Summary
Signal transducer and activator of transcription 5 (STAT5) is crucial for physiological processes that include hematopoiesis, liver metabolism and mammary gland development. However, aberrant continual activity of STAT5 has been causally linked to human leukemias and solid tumor formation. As a regulated transcription factor, precise cellular localization of STAT5 is essential. Conventional nuclear localization signals consist of short stretches of basic amino acids. In this study, we provide evidence that STAT5 nuclear import is dependent on an unconventional nuclear localization signal that functions within the conformation of an extensive coiled-coil domain. Both in vitro binding and in vivo functional assays reveal that STAT5 nuclear import is mediated by the importin-α3/β1 system independently of STAT5 activation by tyrosine phosphorylation. The integrity of the coiled-coil domain is essential for STAT5 transcriptional induction of the β-casein gene following prolactin stimulation as well as its ability to synergize with the glucocorticoid receptor. The glucocorticoid receptor accumulates in the nucleus in response to prolactin and this nuclear import is dependent on STAT5 nuclear import. STAT5 continually shuttles in and out of the nucleus and live cell imaging demonstrates that STAT5 nuclear export is mediated by both chromosome region maintenance 1 (Crm1)-dependent and Crm1-independent pathways. A Crm1-dependent nuclear export signal was identified within the STAT5 N-terminus. These findings provide insight into the fundamental mechanisms that regulate STAT5 nuclear trafficking and cooperation with the glucocorticoid receptor and provide a basis for clinical intervention of STAT5 function in disease.
Key words: STAT5, Nuclear import, Nuclear export
Introduction
The rapid response of cells to external stimuli depends on molecules that can quickly transmit signals from the plasma membrane into the nucleus. The family of signal transducers and activators of transcription (STATs) exemplifies signaling molecules that can sense changes at the plasma membrane and subsequently redirect gene expression (Darnell et al., 1994; Levy and Darnell, 2002; Schindler et al., 2007). STATs are activated by tyrosine phosphorylation in response to Janus kinases (JAKs) associated with cytokine receptors, and this phosphorylation promotes a dimer conformation with the ability to bind specific DNA targets. Proper cellular localization of STATs is thereby critical for their normal biological functions.
Two of the STAT genes, STAT5a and STAT5b, encode proteins that are more than 95% identical, and murine gene knockout studies indicate they have both distinct and redundant functions (Liu et al., 1995; Liu et al., 1997; Moriggl et al., 1999; Nevalainen et al., 2000; Udy et al., 1997; Yao et al., 2006). A double knockout of both STAT5 genes is perinatal lethal, however individual gene knockouts reveal their preferential role in response to cytokines such as prolactin, interleukin-2, erythropoietin, and growth hormone, and consequent effects on biological processes such as mammary gland development, blood cell differentiation, and liver metabolism. Loss of function studies confirm the requirement of STAT5 in normal development, but in contrast, aberrant persistent activation of STAT5 can promote oncogenesis (Cotarla et al., 2004; Hantschel et al., 2012; Hayakawa et al., 1998; Schwaller et al., 2000). Studies in human and murine systems indicate a causal link between tyrosine phosphorylated STAT5 and development of leukemias and mammary tumors. Understanding the mechanisms that regulate STAT5 nuclear trafficking and its consequent impact on gene expression can provide a basis to develop methods to target STAT5 activity in human disease.
The transport of proteins between nucleus and cytoplasm is a tightly regulated process. Movement of molecules into the nucleus is gated through nuclear pore complexes (NPC) that allow passive diffusion of small molecules, but restrict passage of large molecules to those that possess a nuclear localization signal (NLS) (Chook and Blobel, 2001; Cokol et al., 2000; Görlich and Kutay, 1999; Macara, 2001; Mattaj and Englmeier, 1998; Pemberton and Paschal, 2005; Rout and Aitchison, 2001). The best defined NLSs consist of a monopartite or bipartite stretch of basic amino acids, particularly lysine or arginine. In classical transport, the NLS-containing cargo is carried through the NPC by a dimeric importin-α/importin-β1 transporter complex. The NLS is recognized directly by one of six characterized importin-α adapter molecules bound to the importin-β1 protein (Köhler et al., 1999). Importin-β1 facilitates transport through the NPC. In the nucleus, importin-β1 binds to Ran-GTP and releases importin-α and cargo. In this report we identify an unconventional NLS of STAT5 that is constitutively active, independent of tyrosine phosphorylation. The NLS function is required for gene induction by STAT5 and for its ability to cooperate with the glucocorticoid receptor.
Nuclear export of macromolecules is also mediated by transport carriers that recognize a nuclear export signal (NES) in cargo and facilitate transport through the NPC. The best characterized NES consensus comprises a leucine-rich hydrophobic sequence that can be recognized by the exportin chromosome region maintenance 1 (Crm1) in complex with Ran-GTP (Fornerod et al., 1997; Ullman et al., 1997; Wen et al., 1995). Following transport to the cytoplasm, nucleotide exchange of Ran-GTP to Ran-GDP leads to the dissociation of the exportin-cargo complex. A specific inhibitor of Crm1, leptomycin B (LMB), has been a useful tool to determine the role of Crm1 in export of various proteins (Kudo et al., 1998). Recent studies have also identified additional exportins, including exportin-4, 6, 7, importin-13, and calreticulin (Güttler and Görlich, 2011; Pemberton and Paschal, 2005). In this study we demonstrate that continuous nuclear export of STAT5 is mediated by both Crm1-dependent and Crm1-independent mechanisms.
Results
The coiled-coil domain of STAT5a functions as an unconventional NLS
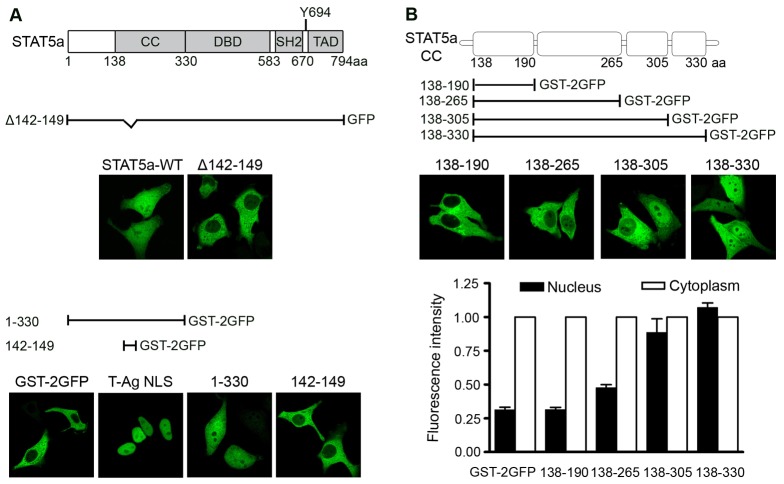
Fluorescence imaging of STAT5a tagged with monomeric EGFP (STAT5a-GFP) was used to investigate the functional NLS in STAT5. Our previous study indicated the constitutive nuclear presence of STAT5a-GFP was independent of tyrosine phosphorylation (Fig. 1A, top) (Iyer and Reich, 2008). In addition, a small deletion in the coiled-coil domain of STAT5a (142–149 a.a.) was found to abrogate the NLS function.
Fig. 1.
STAT5a coiled-coil domain and nuclear import. (A) Top: Linear diagram of STAT5a functional domains including the coiled-coil domain (CC), DNA-binding domain (DBD), Src homology 2 (SH2) domain, transcriptional activation domain (TAD) and specific phosphorylated tyrosine residue 694 (Y694) (Tan and Nevalainen, 2008). Beneath is a linear depiction of STAT5a 142–149 a.a. deletion construct linked to GFP. Images show cellular localization of STAT5a full-length (WT) and deletion construct, as visualized by fluorescence microscopy. Bottom: Linear depictions and fluorescence images of STAT5a fragments linked to GST-2GFP. Numbers correspond to STAT5a amino acids. Images of GST-2GFP and SV40 T antigen NLS linked to GST-2GFP (T-Ag NLS) are also shown. (B) Diagram of STAT5a coiled-coil domain with four α-helices and constructs used in analyses. Representative fluorescent images are shown. Bar graphs show statistical analyses of nuclear and cytoplasmic fluorescence intensity of STAT5a proteins expressed in cells. Fluorescence intensity was quantified in the nucleus and cytoplasm of 10 cells expressing individual constructs by LSM Image Browser program and analyzed statistically by two-tailed tests. Nuclear fluorescence (black bar) was normalized to the cytoplasmic fluorescence (white bar).
To determine if a peptide corresponding to 142–149 a.a. of STAT5a was sufficient to function as an NLS, this peptide was linked to GFP and evaluated for cellular localization. Since a peptide tagged with GFP is small and can passively diffuse into the nucleus, it cannot be used to analyze active transport. For this reason the peptide was linked to a larger protein encoding glutathione S-transferase (GST) and two tandem repeats of GFP (GST-2GFP). The GST-2GFP protein alone does not possess an NLS and does not enter the nucleus (Fig. 1A, bottom panel). However the monopartite NLS from the well characterized SV40 large T antigen is able to efficiently promote nuclear import of GST-2GFP, therefore the GST-2GFP protein can be used to study nuclear trafficking. The STAT5a N-terminus with the coiled-coil domain (1–330 a.a.) linked to GST-2GFP was found to promote nuclear import of the fusion protein, although not as prominently as the T antigen NLS. The STAT5a peptide encoding only 142–149 a.a. does not confer nuclear localization to GST-2GFP. The results suggest that a.a. 142–149 are needed for STAT5a nuclear import, but are not sufficient to function as an NLS.
Since 142–149 a.a. of STAT5a may be part of a larger domain that serves as an NLS, we evaluated the cellular localization of various fragments of the STAT5a coiled-coil domain tagged with GST-2GFP (Iyer and Reich, 2008; Zeng et al., 2002). The crystal structure of unphosphorylated STAT5a has been solved and indicates that the STAT5a coiled-coil domain consists of four α-helices (Neculai et al., 2005). Amino acids 142–149 of STAT5a are located in the first α-helix of the coiled-coil domain. Each α-helix defined by the crystal structure was tested for its contribution to nuclear import and representative images are shown in Fig. 1B. Fluorescence imaging demonstrates that the first α-helix (138–190 a.a.) containing a.a. 142–149 is not sufficient to direct nuclear import. A fragment containing both the first and second α-helices (138–265 a.a.) also is not sufficient for nuclear import; however a fragment containing the first, second and third α-helices (138–305 a.a.) shows nuclear presence in nearly 50% of the cells in culture. Maximal nuclear import in all of the cells is only achieved with all four α-helices of the coiled-coil domain of STAT5a (138–330 a.a.). Fluorescence intensities of nuclear and cytoplasmic signals were measured with the LSM Image Browser program and statistical analyses by two-tailed tests indicate the entire coiled-coil domain of STAT5a is required for the complete nuclear import function (Fig. 1B, bottom graph). The data suggest the STAT5a NLS does not conform to a conventional monopartite or bipartite basic NLS, but is a larger structural domain.
Identification of essential residues in the STAT5a NLS
To more closely investigate the nature of the STAT5a NLS, the sequence of the coiled-coil domain was evaluated for the presence of classical lysine or arginine-rich basic residues (Lange et al., 2007). Although amino acids 142–149 (LQINQTFE) in the first α-helix are needed for nuclear import, they do not include any basic residues. However, since the charged residue glutamic acid (E) is accessible on the surface of STAT5a, it may be available for recognition by importins and its contribution to nuclear import was evaluated. Mutation of glutamic acid 149 to alanine (E149A) was found to inhibit nuclear import mediated by the coiled-coil domain of STAT5a in more than 80% of the population (Fig. 2A). It therefore appears that E149 plays a major role in STAT5a import.
Fig. 2.
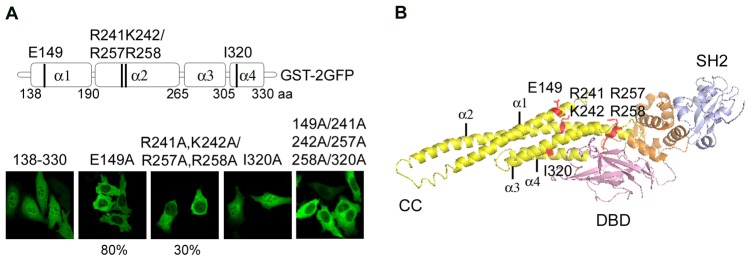
Identification of residues essential for the STAT5a NLS. (A) Location of alanine substitutions in the STAT5a coiled-coil domain. Below are fluorescence images of STAT5a coiled-coil domain tagged with GST-2GFP with the single or combined mutations noted above the image. Images represent the entire population of cells in culture, unless noted as 80% for E149A and 30% for 241A, 242A/257A and 258A. (B) Position of mutated residues in a ribbon diagram of the crystal structure of STAT5a coiled-coil domain (Protein Data Bank ID code 1Y1U). E149A in the first α-helix (α1), basic residues (R241A, K242A/R257A, R258A) in the second α-helix (α2) and I320A in the fourth α-helix (α4) are indicated.
A potential bipartite basic sequence was identified within the second α-helix (R241, K242, R257, R258), and its contribution to STAT5a nuclear import was evaluated by alanine substitutions. The R241A, K242A/R257A, R258A mutant disrupted nuclear import in ∼30% of the cells. It is not clear why the impairment was evident in a subpopulation of the culture, and may indicate this region plays an additive or auxiliary role. Site directed mutation of basic residues in the third and fourth α-helices did not identify an essential amino acid (data not shown), and it is possible that the third and fourth α-helices are needed to maintain conformation of the coiled-coil structure. Comparison of the fourth α-helix of STAT5a with STAT1 and STAT3 revealed a conserved isoleucine in the STAT5a and STAT3 proteins, but not in STAT1. Since nuclear import of STAT5a is independent of tyrosine phosphorylation similar to STAT3 (Liu et al., 2005), whereas STAT1 is dependent on tyrosine phosphorylation (McBride et al., 2002), we evaluated the effect of alanine substitution of this conserved isoleucine 320 (I320A). The I320A mutant alone had no apparent effect on nuclear import, however combined with the other mutations, nuclear import of the coiled-coil domain was completely inhibited in the total population.
The positions of the residues identified to be crucial for nuclear import in the STAT5a coiled-coil domain are indicated in the solved crystal structure of STAT5a (Neculai et al., 2005) (Fig. 2B). The four α-helices of the coiled-coil are shown in yellow and the position of critical a.a. are shown in red. E149 and R241, K242, R257, R258 are exposed on the same surface of the protein. The side chain of I320 in the fourth α-helix is embedded, but may interact with third α-helix and contribute to conformation of the coiled-coil domain.
STAT5a nuclear import is mediated by importin-α/importin-β1 system
Active transport of large proteins is facilitated by the importin-β1/karyopherin-β1 carrier proteins. Most commonly the NLS is recognized by adapter proteins of the importin-α family that dimerize with importin-β1. Importin-β1 mediates transport through the NPC and the importin-α/importin-β1 dimer thereby facilitates nuclear import of cargo. There are six characterized members of the importin-α family that can directly recognize the NLS (Goldfarb et al., 2004; Köhler et al., 1999). To determine if any of the importin-α family members physically interact with STAT5a, in vitro binding assays were performed with importins and STAT5a (Fig. 3A). Mammalian cells were transfected with V5 tagged STAT5a and cellular lysates were used as a source of STAT5a. STAT5a-V5 was immunoprecipitated from the cell lysates using V5 antibody, and incubated with bacterially expressed GST tagged importin family members. STAT5a bound importins were eluted from the beads and analyzed by western blot using anti-GST antibody. Results show STAT5a binding to both importin-α3 and importin-α6. Since importin-α3 is ubiquitously expressed whereas importin-α6 is restricted to the testes, importin-α3 appears to be the primary adaptor that recognizes STAT5a (Köhler et al., 1997; Köhler et al., 1999). To determine if tyrosine-phosphorylated STAT5a has similar importin binding features, an in vitro binding assay was performed with STAT5a isolated from cells treated with epidermal growth factor (EGF) (supplementary material Fig. S1). STAT5a was immunoprecipitated from EGF treated cell lysates, and incubated with GST-importins. STAT5a from EGF-treated cells was found to bind importin-α3, importin-α6, and importin-β1. The binding to importin-β1 may indicate that tyrosine-phosphorylated STAT5a has an additional ability to bind importin-β1.
Fig. 3.
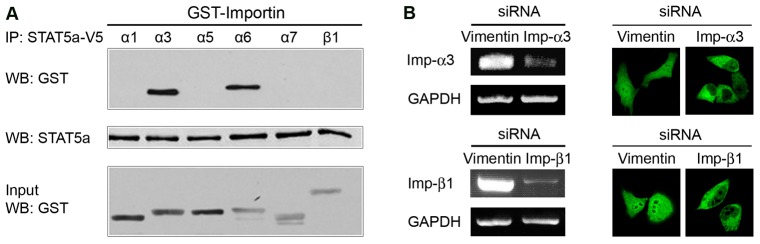
STAT5a nuclear import is mediated by importin-α3/β1 system. (A) STAT5a-V5 expressed in COS-1 cells was immunoprecipitated using protein G agarose beads, and incubated with bacterially expressed GST-importins in vitro. Importins bound to STAT5a were detected by western blot using anti-GST antibody. The input levels of STAT5a are shown with anti-STAT5a antibody. The lower blot shows 10% of purified GST-importin proteins used in the binding assay. (B) Left: The effect of pooled siRNAs on endogenous importin-α3 or importin-β1 mRNA levels detected by RT-PCR. Pooled siRNAs to importin-α3 or importin-β1 reduced the endogenous respective mRNA levels by ∼80%. Vimentin siRNA was used as a control siRNA. The level of GAPDH was quantified as an internal control. Right: The effect of importin-α3 or importin-β1 siRNAs on the cellular localization of STAT5a-GFP. Nuclear accumulation of STAT5a was inhibited in 10–30% of cultures transiently transfected with importin-α3 or importin-β1 pooled siRNAs, whereas there was no effect with vimentin siRNA.
To confirm the functional role of defined importins in STAT5a nuclear import, we evaluated the cellular localization of STAT5a after knockdown of importin-α3 and importin-β1 expression by RNA interference (RNAi; Fig. 3B). siRNA duplexes corresponding to importin-α3, importin-β1 or vimentin control were transfected into cells expressing STAT5a-GFP, and cellular localization of STAT5a-GFP was evaluated by fluorescence microscopy. The knockdown efficiency of corresponding siRNAs in cells was determined by measuring endogenous importin-α3 and importin-β1 mRNA levels. There was a significant inhibition of STAT5a nuclear accumulation if cells were treated with pooled importin-α3 siRNAs, or individual importin-α3 siRNAs (supplementary material Fig. S2). Treatment of cells with pooled importin-β1 siRNAs also decreased STAT5a nuclear import whereas there was no effect of vimentin siRNA. Together with the importin binding assay, results suggest that importin-α3/importin-β1 heterodimer mediates the nuclear translocation of STAT5a.
To understand the interface between STAT5a and importin-α3, specific fragments of importin-α3 able to bind STAT5a were identified. Importin-αs possess an N-terminal importin-β1 binding domain followed by 10 tandem armadillo (ARM) repeats (Cingolani et al., 1999; Conti et al., 1998; Fontes et al., 2000; Herold et al., 1998; Kobe, 1999). Each ARM motif consists of ∼40 a.a. folded into three α-helices. Co-crystal structures of importin-α with conventional NLS peptides indicate a basic NLS can bind to two regions of ARM repeats 2–4 and 6–8. To define the domains of importin-α3 able to directly recognize the unconventional STAT5a NLS, we performed in vitro binding assays with purified proteins from bacteria. Maltose binding protein (MBP) tagged to STAT5a 1–330 a.a. was immobilized on amylose resin and incubated with GST-importin-α3 or GST-importin-α1 as a control (Fig. 4A). Importins bound to STAT5a were detected by western blot, and importin-α3 but not importin-α1, was found to directly bind STAT5a. To further define the region of importin-α3 that binds STAT5a, in vitro binding assays were performed with MBP-STAT5a and GST-importin-α3 deletions. The results showed that importin-α3 can bind to STAT5a through two independent regions, ARM repeats 1–4 and 7–10 (Fig. 4B). Additional deletions of importin-α3 narrowed binding to ARMs 2–4, but maintained binding to a second broader region ARMs 7–10 (supplementary material Fig. S3). From both in vitro binding assays using mammalian and bacterial expression systems and in vivo functional studies using siRNAs, nuclear import of STAT5a appears to be mediated by importin-α3/importin-β1 system.
Fig. 4.
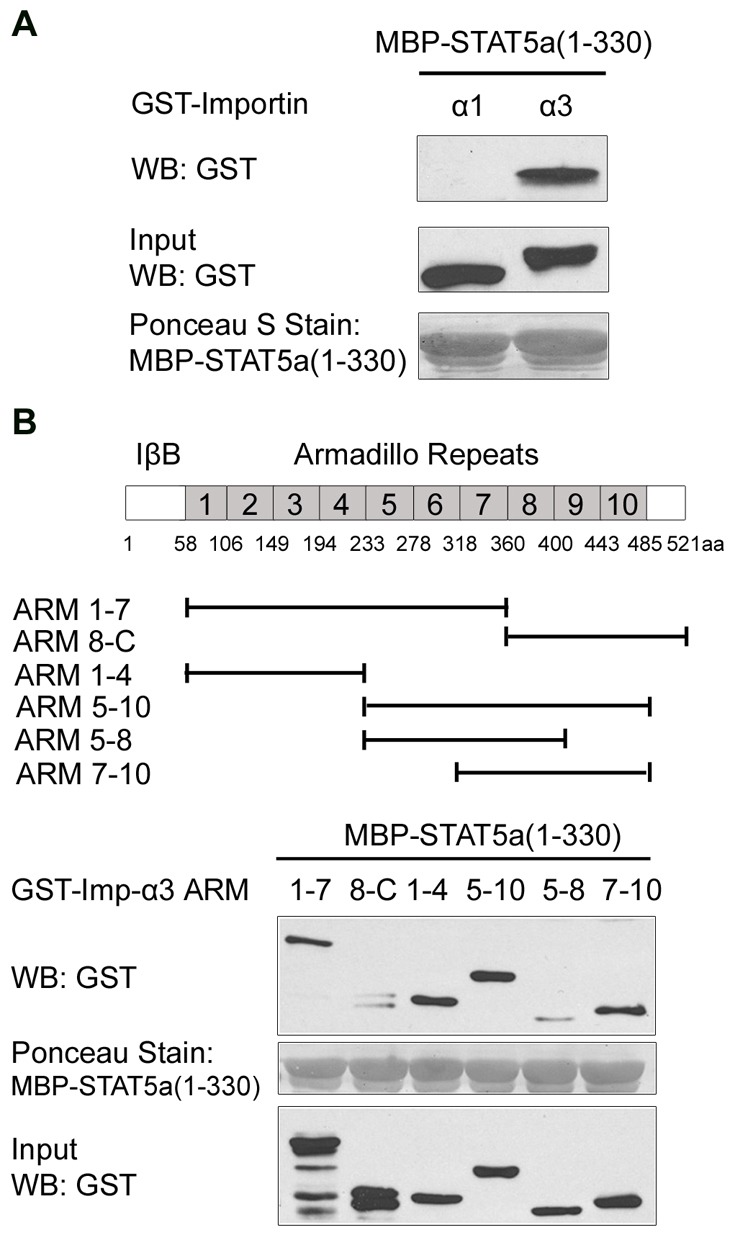
STAT5a directly binds to two independent regions of importin-α3. (A) Bacterially expressed MBP-STAT5a(1–330) was immobilized on the amylose resin and incubated with bacterially purified GST-importin-α3 or importin-α1 as a control. Importins bound to STAT5a were detected by western blot using anti-GST antibody. The protein level of STAT5a bound to resin was examined by Ponceau S Staining. The middle blot shows 10% of purified importin inputs. (B) Top: Linear depiction of importin-α3 motifs including the importin-β binding domain (IβB) and ARM repeats; fragments used in the study are indicated. Bottom: Bacterially expressed MBP-STAT5a (1–330) was immobilized on amylose resin, and incubated with GST-importin-α3 truncations. Importin-α3 deletions bound to STAT5a were identified by western blot using anti-GST antibody. STAT5a bound to resin is shown in the middle blot and 10% of importin input is shown in the lower blot.
STAT5a nuclear import is required for synergy with glucocorticoid receptor and β-casein gene expression
STAT5a has a primary role in mammary epithelial cell differentiation and alveologenesis (Liu et al., 1997). The prolactin (PRL) hormone stimulates the tyrosine phosphorylation of STAT5a during lactation leading to induction of the β-casein gene in concert with the glucocorticoid receptor (Groner, 2002; Happ and Groner, 1993). STAT5a synergizes with the glucocorticoid receptor (GR) for maximal induction of the β-casein gene (Cella et al., 1998; Kabotyanski et al., 2006; Lechner et al., 1997; Stöcklin et al., 1996; Stoecklin et al., 1997; Wyszomierski et al., 1999). The GR is a ligand-dependent transcription factor that is activated by binding glucocorticoid or derivatives such as dexamethasone or hydrocortisone (Funder, 1997; Kumar and Thompson, 1999).
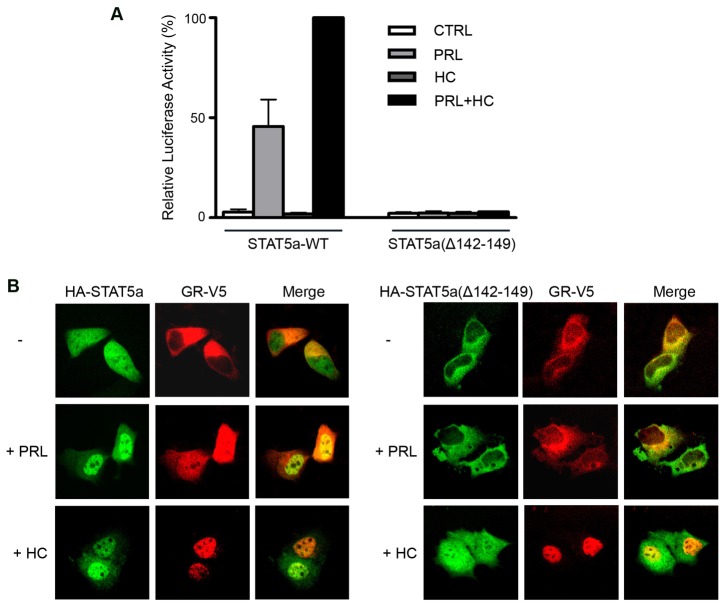
To assess the effect of the STAT5a NLS mutation Δ142–149 on transcriptional induction of the β-casein gene, we evaluated induction of a luciferase reporter gene regulated by the β-casein gene promoter (Fig. 5A). STAT5a wild-type or the NLS mutant Δ142–149 were expressed in a human breast cell line with the β-casein gene reporter, and the cells were stimulated with PRL and/or hydrocortisone (HC). PRL activation of wild type STAT5a induced the β-casein reporter, but activation of the STAT5a NLS mutant did not result in transcriptional induction. The STAT5a NLS mutant Δ142–149 is tyrosine phosphorylated in response to PRL and can bind DNA in vitro (supplementary material Fig. S4) (Iyer and Reich, 2008). To assess the effect of the STAT5a NLS mutant on synergy with the GR, cells expressing STAT5a wild-type or the NLS mutant Δ142–149 were co-treated with hydrocortisone (HC). HC treatment alone had no effect on transcription of the β-casein gene by STAT5a. However co-stimulation with PRL and HC produced the expected synergistic induction of β-casein gene activity with wild-type STAT5a. Cells expressing the STAT5a NLS mutant Δ142–149 did not respond to PRL alone or with HC in this assay. The data indicate that STAT5a nuclear import is required for cooperation with the GR in transcription of β-casein gene.
Fig. 5.
STAT5a nuclear import is required for the transcription of the β-casein promoter and synergy with GR. (A) T47D human breast cells were transfected with luciferase reporter regulated by β-casein gene promoter, β-galactosidase (β-gal) control and STAT5a wild-type or import mutation. After serum starvation, cells were left untreated (CTRL) or treated with prolactin (PRL) or/and hydrocortisone (HC) for 16 hours to activate STAT5a or GR. The level of β-casein gene expression was measured by luciferase assay and normalized to β-gal activity. (B) MCF-7 human breast cells were co-transfected with human prolactin receptor, GR-V5, HA-STAT5a wild type or import mutation and treated with PRL or HC. Immunofluorescence of HA-STAT5a and GR-V5 was detected using either FITC- or Texas Red-conjugated secondary antibody, respectively. Fluorescence intensity measurements are provided in supplementary material Fig. S4. Images represent the majority of transfected cells.
Although STAT5a and GR can synergize to induce the gene expression, the molecular mechanism of their cooperation remains to be completely understood. A physical interaction of STAT5a with GR has been documented (Cella et al., 1998; Stöcklin et al., 1996), and for this reason we investigated the role of the STAT5a NLS on interdependent effects of their nuclear accumulation. Immunofluorescence assays were performed with human breast epithelial cells expressing HA-tagged STAT5a and V5-tagged GR. Cells were treated with PRL or HC for activation of STAT5a or GR, respectively. In the absence of hormone treatment, STAT5a was evident in the nucleus, and GR was primarily resident in the cytoplasm (Fig. 5B, top left panels). After PRL treatment, STAT5a accumulated more prominently in the nucleus as expected due to its induced ability to bind DNA (Iyer and Reich, 2008), and GR also was found to accumulate with STAT5a in the nucleus of a significant percentage of cells. Treatment of cells with HC led to nuclear accumulation of GR as well as STAT5a. The data suggest that activated STAT5a can interact with GR in the cytoplasm and promote the nuclear import of both transcription factors. Similarly, ligand-bound GR can interact with STAT5a in the cytoplasm leading to the nuclear import of GR and STAT5a. Activation of either transcription factor causes the nuclear accumulation of its associated partner.
To determine the influence of the STAT5a NLS on cellular localization in response to PRL or HC, the STAT5a NLS mutant Δ142–149 was evaluated (Fig. 5B, right panel). In the absence of hormone treatment, both STAT5a Δ142–149 and GR resided in the cytoplasm. PRL stimulation did not change the cytoplasmic localization of either factor. However, treatment of cells with HC induced the nuclear accumulation of GR as well as STAT5a Δ142–149. This indicates that ligand-activated GR is able to import STAT5a Δ142–149 into the nucleus even though STAT5a Δ142–149 lacks the independent ability to be imported. Together the results indicate that regulated nuclear accumulation of GR or STAT5a can influence the nuclear localization of its associated factor.
STAT5a has a Crm1-dependent nuclear export signal in the N-terminal domain
Our studies have shown that STAT5a continually shuttles in and out of the nucleus (Iyer and Reich, 2008). The nuclear export of STAT5a can serve both as a mechanism of negative regulation and as a way to recycle the dephosphorylated STAT5a back to the cytoplasm. One of the best characterized nuclear export mechanisms is mediated by Crm1/exportin1 (Fornerod et al., 1997; Ullman et al., 1997). To evaluate the regulation of STAT5a nuclear export by Crm1, cells expressing STAT5a-GFP were treated with the inhibitor leptomycin B (LMB). LMB is an antifungal antibiotic that directly binds to Crm1 and inhibits its export function (Kudo et al., 1998). The nuclear accumulation of STAT5a-GFP dramatically increased in cells treated with leptomycin B, indicating STAT5a export can be mediated by Crm1 (Fig. 6).
Fig. 6.
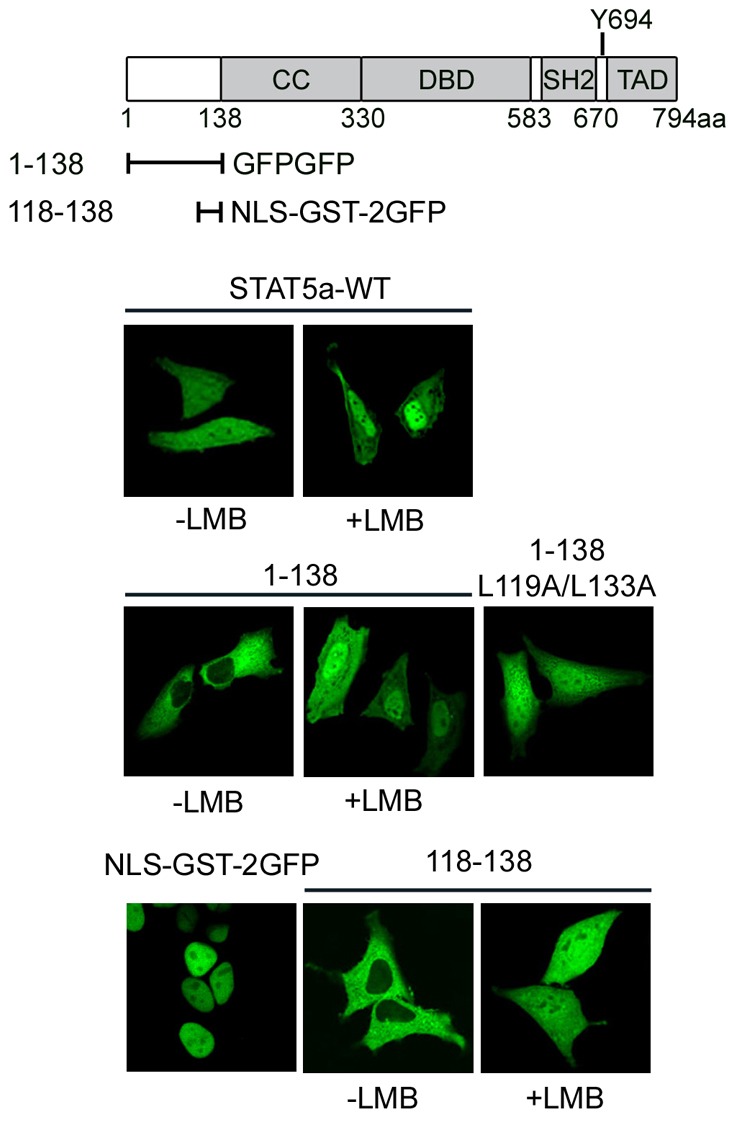
STAT5a has a Crm1-mediated NES in the N-terminal domain. Diagram of STAT5a functional motifs and the STAT5a fragments. Below are shown fluorescence images of wild-type STAT5a-GFP in cells with or without leptomycin B (LMB) treatment; STAT5a (1–138) fragment linked to two tandem repeats of GFP with or without LMB treatment; L119A/L133A double mutation in STAT5a (1–138)-GFPGFP; SV40 large T Ag NLS linked to GST-2GFP (NLS-GST-2GFP); and STAT5a (118–138) linked to GST-2GFP in cells treated with or without LMB.
The cellular localization of STAT5a fragments tagged with GFP was analyzed to identify the Crm1-mediated NES in STAT5a. We found that an N-terminal fragment encoding 1–138 a.a. was sensitive to LMB (Fig. 6). STAT5a 1–138 a.a. localized in the cytoplasm unless nuclear export was inhibited with LMB and then it accumulated in the nucleus. The NES consensus that is recognized by Crm1 is a leucine-rich hydrophobic sequence (Fornerod et al., 1997; Güttler and Görlich, 2011; Wen et al., 1995). Alanine substitution of leucine residues in this region of STAT5a, L119A and L133A, impaired the nuclear export of STAT5a 1–138 a.a.. To determine if a peptide containing this sequence was sufficient to mediate export of a protein containing a characterized NLS, 118–138 a.a was linked to a prominently nuclear protein corresponding to the NLS of the SV40 large T antigen tagged with GST-2GFP (NLS-GST-2GFP). The effective nuclear export function of a.a. 118–138 was evident in its ability to maintain cytoplasmic residence of the NLS-containing protein (Fig. 6, lower panels). The NES function was sensitive to LMB, identifying a functional Crm1-mediated NES in the N-terminus of STAT5a. The function of this NES was also demonstrated in full length STAT5a by introduction of the internal deletion of 118–138 a.a. or the targeted mutation L119A/L133A (supplementary material Fig. S5).
The directional transport of proteins through the NPC is regulated by a gradient of Ran-GTP. The higher relative levels of GTP-bound Ran in the nucleus facilitate the binding of Crm1 to the NES for export, and the GTP to GDP nucleotide exchange in the cytoplasm allows the dissociation of Crm1 from NES (Cook et al., 2007; Yudin and Fainzilber, 2009). To investigate Ran requirement for STAT5a nuclear export, cellular localization of STAT5a was evaluated in the presence of wild type Ran or a constitutively active Ran mutant Q69L that remains in a GTP-bound state (Bischoff et al., 1994; Klebe et al., 1995). Following co-transfection of STAT5a-YFP with CFP-Ran, cellular localization of STAT5a was examined. Expression of Ran Q69L influenced STAT5a to become predominantly cytoplasmic indicating a role of Ran in STAT5a nuclear trafficking (supplementary material Fig. S6).
Evidence supporting the existence of an additional exportin
Photobleaching techniques with live cell imaging can provide information on the kinetics of protein movement within and between nuclear and cytoplasmic compartments of the cell. To assess the kinetics of STAT5a nuclear export, we used cytoplasmic Fluorescence Loss in Photobleaching (cFLIP). With this technique a small region in the cytoplasm of cells expressing STAT5a-GFP was subjected to a continuous high intensity laser (Fig. 7). Cytoplasmic fluorescence was rapidly lost due to the bleaching of STAT5a-GFP as it passed through the path of the laser, indicating rapid continuous movement of STAT5a within the cytoplasm. With additional time of photobleaching, fluorescence was also completely lost from the nucleus by 40 minutes. This indicates STAT5a-GFP is exported from the nucleus and photobleached in the cytoplasm. To test the effect of LMB on the kinetics of STAT5a nuclear export, we performed cFLIP on cells treated with LMB. With the LMB treatment, the rate of nuclear export was delayed as expected, but STAT5a-GFP nuclear fluorescence was still lost by 50 minutes. Curve fitting analyses of multiple cells were used to calculate the half-time loss of nuclear fluorescence in the presence or absence of LMB, and LMB was found to delay the half-time of nuclear fluorescence decay by 1.8-fold (Fig. 7, bottom panel). The similar reduced nuclear export of the STAT5a Δ118–138 NES mutant is shown in supplementary material Fig. S7, as well as a comparative nuclear control with T-Ag-NLS-GST-2GFP protein. Taken together, the live cell imaging data indicate a role of Crm1 in STAT5a nuclear export, but also provide evidence for the function of export that is independent of Crm1 since export occurs in the presence of LMB.
Fig. 7.
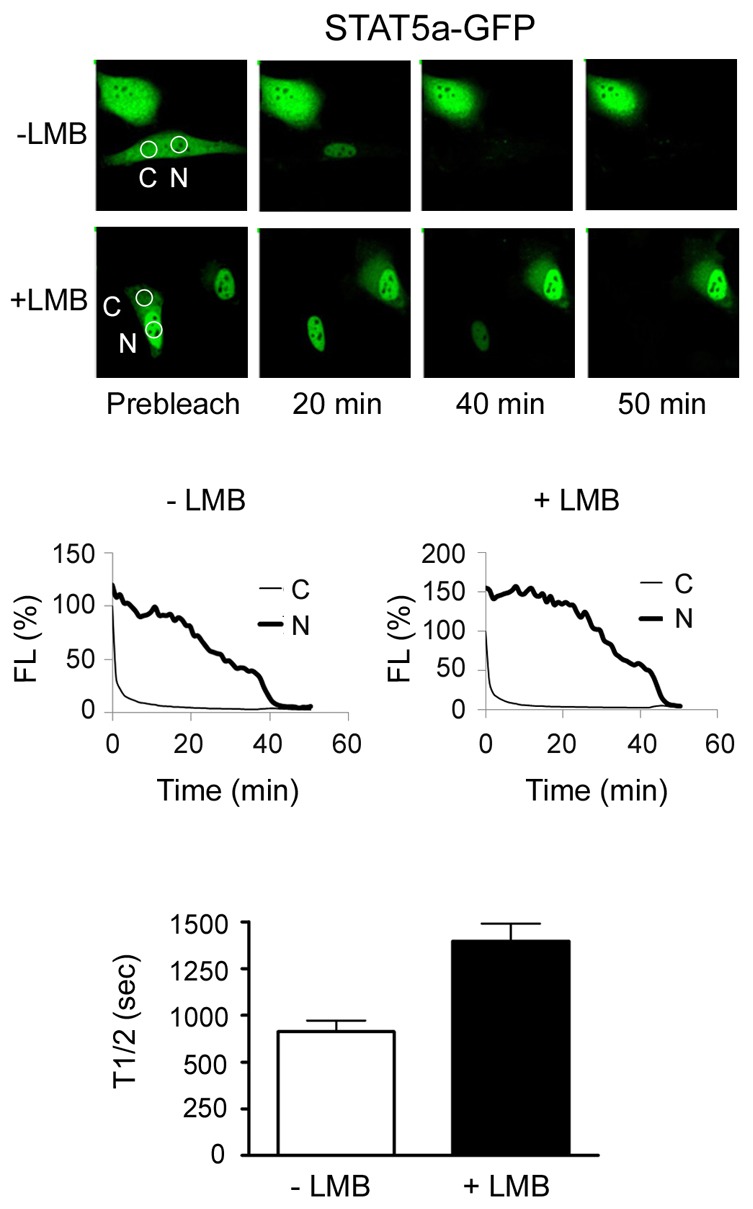
Evidence for additional nuclear export signal in STAT5a. Cytoplasmic fluorescence loss in photobleaching (FLIP). A small region in the cytoplasm of cells expressing STAT5a-GFP untreated or treated with LMB was subjected to a continuous high intensity laser. Time-lapse images capture fluorescence in cytoplasm (C) and nucleus (N). Graphs show loss of fluorescence intensity (FL%) quantified by LSM Image Browser in cytoplasmic and nuclear compartments of one photobleached cell plotted against time. The bar graphs show the half-time (T1/2) of nuclear fluorescence decay, as calculated by curve-fitting analysis for multiple LMB-treated and untreated cells at baseline of cytoplasmic fluorescence.
To investigate the Crm1-independent NES of STAT5a, additional fragments of STAT5a were evaluated for their cellular localization (Fig. 8). The 1–330 a.a. that contains both the Crm1-dependent NES and the NLS within the coiled-coil domain has clear nuclear presence indicating dominance of the STAT5a NLS at steady state. The carboxyl terminal fragment 331–794 a.a of STAT5a localized in the cytoplasm, and this could be due to either the lack of an NLS or the function of an NES. To determine if there is a functional NES in this domain, fragments of this region were linked to the NLS of SV40 T antigen (NLS-GST-2GFP). NLS-GST-2GFP localizes prominently in the nucleus, but when linked to STAT5a 331–583 a.a. or 403–474 a.a. of STAT5a, the protein is exported into the cytoplasm (Fig. 8). The nuclear export mediated by this sequence is independent of Crm1 since LMB did not have any effect on export. The results indicate that STAT5a has a Crm1-independent NES in the DNA-binding domain as well as a Crm1-dependent NES in the N-terminus. It is not clear if DNA binding may inhibit the function of the NES positioned in the DNA-binding domain.
Fig. 8.
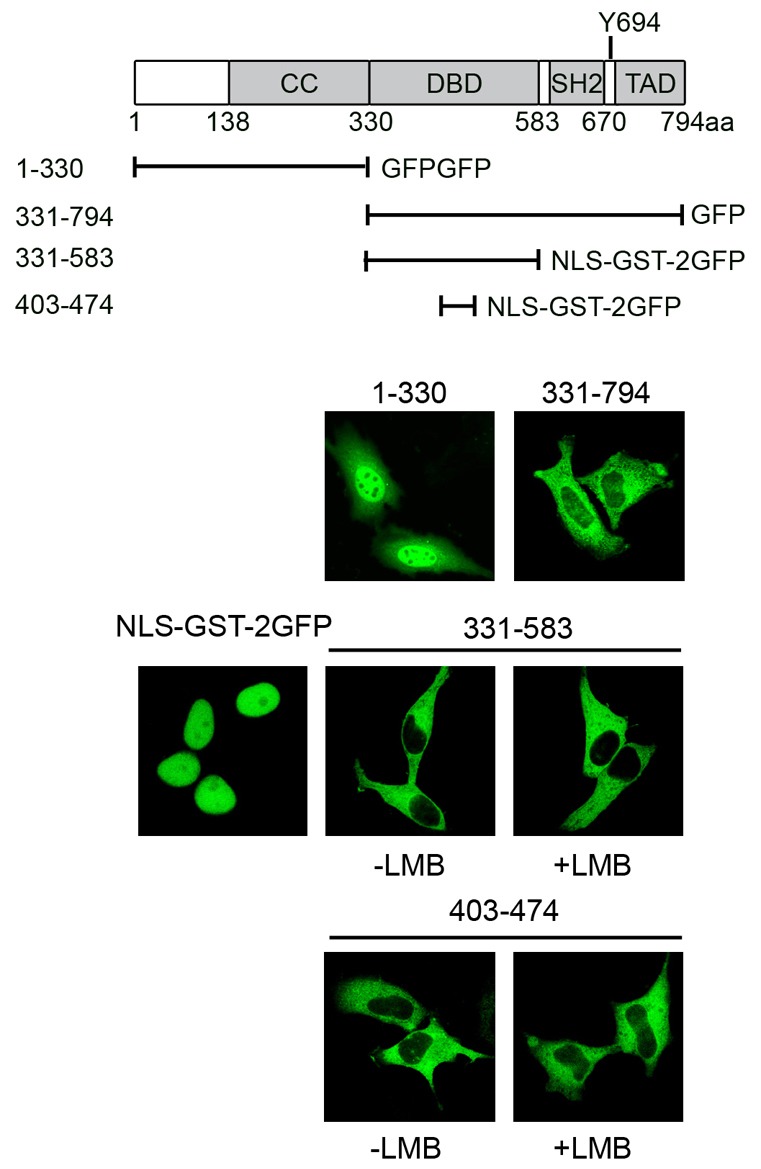
STAT5a has a Crm1-independent NES in the DNA-binding domain. Diagram of STAT5a functional motifs and STAT5a constructs. Below are fluorescence images of STAT5a fragments linked to GFP or to SV40 large T Ag NLS linked to GST-2GFP (NLS-GST-2GFP) with or without LMB treatment.
Discussion
STAT factors classically can sense cytokine and growth factor signals in the cytoplasm and deliver those signals to responsive genes in the nucleus (Levy and Darnell, 2002; Schindler et al., 2007). Movement between cytoplasmic and nuclear compartments is thereby central to their biological function. Although tyrosine phosphorylation of STATs is known to promote the formation of dimers that can bind to specific DNA targets, an increasing number of studies indicate unphosphorylated STATs function in nuclear gene expression (Chatterjee-Kishore et al., 2000; Cui et al., 2007; Yang et al., 2005; Yang and Stark, 2008). In addition, nuclear import of STAT3, STAT5, and STAT6 has been shown to be independent of tyrosine phosphorylation (Chen and Reich, 2010; Cimica et al., 2011; Iyer and Reich, 2008; Liu et al., 2005; Meyer and Vinkemeier, 2004; Zeng et al., 2002). For these reasons, accurate nuclear trafficking is key to the function of both unphosphorylated and tyrosine phosphorylated STATs.
In this study, we describe the novel finding of an unconventional NLS in STAT5a within the conformation of an extensive coiled-coil domain. By analyzing deletion mutants, we previously identified a short stretch of amino acids in the coiled-coil domain to be required for STAT5a nuclear import (142–149a.a.) (Iyer and Reich, 2008). However, this sequence was not sufficient to mediate nuclear import, and so larger fragments of the STAT5a coiled-coil domain were evaluated (Fig. 1). The results indicated that all four α-helices of the coiled-coil domain are required for effective nuclear localization. Although the conformation of this entire domain appears necessary for STAT5a NLS function, several residues were identified to be critical by targeted mutation. Glutamic acid 149 in the first α-helix plays a major role in NLS function, and lesser roles were found for a bipartite basic sequence in the second α-helix and an isoleucine 320 in the fourth α-helix (Fig. 2). The effect of the isoleucine mutation is only evident in combination with other mutations.
The coiled-coil domain of both unphosphorylated and tyrosine phosphorylated STAT5a is expected to be accessible for recognition by importin carrier proteins. The crystal structure of unphosphorylated STAT5a has been solved, and similar to unphosphorylated STAT1, STAT5a appears to form an anti-parallel homodimer (Mao et al., 2005; Neculai et al., 2005). The interaction between monomers is stabilized primarily by the β-barrel of the DNA-binding domains, but also by weak hydrogen bonds with α-helices 1 and 3. Following tyrosine phosphorylation, STAT5a is expected to form a parallel homodimer via reciprocal SH2 domain and phosphotyrosine interactions similar to that of STAT1 and STAT3 (Becker et al., 1998; Chen et al., 1998). The coiled-coil domain of STAT5a should thereby be accessible in both unphosphorylated and tyrosine-phosphorylated dimeric forms for potential interaction with importin transporter proteins. Detection of in vivo interactions between NLS-containing proteins and importins is technically challenging due to the transient interaction of importins with thousands of proteins, and so to evaluate STAT5a recognition by importins we developed in vitro binding assays (Fig. 3). Assays with STAT5a from either mammalian cell lysates or as bacterially purified protein revealed importin-α3 as a primary binding adaptor (Figs 3, 4). More importantly, silencing expression of endogenous importin-α3 or importin-β1 with siRNA caused a significant inhibition of STAT5a nuclear import, indicating a functional requirement of importin-α3 and importin-β1. Our results do not support a previous suggestion that STAT5 is imported independent of importin carriers (Marg et al., 2004). Another study suggested that nuclear import of tyrosine-phosphorylated STAT5 is mediated by a Rac1 GTPase activating protein and is inhibited by a dominant negative N17Rac1 (Kawashima et al., 2009). We have found no evidence for a negative effect of N17Rac1 on nuclear import of either unphosphorylated or tyrosine phosphorylated STAT5a (unpublished observations, NCR).
The requirement of the entire coiled-coil domain of STAT5a for nuclear import suggests the possibility of a significant interface between STAT5a and importins. To determine the domain(s) of importin-α3 that binds STAT5a, we evaluated fragments containing various ARM repeats (Fig. 4; supplementary material Fig. S3). Two regions of importin-α3 were found to bind STAT5a independently, ARMs 2–4 and ARMs 7–10. Co-crystal structures of conventional single or bipartite basic NLS peptides with importin-α have demonstrated NLS binding in an antiparallel configuration to a major site within ARMs 2–4 and a minor site within ARMs 6–8 (Conti et al., 1998; Fontes et al., 2003). Although the STAT5a NLS is not a conventional basic peptide, it does bind importin-α ARMs 2–4. The second binding site in importin-α appears to be more extensive and include ARMs 7–10. It is possible that this reflects an extensive interface between STAT5a and importin-α, or it may indicate the ARM repeats in this region contribute to the structure of the binding site and do not directly interact with STAT5a. Future studies with targeted mutations of asparagine and tryptophan residues of importin-α known to interact with basic NLS peptides may provide clarification. Since two independent regions of importin-α3 can bind STAT5a, it could suggest that one importin protein can bind two STAT5a proteins either as monomers, unphosphorylated dimers, or tyrosine-phosphorylated dimers. Solving a co-crystal structure will provide a more definitive understanding of STAT5-importin interactions.
Functional specificity of STAT5 is in part a consequence of its ability to synergize with other transcription factors, most notably the glucocorticoid receptor (GR) (Hennighausen and Robinson, 2008). In hepatocytes, STAT5b interaction with the glucocorticoid receptor (GR) contributes to normal growth and sexual maturation (Engblom et al., 2007). In the mammary gland, STAT5a interaction with the GR enhances development and expression of the β-casein gene (Liu et al., 1997; Stöcklin et al., 1996). We found the STAT5a NLS mutant (142–149 a.a. deletion) was not able to induce the β-casein gene promoter with or without synergy by the GR in breast cancer cells. Although the precise mechanisms of GR transport remain to be determined, the GR is known to exist in a multimeric chaperone complex and a conformational change occurs following ligand binding to expose an NLS (Vandevyver et al., 2012). Previous studies demonstrated a physical interaction of GR with STAT5a, and transcriptional enhancement of the β-casein gene by GR was dependent on STAT5a binding to its DNA target (Stoecklin et al., 1997; Wyszomierski et al., 1999). However it was not clearly understood if GR-STAT5a interaction takes place in the nucleus or in the cytoplasm. Our study demonstrates that ligand-bound GR can bring the STAT5a NLS mutant (142–149a.a. deletion) into the nucleus, indicating that these factors can interact in the cytoplasm to effect partner import (Fig. 5).
Both positive and negative regulation of STAT5 is necessary to maintain normal physiological processes of blood cell development, mammary development, and body growth. Negative regulation of STAT5 is known to be mediated by tyrosine phosphatases and suppressors of cytokine signaling (SOCS) (Aoki and Matsuda, 2000; Aoki and Matsuda, 2002; Chen et al., 2004; Cornish et al., 2003; Hoyt et al., 2007; Martens et al., 2005). Another possible means of negative regulation is STAT5 export from the nucleus. Our studies indicate that there are two regulatory signals in STAT5a that effect export, a Crm1-dependent NES in the N-terminus, and a Crm1-independent NES within the DNA-binding domain (Figs 6–8). Although Crm1 is a common exportin, other exportins and chaperones have been identified that mediate export of translation and transcription factors (Güttler and Görlich, 2011; Pemberton and Paschal, 2005). Future studies are needed to identify the non-Crm1 exportin for STAT5a. The continuous nuclear import of STAT5a may facilitate its rapid response to activators, and continuous nuclear export may facilitate either recycling of STAT5a or signal termination.
The critical role of negative modulation subsequent to STAT5 activation is apparent. Accumulating evidence indicates continuous STAT5 activity promotes leukemias, myeloproliferative disorders, and solid tumors. Since nuclear trafficking is required for STAT5 to regulate gene expression, it is a putative target for disease intervention. Therefore knowledge of the interface between STAT5 and importins may support the development of import inhibitors. Although a co-crystal structure of STAT5 with importin remains to be solved, our results provide a fundamental understanding of the dynamics and molecular mechanisms of STAT5a nuclear trafficking.
Materials and Methods
Cell culture and reagents
HeLa and COS-1 cells were obtained from American Type Culture Collection (ATCC), and were grown in Dulbecco's modified Eagle's medium (DMEM) with 8% fetal bovine serum (FBS). T47D cells (ATCC) were maintained in RPMI with 10% FBS and MCF-7 cells (a gift from Todd Miller, Stony Brook University, Stony Brook, NY) were cultured in DMEM with 10% FBS. DNA transfections were performed with TransIT-LT1 transfection reagent (Mirus, Madision, WI) according to the manufacturer's instructions. Cells were treated with 10 nM leptomycin B (LMB; a gift from Barbara Wolff-Winiski, Novartis Research Institute, Vienna, Austria). After serum starvation, T47D and MCF-7 cells were stimulated with human recombinant prolactin (PRL; PBL Biomedical Laboratories, New Brunswick, NJ) and hydrocortisone (HC; Sigma-Aldrich, St. Louis, MO) at 1 µg/ml each.
Plasmid constructs
Human full-length STAT5a cDNA and deletion mutants generated by polymerase chain reaction (PCR) were cloned into pcDNA3 (Invitrogen, Carlsbad, CA), pEF1/V5-His (Invitrogen), pCGN (Addgene, Cambridge, MA), or pMAL-c4X (New England Biolabs, Ipswitch, MA) to express STAT5a, or STAT5a proteins tagged with V5, HA or maltose-binding protein (MBP). A monomeric form of enhanced green fluorescent protein (GFP) (Chen and Reich, 2010), glutathione S-transferase (GST)-2GFP, or SV40 large T antigen NLS-GST-2GFP (Kalderon et al., 1984; Liu et al., 2005) were linked to full length or deletion mutants of STAT5a. Site-directed mutagenesis of STAT5a was performed using pfu Turbo DNA polymerase (Stratagene, La Jolla, Ca) with targeted oligonucleotides. Human importin-α or β1 deletion constructs lacking the importin-β1 binding (IBB) domain were subcloned into pGEX-KG for bacterial expression and purification as reported previously (Liu et al., 2005). The β-casein gene promoter responsive luciferase reporter gene was a gift from David Waxman (Boston University School of Medicine, Boston, MA), and the β-galactosidase gene was obtained from Promega. Human full-length GR (Origene, Rockville, MD) was PCR amplified and cloned into pEF1/V5-His (Invitrogen) to generate V5 tagged GR.
Confocal microscopy
Cells were plated on glass coverslips and transfected with STAT5a constructs. After 24 hours of serum starvation, cells were treated with or without LMB for 1 hour. Cells were washed with PBS and fixed with 4% paraformaldehyde for 10 minutes. GFP-tagged protein was visualized with a Zeiss LSM 5 pascal confocal microscope using a 40× oil objective. GFP was excited at 488 nm using an argon laser, and emission was collected using a 505–530 nm filter. Images were captured using Zeiss LSM image browser and presented using Adobe Photoshop.
Bacterial protein expression and purification
Human recombinant importin-α or β1 proteins tagged with GST were bacterially expressed and purified by binding and elution from glutathione agarose beads (Sigma) as reported previously (Liu et al., 2005).
In vitro importin binding assay
COS-1 cells expressing STAT5a-V5 were lysed with cold lysis buffer [50 mM Tris-HCl pH 8.2, 5 mM EDTA, 280 mM NaCl, 0.5% Nonidet P-40, 1 mM PMSF, 1× mammalian protease inhibitor cocktails (Sigma)], and 500 µg of cellular proteins were used for each assay. STAT5a-V5 was captured with anti-V5 antibody, immobilized to protein G beads, and incubated with 15 µg of purified GST-importin-α or β1 proteins. Immunocomplexes were eluted with SDS sample buffer and separated on 10% SDS-PAGE. Proteins were transferred to nitrocellulose membrane (Pierce Biotechnology, Rockford, IL), and detected with anti-GST and anti-STAT5a for western blots. Bacterially expressed MBP-tagged STAT5a was immobilized on amylose resin (New England Biolabs) in cold Column Buffer [20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, and 1 mM DTT, 1 mM PMSF, and 1× bacterial protease inhibitor cocktails (Sigma)] with 0.1% CHAPS (Sigma). Purified GST-importin-α3 and deletions were incubated with STAT5a protein immobilized beads and bound protein complexes were detected by western blot using anti-GST antibody. The amount of STAT5a bound to resin was examined by Ponceau S staining.
Antibodies
Western blots were performed using rabbit anti-STAT5a antibody (sc-1081, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-GST antibody (sc-33613, Santa Cruz Biotechnology). Alexa Fluor 680-labeled (A21109, Invitrogen) or horseradish peroxidase-conjugated (NA934V, Amersham Bioscience, Piscataway, NJ) anti-rabbit IgG was used as secondary antibody for western blots. Reactive signals were detected with an enhanced chemiluminescence system or Odyssey infrared imaging system (Li-COR Bioscience, Lincoln, NE). Two micrograms of anti-V5 antibody (R960-25, Invitrogen) was used for immunoprecipitation. For the immunofluorescence assay, anti-HA (Santa Cruz) and anti-V5 (Santa Cruz) were used as primary antibodies. Anti-rabbit conjugated with FITC (Molecular Probes, Carlsbad, CA) and anti-mouse conjugated with Texas Red (Molecular Probes) were used as secondary antibodies.
RNA interference
Short interfering RNAs (siRNA) specific for human importin-α or β1 (Qiagen Inc., Valencia, CA) were transfected with X-tremeGENE siRNA transfection reagent (Roche, Indianapolis, IN). Vimentin siRNA was used as a negative control. After 24 hours of siRNA transfection, cells were transfected with STAT5a-GFP. Cellular localization of STAT5a-GFP was evaluated after 24 hours by confocal microscopy. Isolation of total RNA was performed with TRIzol reagent (Invitrogen), and cDNA was synthesized with M-MLV reverse transcriptase (Promega, Madison, WI). RT-PCR was carried out using specific primers for importin-α3, β1 or GAPDH as an internal control. Primer sequences for importin-α3 and β1 were reported previously (Chen and Reich, 2010; Liu et al., 2005). ImageJ software was used for quantification of endogenous importin-α3 or importin-β1 levels.
Luciferase reporter assay
T47D human breast cells that express endogenous glucocorcoid receptor and prolactin receptor (PRL-R) were co-transfected with β-casein-luciferase, β-galactosidase (Promega), and pcDNA3-STAT5a wild type or import mutant. After 24 hours of serum starvation, cells were treated with 1 µg/ml of prolactin (PRL) or/and hydrocortisone (HC) for 16 hours. Firefly luciferase (Promega) and luminescent β-galactosidase (Clontech, Mountain View, CA) were measured according to the manufacturer's instructions. Firefly luciferase values were normalized to luminescent β-galactosidase values to eliminate variations of transfection efficiency.
Immunofluorescence assay
MCF-7 human breast cells were seeded on the glass coverslips and co-transfected with GR-V5, hPRL-R, HA-STAT5a or HA-STAT5a(Δ142–149). After 24 hours of serum starvation, cells were treated with or without PRL or HC for an hour. Cells were rinsed with cold PBS and fixed with 4% paraformaldehyde. Following the permeabilization in 0.5% Triton X-100, cells were blocked in 3% BSA, incubated with primary antibodies for 3 hours followed by secondary antibodies conjugated with FITC or Texas Red. Immunofluorescence of cells was visualized by a Zeiss LSM 5 pascal confocal microscope using a 40× oil objective. Images were obtained using Zeiss LSM image browser program and presented using Adobe Photoshop.
Live cell imaging
HeLa cells were seeded on 35 mm glass-bottom tissue culture dishes (Mattek Corporation, Ashland, MA), and transfected with STAT5a-GFP constructs. After serum starvation, cells were treated with or without LMB for an hour. During live cell imaging, cells were maintained at 37°C and 5% CO2 using the Zeiss Tempcontrol 37-2 Digital and CTI Controller 3700. Fluorescence loss in photobleaching (FLIP) with STAT5a-GFP was performed by bleaching the region of interest (ROI) in the cytoplasm every 60 seconds with 100% power of an argon laser at 488 nm for 50 minutes. The time series images were obtained with the Zeiss LSM 510 META NLO two-photon laser scanning microscope system using a 63× oil objective. The excitation wavelength of GFP was 488 nm and emission was detected with a 505 nm filter. Images were obtained using LSM image browser and presented with Adobe Photoshop. Nuclear and cytoplasmic fluorescence intensities of target cells were quantified using LSM Image Browser and graphically plotted using GraphPad Prism software.
Supplementary Material
Acknowledgments
We thank the current and past members of the laboratory for their support and helpful advice, especially Janaki Iyer, Hui-Chen Chen, and Velasco Cimica. We appreciate the support of Guo-Wei Tian and Vitaly Citovsky with imaging analyses.
Footnotes
Author contributions
H.Y.S. and N.C.R. designed experiments and wrote the manuscript. H.Y.S. performed the experiments.
Funding
These studies were supported by the National Institutes of Health (NIH) [grant numbers R56AI095268, RO1CA122910]; a Carol M. Baldwin Breast Cancer Research Award; and a Walk-for-Beauty Foundation Research Award (to N.C.R.). Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.123042/-/DC1
References
- Aoki N., Matsuda T. (2000). A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J. Biol. Chem. 275, 39718–39726 10.1074/jbc.M005615200 [DOI] [PubMed] [Google Scholar]
- Aoki N., Matsuda T. (2002). A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol. Endocrinol. 16, 58–69 10.1210/me.16.1.58 [DOI] [PubMed] [Google Scholar]
- Becker S., Groner B., Müller C. W. (1998). Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 394, 145–151 10.1038/28101 [DOI] [PubMed] [Google Scholar]
- Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91, 2587–2591 10.1073/pnas.91.7.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella N., Groner B., Hynes N. E. (1998). Characterization of Stat5a and Stat5b homodimers and heterodimers and their association with the glucocortiocoid receptor in mammary cells. Mol. Cell. Biol. 18, 1783–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee-Kishore M., Wright K. L., Ting J. P., Stark G. R. (2000). How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 19, 4111–4122 10.1093/emboj/19.15.4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. C., Reich N. C. (2010). Live cell imaging reveals continuous STAT6 nuclear trafficking. J. Immunol. 185, 64–70 10.4049/jimmunol.0903323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J. E., Jr, Kuriyan J. (1998). Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 10.1016/S0092-8674(00)81443-9 [DOI] [PubMed] [Google Scholar]
- Chen J., Yu W. M., Bunting K. D., Qu C. K. (2004). A negative role of SHP-2 tyrosine phosphatase in growth factor-dependent hematopoietic cell survival. Oncogene 23, 3659–3669 10.1038/sj.onc.1207471 [DOI] [PubMed] [Google Scholar]
- Chook Y. M., Blobel G. (2001). Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 10.1016/S0959-440X(01)00264-0 [DOI] [PubMed] [Google Scholar]
- Cimica V., Chen H. C., Iyer J. K., Reich N. C. (2011). Dynamics of the STAT3 transcription factor: nuclear import dependent on Ran and importin-β1. PLoS ONE 6, e20188 10.1371/journal.pone.0020188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Petosa C., Weis K., Müller C. W. (1999). Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399, 221–229 10.1038/20367 [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair R., Rost B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411–415 10.1093/embo-reports/kvd092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998). Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94, 193–204 10.1016/S0092-8674(00)81419-1 [DOI] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., Conti E. (2007). Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 10.1146/annurev.biochem.76.052705.161529 [DOI] [PubMed] [Google Scholar]
- Cornish A. L., Chong M. M., Davey G. M., Darwiche R., Nicola N. A., Hilton D. J., Kay T. W., Starr R., Alexander W. S. (2003). Suppressor of cytokine signaling-1 regulates signaling in response to interleukin-2 and other gamma c-dependent cytokines in peripheral T cells. J. Biol. Chem. 278, 22755–22761 10.1074/jbc.M303021200 [DOI] [PubMed] [Google Scholar]
- Cotarla I., Ren S., Zhang Y., Gehan E., Singh B., Furth P. A. (2004). Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int. J. Cancer 108, 665–671 10.1002/ijc.11619 [DOI] [PubMed] [Google Scholar]
- Cui X., Zhang L., Luo J., Rajasekaran A., Hazra S., Cacalano N., Dubinett S. M. (2007). Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene 26, 4253–4260 10.1038/sj.onc.1210222 [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Jr, Kerr I. M., Stark G. R. (1994). Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 10.1126/science.8197455 [DOI] [PubMed] [Google Scholar]
- Engblom D., Kornfeld J. W., Schwake L., Tronche F., Reimann A., Beug H., Hennighausen L., Moriggl R., Schütz G. (2007). Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 21, 1157–1162 10.1101/gad.426007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes M. R., Teh T., Kobe B. (2000). Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297, 1183–1194 10.1006/jmbi.2000.3642 [DOI] [PubMed] [Google Scholar]
- Fontes M. R., Teh T., Jans D., Brinkworth R. I., Kobe B. (2003). Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 278, 27981–27987 10.1074/jbc.M303275200 [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Funder J. W. (1997). Glucocorticoid and mineralocorticoid receptors: biology and clinical relevance. Annu. Rev. Med. 48, 231–240 10.1146/annurev.med.48.1.231 [DOI] [PubMed] [Google Scholar]
- Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004). Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 10.1016/j.tcb.2004.07.016 [DOI] [PubMed] [Google Scholar]
- Görlich D., Kutay U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Groner B. (2002). Transcription factor regulation in mammary epithelial cells. Domest. Anim. Endocrinol. 23, 25–32 10.1016/S0739-7240(02)00142-X [DOI] [PubMed] [Google Scholar]
- Güttler T., Görlich D. (2011). Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 30, 3457–3474 10.1038/emboj.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O., Warsch W., Eckelhart E., Kaupe I., Grebien F., Wagner K. U., Superti-Furga G., Sexl V. (2012). BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia. Nat. Chem. Biol. 8, 285–293 10.1038/nchembio.775 [DOI] [PubMed] [Google Scholar]
- Happ B., Groner B. (1993). The activated mammary gland specific nuclear factor (MGF) enhances in vitro transcription of the beta-casein gene promoter. J. Steroid Biochem. Mol. Biol. 47, 21–30 10.1016/0960-0760(93)90053-Y [DOI] [PubMed] [Google Scholar]
- Hayakawa F., Towatari M., Iida H., Wakao H., Kiyoi H., Naoe T., Saito H. (1998). Differential constitutive activation between STAT-related proteins and MAP kinase in primary acute myelogenous leukaemia. Br. J. Haematol. 101, 521–528 10.1046/j.1365-2141.1998.00720.x [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G. W. (2008). Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 22, 711–721 10.1101/gad.1643908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A., Truant R., Wiegand H., Cullen B. R. (1998). Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 143, 309–318 10.1083/jcb.143.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt R., Zhu W., Cerignoli F., Alonso A., Mustelin T., David M. (2007). Cutting edge: selective tyrosine dephosphorylation of interferon-activated nuclear STAT5 by the VHR phosphatase. J. Immunol. 179, 3402–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer J., Reich N. C. (2008). Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 22, 391–400 10.1096/fj.07-8965com [DOI] [PubMed] [Google Scholar]
- Kabotyanski E. B., Huetter M., Xian W., Rijnkels M., Rosen J. M. (2006). Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers. Mol. Endocrinol. 20, 2355–2368 10.1210/me.2006-0160 [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984). A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 10.1016/0092-8674(84)90457-4 [DOI] [PubMed] [Google Scholar]
- Kawashima T., Bao Y. C., Minoshima Y., Nomura Y., Hatori T., Hori T., Fukagawa T., Fukada T., Takahashi N., Nosaka T. et al. (2009). A Rac GTPase-activating protein, MgcRacGAP, is a nuclear localizing signal-containing nuclear chaperone in the activation of STAT transcription factors. Mol. Cell. Biol. 29, 1796–1813 10.1128/MCB.01423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C., Bischoff F. R., Ponstingl H., Wittinghofer A. (1995). Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34, 639–647 10.1021/bi00002a031 [DOI] [PubMed] [Google Scholar]
- Kobe B. (1999). Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6, 388–397 10.1038/7625 [DOI] [PubMed] [Google Scholar]
- Köhler M., Ansieau S., Prehn S., Leutz A., Haller H., Hartmann E. (1997). Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417, 104–108 10.1016/S0014-5793(97)01265-9 [DOI] [PubMed] [Google Scholar]
- Köhler M., Speck C., Christiansen M., Bischoff F. R., Prehn S., Haller H., Görlich D., Hartmann E. (1999). Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19, 7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N., Wolff B., Sekimoto T., Schreiner E. P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242, 540–547 10.1006/excr.1998.4136 [DOI] [PubMed] [Google Scholar]
- Kumar R., Thompson E. B. (1999). The structure of the nuclear hormone receptors. Steroids 64, 310–319 10.1016/S0039-128X(99)00014-8 [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007). Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282, 5101–5105 10.1074/jbc.R600026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Welte T., Doppler W. (1997). Mechanism of interaction between the glucocorticoid receptor and Stat5: role of DNA-binding. Immunobiology 198, 112–123 10.1016/S0171-2985(97)80032-0 [DOI] [PubMed] [Google Scholar]
- Levy D. E., Darnell J. E., Jr (2002). Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- Liu X., Robinson G. W., Gouilleux F., Groner B., Hennighausen L. (1995). Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl. Acad. Sci. USA 92, 8831–8835 10.1073/pnas.92.19.8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Robinson G. W., Wagner K. U., Garrett L., Wynshaw-Boris A., Hennighausen L. (1997). Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 11, 179–186 10.1101/gad.11.2.179 [DOI] [PubMed] [Google Scholar]
- Liu L., McBride K. M., Reich N. C. (2005). STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. USA 102, 8150–8155 10.1073/pnas.0501643102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I. G. (2001). Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570–594 10.1128/MMBR.65.4.570-594.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Ren Z., Parker G. N., Sondermann H., Pastorello M. A., Wang W., McMurray J. S., Demeler B., Darnell J. E., Jr, Chen X. (2005). Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17, 761–771 10.1016/j.molcel.2005.02.021 [DOI] [PubMed] [Google Scholar]
- Marg A., Shan Y., Meyer T., Meissner T., Brandenburg M., Vinkemeier U. (2004). Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 165, 823–833 10.1083/jcb.200403057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens N., Uzan G., Wery M., Hooghe R., Hooghe-Peters E. L., Gertler A. (2005). Suppressor of cytokine signaling 7 inhibits prolactin, growth hormone, and leptin signaling by interacting with STAT5 or STAT3 and attenuating their nuclear translocation. J. Biol. Chem. 280, 13817–13823 10.1074/jbc.M411596200 [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Englmeier L. (1998). Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67, 265–306 10.1146/annurev.biochem.67.1.265 [DOI] [PubMed] [Google Scholar]
- McBride K. M., Banninger G., McDonald C., Reich N. C. (2002). Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21, 1754–1763 10.1093/emboj/21.7.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T., Vinkemeier U. (2004). Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J. Biochem. 271, 4606–4612 10.1111/j.1432-1033.2004.04423.x [DOI] [PubMed] [Google Scholar]
- Moriggl R., Topham D. J., Teglund S., Sexl V., McKay C., Wang D., Hoffmeyer A., van Deursen J., Sangster M. Y., Bunting K. D. et al. (1999). Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity 10, 249–259 10.1016/S1074-7613(00)80025-4 [DOI] [PubMed] [Google Scholar]
- Neculai D., Neculai A. M., Verrier S., Straub K., Klumpp K., Pfitzner E., Becker S. (2005). Structure of the unphosphorylated STAT5a dimer. J. Biol. Chem. 280, 40782–40787 10.1074/jbc.M507682200 [DOI] [PubMed] [Google Scholar]
- Nevalainen M. T., Ahonen T. J., Yamashita H., Chandrashekar V., Bartke A., Grimley P. M., Robinson G. W., Hennighausen L., Rui H. (2000). Epithelial defect in prostates of Stat5a-null mice. Lab. Invest. 80, 993–1006 10.1038/labinvest.3780105 [DOI] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. (2005). Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6, 187–198 10.1111/j.1600-0854.2005.00270.x [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D. (2001). The nuclear pore complex as a transport machine. J. Biol. Chem. 276, 16593–16596 10.1074/jbc.R100015200 [DOI] [PubMed] [Google Scholar]
- Schindler C., Levy D. E., Decker T. (2007). JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 282, 20059–20063 10.1074/jbc.R700016200 [DOI] [PubMed] [Google Scholar]
- Schwaller J., Parganas E., Wang D., Cain D., Aster J. C., Williams I. R., Lee C. K., Gerthner R., Kitamura T., Frantsve J. et al. (2000). Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell 6, 693–704 10.1016/S1097-2765(00)00067-8 [DOI] [PubMed] [Google Scholar]
- Stöcklin E., Wissler M., Gouilleux F., Groner B. (1996). Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383, 726–728 10.1038/383726a0 [DOI] [PubMed] [Google Scholar]
- Stoecklin E., Wissler M., Moriggl R., Groner B. (1997). Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 17, 6708–6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. H., Nevalainen M. T. (2008). Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr. Relat. Cancer 15, 367–390 10.1677/ERC-08-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy G. B., Towers R. P., Snell R. G., Wilkins R. J., Park S. H., Ram P. A., Waxman D. J., Davey H. W. (1997). Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl. Acad. Sci. USA 94, 7239–7244 10.1073/pnas.94.14.7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman K. S., Powers M. A., Forbes D. J. (1997). Nuclear export receptors: from importin to exportin. Cell 90, 967–970 10.1016/S0092-8674(00)80361-X [DOI] [PubMed] [Google Scholar]
- Vandevyver S., Dejager L., Libert C. (2012). On the trail of the glucocorticoid receptor: into the nucleus and back. Traffic 13, 364–374 10.1111/j.1600-0854.2011.01288.x [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth J. L., Tsien R. Y., Taylor S. S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473 10.1016/0092-8674(95)90435-2 [DOI] [PubMed] [Google Scholar]
- Wyszomierski S. L., Yeh J., Rosen J. M. (1999). Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol. Endocrinol. 13, 330–343 10.1210/me.13.2.330 [DOI] [PubMed] [Google Scholar]
- Yang J., Stark G. R. (2008). Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 10.1038/cr.2008.41 [DOI] [PubMed] [Google Scholar]
- Yang J., Chatterjee-Kishore M., Staugaitis S. M., Nguyen H., Schlessinger K., Levy D. E., Stark G. R. (2005). Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 65, 939–947 [PubMed] [Google Scholar]
- Yao Z., Cui Y., Watford W. T., Bream J. H., Yamaoka K., Hissong B. D., Li D., Durum S. K., Jiang Q., Bhandoola A. et al. (2006). Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA 103, 1000–1005 10.1073/pnas.0507350103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D., Fainzilber M. (2009). Ran on tracks—cytoplasmic roles for a nuclear regulator. J. Cell Sci. 122, 587–593 10.1242/jcs.015289 [DOI] [PubMed] [Google Scholar]
- Zeng R., Aoki Y., Yoshida M., Arai K., Watanabe S. (2002). Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J. Immunol. 168, 4567–4575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.