in mice. Exp. Parasitol. 1982;54:296–302. doi: 10.1016/0014-4894(82)90038-8. [DOI] [PubMed] [Google Scholar]
- Miller LH, Good MF, Milon G. Malaria pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- Nash T. Surface antigen variability and variation in Giardia lamblia. Parasitol. Today. 1992;8:229–234. doi: 10.1016/0169-4758(92)90119-m. [DOI] [PubMed] [Google Scholar]
- Newbold CI, Pinches R, Roberts DJ, Marsh K. Plasmodium falciparum: the human agglutinating antibody response to the infected red cell surface is predominantly variant specific. Exp. Parasitol. 1992;75:261–292. doi: 10.1016/0014-4894(92)90213-t. [DOI] [PubMed] [Google Scholar]
- Ockenhouse CF, Ho M, Tandon NN, van Seventer GA, Shaw S, White NJ, Jamieson GA, Chulay JD, Webster HK. Molecular basis of sequestration in severe and uncomplicated Plasmodium falciparum malaria: differential adhesion of infected erythrocytes to CD36 and ICAM-1. J. Infect. Dis. 1991;164:163–169. doi: 10.1093/infdis/164.1.163. [DOI] [PubMed] [Google Scholar]
- Ockenhouse CF, Tegoshi T, Maeno Y, Benjamin C, Ho M, Kan KE, Thway Y, Win K, Aikawa M, Lobb RR. Human vascular endothelial cell adhesion receptors for Plasmodium falciparum-infected erythrocytes: roles for endothelial leukocyte adhesion molecule 1 and vascular cell adhesion molecule 1. J. Exp. Med. 1992;176:1183–1189. doi: 10.1084/jem.176.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlik O, Altener C. Modifications of hybridoma technology which improve the yield of monoclonal antibody producing cells. J. Immunol. Meth. 1988;115:55–59. doi: 10.1016/0022-1759(88)90309-2. [DOI] [PubMed] [Google Scholar]
- Pasvol G, Wilson RJM, Smalley ME, Brown J. Separation of viable schizont-infected cells of Plasmodium from human blood. Ann. Trop. Med. Parasitol. 1978;72:87–88. doi: 10.1080/00034983.1978.11719283. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Miller LH, Wellems TE. Multiple sequences in the Plasmodium falciparum genome encode conserved domains homologous to those in erythrocyte-binding proteins. Proc. Natl. Acad. Sci. 1995;92 doi: 10.1073/pnas.92.15.7100. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preer JR., Jr. Surface Antigens of Paramecium. Academic Press; New York: 1986. [Google Scholar]
- Roberts DD, Sherwood JA, Spitalnik SL, Panton LJ, Howard RJ, Dixit VM, Frazier WA, Miller LH, Ginsburg V. Thrombospondin binds falciparum malaria parasitized erythrocytes and may mediate cytoadherence. Nature. 1985;318:64–66. doi: 10.1038/318064a0. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI. Rapid switching to multiple antigenie and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DJ, Biggs BA, Brown G, Newbold CI. Protection, pathogenesis and phenotypic plasticity in Plasmodium falciparum malaria. Parasitol. Today. 1993;9:281–286. doi: 10.1016/0169-4758(93)90121-u. [DOI] [PubMed] [Google Scholar]
- Robson KJH, Jennings MW. The structure of the calmodulin gene from Plasmodium falciparum. Mol. Biochem. Parasitol. 1991;46:19–34. doi: 10.1016/0166-6851(91)90195-c. [DOI] [PubMed] [Google Scholar]
- Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- Slutsky B, Buffo J, Soll D. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- Su XZ, Heatwole V, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum–infected erythrocytes. Cell. 1995;82(this issue) doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Turner CMR, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infection. Parasitology. 1989;99:67–75. doi: 10.1017/s0031182000061035. [DOI] [PubMed] [Google Scholar]
- Turner GDG, Morrison H, Jones M, Davis TME, Looareesuwan S, Buley ID, Gatter KC, Newbold CI, Pukritayakamee S, Nagachinta B, White NJ, Berendt AR. An immunohistochemical study of the pathology of fatal malaria: evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- Udeinya IJ, Miller LH, McGregor IA, Jensen JB. Plasmodium falciparum strain-specific antibody blocks binding of infected erythrocytes to amelanotic melanoma cells. Nature. 1983;303:429–431. doi: 10.1038/303429a0. [DOI] [PubMed] [Google Scholar]
- van Schravendijk MR, Pasloske BL, Baruch DI, Hendunnetti SM, Howard RJ. Immunochemical characterization and differentiation of two ~ 300-kD erythrocyte membrane–associated proteins of Plasmodium falciparum, PfEMP1 and PfEMP3. Am. J. Trop. Med. Hyg. 1993;49:552–565. doi: 10.4269/ajtmh.1993.49.552. [DOI] [PubMed] [Google Scholar]
- Warrell DA, Molyneux ME, Beales PF. Severe and complicated malaria. Trans. R. Soc. Trop. Med. Hyg. 1990;84(Suppl. 2):1–65. [PubMed] [Google Scholar]
- Warrell DA. Clinical Features of Malaria. Edward Arnold; Boston: 1993. [Google Scholar]
- Wynn TA, Eltoum I, Cheever AW, Lewis FA, Gause WC, Sher A. Analysis of cytokine mRNA expression during primary granuloma formation induced by eggs of Schistosoma mansoni. J. Immunol. 1993;151:1430–1440. [PubMed] [Google Scholar]