Summary
Basal bodies and centrioles are conserved microtubule-based organelles the improper assembly of which leads to a number of diseases, including ciliopathies and cancer. Tubulin family members are conserved components of these structures that are integral to their proper formation and function. We have identified the ε-tubulin gene in Tetrahymena thermophila and detected the protein, through fluorescence of a tagged allele, to basal bodies. Immunoelectron microscopy has shown that ε-tubulin localizes primarily to the core microtubule scaffold. A complete genomic knockout of ε-tubulin has revealed that it is an essential gene required for the assembly and maintenance of the triplet microtubule blades of basal bodies. We have conducted site-directed mutagenesis of the ε-tubulin gene and shown that residues within the nucleotide-binding domain, longitudinal interacting domains, and C-terminal tail are required for proper function. A single amino acid change of Thr150, a conserved residue in the nucleotide-binding domain, to Val is a conditional mutation that results in defects in the spatial and temporal assembly of basal bodies as well as their stability. We have genetically separated functions for the domains of ε-tubulin and identified a novel role for the nucleotide-binding domain in the regulation of basal body assembly and stability.
Key words: Basal body, Epsilon tubulin, Microtubule organizing center, Tetrahymena thermophila
Introduction
Microtubules are components of the cytoskeleton that perform roles in essential functions, including cellular transport, cell motility and mitosis. Heterodimers of α- and β-tubulin polymerize to form the protofilaments of microtubules. Since the initial discovery of microtubules and α- and β-tubulin (Borisy and Taylor, 1967), three additional tubulin superfamily members have been identified within higher eukaryotes. These family members are γ-, ε- and δ-tubulin. γ-Tubulin was the first of these three to be identified and has received much attention over the years (Oakley and Oakley, 1989). Currently, γ-tubulin is known to nucleate microtubules at the (−)-end and is present at the two major microtubule organizing centers (MTOCs): centrosomes and basal bodies (Wiese and Zheng, 2000). Much less is known about the function of ε- and δ-tubulin, but it appears that they are organelle specific and localize solely to centrosomes and basal bodies (Chang et al., 2003; Chang and Stearns, 2000; Dupuis-Williams et al., 2002; Dutcher et al., 2002; Dutcher and Trabuco, 1998).
The centrosome is composed of two orthogonal centrioles and the surrounding pericentriolar material, and is critical for separating chromosomes during mitosis. Basal bodies, which are responsible for nucleating cilia and flagella, are composed of a single centriole. In recent years, the importance of cilia function and proper assembly of cilia has become apparent (Marshall and Nonaka, 2006). There is a specific class of diseases, known as ciliopathies, which have all been linked to ciliary dysfunction and result in highly pleiotropic phenotypes (Badano et al., 2006). Basal bodies are essential for proper cilia formation and, as such, it is important to understand their proper assembly.
Basal bodies are unique microtubule-based structures, characterized by a ninefold symmetrical organization of triplet microtubules blades. Triplet microtubules (designated A, B and C) form the core structural component of basal bodies. The C-tubule terminates at the distal end of the basal body and the A- and B-tubule are continuous with the nine outer doublets of the axoneme. Unlike dynamic cytoplasmic microtubules, the microtubules of the basal body are hyper-stabilized (Bobinnec et al., 1998; Pearson et al., 2009a). Within each triplet there is only one complete 13-protofilament microtubule, the A-tubule. The B- and C-tubules form incomplete tubules that share protofilaments with the preceding tubule to complete the triplet (Li et al., 2012). This unique microtubule environment suggests that there are novel proteins required to form and maintain these structures.
ε-tubulin was initially identified through a database search of the human genome. It was found to localize to centrosomes and be essential for centriole duplication in Xenopus extracts (Chang et al., 2003; Chang and Stearns, 2000). Work in Chlamydomonas reinhardtii and Paramecium terauralia identified ε-tubulin as an essential component of basal bodies (Dupuis-Williams et al., 2002; Dutcher et al., 2002). A mutant isolated in Chlamydomonas resulted in singlet, rather than triplet, microtubules in the basal body, and supplied the first indication that ε-tubulin is important for proper triplet microtubule assembly and/or maintenance (Dutcher et al., 2002). A subsequent study in Paramecium used RNAi to deplete ε-tubulin and reported degradation of the microtubule triplets, reaffirming its importance at the basal body (Dupuis-Williams et al., 2002). Thus far, studies have not clearly distinguished between a role in basal body assembly as opposed to maintenance, nor have they determined how ε-tubulin might carry out these functions.
Tetrahymena thermophila is a powerful model organism to study basal body biogenesis. Electron microscopy studies have determined that the core basal body structure is conserved from humans and also established a well-defined structural assembly pathway (Allen, 1969). A number of the molecular components are conserved as well (Kilburn et al., 2007; Li et al., 2004). Tetrahymena has been utilized to assay for the function of basal body components in both assembly and stability, independently, as well as orientation of new basal body assembly and cortical patterning (Culver et al., 2009; Pearson et al., 2009b; Stemm-Wolf et al., 2005; Vonderfecht et al., 2011).
Here we have identified the ε-tubulin homologue in Tetrahymena and found that it is essential for the assembly and stability of basal bodies. ε-tubulin localizes predominately to the ninefold microtubule triplet array that forms the core structure of basal bodies and is essential for its stability. A directed alanine-scanning mutagenesis screen identified a conditional mutant in the nucleotide-binding domain of ε-tubulin. The mutation causes defects in the proper spatial and temporal regulation of basal body assembly, as well as basal body stability. This work highlights the first structure–function relationship established in ε-tubulin and identifies a novel role for the nucleotide-binding domain.
Results
Identification of the Tetrahymena thermophila ε-tubulin homologue
ε-tubulin (ETU1) was initially identified in the T. thermophila macronuclear genome sequencing project (Eisen et al., 2006). We confirmed the identification of a single gene (ETU1) encoding an ε-tubulin homologue with BLAST (basic local alignment search tool) of the T. thermophila genome using the published human ε-tubulin sequence. Analysis of the coding sequence of ETU1 by RT-PCR revealed that the predicted amino acid sequence incorrectly included an 18 amino acid insertion after T279, which is absent in the sequence of the cDNA clone of ETU1. The removal of this sequence also alters the succeeding two amino acids, S298 and S299, in the predicted sequence. The missing insertion changes these amino acids to C280 and P281 (supplementary material Fig. S1). The correct protein is 480 amino acids long with a predicted molecular mass of 54.9 kDa and a predicted pI of 6.25. Alignment of the ETU1-derived amino acid sequence with other published and predicted sequences shows that this gene is indeed an ε-tubulin homologue and not another member of the tubulin superfamily (supplementary material Fig. S1, Fig. S2A,B). Tetrahymena ETU1 is 49% identical and 71% similar to human ε-tubulin (TUBE1; supplementary material Fig S1, Fig. S2B). Comparison of ETU1 with ATU1 (α-tubulin) and BTU1 (β-tubulin) of T. thermophila shows 30%/48% and 32%/53% identity/similarity, respectively (supplementary material Fig. S2A). These results are consistent with the sequence analysis of other tubulin family members (Inclán and Nogales, 2001).
ε-tubulin is a component of basal bodies and localizes specifically to the microtubule scaffold
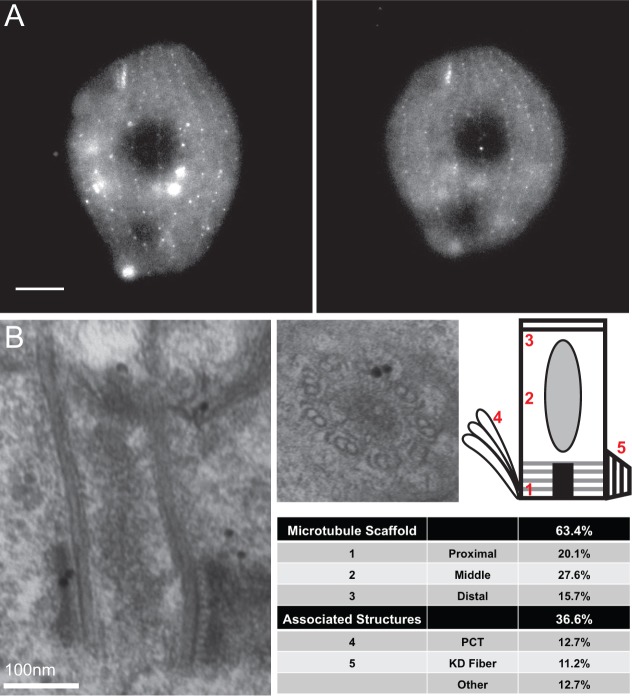
To confirm that ε-tubulin is a component of basal bodies, we constructed an N-terminal GFP-tagged version of ETU1 (GFP-ETU1) under the transcriptional control of the cadmium responsive MTT1 (metallothionein) promoter (Shang et al., 2002). GFP–Etu1p localizes to the basal bodies (punctate signal) and cytoplasm in Tetrahymena (Fig. 1A). The localization of GFP–Etu1p to the cytoplasm is consistent with the findings of a cytoplasmic pool in tissue culture (Chang and Stearns, 2000).
Fig. 1.
GFP–Etu1p localizes to the microtubule scaffold of basal bodies. (A) GFP-ETU1 was induced with 0.1 µg/ml CdCl2 and localized to basal bodies as well as the cytoplasm. GFP–Etu1p (white) localizes to the basal bodies of the cortical rows as well as the oral apparatus. Scale bar: 10 µm. (B) GFP–Etu1p was detected in the ultrastructure of basal bodies using immuno-electron microscopy. A secondary antibody conjugated to 10 nm gold was used to identify the primary antibody against GFP. Gold particles, as seen in B, were mapped from 85 basal bodies and tabulated. The number of gold particles = 134.
We then conducted immunogold electron microscopy against GFP–Etu1p to determine the ultrastructural localization of Etu1p (Kilburn et al., 2007). The majority of the GFP–Etu1p signal (63.4%) localized to the microtubule scaffold of basal bodies (n = 134 gold particles on 85 basal bodies). GFP–Etu1p was evenly distributed along the length of the basal body microtubule scaffold with a slightly larger population localizing to the middle portion (proximal: 15.7%; middle: 27.6%; distal: 20.1%). The remainder of the labeling was found on the accessory structures associated with basal bodies (Fig. 1B). The ultrastructural localization of GFP–Etu1p is consistent with ETU1 functioning at the microtubule triplets.
ε-tubulin is an essential protein in Tetrahymena thermophila
To determine the function of ETU1 in Tetrahymena, we constructed a complete knockout of the gene using homologous recombination to delete ETU1 (entire open reading frame) from the micronucleus (germline) of Tetrahymena. Once two strains of different mating types were constructed, they were mated and the subsequent progeny lacked ETU1 in the macronucleus (somatic nucleus), thus creating the etu1Δ strain. Staining of basal bodies using centrin 1 (Cen1), a basal body marker (Stemm-Wolf et al., 2005), revealed a decrease in the basal body density within cortical rows upon removal of the ETU1 gene product (supplementary material Fig. S3A–C). Concurrently, the cortical rows also became disorganized. These phenotypes were exacerbated with time. These phenotypes preceded the eventual death of etu1Δ cells after ∼3 days.
ε-tubulin is required for the proper assembly and maintenance of basal bodies
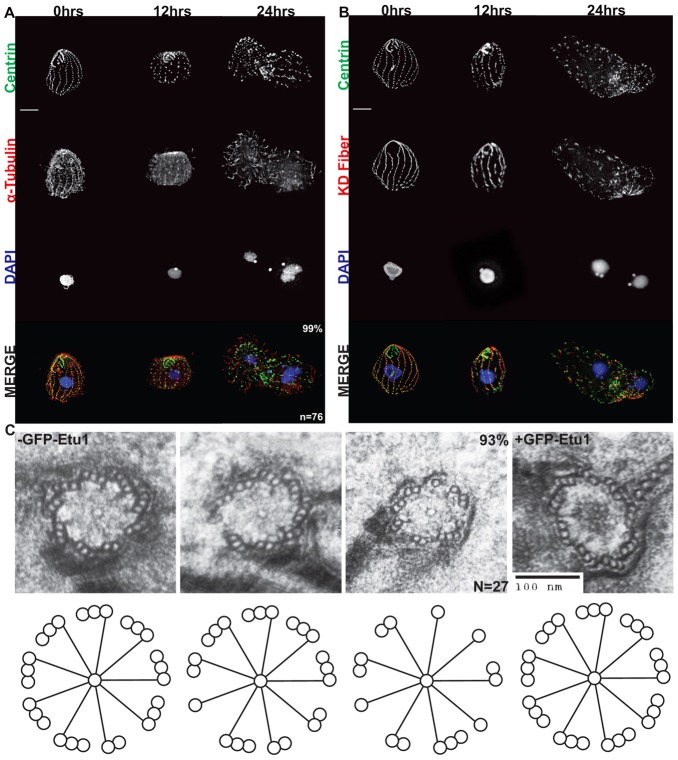
The function of ETU1 was also addressed by depleting it from cells. The etu1Δ strain was rescued with cadmium inducible GFP-ETU1. These cells grow in the presence of cadmium (1 µg/ml) and removal of cadmium from the medium leads to depletion of GFP–Etu1p (supplementary material Fig. S4). After depletion of GFP–Etu1p, the cells stopped growing and displayed phenotypes similar to the etu1Δ strain (Fig. 2A,B). After 24 hours of GFP–Etu1p depletion, Cen1 staining revealed a loss of basal bodies within the cortical rows and loss of overall cortical row organization (Fig. 2A,B). A basal body accessory structure, called the kinetodesmal (KD) fiber, is located adjacent to cortical row basal bodies in wild-type cells and points toward the anterior of the cell. In etu1Δ::GFP-ETU1 cells depleted of GFP–Etu1p, the KD fibers are still present, but no longer show the characteristic organization and are pointing in all directions (Fig. 2B). The cells were also stained for α-tubulin (Atu1; Fig. 2A). In wild-type cells, this antibody will label cilia as well as the underlying cortical microtubules. Labeling of etu1Δ::GFP-ETU1 cells showed that they lose the underlying cortical microtubules, and in some cases cilia, upon depletion of GFP–Etu1p (Fig. 2A).
Fig. 2.
ETU1 is essential for proper assembly and maintenance of basal bodies. The etu1Δ::MTT-GFP-ETU1 strain was depleted for GFP–Etu1p by removal of CdCl2 from the medium. Cells were fixed at the indicated times after GFP–Etu1p depletion. (A) Staining for Cen1, DNA (DAPI), and α-tubulin. (B) As in A except an antibody staining the KD fiber was used instead of the one for α-tubulin. Scale bars: 10 µm. (C) Basal bodies in cells after 12 hours of depletion commonly contained incomplete microtubule triplets (93%). n = 27 basal bodies. Scale bar: 100 nm.
Removal of cadmium from the medium and subsequent depletion of GFP–Etu1p in the etu1Δ::GFP-ETU1 strain also leads to a significant decrease in basal body density at 24 hours. In the presence of GFP–Etu1p, the cells maintain and average basal body density of 0.43±.08 basal bodies/µm2. Upon depletion of GFP–Etu1p for 24 hours, the basal body density decreases to 0.27±.05 basal bodies/µm2. These data are consistent with ETU1 being essential for basal body assembly and/or stability.
The immunofluorescence analysis demonstrated a reduction in basal body number, but did not address basal body structural phenotypes. We conducted electron microscopy of cells after 12 hours of depletion. The basal bodies of these cells displayed consistent defects in the microtubule triplets. The presence of a combination of singlet, doublet and triplet microtubule blades was observed in a majority of the basal bodies and a complete set of ninefold triplets was only seen in two instances (Fig. 2C). These data are consistent with ETU1 playing an essential role in the maintenance of the microtubule triplets.
Defects due to ε-tubulin depletion are reversible
The regulatable system we used to analyze the depletion of GFP–Etu1p can also be used to analyze the recovery of basal bodies and cortical organization upon re-introduction of GFP–Etu1p. After depletion of GFP–Etu1p for 24 hours, the defects can be rescued by re-introduction of cadmium (1 µg/ml), which induces the expression of GFP-ETU1. Twenty-four hours after cadmium induction the cells are nearly recovered to wild-type basal body and cortical organization (supplementary material Fig. S5A,B). We conclude from these data that ETU1 is essential for the assembly of basal bodies.
Alanine-scanning mutagenesis of ε-tubulin
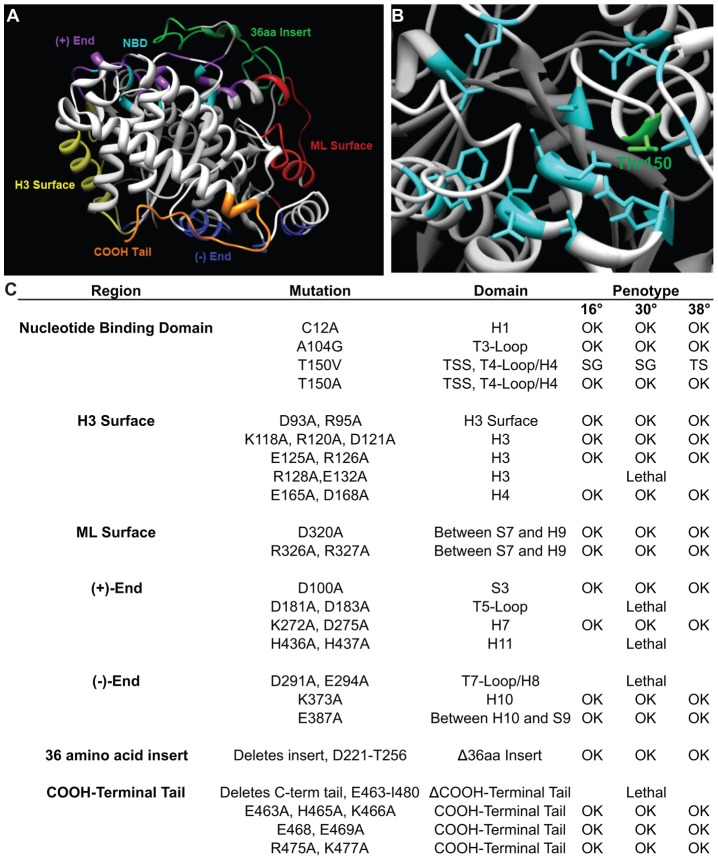
ε-tubulin clearly plays an essential role in basal body assembly and stability, but it is unclear how each domain of ε-tubulin contributes to these functions. We undertook a systematic mutagenesis screen of each of the domains of ε-tubulin to shed light on their individual functions. Previous mutagenic analysis of α- and β-tubulin utilized a systematic alanine scanning approach regardless of any structural features (Richards et al., 2000). We focused our mutagenesis by limiting it to areas within characterized domains of the tubulin family, as defined in a previous human sequence and structural analysis (Inclán and Nogales, 2001). Etu1p is predicted to have the six canonical tubulin domains: (1) nucleotide-binding domain (NBD); (2) (+)-end; (3) (−)-end; (4) H3 surface; (5) ML surface; and (6) C-terminal tail (Fig. 3A). There are also two unique and conserved amino acid inserts in ε-tubulin. The first is a 5 amino acid insert at amino acid 60, and the second is a 36 amino acid insert at amino acid 221 (supplementary material Fig. S2B). The position of these inserts is conserved among ε-tubulins, but the size and sequence of the inserts are not conserved. The small insert is predicted to be within the ML surface and may be involved in unique lateral interactions of Etu1p with possible binding partners. The larger insert is located within the (+)-end of Etu1p and most likely prevents the inter- and intra-dimer longitudinal interactions seen with α- and β-tubulin, but may provide a unique surface for association with a yet unidentified binding partner.
Fig. 3.
Directed mutagenesis of ETU1. The domains of Etu1p were identified based on Inclan and Nogales (Inclan and Nogales, 2001). (A) The predicted tertiary structure of Etu1p. The domains have been colored and labeled. (B) The residues that are involved in GTP hydrolysis and make up the nucleotide-binding domain (NBD) have been highlighted. Thr150 is highlighted in green. (C) ETU1 mutants created in this study. TSS, tubulin signature sequence; SG, slow growth; TS, temperature sensitive.
We mutagenized each domain using an alanine-scanning approach, i.e. changing the charged residues within a five amino acid window to alanines (Cunningham and Wells, 1989). The only domains for which this approach was not applied was the NBD (Fig. 3B), which does not possess many charged residues, and the large ε-tubulin specific insertion, which was removed. Conserved residues within the NBD, shown previously to be involved with nucleotide binding (Löwe et al., 2001), were mutated to alanine, valine, or glycine, similar to mutations made in γ-tubulin (Shang et al., 2005). In all, 23 different mutations were made in the domains of Etu1p (Fig. 3C), and each allele was screened for its ability to rescue etu1Δ cells. In total, 5 of the 23 mutant alleles were unable to restore viability to the etu1Δ strain, and were determined to be lethal after two transformation attempts. The lethal mutations were dispersed across nearly all of the domains of Etu1p, including the H3 surface (R128A), the (+)-end (D181A and H436A), the (−)-end (D291A), and the C-terminal tail (E463-I480).
Next, we wished to determine whether any of the 18 viable mutants displayed conditional phenotypes. Each mutant strain was subjected to temperature shifts from the permissive temperature, 30°C, to 16°C or 38°C (supplementary material Fig. S6A–C). The cells were then assayed for growth, cell size and overall cellular morphology. Of the 18 mutants that were able to rescue etu1Δ, 17 displayed wild-type growth and cellular morphology at the temperatures tested. Of note, the conserved large insert is not essential and was removed with no apparent phenotype. We had hypothesized that mutations within the conserved NBD would be lethal, but we were able to isolate three viable mutations in conserved residues within the NBD (C12A, A104G and T150A). Interestingly, the only conditional mutation isolated that affected growth rate is in the nucleotide-binding domain, Thr150Val (Fig. 3B). Thr150 is a conserved residue within the tubulin signature sequence (GGTGSG) that has been implicated in GTP binding in α- and β-tubulin (Löwe et al., 2001).
Thr150 is required for proper regulation of basal body duplication
The etu1-T150V mutation led to severe morphological defects at the non-permissive temperature of 38°C. Immunofluorescence of etu1-T150V containing cells using antibodies to Cen1 and either Atu1 (α-tubulin) or the KD fiber showed discrete Cen1 foci indicating the presence of basal bodies, or some residual basal body structure (Fig. 4). The cells maintained cilia, but showed a high level of cortical disorganization as indicated by the KD fiber staining (Fig. 4). In addition, Etu1p-T150V cells revealed a significant increase in basal body (bb) density at 38°C as measured by Cen1 foci (0.33±.08 bb/µm2) compared to the wild-type control (0.24±.03 bb/µm2) and Etu1p-T150V 30°C control (0.28±.06 bb/µm2; Fig. 4). Wild-type cells normally display a decreased basal body density upon temperature shift from 30°C to 38°C, possibly due to an increase in cell size, but Etu1p-T150V cells actually display an increase in basal body density as well as an increase in cell size (supplementary material Fig. S6B,C). The increase in basal body density in Etu1p-T150V cells indicates a loss of the temporal regulation of basal body assembly because it is a highly regulated process (Nanney et al., 1978). Therefore, we conclude that the NBD of Etu1p is essential for the proper regulation of basal body assembly.
Fig. 4.
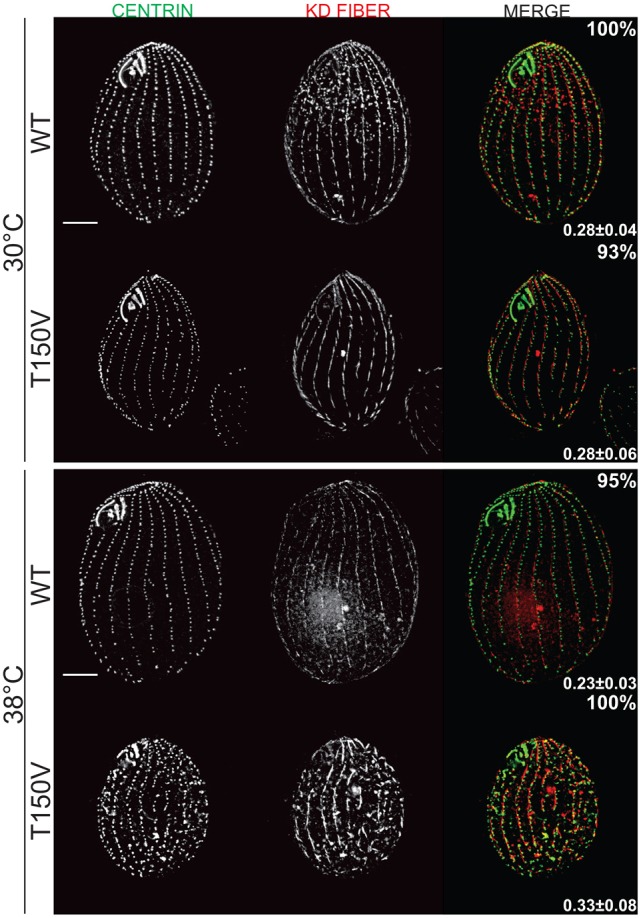
Thr150 is essential for proper regulation of basal body assembly. Thr150 in the NBD was mutated to Val (Etu1p-T150V). (A) Staining for Cen1 and the KD fiber at 30°C (n = 98 and n = 86, WT and mutant respectively), and at 38°C (n = 76 and n = 87, WT and mutant, respectively). Basal body density for each strain was measured by counting Cen1 foci and is indicated in the lower right corner of each sample set (n = 25). Etu1p-T150V cells show a significant increase (P<0.01) in basal body density as compared to WT at 38°C and Thr150Val at 30°C. Percentages indicate the frequency of the observed phenotype. Scale bars: 10 µm.
Thr150 of ε-tubulin is essential for basal body stability
In addition to the effect of etu-T150V on basal body duplication, we wanted to determine whether the NBD is also involved in basal body stability. We arrested Etu1p-T150V cells in G1, which halts basal body duplication, at 30°C and 38°C for 48 hours and performed immunofluorescence using an antibody to Cen1 (Fig. 5A). Basal body density in Etu1p-T150V cells is significantly decreased at restrictive temperature (0.36±0.06 bb/µm2) compared to those arrested at permissive temperature (0.46±0.08 bb/µm2). In contrast, the wild-type control did not show a significant change in basal body density at the two temperatures (30°C: 0.54±0.07; 38°C: 0.49±0.08 bb/µm2; Fig. 5B). We conclude that the NBD of Etu1p is essential for the stability of basal bodies. Interestingly, the orientation of the KD fiber shows that the cortical organization is preserved when basal bodies are not duplicating (supplementary material Fig. S7). These results suggest that the disorganization we observe in cycling cells is likely due to new basal bodies being improperly oriented during assembly as well as overall loss of cortical organization.
Fig. 5.
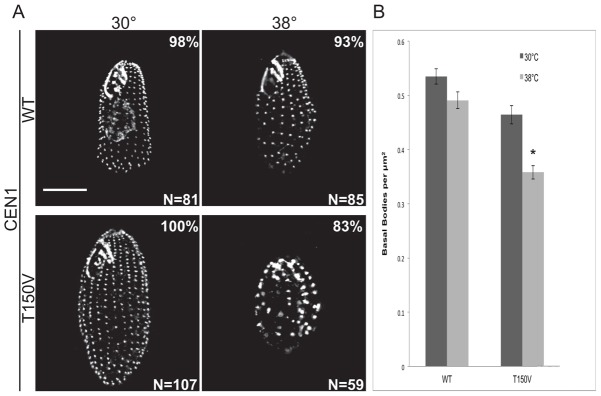
Thr150 is required for basal body stability. Strains were arrested in G1 for 48 hours at 30°C and 38°C. (A) Immunofluorescent images of cells stained for Cen1. Scale bar: 10 µm. (B) Quantification of basal body density. The basal body density in Etu1p-T150V cells at 38°C is significantly different from Etu1p-T150V at 30°C and wild-type at both temperatures (*P<0.01). Error bars indicate the s.e.m.
Mutation of ε-tubulin Thr150 results in misorientation of newly assembled basal bodies
In cycling Etu1p-T150V cells there are a disproportionate number of basal bodies mislocalized from the cortical rows. We asked directly whether new basal bodies were being assembled off the axis of the cortical row. Etu1p-T150V cells were arrested in G1 at 30°C overnight then released into growth conditions at 30°C and 38°C to allow new basal body assembly. We performed immunofluorescence with antibodies against Cen1, which labels all basal bodies, and K-like antigen, which will label only mature basal bodies (Shang et al., 2005; Williams et al., 1990). These two antibodies allow us to distinguish between newly assembled basal bodies (− K-like antigen) and older basal bodies (+ K-like antigen; Fig. 6A). To measure the angle of newly assembled basal bodies a line was drawn connecting two basal bodies within a cortical row (Fig. 6A, centrin panels, arrows). Then a line was drawn to a third, newly assembled, basal body (arrowheads) and the angle created by these two lines was measured (Fig. 6B).
Fig. 6.
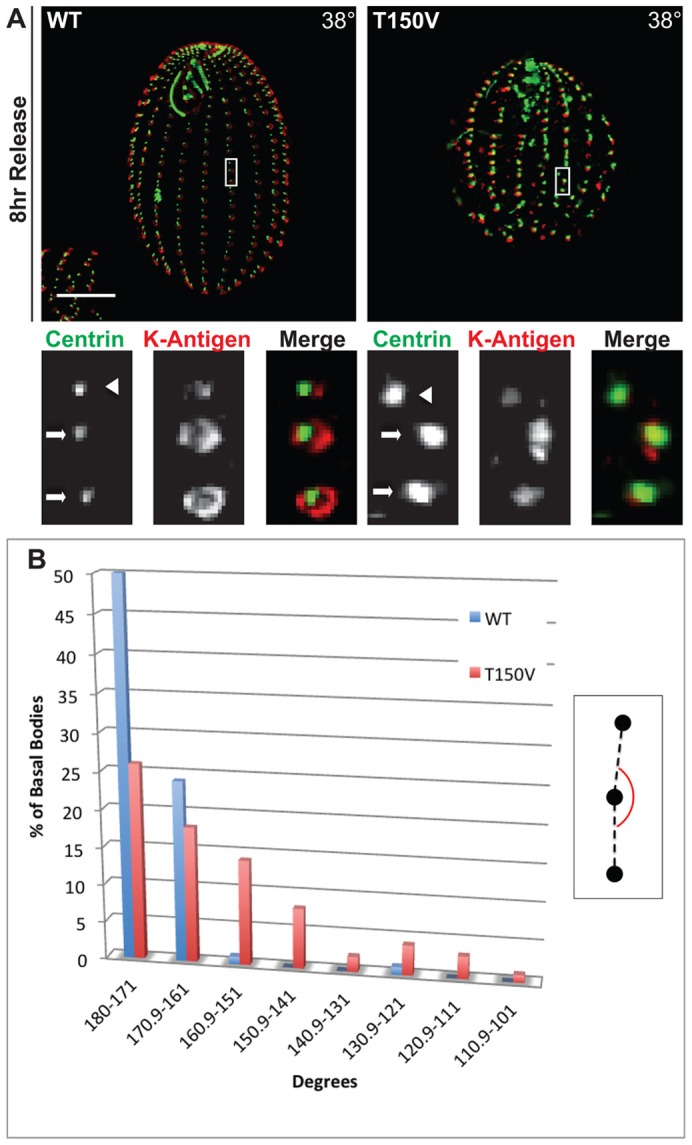
Thr150 is required for proper orientation of newly assembled basal bodies. Cells were arrested in G1 at 30°C for 18 hours to halt new basal body assembly then released into growth medium at 38°C to initiate basal body assembly. (A) The angle of basal body assembly was calculated by measuring the angle between two mature basal bodies within the cortical row (arrows) and the newly assembled basal body (arrowhead), based on lack of staining for K-like antigen. Also see inset in Fig. 9B. (B) The inset shows the measurement of the new basal body angle. The plot shows the frequency of new basal body angle for cells grown at 38°C. n = 76 basal bodies.
The etu1-Thr150Val mutation resulted in significant orientation defects at restrictive temperature. Wild-type control cells (Fig. 6B, blue bar) assembled basal bodies mostly within the two bins that correspond most closely to assembly in a straight line, i.e. within the cortical rows. In contrast, the Etu1p-T150V strain (Fig. 6B, red bar) showed a wide distribution of basal body assembly angles, indicating that the NBD is essential for proper orientation of newly assembled basal bodies. These data most likely underrepresent the actual orientation defects, as we were unable to count a large number of newly assembled basal bodies because they could not be unambiguously linked to a parent basal body. This may be due to de novo basal body assembly or deregulation of the temporal and spatial control of basal body.
Electron microscopy of cycling and arrested Etu1p-T150V cells reveals defects in basal body spacing and triplet microtubule stability
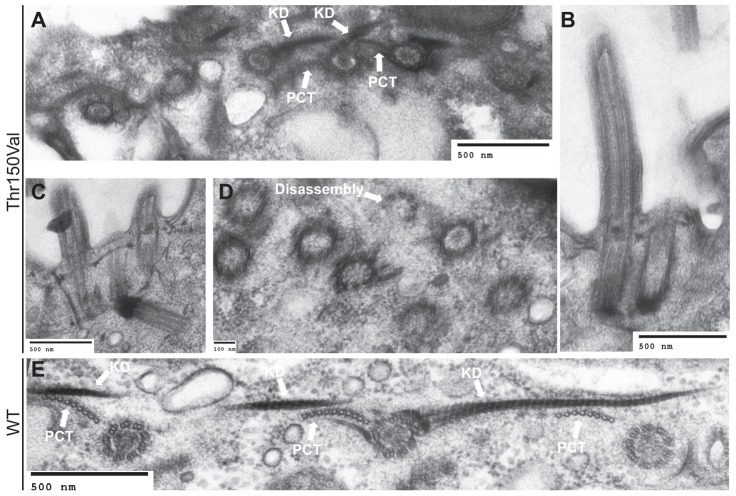
In order to assess whether there are ultrastructural defects associated with the etu1-Thr150Val allele, we conducted electron microscopy on the mutant cells after growth at 38°C for 24 hours. Our analysis revealed that there are gross defects in basal body assembly consistent with the immunofluorescence data. Mature basal bodies [identified by the KD fiber and post-ciliary microtubules (PCT)] are unusually close to each other within the cortical rows (Fig. 7A–C) as compared to wild-type (Fig. 7E). Furthermore, some basal bodies were observed to have been aberrantly nucleated, such that they were oriented away from the plasma membrane towards the depths of the cytoplasm (Fig. 7C). It is also possible that these basal bodies were properly oriented during assembly, but ultimately unable to remain at the cell cortex, thus leading to the improper orientation. We also observed cilia in very close proximity to each other, which is uncharacteristic in wild-type cells (Fig. 7B,C). High density, disorganized aggregates of basal bodies were also found in the mutant cells (Fig. 7D). Basal bodies within these aggregates are often within 100 nm of each other (Fig. 7D), which is beyond the resolution of our fluorescence microscope. These observations suggest that our immunofluorescence analysis most likely underestimates the number of basal bodies in the mutant cells. Within the mutant cells, we also observed a basal body that appears to be disassembling from the distal end (Fig. 7D). These data are consistent with the immunofluorescence data of the cycling Etu1p-Thr150Val mutant cells and further confirmed the role of ETU1 in proper coordination of basal body assembly and stability.
Fig. 7.
Thr150 is required for proper separation and organization of basal bodies. Etu1p-T150V and WT cells were grown at 38°C for 24 hours then analyzed by transmission electron microscopy. (A) Mature basal bodies [indicated by the presence of the KD fiber (KD) and post-ciliary microtubule (PCT)] are present in abnormally close proximity. (B) Two basal bodies have assembled and inserted into the membrane abnormally close to each other. (C) Basal bodies are unusually close, and a basal body is misoriented in the cytoplasm. (D) Basal bodies have assembled unusually close to each other with no apparent organization and there is also a basal body that is disassembling from the distal end. (E) Basal bodies within a cortical row with proper spacing and organization in a wild-type cell.
The ultrastructural consequences of the basal body stability defects we observed using immunofluorescence were examined by electron microscopy on G1 arrested mutant cells at non-permissive temperature. We observed basal bodies with incomplete microtubule triplets (Fig. 8A–C), most commonly a mix of doublet and triplet microtubules, which we do not observe in wild-type cells (Fig. 8D). We also observed complete loss of a triplet set of microtubules (Fig. 8B) (see also Pearson et al., 2009b; Vonderfecht et al., 2011). This analysis demonstrates that a mutation within the NBD of ε-tubulin can have drastic effects on the triplet microtubule integrity within the basal body.
Fig. 8.
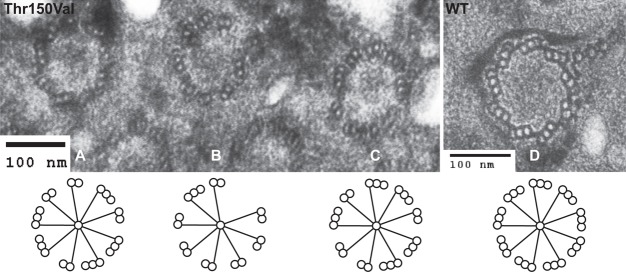
Thr150 is required for basal body stability. Etu1p-T150V and wild-type cells were starved for 24 hours at 38°C and then analyzed by transmission electron microscopy. Many basal bodies lack complete microtubule triplets. (A,C) These basal bodies show a combination of triplet and doublet microtubules. (B) This basal body is missing an entire triplet and the remainder are a combination of doublet and triplet microtubules. (D) A wild-type (WT) basal body with a complete set of microtubule triplets.
Discussion
ε-tubulin is a member of the tubulin superfamily and is an essential component of basal bodies and centrioles. Our work shows that it is primarily a component of the ninefold symmetric microtubule triplet scaffold that forms the canonical basal body structure and is essential for the assembly and stability of basal bodies. Mutagenic analysis of ε-tubulin revealed a novel role for the nucleotide-binding domain. We found that the NBD is necessary for basal body stability, but not required for new basal body assembly. However, it is essential for the proper spatial and temporal regulation of basal body assembly.
Conservation of ε-tubulin with basal body structure
Basal body structure and assembly have been well characterized morphologically. Previous electron microscopy studies have defined the structural intermediates of basal body assembly (Allen, 1969; Dippell, 1968). One of the canonical structural features of basal bodies is the ninefold microtubule triplets, which are a conserved feature across almost all eukaryotes. The microtubules within the triplets form a unique microtubule environment because only the A-tubule is a complete 13-protofilament microtubule. The subsequent B- and C-tubules each share protofilaments with the adjacent tubule and are predicted to have 15 protofilaments in total (Li et al., 2012). Cryo-electron tomography of basal bodies in Chlamydomonas has determined that there are proteins in addition to α- and β-tubulin in and around the B- and C-tubules (Li et al., 2012). Therefore, proteins that primarily localize to the microtubule scaffold may be important components for the assembly and stability of the microtubule triplets.
ε-tubulin is a widely conserved component of basal bodies and centrioles. After its initial identification (Chang and Stearns, 2000), homologues were identified in nearly all organisms that possess basal bodies with ninefold triplet microtubule organization (Carvalho-Santos et al., 2011; Hodges et al., 2010). Notably, ε-tubulin has not been found in organisms that lack the canonical triplet microtubule organization, C. elegans, D. melanogaster and S. cerevisiae. C. elegans centrioles are composed of singlet microtubules and are 100 nm in diameter and 150 nm in length (Pelletier et al., 2006). Drosophila centrioles consist of doublet microtubules and are 80 nm in diameter by 160 nm in length (Vidwans et al., 2003). The one exception is the Drosophila spermatid, which forms basal bodies that are ten times larger than their somatic counterparts and are composed of ninefold microtubule triplets (González et al., 1998). The mechanism by which they are formed is unclear. Finally, the yeast spindle pole body is a more divergent microtubule organizing center, which has many conserved components but varies greatly in structure (Jaspersen and Winey, 2004). In contrast to these organisms, Tetrahymena, and most other eukaryotes, have basal bodies composed of microtubule triplets that are ∼500 nm in length and 200 nm in diameter. The difference in size and composition of basal bodies supports the hypothesis that there are distinct molecular components conserved with this structure.
The specific structural elements required for triplet microtubule formation remain unknown, but the presence of unique proteins to facilitate the formation of these structures is logical. Phylogenetic and mutational data on ε-tubulin supports the notion that ε-tubulin is likely one of these factors. Our study and others have clearly established an important role for ε-tubulin in assembly and stability of the microtubule triplets, but the mechanism is still unclear. ε-tubulin localizes to the microtubule scaffold, but whether it is integrated into the tubules themselves or merely associated with the scaffold remains to be determined. Further work to identify ε-tubulin binding partners and high-resolution electron tomography of basal bodies may resolve this question.
Novel role of NBD of ε-tubulin at the basal body
Previous studies on ε-tubulin have been unable to resolve whether this tubulin isoform is required for assembly of basal bodies, stability of basal bodies, or both. The phenotypes observed in Chlamydomonas and Paramecium could be a result of either phenomenon (Dupuis-Williams et al., 2002; Dutcher et al., 2002), but from our work we have concluded that ε-tubulin is indeed involved in both processes.
Our directed mutagenic analysis, which is the most comprehensive to date, has revealed a separation of function between domains within ε-tubulin. Our initial screening of the mutants identified five lethal mutations. The lethal mutations were in the lateral interacting H3 surface, the two longitudinal interacting surfaces, and the C-terminal tail (Fig. 3A). In α- and β-tubulins these domains are important for protein–protein interactions and may also perform the same function in ε-tubulin. In α- and β-tubulins the H3 and ML surfaces are responsible for protofilament to protofilament interactions within the microtubule, in which the H3 surface of a tubulin subunit would interact with the corresponding ML surface on the adjacent tubulin subunit. The (+) and (−) ends, the longitudinal interacting surfaces, are responsible for inter- and intra-tubulin dimer interactions. The remaining domain, the C-terminal tail, is essential as a whole, but individual mutations are viable. In α- and β-tubulin this tail is the site of most post-translational modifications, is negatively charged, and provides a docking surface for microtubule-associated proteins (Duan and Gorovsky, 2002; Garnham and Roll-Mecak, 2012; Wloga and Gaertig, 2010). Unlike the tails of α- and β-tubulin, with pIs of 3.41 and 3.25 respectively, the C-terminal tail of Etu1p tail has a near neutral charge with a pI of 6.86. The lack of charge on the Etu1p tail implies a unique function for this domain. This raises the question of what essential function this domain is performing and will require further investigation.
We identified a number of viable mutants across all of the domains in ε-tubulin. Although these mutants were viable, thirteen showed a significant cell size change at one of the temperatures tested, which may indicate more subtle defects. Surprisingly, deletion of the large conserved insert at amino acid 221 resulted in viable cells with no apparent phenotype. We hypothesized that this insert was essential to a novel function of ε-tubulin, but our results indicate this is not the case and raise questions about the function of this domain.
Our study isolated one strongly conditional mutant, etu1-Thr150Val, in the NBD of ε-tubulin. This conditional mutant has allowed us to establish the first structure function relationship in ε-tubulin. The NBD of ε-tubulin is essential for proper temporal and spatial regulation of basal body assembly as well as basal body stability. Unlike depletion of ε-tubulin, we did not observe any defects in cell division (cytokinesis defects or multinucleate cells), and cells were able to assemble new basal bodies. This separation of function indicates that other domains are important for basal body assembly. The new insights we have uncovered regarding the NBD of ε-tubulin provide strong evidence for the importance of GTP binding in the function of ε-tubulin, although we have not conclusively ruled out that the mutant allele is improperly folded (Zabala et al., 1996). The dissimilar, in fact nearly opposite, phenotypes observed in the null/depletion experiments versus experiments with the T150V allele strongly suggest that the T150V phenotypes (such as off axis basal body assembly) arise from aberrant function as opposed to loss of the protein, which would mimic the null or depletion alleles. To date, there is no published data on the binding of GTP by ε-tubulin, but it would be very surprising if a member of the tubulin superfamily did not bind GTP. Assuming ε-tubulin does bind GTP, our data would be consistent with GTP binding by ε-tubulin playing an essential role in basal body stability and temporal and spatial control of assembly.
We did uncover an unexpected function of the NBD. etu1-Thr150Val results in a significant increase in basal body density implying that the NBD is part of mechanism to suppress over-duplication of basal bodies. Previous mutational analysis of γ-tubulin isolated a mutant of the corresponding residue in the NBD that showed a very similar phenotype (Shang et al., 2005). Two mutations within the NBD of γ-tubulin resulted in over-duplication and misorientation of newly formed basal bodies, but no defect in basal body stability as we observed for ε-tubulin. This group postulated a model whereby the NBD of γ-tubulin acts as part of a network suppressing un-licensed basal body duplication. This conclusion is consistent with the role of γ-tubulin in microtubule nucleation, which one would presume is essential for basal bodies, but the role of ε-tubulin is less clear. The binding of GTP through the NBD of ε-tubulin may act as another step to regulate basal body assembly by suppressing over-duplication and ensuring proper orientation and stability. The combination of these studies has begun to establish another level of basal body regulation that has not been well defined and will surely require further refinement as novel discoveries are made in the field.
Materials and Methods
T. thermophila cell culture
T. thermophila cells were grown at 30°C (unless otherwise indicated) in 2% SPP (2% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% FeEDTA). To arrest the cells in G1 and also for starvation conditions, cells were washed and resuspended in 10 mM Tris pH 7.4. Cell densities were determined with a Z2 Coulter counter (Beckman Coulter) or a hemacytometer.
Identification and alignment of ETU1
ETU1 was initially identified during the sequencing of the T. thermophila macronuclear genome (Eisen et al., 2006). BLAST search of the Tetrahymena genome using the human ε-tubulin confirmed a single homologue in Tetrahymena. The Tetrahymena ETU1 cDNA was isolated with the Superscript® II One-Step RT-PCR System (Invitrogen). The cDNA was cloned into pBluescript KS(−) and confirmed by sequencing. The alignment of Etu1p with its homologues was done with ClutalW 2.0 (Larkin et al., 2007). Phylogenetic analysis of ETU1 was done with MUSCLE (Edgar, 2004) and PhyML (Guindon et al., 2010) using Phylogeny.fr (Dereeper et al., 2008). The specific isoforms for α-, β- and γ-tubulin used for each organism were: Homo sapiens (Hs)-TUBA1B, TUBB1, TUBG1; Mus musculus (Mm)-TUBA1A, TUBB1, TUBG1; Drosophila melanogaster (Ds)-TUBA1, TUBB2C, TUBG1; Xenopus laevis (Xl)-TUBA1B, TUBB, TUBG1; Chlamydomonas reinhardtii (Cr)-TUA, TUBB1, TUG; T. thermophila (Tt)-ATU1, BTU1, GTU1.
Plasmids
etu1Δ::NEO2 was constructed from the plasmid p4T2-1 (Gaertig et al., 1994). The 1.4 kb upstream of ETU1 was amplified by PCR and flanked with KpnI and XhoI. The 1.3 kb downstream of ETU1 was similarly amplified by PCR and flanked by BamH1 and SacII. Each fragment was then digested and ligated into p4T2-1.
GFP-ETU1 was constructed by cloning ETU1 with flanking KpnI and XhoI sites into the pENTR 4 Dual Selection vector (Invitrogen). ETU1 was then subcloned into pBSMTTGFPgtw (provided by Doug Chalker, Washington University, St. Loius, MO) using the Gateway system (Invitrogen) to create pBSMTTGFPETU1.
ETU1WFpBS was constructed using the wild-type locus. The ETU1 locus including 527 base pairs upstream, the ORF, and the 867 base pairs downstream were amplified by PCR and flanked with SpeI, which is in the upstream region, and NotI. The DNA was subsequently digested and ligated into pBluescript. ETU1WFpBS-XL was constructed from ETU1-WFpBS by subcloning the upstream flanking sequence from etu1Δ::NEO2. etu1Δ::NEO2 was digested with KpnI and SpeI and the resulting 864 bp fragment was purified and ligated into ETU1WFpBS.
The ETU1 mutant plasmids were constructed using either overlap PCR or inverse PCR techniques to mutate either ETU1WFpBS or ETU1WFpBS-XL. Each mutant was then confirmed by sequencing (Macrogen, Rockland, MD).
Construction of the ETU1 knockout strain
etu1Δ::NEO2 was digested with KpnI and SacII to release the NEO2 cassette and introduced into the micronucleus of conjugating cells (B2086 and CU427) using biolistic bombardment (Cassidy-Hanley et al., 1997) and removed the entire ETU1 gene. The integration of the cassette was confirmed by PCR. Genetic crosses were performed to create knockout heterokaryons as previously described (Hai et al., 2000). Complete germline knockout cells were created by mating the knockout heterokaryons to star strains. Upon mating of the germline knockouts, ETU1 is eliminated from the somatic nucleus (macronucleus) to create etu1Δ. Elimination of ETU1 was validated by PCR, which confirmed the correct integration of the NEO2 cassette. etu1Δ was also rescued with ETU1WFpBS and pBSMTTGFPETU1, which both re-introduced ETU1.
Analysis of the ETU1 knockout strain
The two germline knockout strains, EPMS1 and EPMS10, and CU427 (WT) were grown in SPP medium and then starved overnight in 10 mM Tri pH 7.4. Two matings were setup, EPMS1+EPMS10 and EPMS1+CU427, to create etu1Δ and ETU1, respectively. Cells were mixed at equal numbers in 10 mM Tris pH 7.4 and incubated at 30°C for 10 hours to allow for conjugation. An equal volume of 2× SPP medium was then added and the cells were allowed to grow for another 7 hours. 100 µg/ml paromomycin was added to the cultures, designating the 0 hour timepoint. Modified from Bayless et al. (Bayless et al., 2012).
Macronuclear transformation
The GFP-tagged allele of ETU1 was transformed into the macronucleus of starved cells (B2086) by biolistic transformation (Cassidy-Hanley et al., 1997). Transformants were selected by growth in 2% SPP with 7.5 µg/ml cycloheximide. After initial selection, transformants were grown in 2% SPP with 15 µg/ml cycloheximide. Transformation was also confirmed by fluorescence and western blot of the fusion protein.
Immunofluorescence microscopy
Imaging was performed on a Nikon Ti-Eclipse inverted microscope (Nikon, Tokyo, Japan) with a 60× Plan Apo VC NA 1.40 objective and a charge-coupled device camera (Cool-SNAP hq2, Photometrics, Tucson, AZ). All imaging was done at room temperature. Metamorph Imaging Software (Molecular Devices, Sunnyvale, CA) was used to acquire all images. Exposure times were between 10 and 500 ms. Images were deconvolved using the nearest-neighbors algorithm and subjected to maximum projection.
Live-cell imaging was used to image cells expressing GFP–Etu1p. Cells were washed into 10 mM Tris pH 7.4 and then placed on glass microscope slides with a coverslip. Cells were prepared for immunofluorescence by chemical fixation. Cells were fixed with either a paraformaldehyde/ethanol fix (Stuart and Cole, 2000) or an ethanol fix. For the ethanol fix cells were washed with 10 mM Tris pH 7.4 and resuspended in 70% ethanol with 0.1% Triton X-100 and incubated on ice for 5 minutes, after which the protocol is the same as the paraformaldehyde/ethanol protocol. All primary antibodies were diluted in 1% BSA in phosphate buffered saline (PBS). The Tetrahymena Cen1 antibody (Stemm-Wolf et al., 2005) was diluted 1:2000. The monoclonal Atu1 antibody (provided by Joe Frankel, University of Iowa, Iowa City, IA) was diluted 1:50. The monoclonal KD-fiber antibody (provided by Joe Frankel) was diluted 1:250. The monoclonal K-like antigen antibody (provided by Joe Frankel) was diluted 1:50. All incubations were either done overnight at 4°C or at room temperature for 1 hour. Cells were then washed three times with 0.1% BSA in PBS. Secondary antibodies were diluted in 1% BSA in PBS. The secondary antibodies used were anti-rabbit Alexa Fluor 488 (Invitrogen/Molecular Probes, Carlsbad, CA), anti-mouse Texas Red (Jackson ImmunoResearch Labs, West Grove, PA). Cells were then washed three times in 0.1% BSA in PBS and mounted with Citifluor (Citifluor, London, UK).
Rescue of etu1Δ and depletion of GFP–Etu1p
The etu1Δ heterokaryon strains were starved 18–24 hours and mated in 10 mM Tris pH 7.4. After 24 hours post-mixing, pBSMTTGFPETU1 was transformed into cells using biolistic bombardment. The cells were then allowed to recover in 2% SPP with 1 µg/ml CdCl2. In order to shut off GFP-ETU1 expression, cells were washed with 10 mM Tris pH 7.4 and resuspended in 2% SPP without CdCl2. Shutoff was assessed by phenotypic analysis and western blot.
Electron microscopy
Cells were prepared by high-pressure freezing followed by freeze-substitution (Giddings et al., 2010; Meehl et al., 2009). Thin sections from these samples were immunolabeled as described in Meehl et al. using a rabbit polyclonal antibody (generous gift of Chad Pearson, University of Colorado, Denver) (Meehl et al., 2009). Imaging analysis was done on either a Phillips CM10 or CM100 electron microscope (FEI, Inc., Hillsboro, OR). Structural domains within basal bodies were identified as previously described (Kilburn et al., 2007).
Western blot analysis
Tetrahymena whole-cell extracts were prepared by spinning down cells and washing with 10 mM Tris pH 7.4. Cells were then resuspended in reducing sample buffer (20% glycerol, 2% SDS, 125 mM Tris, pH 6.8, 5% β-mercaptoethanol) and boiled at 95°C for 5 minutes. 5,000 cells were loaded per lane onto a 10% SDS-PAGE gel. The protein was then transferred to an Immobilon PVDF membrane (Millipore, Billerica, MA) using a Bio-Rad transfer apparatus. The membrane was blocked in 10% milk in Tris-buffered saline with 0.05% TWEEN 20 (TBST) for 30 minutes at room temperature and washed with TBST for 5 minutes. All primary antibodies were diluted in 5% milk and incubated at room temperature for 1 hour. The monoclonal GFP antibody (Covance Inc., Princeton, NJ) was diluted 1:5000. The monoclonal Atu1 antibody (provided by Joe Frankel) was diluted 1:2500. The membrane was washed three times for 5 minutes in TBST. The secondary antibody from LI-COR (anti-mouse IR680) was diluted 1∶10,000 in 5% milk and incubated at room temperature for 1 hour. The membrane was washed three times for 5 minutes with TBST. The protein band intensity was determined with a LI-COR Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE). Since both of the primary antibodies used were monoclonal, the blot was probed for each protein independently. After detecting GFP–Etu1p the membrane was stripped by incubating in 0.25 mM glycine pH 2 with 1% SDS for 40 minutes at room temperature. The membrane was washed twice with PBS for 10 minutes and the once with TBST for 10 minutes. The western blot protocol was then repeated with the Atu1 antibody.
Modeling of Etu1p
The structure of Etu1p was predicted using the I-TASSER server (Roy et al., 2010; Zhang, 2007). The predicted domains of Etu1p are based on previous sequence and structural analysis (Inclán and Nogales, 2001) and the subsequent modeling was done with the UCSF Chimera package (Pettersen et al., 2004).
Etu1p mutant strain analysis
Basal body density
Images were gathered of Etu1p mutant cells that were fixed and labeled for Cen1. ImageJ (National Institutes of Health, Bethesda, MD) was used to calculate cell area and count Cen1 foci to determine basal bodies per square micrometer. In cases where the oral apparatus was present, its area and Cen1 foci were subtracted from the total. Twenty-five cells were counted for each condition.
Assignment of new basal body angle
ImageJ was used to identify the angle of new basal body assembly. Three basal bodies within a row were used to establish an angle. The first two basal bodies were assigned the designation of ‘in a cortical row’ and the third, new basal body was assigned an angle based on the first two (Vonderfecht et al., 2011). A total of 76 basal bodies were counted for each condition.
Supplementary Material
Acknowledgments
We thank Alex Stemm-Wolf, Shelly Jones, Tom Giddings, and Eileen O'Toole for discussion and critique of the work; Chad Pearson for helpful discussion; Dr Joe Frankel for antibodies; and Dr Doug Chalker for plasmids. Protein modeling was performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the National Institutes of Health (National Center for Research Resources grant 2P41RR001081, National Institute of General Medical Sciences grant 9P41GM103311).
Footnotes
Author contributions
I.R. executed all of the experiments, with the exception of EM which was done by C.C. and T.H.G. I.R. and M.W. analyzed the data and prepared the manuscript.
Funding
This work was supported by the National Institute of General Medical Sciences [grant number RO1 GM074746 to M.W.]; and National Institutes of Health [grant number T32 GM08759 to I.R.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.128694/-/DC1
References
- Allen R. D. (1969). The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 40, 716–733 10.1083/jcb.40.3.716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125–148 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- Bayless B. A., Giddings T. H., Jr, Winey M., Pearson C. G. (2012). Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell 23, 4820–4832 10.1091/mbc.E12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L. M., Rieder C. L., Eddé B., Bornens M. (1998). Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143, 1575–1589 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy G. G., Taylor E. W. (1967). The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J. Cell Biol. 34, 525–533 10.1083/jcb.34.2.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B., Bettencourt-Dias M. (2011). Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D., Bowen J., Lee J. H., Cole E., VerPlank L. A., Gaertig J., Gorovsky M. A., Bruns P. J. (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146, 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Stearns T. (2000). δ-tubulin and ε-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2, 30–35 10.1038/71350 [DOI] [PubMed] [Google Scholar]
- Chang P., Giddings T. H., Jr, Winey M., Stearns T. (2003). ε-tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 5, 71–76 10.1038/ncb900 [DOI] [PubMed] [Google Scholar]
- Culver B. P., Meehl J. B., Giddings T. H., Jr and Winey M. (2009). The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol. Biol. Cell 20, 1865–1877 10.1091/mbc.E08-08-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. C., Wells J. A. (1989). High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085 10.1126/science.2471267 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M. et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, Suppl. 2W465–W469 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippell R. V. (1968). The development of basal bodies in paramecium. Proc. Natl. Acad. Sci. USA 61, 461–468 10.1073/pnas.61.2.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Gorovsky M. A. (2002). Both carboxy-terminal tails of α- and β-tubulin are essential, but either one will suffice. Curr. Biol. 12, 313–316 10.1016/S0960-9822(02)00651-6 [DOI] [PubMed] [Google Scholar]
- Dupuis-Williams P., Fleury-Aubusson A., de Loubresse N. G., Geoffroy H., Vayssié L., Galvani A., Espigat A., Rossier J. (2002). Functional role of ε-tubulin in the assembly of the centriolar microtubule scaffold. J. Cell Biol. 158, 1183–1193 10.1083/jcb.200205028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Trabuco E. C. (1998). The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9, 1293–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Morrissette N. S., Preble A. M., Rackley C., Stanga J. (2002). ε-tubulin is an essential component of the centriole. Mol. Biol. Cell 13, 3859–3869 10.1091/mbc.E02-04-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., Badger J. H., Ren Q., Amedeo P., Jones K. M. et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286 10.1371/journal.pbio.0040286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., Gu L., Hai B., Gorovsky M. A. (1994). High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22, 5391–5398 10.1093/nar/22.24.5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham C. P., Roll-Mecak A. (2012). The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton (Hoboken) 69, 442–463 10.1002/cm.21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings T. H., Jr, Meehl J. B., Pearson C. G., Winey M. (2010). Electron tomography and immuno-labeling of Tetrahymena thermophila basal bodies. Methods Cell Biol. 96, 117–141 10.1016/S0091-679X(10)96006-8 [DOI] [PubMed] [Google Scholar]
- González C., Tavosanis G., Mollinari C. (1998). Centrosomes and microtubule organisation during Drosophila development. J. Cell Sci. 111, 2697–2706 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hai B., Gaertig J., Gorovsky M. A. (2000). Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62, 513–531 10.1016/S0091-679X(08)61554-X [DOI] [PubMed] [Google Scholar]
- Hodges M. E., Scheumann N., Wickstead B., Langdale J. A., Gull K. (2010). Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407–1413 10.1242/jcs.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inclán Y. F., Nogales E. (2001). Structural models for the self-assembly and microtubule interactions of γ-, δ- and ε-tubulin. J. Cell Sci. 114, 413–422 [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. (2004). The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20, 1–28 10.1146/annurev.cellbio.20.022003.114106 [DOI] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Jr, Culver B. P., Yates J. R., 3rd, Winey M. (2007). New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178, 905–912 10.1083/jcb.200703109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R. et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Li J. B., Gerdes J. M., Haycraft C. J., Fan Y., Teslovich T. M., May-Simera H., Li H., Blacque O. E., Li L., Leitch C. C. et al. (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–552 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- Li S., Fernandez J. J., Marshall W. F., Agard D. A. (2012). Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 31, 552–562 10.1038/emboj.2011.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J., Li H., Downing K. H., Nogales E. (2001). Refined structure of α β-tubulin at 3.5 A resolution. J. Mol. Biol. 313, 1045–1057 10.1006/jmbi.2001.5077 [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Nonaka S. (2006). Cilia: tuning in to the cell's antenna. Curr. Biol. 16, R604–R614 10.1016/j.cub.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Meehl J. B., Giddings T. H., Jr and Winey M. (2009). High pressure freezing, electron microscopy, and immuno-electron microscopy of Tetrahymena thermophila basal bodies. Methods Mol. Biol. 586, 227–241 10.1007/978-1-60761-376-3_12 [DOI] [PubMed] [Google Scholar]
- Nanney D. L., Chen S. S., Meyer E. B. (1978). Scalar constraints in Tetrahymena evolution. Quantitative basal body variations within and between species. J. Cell Biol. 79, 727–736 10.1083/jcb.79.3.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley C. E., Oakley B. R. (1989). Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338, 662–664 10.1038/338662a0 [DOI] [PubMed] [Google Scholar]
- Pearson C. G., Giddings T. H., Jr and Winey M. (2009a). Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol. Biol. Cell 20, 904–914 10.1091/mbc.E08-08-0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Osborn D. P., Giddings T. H., Jr, Beales P. L., Winey M. (2009b). Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 187, 905–920 10.1083/jcb.200908019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Müller-Reichert T. (2006). Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004). UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Richards K. L., Anders K. R., Nogales E., Schwartz K., Downing K. H., Botstein D. (2000). Structure-function relationships in yeast tubulins. Mol. Biol. Cell 11, 1887–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., Gorovsky M. A. (2002). A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99, 3734–3739 10.1073/pnas.052016199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Tsao C. C., Gorovsky M. A. (2005). Mutational analyses reveal a novel function of the nucleotide-binding domain of γ-tubulin in the regulation of basal body biogenesis. J. Cell Biol. 171, 1035–1044 10.1083/jcb.200508184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Morgan G., Giddings T. H., Jr, White E. A., Marchione R., McDonald H. B., Winey M. (2005). Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell 16, 3606–3619 10.1091/mbc.E04-10-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K. R., Cole E. S. (2000). Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 62, 291–311 10.1016/S0091-679X(08)61538-1 [DOI] [PubMed] [Google Scholar]
- Vidwans S. J., Wong M. L., O'Farrell P. H. (2003). Anomalous centriole configurations are detected in Drosophila wing disc cells upon Cdk1 inactivation. J. Cell Sci. 116, 137–143 10.1242/jcs.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderfecht T., Stemm-Wolf A. J., Hendershott M., Giddings T. H., Jr, Meehl J. B., Winey M. (2011). The two domains of centrin have distinct basal body functions in Tetrahymena. Mol. Biol. Cell 22, 2221–2234 10.1091/mbc.E11-02-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. (2000). A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2, 358–364 10.1038/35014051 [DOI] [PubMed] [Google Scholar]
- Williams N. E., Honts J. E., Kaczanowska J. (1990). The formation of basal body domains in the membrane skeleton of Tetrahymena. Development 109, 935–942 [DOI] [PubMed] [Google Scholar]
- Wloga D., Gaertig J. (2010). Post-translational modifications of microtubules. J. Cell Sci. 123, 3447–3455 10.1242/jcs.063727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala J. C., Fontalba A., Avila J. (1996). Tubulin folding is altered by mutations in a putative GTP binding motif. J. Cell Sci. 109, 1471–1478 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2007). Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 69, Suppl. 8108–117 10.1002/prot.21702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.