Summary
Protrusion formation is the first step that precedes cell movement of motile cells. Spatial control of actin polymerization is necessary to achieve directional protrusion during cell migration. Here we show that the spatial coordinators p190RhoGEF and p190RhoGAP regulate actin polymerization during leading edge protrusions by regulating the actin barbed end distribution and amplitude. The distribution of RhoC activity and proper balance of cofilin activation achieved by p190RhoGEF and p190RhoGAP determines the direction of final protrusive activity. These findings provide a new insight into the dynamic plasticity in the amplitude and distribution of barbed ends, which can be modulated by fine-tuning RhoC activity by upstream GEFs and GAPs for directed cell motility.
Key words: RhoC GTPase, p190RhoGAP, p190RhoGEF, Directional protrusion, Chemotaxis, Actin cytoskeleton
Introduction
Polarized protrusion of the cell toward a source of chemoattractant is the initial motile response of diverse cell types, from Dictyostelium to tumor cells, during chemotaxis (Weiner, 2002). Polarized protrusion at the leading edge is coupled to actin polymerization and is essential for the establishment of directional migration (Insall and Machesky, 2009). Regulators of leading edge protrusion include actin-binding proteins such as cofilin and Arp2/3, and both function synergistically to generate actin-filament-associated free barbed ends (Chan et al., 2000; DesMarais et al., 2004; Oser et al., 2009). The cofilin activity cycle is temporally and spatially regulated to restrict active cofilin at specific locations at the cell membrane, thereby defining the location of actin polymerization and direction of cell motility (Ghosh et al., 2004; Mouneimne et al., 2006). Through phosphorylation at serine 3 (pCofilinS3), cofilin is inactivated and is not able to bind to actin (Van Troys et al., 2008). In tumor cells, phosphorylation of cofilin is regulated by RhoC/ROCK/LIMK pathway (Bravo-Cordero et al., 2011). However, the mechanism of how cofilin activity is spatiotemporally regulated during polarized protrusions of the leading edge is not known. Moreover, there are different models that explain the role of cofilin at the leading edge during actin polymerization and barbed end formation (DesMarais et al., 2005; Pollard and Borisy, 2003). However, neither model explains at a molecular level how spatial control of actin dynamics is achieved during directional cell migration. As motility is a crucial step for multiple processes from development and homeostasis to metastasis, understanding the molecular pathways that drive spatiotemporal control of protrusion formation is a fundamental question to be answered.
The Rho family of p21 small GTPases have been shown to be master regulators of actin dynamics through their ability to interact with many different downstream effectors (Ridley, 2012). Regulation of GTPase signaling pathways involves multiple layers of regulatory molecules including the GEFs, GAPs and GDIs (Ridley, 2012). It has been suggested that the specificity of GTPase signaling cascades rely on spatial and temporal segregation of functions between the specific GEF/GAP modular groups, dictating specific outcomes (Pertz, 2010). Through this spatially and temporally discrete upstream regulatory control, RhoGTPases can be activated/deactivated very rapidly and locally in order to trigger specific signaling pathways. These pathways require precise coordination in time and space of all of the components to generate the final, spatiotemporal output signal/function. However it is not yet well understood how cells spatially integrate the activities of GEFs and GAPs to define the final outputs including actin polymerization and protrusion formation.
Among all the Rho isoforms, RhoC is best known to be essential for metastasis, a process highly dependent on motility mechanisms (Clark et al., 2000). While the importance of RhoC in cell motility has been shown (Vega et al., 2011; Wu et al., 2010), the mechanisms of how it regulates actin polymerization during leading edge protrusions still remain unknown.
RhoGTPases have been shown to localize to dynamic activity zones in different processes. For example, RhoA and Cdc42 localize in concentric rings around wounds in Xenopus oocytes during wound closure (Benink and Bement, 2005); RhoC localizes in areas surrounding invadopodia, actin-rich structures capable of degrading extracellular matrix (Bravo-Cordero et al., 2011); and RhoA, Rac and Cdcd42 localize at the leading edge during lamellipodium formation (El-Sibai and Backer, 2007; El-Sibai et al., 2008; Machacek et al., 2009). These examples highlight the high degree of spatial and temporal regulation of GTPases in different subcellular processes. However, how these activity zones are setup and sustained during polarized protrusions of the leading edge has not been explored. It is likely that GEFs and GAPs are involved in mediating the formation of these ‘activity zones’ but how they spatiotemporally regulate Rho GTPases and ultimately actin dynamics is still unknown.
We show here how p190RhoGEF, p190RhoGAP and RhoC regulate polarized protrusion formation. In our model, the spatially coordinated localization patterns of these proteins regulate RhoC activity within dynamic activity zones at the leading edge protrusion. Focused RhoC activity determines the directionality of leading edge protrusions, by spatially restricting actin polymerization. Interestingly, we have found that p190RhoGEF and p190RhoGAP play an important role in determining the sites of actin polymerization. By interfering with their activities, actin barbed end amplitude can be spatially and temporally modulated. This suggests a dynamic control of barbed end production and protrusion activity that can be finely tuned by the upstream regulators of RhoC GTPase. Overall, these results show that the p190RhoGEF/p190RhoGAP/RhoC signaling pathway has a critical role in achieving cellular asymmetry during cell protrusion and lead to a new model of how actin polymerization sites are generated and sustained during directional cell protrusion.
Results
RhoC is necessary for protrusion formation during chemotaxis
Invasive tumor cells are chemotactic and form lamellipodial protrusions towards EGF (Segall et al., 1996). We sought to test how RhoC is involved in lamellipodium protrusion formation after EGF stimulation. We used the MTLn3 adenocarcinoma cell line because it is well characterized for the formation of broad flat lamellipodia with no ruffles (supplementary material Fig. S16), so there is negligible contribution due to thickness variation to the fluorescence intensity (Bailly et al., 1998a; Bailly et al., 1998b; Chan et al., 1998; Rotsch et al., 2001) facilitating the study of the actin rearrangements in the early stages of protrusion formation after EGF stimulation (Segall et al., 1996). We knocked down RhoC in MTLn3 cells (supplementary material Fig. S1a) and studied how EGF-stimulated protrusions were affected. Results indicate that protrusive activity as well as other protrusion parameters are reduced upon RhoC depletion (Fig. 1A,B; supplementary material Fig. S1b,c; Movies 1 and 2).
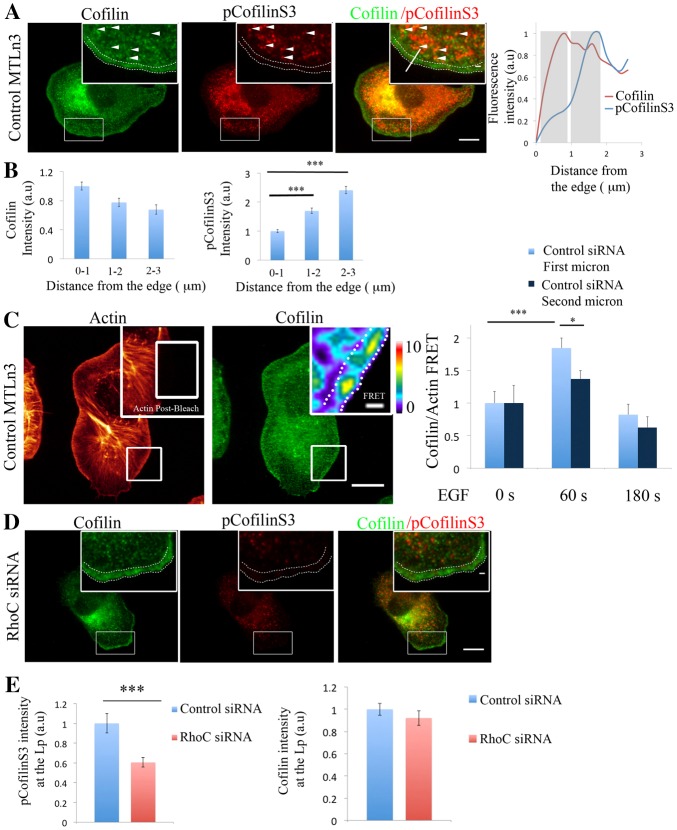
Fig. 1.
RhoC is essential for protrusion formation and directional protrusion during chemotaxis. (A) Still images from time-lapse movies of control- and RhoC-siRNA-treated MTLn3 cells stimulated with EGF. Dotted lines show the cell edge. (B) Quantification of protrusive activity in response to EGF in control- and RhoC-siRNA-treated cells. Membrane protrusion is standardized to time 0. Number of cells analyzed: control siRNA = 26 cells, RhoC siRNA = 28. (C) Still images from time-lapse movies of control- and RhoC-siRNA-treated MTLn3 cells stimulated with an EGF-filled micropipette (positioned at the asterisk). The white arrows indicate the resulting directions of protrusion. (D) Standardized membrane protrusion at the front and the back of control- and RhoC-siRNA-treated MTLn3 cells during the 10 minutes after EGF stimulation. Number of cells analyzed: control siRNA = 23 cells, RhoC siRNA = 18 cells. (E) The average chemotactic index of control- and RhoC-siRNA-treated MTLn3 cells over 10 minutes expressed as the cosine (θ). Number of cells analyzed: control siRNA = 24 cells, RhoC siRNA = 11 cells. (F) The polarity index, which corresponds to the angle at which the maximum protrusion is occurring in reference to the pipette position at each time point. The cosine values of these angles are calculated, and these values represent the polarity index of the cell at each time point after stimulation (see Materials and Methods for a detailed description). The polarity indices from different cells are averaged and then plotted versus time after stimulation. Number of cells analyzed: control siRNA = 20 cells, RhoC siRNA = 24 cells. Scale bars: 10 µm. *P<0.05. Error bars indicate the s.e.m.
Gradient sensing of chemotactic cues is the initial step in chemotaxis. To explore the role of RhoC during gradient sensing, we applied local EGF signals to cells using a pipette. Control MTLn3 cells show newly formed protrusions oriented towards the pipette, which demonstrates their ability to sense the EGF gradient (Fig. 1C,D; supplementary material Fig. S2a; Movie 3). However, cells knocked down for RhoC cannot properly sense the chemotactic gradient, resulting in protrusions with significantly reduced orientational accuracy towards the EGF gradient (Fig. 1C,D,F; supplementary material Fig. S2a,c; Movie 4). Chemotactic index measured as the cosθ (see Materials and Methods and supplementary material Fig. S2b) shows that chemotaxis towards EGF is severely impacted in cells depleted of RhoC (Fig. 1E; supplementary material Fig. S2b). By contrast, cell velocity is unchanged upon RhoC knockdown [control siRNA 0.65±0.07 µm/minute (n = 26 cells); RhoC siRNA 0.67±0.03 µm/minute (n = 24 cells)]. Together, these data show that RhoC is required to form directional protrusions during chemosensing for directional motility.
RhoC activity areas are localized behind the leading edge during cell protrusions
To determine how RhoC regulates leading edge protrusions, we examined RhoC activity using a RhoC FLARE.sc biosensor (Bravo-Cordero et al., 2011; J. Zawistowski, M. Sabouri-Ghomi, G. Danuser, K. M. Hahn and L.H., unpublished.). Using quantitative fluorescence biosensor imaging we observed that RhoC activity is enriched in areas behind the leading edge of the cell. The leading edge is defined as the region that extends 1 µm inward from the cell membrane (DesMarais et al., 2002), while RhoC activity was found between the second (1–2 µm from the cell edge) and third (2–3 µm from the cell edge) micrometer, in an EGF-driven protrusion (Fig. 2A,B; supplementary material Fig. S14).
Fig. 2.

RhoC is activated in areas behind the leading edge of the cell during EGF-stimulated protrusions. (A) Representative image of an MTLn3 cell expressing the RhoC biosensor and stimulated globally with EGF. White arrows point to the areas at the protrusion where RhoC activity is high. (B) Quantification of RhoC activity in areas of 1 µm depth and 10 µm length, starting at the cell edge of the MTLn3 cell stimulated globally with EGF for 1 minute. Values are normalized to the 0–1 µm values. Number of cells = 19. (C) RhoC activity and staining of cofilin in MTLn3 cells expressing RhoC biosensor before and 1 minute after stimulation with an EGF-filled micropipette. Asterisks indicate where the pipette was placed. RhoC activity and cofilin radial sweep of stimulated cell is shown (this sweep is the outermost 4 µm strip of the cell). Vertical white lines in the radial sweep indicate the positions where the front and the back of the cells are located with respect to the pipette. Spatial distances and horizontal line in the radial sweep images highlight the first and the second micrometers (D) RhoC activity and cofilin intensity profiles along the indicated dotted line in the radial-sweep images. Grey rectangles highlight the first and the second micrometer behind the cell edge. (E) Quantification of RhoC activity in an area of 1 µm width, 1 µm behind the cell edge in MTLn3 cells expressing RhoC biosensor and non-stimulated or stimulated for 1 minute with an EGF-filled pipette. Areas were analyzed in the regions facing the pipette and in the mirror back (see Materials and Methods for detailed description and supplementary material Fig. S2d). Scale bars: 10 µm (main image); 1 µm (insert). **P<0.01, ***P<0.001. The pseudocolor scale shows ratio limits of black to red for RhoC activity. Values are normalized to the first micrometer values. Error bars indicate the s.e.m.
Upon local EGF stimulation RhoC activity is polarized and elevated between the second and third micrometer area behind the leading edge (Fig. 2C,D; supplementary material Fig. S14 and Fig. S17). These areas are immediately behind the region where cofilin [a marker for leading edge (Chan et al., 2000)] localizes (Fig. 2D). RhoC is not activated in areas at the back of the cell (Fig. 2C,E; supplementary material Fig. S2d). These results suggest that RhoC activation facilitates directional protrusion towards an EGF source (Fig. 2E).
RhoC activation defines actin polymerization sites during leading edge protrusions
Activation of RhoC increases pCofilinS3 levels in a ROCK-dependent manner (Bravo-Cordero et al., 2011). Indeed, cofilin phosphorylation status is regulated by the RhoC/ROCK/LIMK signaling pathway in invasive tumor cells (Bravo-Cordero et al., 2011) (supplementary material Fig. S3a,b). Phosphorylation of cofilin is increased after 1 minute of EGF stimulation, remains elevated for 10 minutes after EGF stimulation (supplementary material Fig. S4) and gradually returns to unstimulated levels after 30 minutes (Song et al., 2006). This phosphorylation step is dependent on RhoC but not RhoA (supplementary material Fig. S4) and it is necessary to place cofilin activity to the very tip of the leading edge following release from PIP2 binding (Song et al., 2006; van Rheenen et al., 2007) during lamellipodium extension. Inhibiting cofilin activity by highly increasing phosphorylation levels (Zebda et al., 2000) (supplementary material Fig. S3c) prevents lamellipodium protrusions.
During protrusion formation after EGF stimulation, LIMK1 (Song et al., 2006) and pCofilinS3 (Fig. 3A,B; supplementary material Fig. S15) are enriched behind the leading edge. Analysis of the cofilin–actin binding by acceptor photobleaching FRET (van Rheenen et al., 2007) shows that this interaction takes place after 1 minute following the EGF stimulation and is localized within the very first micrometer of the leading edge (Fig. 3C). After RhoC depletion, we found that pCofilinS3 levels decreased in the lamellipodium while total cofilin levels remained constant (Fig. 3D,E). These results demonstrate that RhoC is required for increased pCofilinS3 levels during protrusion. We hypothesized that this effect will have a direct impact on the cofilin/actin binding. Upon RhoC depletion, cofilin and actin interaction, measured by acceptor photobleaching FRET, is increased, and the spatial confinement of cofilin activity at the very first micrometer of the leading edge is lost (Fig. 4A,B; supplementary material Fig. S5). We further confirmed these results by using in situ PLA (proximity ligation assay) to detect the interaction between cofilin and actin (supplementary material Fig. S6).
Fig. 3.
RhoC regulates pCofilinS3 levels in areas behind the leading edge during cell protrusion. (A) Representative images of MTLn3 cells stained for cofilin and pCofilinS3 showing cofilin and pCofilinS3 distribution at EGF-stimulated protrusions. Dotted double lines in all panels delineate the leading edge. White arrowheads indicate areas of colocalization between cofilin and pCofilinS3. Inserts in this and other figures show higher magnifications of the boxed regions. pCofilinS3 and cofilin intensity profiles along the indicated line in the overlay insert image are shown on the right. Grey rectangles highlight the first and the second micrometer behind the cell edge. (B) Quantification of cofilin and pCofilinS3 fluorescence intensity at adjacent regions of 1 µm width and 10 µm length at different distances from the edge, starting at the edge of the cell. Number of cells analyzed = 26 cells. P-values obtained by comparison with the control siRNA value. (C) Representative images of EGF-stimulated MTLn3 cells, fixed and labeled for cofilin (donor; Alexa Fluor 488) and F-actin (acceptor; Rhodamine). Using a high laser power, a region was bleached in the acceptor fluorescence channel (white box in the insert of the actin image). Insert: FRET efficiency image of the bleached area. Right panel: FRET efficiency was calculated in an area of 1 µm depth right at the edge of the cell or 1–2 µm behind the edge, in MTLn3 cells unstimulated and stimulated with EGF for 1 or 3 minutes (n>20 bleach regions per condition from different cells). FRET efficiency is normalized to control time = 0 s. Pseudocolor scale value represent FRET efficiency values. (D) Representative images of RhoC-siRNA-treated MTLn3 cells stained for cofilin and pCofilinS3. (E) Quantification of cofilin and pCofilinS3 fluorescence intensity at the lamellipodium [region of 3 µm from the cell edge to the cell interior (Chan et al., 2000; DesMarais et al., 2002)] in control- and RhoC-siRNA-treated MTLn3 cells stimulated with EGF. Values are normalized to control siRNA. Number of cells analyzed: control siRNA = 26, RhoC siRNA = 26. P-values obtained by comparison with the control siRNA value. Scale bars: 10 µm (main image); 1 µm (insert). *P<0.05, **P<0.01, ***P<0.001. Error bars indicate the s.e.m.
Fig. 4.
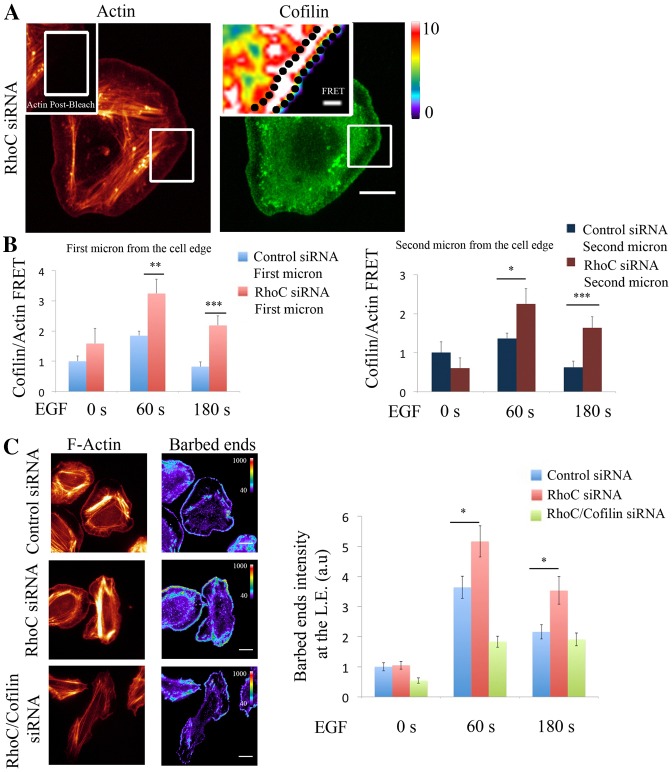
RhoC spatially confines cofilin/actin interaction to regulate cofilin-dependent barbed ends formation during leading edge protrusion. (A) Representative images of EGF-stimulated RhoC-siRNA-treated MTLn3 cells, fixed and labeled for cofilin (donor; Alexa Fluor 488) and F-actin (acceptor; Rhodamine). Insert: FRET efficiency image corresponds to the bleach area. (B) Graphs of FRET efficiency measurements calculated in: an area of 1 µm width right at the edge (left graph) or in an area 1 µm in width and 1 µm behind the cell edge (right graph), for control- and RhoC-siRNA-treated MTLn3 cells unstimulated, and stimulated with EGF for 1 or 3 minutes at each bleached region. n>10 bleach regions per condition from different cells. FRET efficiency is normalized to control time (0 seconds). Pseudocolor scale value represents FRET efficiency values. (C) Representative images of F-actin and barbed ends staining in: control-, RhoC- and RhoC/cofilin-siRNA-treated MTLn3 cells stimulated with EGF for 1 minute. Quantification of barbed end intensity in response to EGF at lamellipodia in control siRNA-treated MTLn3cells, RhoC-siRNA-treated MTLn3cells and RhoC/cofilin-siRNA-treated cells; n = 3 independent experiments with more than 20 cells per group. Scale bars: 10 µm (main images); 1 µm (inserts). P-values obtained by comparison with the control siRNA value; *P<0.05, **P<0.01, ***P<0.001. Error bars indicate the s.e.m.
Cofilin activity in protrusions involves the binding of cofilin to F-actin followed by severing to increase cofilin-dependent free actin barbed ends after 1 minute of EGF stimulation (supplementary material Fig. S7a,b). Based on our results we hypothesized that depletion of RhoC will have an impact on cofilin-generated barbed ends. Indeed, depletion of RhoC significantly increases barbed end formation at the leading edge protrusions in a cofilin-dependent manner (Fig. 4C; supplementary material Fig. S7c).
Collectively, these results demonstrate that cofilin interacts with actin at the leading edge of EGF-driven protrusions and RhoC spatially regulates this interaction as well as cofilin-mediated barbed end formation.
Spatial activation of RhoA regulates lamellar protrusion formation but not lamellipodium protrusion
The closely related isoform RhoA has been previously shown to mediate mDia-dependent actin filament polymerization at the lamellar compartment upon depletion of N-Wasp/Wave2 (Sarmiento et al., 2008). By using a RhoA FLARE.sc biosensor (Pertz et al., 2006) we localized RhoA activity in the first micrometer of the protruding lamellipodium (supplementary material Fig. S8a; Movie 9). Moreover, depletion of RhoA increases protrusive activity without affecting barbed end formation or cofilin phosphorylation levels (supplementary material Fig. S8b–d and Fig. S4). Depletion of N-WASp/Wave2 in cells expressing the RhoA biosensor (supplementary material Fig. S8e; Movie 10) reveals that RhoA is highly activated in the mDia1-dependent protrusions that facilitate lamellar protrusion in absence of the lamellipodium. Moreover, depletion of RhoC did not affect RhoA activation after EGF stimulation or spatial distribution of RhoA activity (supplementary material Fig. S9).
RhoC activity during protrusion is regulated by p190RhoGEF and p190RhoGAP spatial coordinators
We next sought to determine the GEF and GAP responsible for RhoC regulation at the leading edge. p190RhoGEF has been shown to mediate the activation of RhoA, RhoB and RhoC (Bravo-Cordero et al., 2011; Jaiswal et al., 2011; Lim et al., 2008; van Horck et al., 2001) but it is more efficient at catalyzing the nucleotide exchange of RhoC compared to RhoA or RhoB (Jaiswal et al., 2011) by fluorescence spectroscopy measurements. At invadopodia, RhoC activity is spatially regulated by p190RhoGEF. Given the importance of RhoC activity for efficient protrusion formation and actin dynamics in both compartments we tested how p190RhoGEF regulates RhoC at leading edge protrusions.
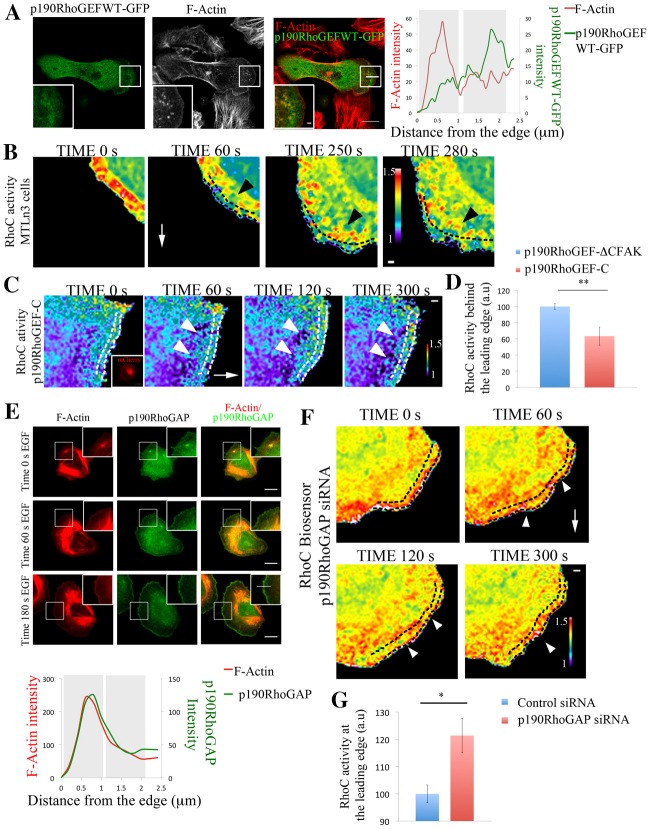
Live-cell imaging of the RhoC biosensor revealed that RhoC activity is highest in areas between 1 and 2 µm behind the cell edge and this is a dynamic process during protrusion formation (Fig. 5B; supplementary material Fig. S14; Movie 5). During protrusion, p190RhoGEF accumulates at discrete areas behind the leading edge, in areas similar to where RhoC is activated (Fig. 5A; supplementary material Fig. S10a,b; Movie 8). To test how RhoC activity is regulated by p190RhoGEF during EGF-induced lamellipodium protrusion, we overexpressed a mutant version of the GEF lacking the catalytic DH-PH domains. This mutant, named p190RhoGEF-C, competes against the endogenous GEF for proper localization through focal adhesion kinase (FAK) binding (due to the native FAK binding site at the C-terminus of the GEF being maintained) to act as a dominant negative but does not bind to GTPases (Lim et al., 2008; Zhai et al., 2003). As a control we used a further truncated version of this mutant lacking this FAK-binding site [referred to in Fig. 5C and throughout Fig. 6 as p190RhoGEF-CΔFAK (control)], containing just a few amino acids of the C-terminal domain of p190RhoGEF attached to mCherry, being diffusible in the cell cytoplasm with no activity (see Materials and Methods) (Zhai et al., 2003). p190RhoGEF-C (dominant negative mutant) affects the spatial activation of RhoC by decreasing RhoC levels in areas 1 µm behind the cell edge where RhoC is observed to be activated in p190RhoGEF-CΔFAK (control; Fig. 5C,D; supplementary material Fig. S14; Movie 6).
Fig. 5.
p190RhoGEF and p190RhoGAP regulate RhoC activity in areas behind the leading edge. (A) Representative images of MTLn3 cells expressing p190RhoGEFWT–GFP stimulated with EGF for 1 minute and stained for F-actin. Right panel: p190RhoGEFWT–GFP and F-actin intensity profiles along the indicated line in overlay image are shown. (B) Still images from a time-lapse movie of MTLn3 cells expressing RhoC biosensor after EGF stimulation. Dashed black lines delineate an area ∼1 µm depth from the cell edge. Black arrowheads point to areas of high RhoC activation behind the leading edge. White arrows point in the direction of the protrusion. Time points are the time after EGF was added. (C) Still images from a time-lapse movie of a MTLn3 cell expressing RhoC biosensor and p190RhoGEF-C–mCherry (inactive mutant; see Materials and Methods for detailed description) after EGF stimulation. Insert in the time 0 s panel shows the fluorescence intensity of p190RhoGEF-C–mCherry. The white arrow points in the direction of the protrusion. White arrowheads point to areas of fluorescence 1 µm behind the cell edge. Dashed white lines represent an area of approximately 1 µm depth from the cell edge. Time points are the times after EGF was added. (D) Quantification of RhoC activity in an area of 1 µm width and 10 µm length, 1 µm behind the cell edge in cells expressing p190RhoGEF-CΔFAK–mCherry (control; see Materials and Methods for detailed description) and p190RhoGEF-C–mCherry (inactive mutant) and stimulated with EGF for 1 minute. Number of cells analyzed: control (p190RhoGEF-ΔCFAK) = 11, p190RhoGEF-C = 11. P-values obtained by comparison with the control siRNA value. Values are normalized to p190RhoGEF-CΔFAK (control). (E) Representative images of MTLn3 cells stained for F-actin and p190RhoGAP at different times after EGF stimulation. Bottom panel: p190RhoGAP and actin intensity profiles along the line shown in the 180 s EGF overlay insert image. (F) Still images from a time-lapse movie of a p190RhoGAP-siRNA-treated MTLn3 cell expressing RhoC biosensor, after EGF stimulation. Time points are the time after EGF was added. The white arrow points in the direction of the protrusion. White arrowheads point to areas at the leading edge showing high RhoC activation. Dashed black lines delineate an area of ∼1 µm depth from the cell edge. (G) Quantification of RhoC activity in an area of 1 µm width and 10 µm length, starting at the cell edge in control- and p190RhoGAP-siRNA-treated MTLn3 cells stimulated with EGF for 1 minute. P-values obtained by comparison with the control siRNA value. Number of cells analyzed: control siRNA = 12, p190RhoGAP siRNA = 24. The pseudocolor scale here shows ratio limits of black to red for RhoC activity. Scale bars: 10 µm (main images); 1 µm (inserts). *P<0.05, **P<0.01. Error bars indicate the s.e.m.
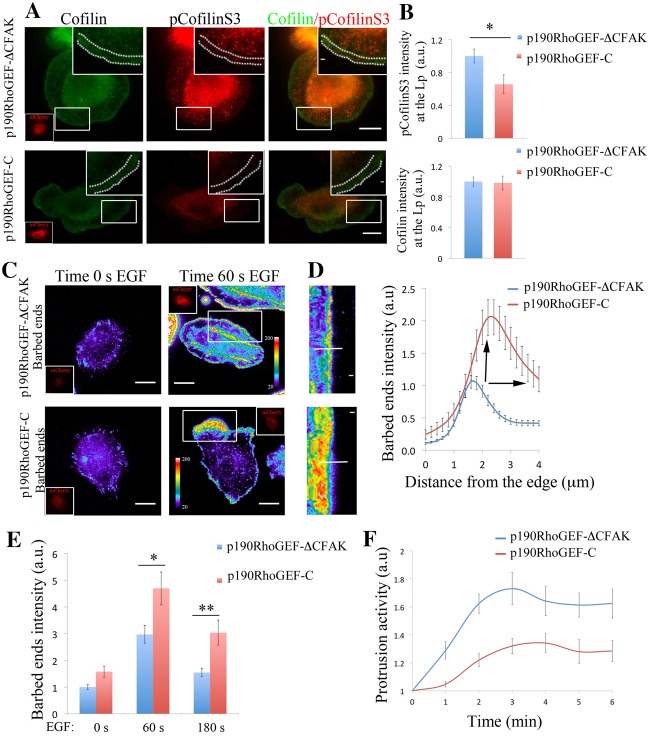
Fig. 6.
p190RhoGEF regulates pCofilinS3 levels, barbed ends amplitude and protrusion formation. (A) Representative images of MTLn3 cells expressing p190RhoGEF-CΔFAK–mCherry (control) and p190RhoGEF-C–mCherry (inactive mutant) stained for cofilin and pCofilinS3 showing cofilin and pCofilinS3 distribution at EGF-stimulated protrusions. Dotted lines in the enlarged image delineate the leading edge. Inserts in cofilin panels show the mCherry fluorescence intensity of p190RhoGEF-CΔFAK–mCherry or p190RhoGEF-C–mCherry. (B) Quantification of cofilin and pCofilinS3 fluorescence intensity at the lamellipodium of MTLn3 cells expressing p190RhoGEF-CΔFAK–mCherry (control) and p190RhoGEF-C–mCherry, and stimulated with EGF. Values are normalized to p190RhoGEF-ΔC–mCherry. Number of cells analyzed: p190RhoGEF-CΔFAK–mCherry = 24, p190RhoGEF-C–mCherry = 11. P-values obtained by comparison with the p190RhoGEF-CΔFAK–mCherry value. (C) Representative images of barbed ends staining after 1 minute EGF stimulation, in MTLn3 cells expressing p190RhoGEF-CΔFAK–mCherry (control) and p190RhoGEF-C–mCherry. (D) Radial sweep of white boxes in panel C. A white line is used to generate the quantification depicted in the right panel. Right panel: fluorescence intensity of barbed end measured from the cell edge (0 µm) into the cell (4 µm) through a region 5 µm wide, in p190RhoGEF-CΔFAK–mCherry (control) and p190RhoGEF-C–mCherry MTLn3 cells. Values are the means of 34 cells (with three regions per cell) in p190RhoGEF-CΔFAK (control) and 30 cells (with three regions per cell) in p190RhoGEF-C–mCherry. Black arrows point to the increase in p190RhoGEF-C–mCherry barbed end amplitude compared with that of the control. (E) Quantification of barbed end intensity in response to EGF at lamellipodia in p190RhoGEF-CΔFAK (control) and p190RhoGEF-C–mCherry MTLn3 cells. n = 3 independent experiments with ∼45 cells per group. (F) Quantification of protrusive activity in response to EGF in MTLn3 cells expressing p190RhoGEF-CΔFAK–mCherry (control) and p190RhoGEF-C–mCherry. Membrane protrusion is standardized to time 0. Number of cells analyzed: p190RhoGEF-CΔFAK–mCherry (control) = 7, p190RhoGEF-C–mCherry = 9. Scale bars: 10 µm (main images); 1 µm (inserts). Pseudocolor scale bar shows the barbed end intensity. *P<0.05, **P<0.01. Error bars indicate the s.e.m.
p190RhoGAP can catalyze the GTP hydrolysis of RhoC (Kusama et al., 2006; Thomas et al., 2011; Wang et al., 2003) regulating RhoC activity (supplementary material Fig. S11). p190RhoGAP was shown to regulate protrusion and polarity through inactivation of RhoA in fibroblasts (Arthur and Burridge, 2001) and RhoC activity at invadopodial protrusions (Bravo-Cordero et al., 2011). In MTLn3 cells, p190RhoGAP is enriched in areas close to the membrane at the leading edge during protrusion formation and present also within the cytoplasm (Fig. 5E; supplementary material Fig. S11) contributing to maintaining the low activity of RhoC within such areas (supplementary material Fig. S11) since depletion of p190RhoGAP spatially disrupted the RhoC activation zones and increased RhoC activity immediately at the leading edge and in other areas (Fig. 5F,G; supplementary material Movie 7; Fig. S14). These results suggest that these two molecules, p190RhoGEF and p190RhoGAP act synergistically as RhoC modulators, spatially defining the areas where RhoC is activated.
p190RhoGEF and p190RhoGAP spatially balance actin barbed end amplitude for efficient protrusion
We next questioned how p190RhoGEF and p190RhoGAP contribute to the pCofilinS3 levels at leading edge protrusions, the balance of actin barbed end formation in time and space and ultimately, how they regulate lamellipodial protrusion. Overexpression of p190RhoGEF-C decreases total pCofilinS3 (supplementary material Fig. S10c) levels and in areas behind the leading edge (Fig. 6A,B) and knocking down p190RhoGAP generates a dramatic redistribution of the pCofilinS3 pool to the leading edge of the cell where pCofilinS3 is normally absent in control cells (Fig. 7A,B). These results show that inhibition of p190RhoGAP displaces pCofilinS3 to the leading edge as it does for RhoC activation, while inhibition of p190RhoGEF activity inhibits cofilin phosphorylation.
Fig. 7.
p190RhoGAP spatially restricts pCofilinS3 and barbed ends amplitude for efficient protrusion formation. (A) Representative images of control- and p190RhoGAP-siRNA-treated MTLn3 cells stained for cofilin and pCofilinS3 showing cofilin and pCofilinS3 distribution at EGF-stimulated protrusions. Dotted lines in the enlarged images delineate the leading edge. Right panel: pCofilin and cofilin intensity profiles along the indicated line in the close up insert on the overlay image. (B) Quantification of cofilin and pCofilinS3 fluorescence intensity at the lamellipodium of control- and p190RhoGAP-siRNA-treated MTLn3 cells, stimulated with EGF. Values are normalized to those of control cells. Number of cells analyzed: control siRNA = 38 cells, p190RhoGAP siRNA = 35 cells. P-values obtained by comparison with the control siRNA value. (C) Representative images of barbed ends and F-actin staining in response to EGF stimulation in control and p190RhoGAP siRNA MTLn3 cells. (D) Radial sweep of white boxes in panel C. A white line is used to generate the quantification depicted in the right panel. Right panel: fluorescence intensity of barbed end measured from the cell edge (0 µm) into the cell (4 µm), through a region 5 µm wide, in control- and p190RhoGAP-siRNA-treated MTLn3 cells. Values are the means of 24 control cells (with three measurements per cell) and 21 (with three measurements per cell) p190RhoGAP siRNA cells. Black arrows point to the decrease in p190RhoGAP siRNA barbed end amplitude compared with that of the control. (E) Quantification of barbed end intensity in response to EGF at lamellipodia in control- and p190RhoGAP-siRNA-treated MTLn3 cells; n = 3 independent experiments with ∼35 cells per group. (F) Quantification of protrusive activity in response to EGF in control- and p190RhoGAP-siRNA-treated MTLn3 cells. Membrane protrusion is standardized to that at time 0. Number of cells analyzed: control siRNA = 26 cells, p190RhoGAP siRNA = 28 cells. Scale bars: 10 µm (main image); 1 µm (insert). Pseudocolor scale bar shows the barbed end intensity **P<0.01, ***P<0.001. Error bars indicate the s.e.m.
By interfering with these two molecules, we spatially modulated the amplitude of the barbed end area at the first phase of actin polymerization during lamellipodium protrusion (Fig. 6C,D; Fig. 7C,D). Cells expressing p190RhoGEF-C (dominant negative) have an increase in barbed ends (i.e. increased amplitude), and the barbed ends are no longer restricted to a very sharp band at the edge of the lamellipodium as it is in control cells (Fig. 6C–E). In contrast, the barbed end amplitude is decreased in cells depleted for p190RhoGAP (Fig. 7C–E).
These results show that the effect on barbed ends is mediated by the regulatory mechanism of these GEF/GAP molecules over the RhoC/cofilin pathway (Fig. 5; Fig. 6A,B; Fig. 7A,B). The activities of p190RhoGEF and p190RhoGAP are crucial for keeping the active versus the inactive cofilin pool spatially separated to properly focus actin barbed end generation during protrusion formation (Fig. 6A,B; Fig. 7A,B).
Interestingly, protrusion activity is affected in the same way when interfering with either of p190RhoGEF or p190RhoGAP (Fig. 6F; Fig. 7F). In either of these cases, the overall protrusion is significantly impaired when p190RhoGAP is depleted or when p190RhoGEF activity is inhibited by expressing the p190RhoGEF-C, suggesting that cells are losing the balance between RhoC activation and deactivation and are not able to form a polarized protrusion.
Other GEFs and GAPs known to regulate Rho GTPases activity include p115RhoGEF, LARG (activated also by integrins as p190RhoGEF) (Dubash et al., 2007; Jaiswal et al., 2011) and DLC-1 GAP (Healy et al., 2008). However, they display different patterns of localization compared to p190RhoGEF or p190RhoGAP during lamellipodium protrusion. While the p115 and LARG localize right at the tip of lamellipodial protrusions, DLC-1 localizes at focal adhesions (supplementary material Fig. S12a; Fig. S13a). None of these molecules affect cofilin phosphorylation on Ser3 (supplementary material Fig. S12d; Fig. S13d). Also none affect lamellipodium protrusion except for LARG (supplementary material Fig. S12c; Fig. S13c) where its knockdown decreased protrusion activity but not affect cofilin phosphorylation.
Discussion
Actin polymerization is a driving force that allows cells to extend the cell membrane in different physiological processes. From Dictyostelium (King and Insall, 2009) to tumor cells (Mouneimne et al., 2006) extension of a protrusion towards a chemotactic signal is the initial response during directional movement. During chemotaxis, actin polymerization takes place at the edge facing the chemotatic source. In this way, highly motile cells like neutrophils and tumor cells respond to chemotactic signals by adopting a polarized morphology. But how do motile cells know where to establish a leading edge? To achieve this feat, motile cells must restrict actin polymerization to the leading edge and this restriction has to be mediated by spatial placement of different components that regulate the actin polymerization machinery. We show here that: (1) spatial regulation of a leading edge protrusion is mediated by RhoC, restricting cofilin activity and actin dynamics to the leading edge of the cell; (2) p190RhoGEF and p190RhoGAP molecules are important regulators of RhoC activation during leading edge protrusion; and (3) barbed end formation and actin polymerization can be spatially modulated by regulating the activity of RhoC through p190RhoGEF and p190RhoGAP activities. Identification of the molecules that carefully fine-tune the areas of actin polymerization introduces a new perspective on how actin polymerization is spatially regulated, and thus how directional cell migration is determined.
Our results support a new model (Fig. 8) for actin polymerization and barbed end generation where the amplitude of barbed ends at leading edge protrusions can be fine-tuned at a molecular level by modulating RhoC GTPase activity, by p190RhoGAP/p190RhoGEF, and ultimately cofilin severing activity. To date it has been an open question as to how cells maintain active cofilin right at the leading edge after it is released from the membrane during protrusion. Our data here shed light onto this question. We show that RhoC activation behind the leading edge determines the localization of active cofilin and the sites of actin polymerization by increasing phosphorylated cofilin behind the leading edge. This spatially distinct RhoC activation sculpts the shape and amplitude of the zone of actin polymerization to define the shape and size of leading edge protrusions. In cells that have high motile activity (such as tumor cells), the ability to dynamically change the amount and distribution of actin barbed ends is a spatially precise and efficient approach to regulate protrusion formation and thus allow the cells to quickly change direction. This RhoC activation pattern supports a barbed end creation mechanism for cofilin (DesMarais et al., 2005). This model is applicable to cells that are highly motile and capable of quickly changing direction in contrast to the classic treadmill model, where cofilin is speculated to only supply actin monomers disassembled from the pointed ends of actin filaments at the rear of the protrusion (Pollard and Borisy, 2003).
Fig. 8.
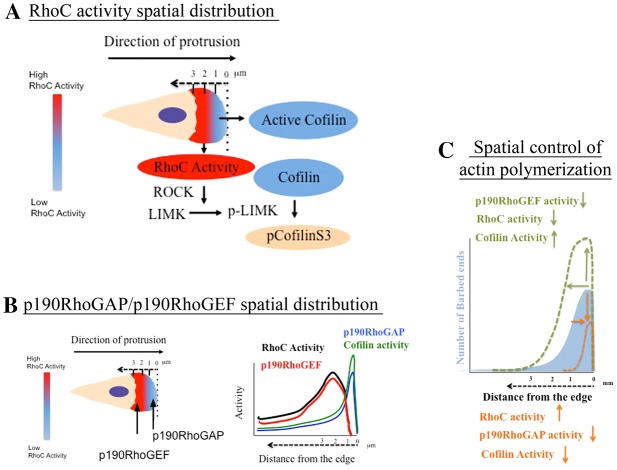
Spatiotemporal control of actin polymerization by p190RhoGEF/p190RhoGAP/RhoC/cofilin during lamellipodium protrusion. (A) During EGF-stimulated lamellipodium protrusions RhoC is activated in areas 1–3 µm behind the cell edge. In those areas, activation of RhoC leads to cofilin phosphorylation by the ROCK/LIMK pathway. Red indicates areas of high RhoC activity behind the leading edge and blue indicates areas of low RhoC activity and also areas of active cofilin. Cream color represents the cytosol. The dashed arrow shows the distance from the edge. Pseudocolor scale bar represents how RhoC activity is distributed at a protruding lamellipodium. (B) Activation of RhoC is confined behind the edge by the action of p190RhoGEF located behind the leading edge and p190RhoGAP located right at the leading edge. In this way, bursts of high RhoC activity are localized to those areas between 1 and 2 µm behind the cell edge. The graph on the left shows where the activities will be located along the lamellipodium. (C) The spatiotemporal regulation of RhoC activity has a direct impact on the distribution of cofilin as a driver of actin polymerization . Our model proposes a new mechanism of barbed end amplitude control where the amount and distribution of RhoC activity by p190RhoGEF and p190RhoGAP molecules dictates the areas of actin polymerization. By modulating the activities of any of these components (p190RhoGAP/p190RhoGEF/RhoC), actin polymerization can be precisely fine-tuned at the leading edge during lamellipodium protrusion, increasing or decreasing the amount of actin polymerization sites. The blue filled area represents barbed end amplitude in control cells. The dotted green line represents barbed end amplitude upon: decrease of p190RhoGEF, decrease of RhoC activity or when cofilin activity is increased. The orange dotted line represents barbed ends amplitude upon: increase RhoC activity, decrease of p190RhoGAP activity or decrease of cofilin activity.
Furthermore, our results suggest a complex relationship between actin polymerization and the leading edge protrusion. More actin polymerization does not necessarily mean more efficient protrusion since the spatial location of the barbed ends needs to be carefully controlled for efficient protrusion and directed cell migration. Here we have shown that the spatial localization of RhoC activity acts as a directional compass to restrict where actin polymerization may occur by confining cofilin-severing activity, which determines where protrusions are formed.
Our study demonstrates the existence of dynamic activity zones of RhoC and RhoA during cell protrusion. While RhoA localizes at the tip of the leading edge and mediates lamellar extension through the activation of a mDia-dependent pathway (Sarmiento et al., 2008), RhoC restricts actin polymerization at the tip of the lamellipodium through regulation of the cofilin pathway. The strong localization of RhoA at the tip of the lamellipodium is involved in inhibiting Rac as previously shown (El-Sibai et al., 2008) and may stabilize microtubules at leading edge focal adhesions through the RhoA–mDia pathway (Yamana et al., 2006). In other cell systems including leukocytes (Worthylake and Burridge, 2003), RhoA is involved in regulation of the membrane protrusions and ROCK/LIMK/cofilin activity but not in invasive tumor cells as shown here. These possible roles of RhoA may explain the increase in protrusive activity after RhoA depletion. As shown here, the GEF LARG localizes at the leading edge where RhoA activity is highly enriched and it is necessary for lamellipodium formation. Therefore, we speculate that LARG mediates the activation of a specific pool of RhoA to facilitate adhesion, and thereby stabilization of the lamellipodium (Dubash et al., 2007). However, it is not involved in cofilin regulation.
How are the RhoC activity zones initiated and sustained? The spatial distribution of p190RhoGEF and p190RhoGAP generate spatially constrained bursts of RhoC activity behind the leading edge that appears to be a major component of the cellular directional sensing during chemotaxis. These two molecules contribute to the spatial distribution and levels of the pCofilinS3 pool. Interfering with any of the components of the axis of p190RhoGEF/p190RhoGAP/RhoC/cofilin affect protrusion formation similarly. The results of studies of the leading edge of locomotory protrusions such as lamellipodia as described here, and invadopodia described previously (Bravo-Cordero et al., 2011) indicate that a common mechanism, the p190RhoGEF/p190RhoGAP/RhoC/cofilin signaling module, is at work in regulating protrusion size and shape.
The next layer of complexity in GTPase signaling is how GEF/GAP activation is regulated in order to control the RhoC-GTPase activity. GEFs are multidomain proteins that can bind to other proteins and lipids (Bos et al., 2007). Based on our results and others, we hypothesize that in the case of p190RhoGEF, FAK could be mediating GEF recruitment to specific areas at the leading edge (Lim et al., 2008). This interaction could localize p190RhoGEF to specific areas of the membrane where latent RhoC is delivered by GDI. Also, p190RhoGEF regulates RhoA downstream of integrins, suggesting that an integrin/FAK pathway may be involved in the activation of p190RhoGEF (Miller et al., 2012). By contrast, an association between p120Ras and p190RhoGAP has been shown to mediate p190RhoGAP localization to the leading edge in fibroblasts (Bradley et al., 2006). We can speculate that similar mechanisms could mediate p190RhoGAP/p190RhoGEF localization in tumor cells but more work needs to be done in order to understand precisely how GEFs and GAPs are recruited to different subcellular locations.
Cell motility is an important component of pathological processes like metastasis, and RhoC has previously been shown to be essential for metastasis (Clark et al., 2000). In addition, the invasive and migratory populations of tumor cells in primary breast tumors overexpress RhoC and some of the genes of the cofilin signaling pathway indicating over activation of the p190RhoGEF/p190RhoGAP/RhoC/cofilin axis (Bravo-Cordero et al., 2011; Wang et al., 2004; Wang et al., 2007). While RhoC depleted cells show 2D locomotion with no change in overall cell velocity, protrusion formation is dramatically inhibited. Recent results from Lauffenburger lab (Meyer et al., 2012) show that acute regulation of protrusion dynamics in 2D (but not other parameters like speed or persistence) reflects long-term 3D migration response and this correlates with the invasive behavior of tumor cells. These results are consistent with our findings and suggest that protrusion formation is a key indicator of migratory behavior of invasive tumor cells in the tumor microenvironment and highlights the importance of elucidating the mechanisms of protrusion formation to understand complex processes like tumor metastasis. Our results explain how RhoC mediates tumor metastasis, by regulating actin dynamics in cell protrusion. This report and our previous work (Bravo-Cordero et al., 2011), showing that RhoC mediates the focused formation of invadopodial protrusions, reinforce the role of RhoC as a key regulator of directed protrusion activity as needed for efficient metastasis. The axis p190RhoGEF/p190RhoGAP/RhoC/cofilin may be one of the predominant pathways that regulate tumor cell metastasis and may be a therapeutic target to prevent metastasis.
Materials and Methods
Cell culture and DNA transfection
For all experiments, MTLn3 cells, derived from the 13762NF rat mammary adenocarcinoma, were cultured in α-MEM supplemented with 5% FBS and antibiotics as described previously (Mouneimne et al., 2004). Transfections were performed 24 hours before each experiment by applying 2 µg DNA mixed with Lipofectamine to MTLn3 cells. The MTLn3 cell lines stably expressing the tet-OFF inducible RhoC FLARE.sc (single chain) and RhoA FLARE.sc biosensors were cultured in the presence of doxycyclin (1 µg/ml) to repress the biosensor expression during normal propagation. The drug was removed 72 hours prior to the experiment as previously described (Bravo-Cordero et al., 2011). Briefly, RhoC FLARE.sc inducible, stable cell line was produced by using retroviral transduction of first, virus containing the expression cassette for the tet-OFF system (Clontech) and the stable selection using G418 (1 mg/ml), followed by a second viral transduction containing the biosensor expression cassette driven by tetracyclin-inducible CMV promoter (Clontech). The transduced cells were selected for stable incorporation of the biosensor expression cassette using puromycin (10 µg/ml). Cells were then FACS sorted to tightly gate for near-identical expression levels of the biosensor, equating to ∼20% of the endogenous protein levels (Bravo-Cordero et al., 2011). In addition, these methods of producing biosensor stable cell lines in order to achieve the low but uniform expression levels are described in detail in the previous publications (Hodgson et al., 2008; Hodgson et al., 2010; Machacek et al., 2009; Pertz et al., 2006).
EGF upshift assays
The EGF upshift assay was performed as described previously (Mouneimne et al., 2004). In brief, cells were starved in L15 medium (GIBCO BRL) supplemented with 0.35% BSA for 3 to 4 hours. Cells were then stimulated with a bath application of 5 nM EGF (Invitrogen), treated at 37°C. Protrusive activity was quantified as an increase in total cell area as described previously (Segall et al., 1996).
Micropipette assays
The micropipette assay was performed as described previously (Mouneimne et al., 2006). Time-lapse series were taken using 20×objectives on a on a wide-field microscope (Inverted Olympus IX70) and analyzed using Image J. Directional protrusion and chemotaxis index was calculated as previously described (Mouneimne et al., 2006). Briefly, front and back protrusions were measured along a line going through the centroid and the tip of the micropipette (supplementary material Fig. S2a): front protrusion is the distance from the centroid of the cell to the point of intersection between the cell perimeter and the line going through the centroid and pipette tip, at the side of the pipette (the front side); back protrusion is the distance from the centroid of the cell to the point of intersection but at the other side of the cell (the back side). Protrusion values were standardized over the corresponding value at time 0. Chemotaxis index is the cosine of the angle between the line formed by the initial centroid (before the micropipette introduction) and the last centroid (after 10 minutes). For the polarity index measurements a custom ImageJ macro was written, which calculates the distance between the cell centroid and the cell edge at every 22.5° for every time point. The angle 0° was defined as the line connecting the cell centroid to the pipette tip. The angle that is protruding more at each time point is annotated and cosine of that angle is calculated. Micropipette biosensor imaging experiments were performed in a similar way and cells were fixed and stained once the experiment is done. Regions were selected as shown in supplementary material Fig. S2d and average intensity per unit area was measured for RhoC activity.
The front of the cell is defined as the region situated within a sector that is at a 35° angle (∼10 µm arc length) in reference to the vector linking the centroid and the pipette tip because this is the range of angles of protrusions exhibited by cells during chemoattractant sensing. Leading edge was defined as the region that extends 1 µm inward from the cell membrane where most of actin polymerization occurs (Chan et al., 2000; DesMarais et al., 2002). The analysis in each region is done by using a custom computer algorithm (detailed in leading edge analysis section) and the average intensity per unit area was measured for RhoC activity.
p190RhoGEF constructs
A mutant version of p190RhoGEF in which the C-terminal portion of the GEF containing the FAK-binding site (p190RhoGEF-C) was overexpressed as the functional competitor to the endogenous GEF, was produced by first PCR amplifying the fragment of p190RhoGEF (3839–4742 base pairs) using a primer pair: 5′-gatatatatttaattatgaattcgagagtctgcaagtggctgtgaagg-3′ and 5′-ctccctcgtaattatataaattattctcgagtcatgatgggtttggtttattggacgtg-3′, encoding EcoRI and XhoI restriction sites. The resulting fragment was spliced into the pTriEX-His-Myc-4 backbone at the EcoRI/XhoI sites. This construct further contained a mCherry at the NcoI/EcoRI restriction sites. A control version of this mutant GEF (p190RhoGEF-CΔFAK) in which the C-terminal portion of the GEF did not contain the FAK-binding site, was produced by PCR amplifying the fragment of p190RhoGEF (3902–4742 base pairs) using a primer pair: 5′-gatatatatttaattatgaattcggagggactgtcctgatggacacac-3′ and 5′-ctccctcgtaattatataaattattctcgagtcatgatgggtttggtttattggacgtg-3′, encoding EcoRI and XhoI restriction sites. The resulting fragment was spliced into the pTriEX-His-Myc-4 backbone at the EcoRI/XhoI sites. This construct further contained a mCherry at the NcoI/EcoRI restriction sites.
Immunofluorescence
All experiments in this study were performed using MTLn3 cells cultured on glass. 100,000 MTLn3 cells were plated 16 hours before fixation. The cells were fixed and immunofluorescence was performed as described previously (Eddy et al., 2000). The fixed cell experiments were performed on a wide-field microscope (Inverted Olympus IX70) and images were acquired with a cooled CCD camera (Sensicam QE cooled CCD camera) with a 60× NA = 1.4 oil immersion objective using IP Laboratory 4.0 software or on a Confocal SP5 AOBS with a 63× 1.4 NA oil-immersion objective (Leica, Mannheim, Germany) using Leica confocal software (LCS) for acquisition of images (Fig. 5A). For leading edge measurements of fluorescence intensity of cofilin and pCofilinS3 MTLn3 cells were stimulated with EGF at different time points, fix and stained with specific antibodies. At least three different regions of 10 µm in length were selected per cell in at least three different experiments. Using a custom written ImageJ macro, a region width of 1 or 3 µm in width (for lamellipodium measurements) right at the leading edge and 1, 2 and 3 µm behind was highlighted and average intensity per unit area was measured for cofilin and pCofilinS3.
Acceptor photobleaching experiments
Acceptor photobleaching experiments were performed and analyzed as previously described (Oser et al., 2009; van Rheenen et al., 2007). Briefly, donor and acceptor confocal images were collected before and after photobleaching of the acceptor on a laser scanning microscope (LSM 5 LIVE DuoScan; Carl Zeiss, Inc.). All images were corrected for laser fluctuations, overall sample bleaching and background. The post-bleached donor image was corrected for imaging bleaching and the gain in donor and the loss in acceptor fluorescence were determined at different areas. FRET was calculated by relating the gain of donor fluorescence to the total amount post-bleach acceptor fluorescence. All FRET calculations were performed on the mean values of regions of interest of areas of 1 µm depth right at the cell periphery or 1 µm inside. No significant photo conversion of the acceptor was observed during photobleaching. The formula used to calculate FRET efficiency is: (DonorPost−DonorPre)/DonorPost.
Barbed ends assay
The actin barbed end assay was performed using biotin–actin as described previously (Sarmiento et al., 2008). For barbed ends quantifications cells were traced and a custom designed macro automated the collection of pixel intensity in a perimeter of the cell starting at the cell edge and extending to 6 µm inside the cell in 0.2 µm steps. The averaged fluorescence intensity of the first 1 µm inside the cell was used to represent the barbed ends.
Reagents and primary antibodies
The antibodies used were as follows: p190RhoGAP (610149) mouse monoclonal antibody against p190RhoGAP was purchased from BD Laboratories. AE441 is an affinity purified rabbit polyclonal antibody raised against an 11 amino acid cofilin-derived peptide containing a phosphorylated serine residue at position 3 and recognizes phosphorylated cofilin (pCofilinS3) exclusively, as shown by isoelectric focusing gel analysis. AE774 is a chicken IgY antibody raised against purified recombinant full-length rat cofilin that recognizes both serine-3-phosphorylated and dephosphorylated cofilin (Song et al., 2006). AC-15 is a mouse monoclonal IgG1 antibody that recognizes the β-isoform of actin (Sigma-Aldrich, St. Louis, MO, USA). RhoC (D40E4) (Cell Signaling Technology, Inc., Danvers, MA). RhoA (26C4) (sc-418), LARG (H-70; sc-25638) a rabbit polyclonal antibody against LARG and DLC-1 (C-12; sc-271915) a mouse monoclonal against DLC-1 were purchased from Santa Cruz Biotechnology. p115RhoGEF rabbit monoclonal (D25D2) against p115RhoGEF was purchased from Cell Signaling. Wave2 and N-WASp rabbit polyclonal antibodies were used as previously described (Sarmiento et al., 2008). DiI lipophilic tracer dye was purchased from Invitrogen and used following manufacturer's instructions.
Live-cell imaging of RhoC and RhoA biosensors
Live-cell imaging of RhoC and RhoA FLARE.sc biosensors was done as previously described (Bravo-Cordero et al., 2011) (Hodgson et al., 2008; Hodgson et al., 2010; Machacek et al., 2009; Pertz et al., 2006). Briefly, MTLn3 cells stably incorporating RhoC or RhoA biosensor were starved in L15 medium (Invitrogen, Carlsbad, CA) supplemented with 0.35% BSA (starvation medium), for 3–4 hours. To stimulate cells, MTLn3 cells were treated with 5 nM epidermal growth factor (EGF); (Invitrogen, Carlsbad, CA) for various times. Imaging was performed in imaging medium [Ham's F-12K without phenol red (Biosource, Camarillo, CA), 10 mM Hepes and 10 µg/ml Oxyfluor reagent (Oxyrase Inc., Mansfield, OH) with 0.35% BSA] in a heated closed chamber.
Because the RhoA/C FLARE.sc biosensors are based on single-chain designs in which the FRET donor and acceptor halves are linked into a single molecule via an optimized linker, the local concentrations of the FRET donor and acceptor molecules are always equimolar (Hodgson et al., 2010; Pertz et al., 2006). Because of this specific feature, we are able to make the simplifying assumption that the ratio of FRET to the donor emission would be sufficient to describe the donor occupancy states (Zal and Gascoigne, 2004) and the resulting ratiometric data are useful for making relativistic comparisons of the GTPase-occupancy states. Thus, activation levels of Rho biosensors were measured in living cells by monitoring the ratio of the FRET between ECFP to Citrine-YFP and the donor ECFP intensities for RhoA sensor and monomeric Cerulean to monomeric Venus FRET and Cerulean intensities for the RhoC biosensor. Time-lapse sequences were acquired on a custom Olympus IX81ZDC inverted microscope with a beam splitter which enabled simultaneous acquisitions of both FRET and CFP channels using two Coolsnap ESII CCD cameras (Roper Photometrics, Tucson, AZ) mounted on the left-side 100% throughput port via an Olympus beamsplitters module, and a set of excitation/emission filterwheels to direct the DIC and the mCherry emission to a third camera (CoolsnapHQII; Roper Photometrics, Tucson, AZ) mounted on the bottom 100% throughput port of the microscope (Spiering and Hodgson, 2012). Images were obtained using an Olympus 60× PlanApoN 1.45 NA UIS2 DIC lens and the Metamorph software. The filter sets used for ratiometric imaging were (Excitation, emission, respectively): ECFP or mCerulean: ET436/20X, ET480/40M (Chroma Technology, Rockingham, VT); FRET: ET436/20X, ET535/30M (Chroma Technology). The main fluorescence turret utilized a 10/90 (reflection/transmittance) mirror (Olympus, Center Valley, PA) that provided compatibility with all of the band pass filters used. Additionally, a 575DCXR dichroic mirror (Chroma Technology, Rockingham, VT) was installed to replace the internal prism of the microscope, enabling beamsplitting of CFP–YFP FRET emissions to the left port of the microscope and all the longer wavelengths to the bottom port of the microscope. 505DCXRU dichroic mirror (Chroma Technology, Rockingham, VT) was used to split the CFP- and YFP-FRET emissions in the external beamsplitter attached to the left side port of the microscope. Cells were illuminated with a 100 W Hg arc lamp through a 10% transmittance neutral density filter. At each time point, CFP and FRET images were recorded simultaneously for 700 mseconds exposure time with binning 2×2, using the two side-mounted cameras. The images were taken at 10 second intervals for the total duration of up to 80 consecutive frames.
Image processing and analysis
Metamorph, version 7.8.0 (Molecular Devices, Sunnyvale, CA) was used to perform the image processing and data analysis. All images were corrected for the readout/dark-current noise, shading corrected, and background subtracted, as previously described (Hodgson et al., 2010). Because the dual-camera system was used (Spiering and Hodgson, 2012), fields of view in CFP and FRET channels were a priori calibrated using custom multispectral beads (Bangs Laboratories) and a morphing algorithm based on multi-order Legendre polynomials (Danuser, 1999) to achieve 1/10th of pixel accuracy in pixel-by-pixel matching. This technology corrected for any misalignments within the field of view including camera mounting angle differences, field shear and lateral chromatic dispersion differences within the microscope objective lens. Following the morphing, CFP/FRET images were manually thresholded based on the foreground signal intensities to generate corresponding binary masks with a value of zero outside the cell and a value of one inside the cell. Here, the edges of the cells were carefully analyzed in pseudocolor display to ascertain that we did not over or under-select the regions of the edge, especially where the leading edge structures were very thin. The morphed and corrected image sets were multiplied by the corresponding binary masks to set areas outside the cell uniformly zero to minimize noise and other artifacts associated with the ratiometric calculations (Hodgson et al., 2006). These images were then X–Y translation aligned automatically based on the global optimal cross-correlation analysis (Hodgsonet al., 2010; Shen et al., 2008) to ascertain registration with subpixel accuracy. Masked FRET image was divided by the masked CFP image to yield a ratio, reflecting RhoC activity throughout the cell. For image visualization purposes only, a linear pseudocolor lookup table (LUT) was applied to the ratiometric images and the ratio images were normalized to the lower scale value. This application of the LUT and visual scaling was for data representation only; thus the original raw ratio data values were unmodified by this process, important for the later analysis of the ratio values. In every data set, raw fluorescent images were carefully inspected to verify that all portions used to create the ratio image had a high enough signal to noise ratio, typically this equated to the signal to noise ratio of at least 2∶1 at the thinnest region of the cell (Hodgson et al., 2010). In time-lapse experiments, the ratio was corrected for photobleaching using a previously published method (Hodgson et al., 2006). The representative images shown in Fig. 5 and the movies represent a running sum of three sequential frames.
Leading edge analysis
Leading edge was defined as the region that extends 1 µm inward from the cell membrane where most of actin polymerization occurs (Chan et al., 2000; DesMarais et al., 2002). Leading edge analysis in fixed samples was performed as follows: We stimulated MTLn3 cells with 5 nM EGF, fixed and stained with appropriate antibodies to identify the leading edge using features including cofilin, cortactin or F-Actin staining. MTLn3 cells expand uniformly after EGF stimulation as seen in supplementary material Movie 1. Images of fixed cells were taken by using a wide-field Olympus inverted microscope with a 60× 1.4 NA oil immersion objective lens. For the leading edge measurements, we divided the cell into three regions of 120° each. We chose three regions of 10 µm long arc-length segments along the cell edge in those regions to mimic the areas used in the pipette stimulated experiments. Regions that protruded can be easily identified as flat protrusion extending outside of the cell containing cofilin, cortactin and F-actin at the very first micrometer. A computer algorithm (freely available upon request) was used to produce accurate regions of interest with the user-drawn 10 µm cell edge arc-length segment as the starting point from which a user-specified distance from the leading edge toward the inside of the cell (1 µm, 2 µm or 3 µm) was selected as the new regions of interest. Intensity measurements are made in these regions and intensity per unit area in each particular 1 µm depth segment was calculated.
Colocalization analysis
MTLn3 cells were fixed and stained for cofilin and pCofilinS3. The images were then analyzed for colocalization (Pearson correlation) using the Jacop colocalization plugin for ImageJ.
Proximity ligation assay
The cofilin and actin interaction was detected in situ using Duolink PLA Kits (Olink Bioscience, Sweden: Duolink II Detection Reagents Far Red fit, Duolink II Probemaker MINUS kit for making cofilin PLA probe by conjugation of the MINUS oligonucleotide with the anti-cofilin chicken IgY and Duolink II Probemaker PLUS kit for making β-actin PLA probe by conjugation of the PLUS oligonucleotide with the anti β-actin mouse IgG) according to manufacturer's instructions. Briefly, fixed cells were blocked in a pre-heated humidity chamber for 30 minutes at 37°C. PLA probes (Cofilin PLA MINUS and β-actin PLA PLUS) mixture were used at a 1∶400 dilution of each probe and incubated for 1 hour at 37°C. Nuclei were stained with Duolink II mounting medium containing DAPI. Images were acquired on epi-fluorescence Deltavision Core Microscope (Applied Precision, LLC), with 60×, 1.42 NA objective and analyzed using a custom-written ImageJ macro that calculates the number of PLA dots.
RNAi
The following rat siRNA reagents from Dharmacon (Thermo Scientific, Lafayette, CO, USA) were used: RhoC (L-089673), p190RhoGAP (M-088596), p115RhoGEF (L-099751), LARG (L-101376), DLC-1 (L-088843), RhoA L-095222 siRNA. siRNA duplexes were designed against the following gene sequences: Wave2 (5′-AAACCTATAACAGTGTGACG-3′) and N-WASP (5′-AAGACGAGATGCTCCAAATGG-3′) (Sarmiento et al., 2008). siRNA were transfected into MTLn3 cells at 100 nM each using Oligofectamine (Invitrogen, Carlsbad, CA) (Bravo-Cordero et al., 2011). The transfection was terminated after 4 hours by addition of 2×serum-containing medium. All experiments were performed 48 hours after transfection with more than 95% knockdown efficiency for the siRNA used (Bravo-Cordero et al., 2011). Control scrambled siRNA and cofilin siRNA (5′-AAGGTGTTCAATGACATGAAA-3′) sequences were described previously (Sidani et al., 2007).
Western blotting
For western blot analysis, whole-cell lysates were prepared by washing twice with ice-cold PBS before direct extraction with Laemmli sample buffer supplemented with the phosphatase inhibitors 50 mM NaF, 1 mM Na3VO4 and protease inhibitor cocktail set III (EMD Biosciences, San Diego, CA). Western blots were performed as follows: the samples were resolved by SDS-PAGE, transferred to nitrocellulose, blocked for 1 hour in Odyssey blocking solution (LI-COR, Lincoln, NE), incubated with primary antibodies overnight at 4°C. The appropriate secondary antibodies (mouse 680, rabbit 800; LI-COR, Lincoln, NE); chicken 800 (Rockland, Gilbertsville, PA) were incubated for 1 hour at RT and analyzed using the Odyssey Infrared Imaging System (LI-COR, Lincoln, NE). For quantification, the median gray value of the band was calculated after subtraction of the background.
RhoA pull-down assays
GST-PBD binding assay was done as described previously (Sarmiento et al., 2008). GST–Rhotekin–RBD was expressed in Escherichia coli and purified using glutathione–Sepharose beads. Cell lysates were incubated with GST–RBD protein for 1 hour at 4°C while rotating. Bound proteins were collected by centrifugation and resuspended in Laemmli buffer. RhoA was detected by western blotting with an anti-RhoA antibody.
Statistical analysis
Statistical significance was calculated using unpaired, two-tailed t-test. Values were considered statistically significant if the P<0.05. For all figures, * indicates P<0.05; ** indicates P<0.01; and *** indicates P<0.001. Error bars indicate the standard error of the mean (s.e.m.).
Supplementary Material
Acknowledgments
We thank the Condeelis, Cox, Hodgson and Segall laboratories for helpful discussions; the Analytical Imaging Facility of the Gruss Lipper Biophotonics Center at Albert Einstein College of Medicine; and the AECOM Flow Cytometry facility for technical help. The authors acknowledge Dr Keith Burridge (University of North Carolina at Chapel Hill) for providing the expression constructs for p115RhoGEF, LARG; and Dr Klaus M. Hahn (University of North Carolina at Chapel Hill) for providing the RhoC FLARE.sc biosensor.
Footnotes
Author contributions
J.J.B.-C., L.H. and J.C. designed the study; J.J.B.-C. performed the experiments that constitute the main body of this work; M.R.-J., V.P.S., X.C. and R.E. designed and performed additional experiments; J.C. and L.H. provided expertise and intellectual input, assisted in interpreting the data and coordinated the project; J.J.B.-C. took a leadership role in writing the manuscript; and J.C., L.H., M.R.-J. and V.P.S. edited the manuscript.
Funding
This work was funded by the National Institutes of Health [grant numbers GM064346 to J.J.B.-C.; GM093121 to L.H. and J.J.B.-C.; CA150344 to J.C., J.J.B.-C. and R.E.; CA100324 to X.C.; CA159663 to M.R.-J.]; a Sinsheimer Foundation Young Investigator Award [to L.H.]; and a postdoctoral fellowship from Susan G. Komen for the Cure© [grant number KG111405 to V.P.S.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.123547/-/DC1
References
- Arthur W. T., Burridge K. (2001). RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol. Biol. Cell 12, 2711–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly M., Condeelis J. S., Segall J. E. (1998a). Chemoattractant-induced lamellipod extension. Microsc. Res. Tech. 43, 433–443 [DOI] [PubMed] [Google Scholar]
- Bailly M., Yan L., Whitesides G. M., Condeelis J. S., Segall J. E. (1998b). Regulation of protrusion shape and adhesion to the substratum during chemotactic responses of mammalian carcinoma cells. Exp. Cell Res. 241, 285–299 10.1006/excr.1998.4031 [DOI] [PubMed] [Google Scholar]
- Benink H. A., Bement W. M. (2005). Concentric zones of active RhoA and Cdc42 around single cell wounds. J. Cell Biol. 168, 429–439 10.1083/jcb.200411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H., Wittinghofer A. (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Bradley W. D., Hernández S. E., Settleman J., Koleske A. J. (2006). Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol. Biol. Cell 17, 4827–4836 10.1091/mbc.E06-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J. J., Oser M., Chen X., Eddy R., Hodgson L., Condeelis J. (2011). A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr. Biol. 21, 635–644 10.1016/j.cub.2011.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. Y., Raft S., Bailly M., Wyckoff J. B., Segall J. E., Condeelis J. S. (1998). EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111, 199–211 [DOI] [PubMed] [Google Scholar]
- Chan A. Y., Bailly M., Zebda N., Segall J. E., Condeelis J. S. (2000). Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148, 531–542 10.1083/jcb.148.3.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E. A., Golub T. R., Lander E. S., Hynes R. O. (2000). Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532–535 10.1038/35020106 [DOI] [PubMed] [Google Scholar]
- Danuser G. (1999). Photogrammetric calibration of a stereo light microscope. J. Microsc. 193, 62–83 10.1046/j.1365-2818.1999.00425.x [DOI] [PubMed] [Google Scholar]
- DesMarais V., Ichetovkin I., Condeelis J., Hitchcock-DeGregori S. E. (2002). Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115, 4649–4660 10.1242/jcs.00147 [DOI] [PubMed] [Google Scholar]
- DesMarais V., Macaluso F., Condeelis J., Bailly M. (2004). Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 117, 3499–3510 10.1242/jcs.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Ghosh M., Eddy R., Condeelis J. (2005). Cofilin takes the lead. J. Cell Sci. 118, 19–26 10.1242/jcs.01631 [DOI] [PubMed] [Google Scholar]
- Dubash A. D., Wennerberg K., García-Mata R., Menold M. M., Arthur W. T., Burridge K. (2007). A novel role for Lsc/p115 RhoGEF and LARG in regulating RhoA activity downstream of adhesion to fibronectin. J. Cell Sci. 120, 3989–3998 10.1242/jcs.003806 [DOI] [PubMed] [Google Scholar]
- Eddy R. J., Pierini L. M., Matsumura F., Maxfield F. R. (2000). Ca2+-dependent myosin II activation is required for uropod retraction during neutrophil migration. J. Cell Sci. 113, 1287–1298 [DOI] [PubMed] [Google Scholar]
- El-Sibai M., Backer J. M. (2007). Phospholipase C gamma negatively regulates Rac/Cdc42 activation in antigen-stimulated mast cells. Eur. J. Immunol. 37, 261–270 10.1002/eji.200635875 [DOI] [PubMed] [Google Scholar]
- El-Sibai M., Pertz O., Pang H., Yip S. C., Lorenz M., Symons M., Condeelis J. S., Hahn K. M., Backer J. M. (2008). RhoA/ROCK-mediated switching between Cdc42- and Rac1-dependent protrusion in MTLn3 carcinoma cells. Exp. Cell Res. 314, 1540–1552 10.1016/j.yexcr.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D. S., Condeelis J. S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746 10.1126/science.1094561 [DOI] [PubMed] [Google Scholar]
- Healy K. D., Hodgson L., Kim T. Y., Shutes A., Maddileti S., Juliano R. L., Hahn K. M., Harden T. K., Bang Y. J., Der C. J. (2008). DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol. Carcinog. 47, 326–337 10.1002/mc.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson L., Nalbant P., Shen F., Hahn K. (2006). Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol. 406, 140–156 10.1016/S0076-6879(06)06012-5 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Pertz O., Hahn K. M. (2008). Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 85, 63–81 10.1016/S0091-679X(08)85004-2 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Shen F., Hahn K. (2010). Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. 46, 14.11.1–14.11.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R. H., Machesky L. M. (2009). Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17, 310–322 10.1016/j.devcel.2009.08.012 [DOI] [PubMed] [Google Scholar]
- Jaiswal M., Gremer L., Dvorsky R., Haeusler L. C., Cirstea I. C., Uhlenbrock K., Ahmadian M. R. (2011). Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG). J. Biol. Chem. 286, 18202–18212 10.1074/jbc.M111.226431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. S., Insall R. H. (2009). Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. 19, 523–530 10.1016/j.tcb.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Kusama T., Mukai M., Endo H., Ishikawa O., Tatsuta M., Nakamura H., Inoue M. (2006). Inactivation of Rho GTPases by p190 RhoGAP reduces human pancreatic cancer cell invasion and metastasis. Cancer Sci. 97, 848–853 10.1111/j.1349-7006.2006.00242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Lim S. T., Tomar A., Gardel M., Bernard-Trifilo J. A., Chen X. L., Uryu S. A., Canete-Soler R., Zhai J., Lin H. et al. (2008). PyK2 and FAK connections to p190Rho guanine nucleotide exchange factor regulate RhoA activity, focal adhesion formation, and cell motility. J. Cell Biol. 180, 187–203 10.1083/jcb.200708194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009). Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. S., Hughes-Alford S. K., Kay J. E., Castillo A., Wells A., Gertler F. B., Lauffenburger D. A. (2012). 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J. Cell Biol. 197, 721–729 10.1083/jcb.201201003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N. L., Lawson C., Chen X. L., Lim S. T., Schlaepfer D. D. (2012). Rgnef (p190RhoGEF) knockout inhibits RhoA activity, focal adhesion establishment, and cell motility downstream of integrins. PLoS ONE 7, e37830 10.1371/journal.pone.0037830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S. C., Ghosh M., Eddy R., Backer J. M., Condeelis J. (2004). Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166, 697–708 10.1083/jcb.200405156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. (2006). Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16, 2193–2205 10.1016/j.cub.2006.09.016 [DOI] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C. C., Bravo-Cordero J. J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A. J., Condeelis J. (2009). Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186, 571–587 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O. (2010). Spatio-temporal Rho GTPase signaling – where are we now? J. Cell Sci. 123, 1841–1850 10.1242/jcs.064345 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006). Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Ridley A. J. (2012). Historical overview of Rho GTPases. Methods Mol. Biol. 827, 3–12 10.1007/978-1-61779-442-1_1 [DOI] [PubMed] [Google Scholar]
- Rotsch C., Jacobson K., Condeelis J., Radmacher M. (2001). EGF-stimulated lamellipod extension in adenocarcinoma cells. Ultramicroscopy 86, 97–106 10.1016/S0304-3991(00)00102-9 [DOI] [PubMed] [Google Scholar]
- Sarmiento C., Wang W., Dovas A., Yamaguchi H., Sidani M., El-Sibai M., Desmarais V., Holman H. A., Kitchen S., Backer J. M. et al. (2008). WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J. Cell Biol. 180, 1245–1260 10.1083/jcb.200708123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. E., Tyerech S., Boselli L., Masseling S., Helft J., Chan A., Jones J., Condeelis J. (1996). EGF stimulates lamellipod extension in metastatic mammary adenocarcinoma cells by an actin-dependent mechanism. Clin. Exp. Metastasis 14, 61–72 10.1007/BF00157687 [DOI] [PubMed] [Google Scholar]
- Shen C. H., Chen H. Y., Lin M. S., Li F. Y., Chang C. C., Kuo M. L., Settleman J., Chen R. H. (2008). Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res. 68, 7779–7787 10.1158/0008-5472.CAN-08-0997 [DOI] [PubMed] [Google Scholar]
- Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J. M. et al. (2007). Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 179, 777–791 10.1083/jcb.200707009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Chen X., Yamaguchi H., Mouneimne G., Condeelis J. S., Eddy R. J. (2006). Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 119, 2871–2881 10.1242/jcs.03017 [DOI] [PubMed] [Google Scholar]
- Spiering D., Hodgson L. (2012). Multiplex imaging of Rho family GTPase activities in living cells. Methods Mol. Biol. 827, 215–234 10.1007/978-1-61779-442-1_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Overdevest J. B., Nitz M. D., Williams P. D., Owens C. R., Sanchez-Carbayo M., Frierson H. F., Schwartz M. A., Theodorescu D. (2011). Src and caveolin-1 reciprocally regulate metastasis via a common downstream signaling pathway in bladder cancer. Cancer Res. 71, 832–841 10.1158/0008-5472.CAN-10-0730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Horck F. P., Ahmadian M. R., Haeusler L. C., Moolenaar W. H., Kranenburg O. (2001). Characterization of p190RhoGEF, a RhoA-specific guanine nucleotide exchange factor that interacts with microtubules. J. Biol. Chem. 276, 4948–4956 10.1074/jbc.M003839200 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S. C., Backer J. M., Eddy R. J., Condeelis J. S. (2007). EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179, 1247–1259 10.1083/jcb.200706206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008). Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649–667 10.1016/j.ejcb.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Vega F. M., Fruhwirth G., Ng T., Ridley A. J. (2011). RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J. Cell Biol. 193, 655–665 10.1083/jcb.201011038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang L., Luo Y., Zheng Y. (2003). A novel strategy for specifically down-regulating individual Rho GTPase activity in tumor cells. J. Biol. Chem. 278, 44617–44625 10.1074/jbc.M308929200 [DOI] [PubMed] [Google Scholar]
- Wang W., Goswami S., Lapidus K., Wells A. L., Wyckoff J. B., Sahai E., Singer R. H., Segall J. E., Condeelis J. S. (2004). Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 64, 8585–8594 10.1158/0008-5472.CAN-04-1136 [DOI] [PubMed] [Google Scholar]
- Wang W., Wyckoff J. B., Goswami S., Wang Y., Sidani M., Segall J. E., Condeelis J. S. (2007). Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 67, 3505–3511 10.1158/0008-5472.CAN-06-3714 [DOI] [PubMed] [Google Scholar]
- Weiner O. D. (2002). Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr. Opin. Cell Biol. 14, 196–202 10.1016/S0955-0674(02)00310-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake R. A., Burridge K. (2003). RhoA and ROCK promote migration by limiting membrane protrusions. J. Biol. Chem. 278, 13578–13584 10.1074/jbc.M211584200 [DOI] [PubMed] [Google Scholar]
- Wu M., Wu Z. F., Rosenthal D. T., Rhee E. M., Merajver S. D. (2010). Characterization of the roles of RHOC and RHOA GTPases in invasion, motility, and matrix adhesion in inflammatory and aggressive breast cancers. Cancer 116, Suppl, 2768–2782 10.1002/cncr.25181 [DOI] [PubMed] [Google Scholar]
- Yamana N., Arakawa Y., Nishino T., Kurokawa K., Tanji M., Itoh R. E., Monypenny J., Ishizaki T., Bito H., Nozaki K. et al. (2006). The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol. Cell. Biol. 26, 6844–6858 10.1128/MCB.00283-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T., Gascoigne N. R. (2004). Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 86, 3923–3939 10.1529/biophysj.103.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebda N., Bernard O., Bailly M., Welti S., Lawrence D. S., Condeelis J. S. (2000). Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151, 1119–1128 10.1083/jcb.151.5.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Lin H., Nie Z., Wu J., Cañete-Soler R., Schlaepfer W. W., Schlaepfer D. D. (2003). Direct interaction of focal adhesion kinase with p190RhoGEF. J. Biol. Chem. 278, 24865–24873 10.1074/jbc.M302381200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.