Summary
Sister chromatid cohesion relies on cohesin, a complex comprising a tri-partite ring and a peripheral subunit Scc3, which is found as two related isoforms SA1 and SA2 in vertebrates. There is a division of labor between the vertebrate cohesin complexes; SA1-cohesin is required at telomeres and SA2-cohesin at centromeres. Depletion of SA1 has dramatic consequences for telomere function and genome integrity, but the mechanism by which SA1-cohesin mediates cohesion at telomeres is not well understood. Here we dissect the individual contribution of SA1 and the ring subunits to telomere cohesion and show that telomeres rely heavily on SA1 and to a lesser extent on the ring for cohesion. Using chromatin immunoprecipitation we show that SA1 is highly enriched at telomeres, is decreased at mitosis when cohesion is resolved, and is increased when cohesion persists. Overexpression of SA1 alone was sufficient to induce cohesion at telomeres, independent of the cohesin ring and dependent on its unique (not found in SA2) N-terminal domain, which we show binds to telomeric DNA through an AT-hook motif. We suggest that a specialized cohesion mechanism may be required to accommodate the high level of DNA replication-associated repair at telomeres.
Key words: SA1, Cohesion, Telomeres
Introduction
Sister chromatid cohesion ensures the faithful distribution of sister chromatids in mitosis and provides a template for homology directed repair and recombination in late S and G2 phase of the cell cycle. Cohesion is mediated by cohesin, a ring complex comprising Smc1, Smc3, and Scc1(Anderson et al., 2002; Haering et al., 2002), and a peripheral subunit Scc3, that in vertebrates exists as two closely related isoforms SA1 and SA2 (Losada et al., 2000; Sumara et al., 2000). A cohesin complex contains either SA1 or SA2. In somatic cells SA2 is found in great excess (12- to 15-fold) over SA1 (Holzmann et al., 2011). Precisely how the ring works in vivo to cohere sisters is not known, but models range from the ring encircling the sisters to hold them together (Gruber et al., 2003) to rings on each sister that are held together by a protein bridge (Chang et al., 2005; Zhang et al., 2008).
The best-understood and most fundamental role of cohesin is at centromeres, where it concentrates to hold sister chromatids together against the spindle forces until the metaphase to anaphase transition (Tanaka et al., 2000). Cohesin is also distributed along chromosome arms, albeit at a much lower frequency; the average distance between two cohesin sites is ∼340 kb in human cells (Wendt et al., 2008). Cohesin binds to the same sites as the CCCTC-binding factor CTCF (Parelho et al., 2008; Rubio et al., 2008; Stedman et al., 2008; Wendt et al., 2008) a zinc finger DNA-binding protein that acts as a transcriptional activator/repressor and insulator-binding protein (Wallace and Felsenfeld, 2007), indicating a role for cohesin in the organization of interphase chromatin. Cohesin is also required for DNA repair; it is recruited to sites of double strand breaks to hold the sisters in close proximity for repair (Ström et al., 2004; Ünal et al., 2004). The observation that cohesin needs to be recruited to sites of damage, suggests that normally there is insufficient cohesion to hold the sisters in the close proximity needed for DNA repair.
Telomeres are unique heterochromatic structures that contain TTAGGG repeats and shelterin, a six-subunit complex comprising TRF1, TRF2, TIN2, RAP1, TPP1 and POT1 (de Lange, 2005). Shelterin function is regulated by transiently associating proteins, such as tankyrase 1, a poly(ADP-ribose) polymerase that PARsylates TRF1 (Smith et al., 1998) and can influence association of TRF1 and its shelterin binding partner TIN2 with telomeres (Houghtaling et al., 2004; Smith and de Lange, 2000). Telomeres rely on specialized mechanisms for their replication (Gilson and Géli, 2007; Stewart et al., 2012), protection (Palm and de Lange, 2008), and cohesion. The first indication for a distinct cohesion mechanism came with the observation that sister telomeres remained cohered at mitosis in normal human cells approaching senescence (Ofir et al., 2002; Yalon et al., 2004). A subsequent study in HeLa cells showed that depletion of tankyrase 1 led to persistent telomere (but not arm or centromere) cohesion at mitosis (Dynek and Smith, 2004). Subsequently, the shelterin subunits TIN2 and TRF1, were found to be associated with SA1-cohesin, but not SA2-cohesin (Canudas et al., 2007). In cells depleted of TIN2 or SA1, but not SA2, telomere cohesion was not established in S phase (Canudas and Smith, 2009). Conversely, depletion of SA2, but not SA1 or TIN2, led to a defect in centromere cohesion (Canudas and Smith, 2009). Additional studies showed that the heterochromatin protein HP1γ associated with TIN2 and was required for cohesion at telomeres, but not at centromeres (Canudas et al., 2011).
The studies described above indicate a unique mode of telomere cohesion in human cells that relies exclusively upon SA1-cohesin. Similar results were obtained in mouse cells deleted for SA1 (Remeseiro et al., 2012a). The need for specialized cohesion at telomeres is likely due to the repetitive G-rich structure, which poses problems for replication fork progression, leading to nicks and gaps that must be repaired prior to mitosis. Consistent with this notion, depletion of SA1 led to the inability to repair breaks in G2 and sister telomere loss in human cells (Canudas and Smith, 2009), and to fragile telomeres in mouse cells (Remeseiro et al., 2012a). Despite its importance in genome integrity, the mechanism by which SA1-cohesin mediates sister telomere cohesion is not well understood.
Here we dissect the individual contribution of SA1 and the ring subunits to telomere cohesion. Surprisingly, we find that while depletion of the ring subunits dramatically affects centromere cohesion, it only minimally affects cohesion at telomeres. Instead, we show that SA1, through its unique AT-hook, is the major driving force for cohesion at telomeres. Hence, telomeres employ (in addition to the canonical ring-mediated cohesion that occurs along chromosome arms) a specialized SA1-mediated mechanism for cohesion. We suggest that the difficulty to replicate G-rich DNA at telomeres demands a specialized (more intimate) association between sister chromatids to allow continual surveillance and repair during and after DNA replication.
Results
Resolution of telomere cohesion occurs in G2/M and is dependent on PARP-active tankyrase 1
Telomere cohesion (unlike centromere cohesion) cannot be analyzed by metaphase-spread analysis; telomere cohesion is sensitive to the hypotonic swelling conditions used during metaphase-spread preparation (Canudas et al., 2007; Dynek and Smith, 2004). Hence, for all the experiments described herein, in order to preserve the protein-protein interactions that mediate sister telomere cohesion, live cells are fixed immediately (without pretreatment) in methanol-acetic acid and subjected directly to fluorescent in situ hybridization (FISH).
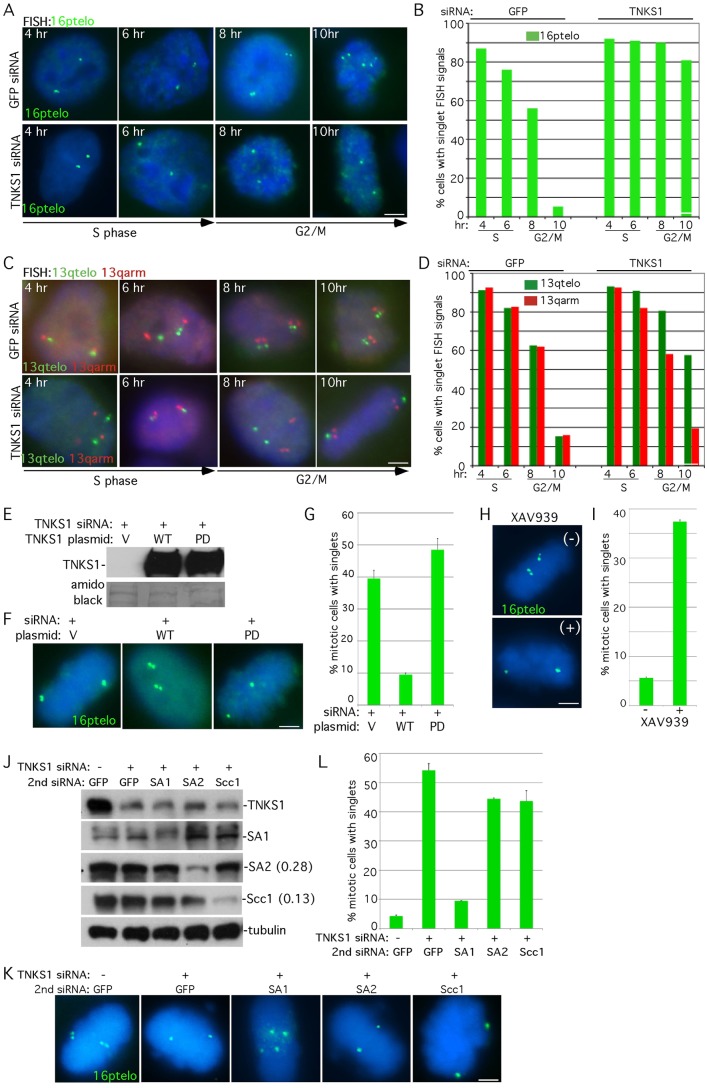
We showed previously (using chromatin immunoprecipitation (ChIP) analysis across the cell cycle) that tankyrase 1 localized to telomeres in G2/M (Bisht et al., 2012). To determine if this localization coincided temporally with resolution of telomere cohesion, we performed FISH analysis with a chromosome-specific subtelomere probe across the cell cycle. Cells were treated with TNKS1 siRNA or GFP as a control, synchronized by a double thymidine block, released for 4, 6, 8 and 10 hours and analyzed by FISH with a 16ptelo probe (Fig. 1A,B). In control cells in S phase (4–6 hours after release) the majority of telomeres appeared as singlets indicating that they were cohered. As cells entered G2/M (8–0 hours after release), telomere cohesion was resolved (Fig. 1A, upper panel and Fig. 1B, left side), coinciding precisely with the time when tankyrase 1 localizes to telomeres (Bisht et al., 2012). In cells depleted of tankyrase 1, telomeres remained as singlets throughout the cell cycle (Fig. 1A, bottom panel and Fig. 1B, right side). To determine if resolution of telomeres and arms was coincident across the cell cycle, double FISH was performed with probes against the telomere and arm of the same chromosome (13qtelo 13qarm). In control cells resolution of telomere and arm cohesion occurred simultaneously across the cell cycle (Fig. 1C, upper panel and Fig. 1D, left side). By contrast, in tankyrase-1-depleted cells arm, but not telomere, cohesion was resolved in G2/M (Fig. 1C, lower panel and Fig. 1D, right side).
Fig. 1.
Resolution of telomere cohesion occurs in G2/M and is dependent on PARP-active tankyrase 1. (A–D) Cell cycle analysis of cohesion. HeLa.I.2.11 cells were synchronized by a double thymidine block, treated with control (GFP) or TNKS1 siRNA, released at 4, 6, 8 and 10 hours, and analyzed by FISH with (A) a 16ptelo (green) or (C) a 13qtelo (green) and 13qarm (red) probe. (B,D) Graphical representation of the frequency of cells with singlet FISH signals from (A; n = 250 cells or more each) and (C; n = 150 cells each), respectively. (E–I) Resolution of sister telomere cohesion requires the PARP activity of tankyrase 1. (E–G) Supertelomerase HeLa cells were synchronized by a double thymidine block, co-transfected with tankyrase 1 siRNA and a control vector (V) or a plasmid containing tankyrase 1 wild type (WT) or PARP-Dead (PD), released for 10 hours, isolated by mitotic shake-off, and analyzed by (E) immunoblot or by (F) FISH with a 16ptelo probe. (G) Graphical representation of the frequency of mitotic cells with singlet FISH signals derived from two independent experiments. Values are means ± s.e.m. (n = 20 cells or more each). (H,I) HeLa1.2.11 cells were synchronized by a double thymidine block, released for 10 hours in the absence (−) or presence (+) of the tankyrase PARP inhibitor XAV939 (1 µm), isolated by mitotic shake-off and analyzed by (H) FISH with a 16ptelo probe. (I) Graphical representation of the frequency of mitotic cells with singlet FISH signals. Values are means ± s.d. from three independent experiments (n = 99 cells or more each). (J–L) Tankyrase 1-induced persistent telomere cohesion is rescued by depletion of SA1, but not by depletion of SA2 or the cohesin ring. HeLaI.2.11 cells were transfected without (−) or with (+) TNKS1 siRNA along with a second siRNA against GFP, SA1, SA2 or Scc1 for 48 hours and analyzed by (J) immunoblot (protein levels relative to α-tubulin and normalized to the GFP siRNA control are indicated next to the blots) or isolated by mitotic shake-off, and analyzed by (K) FISH using a 16ptelo probe (green). (L) Graphical representation of the frequency of mitotic cells with unseparated telomeres in mitosis. Values are means ± s.e.m. from two independent experiments (n = 120 cells or more each). In A, C, F, H, and K DNA was stained with DAPI (blue). Scale bars: 5 µm.
To demonstrate the specificity of resolution of telomere cohesion, we asked if it required the catalytic activity of tankyrase 1. Cells were treated with TNKS1 siRNA and co-transfected with siRNA resistant plasmids encoding wild type or PARP-dead tankyrase 1. Cells were synchronized by a double thymidine block, released for 10 hours, isolated by mitotic shake-off, and analyzed by immunoblot (Fig. 1E) and by FISH with a 16ptelo probe (Fig. 1F,G). Wild type tankyrase 1, but not PARP-dead, rescued the persistent sister telomere cohesion. Consistent with the notion that the catalytic activity of tankyrase 1 was required for resolution of telomere cohesion, treatment of cells with the tankyrase-specific PARP inhibitor XAV939 (Huang et al., 2009) led to persistent sister telomere cohesion (Fig. 1H,I). Thus, the PARP activity of tankyrase 1 is required to disrupt cohesion at telomeres.
We showed previously that persistent telomere cohesion induced by tankyrase 1 depletion could be rescued by depletion of SA1, but not SA2 (Canudas et al., 2007) and see Fig. 1J–L. We now sought to determine if depletion of the cohesin ring would similarly rescue the tankyrase 1-induced persistent cohesion. Cells were subjected to double siRNA treatment with TNKS1 and individual cohesin subunits, analyzed by immunoblot (Fig. 1J), and by FISH with a 16ptelo probe following mitotic shake-off (Fig. 1K,L). Depletion of SA1 led to a 6-fold reduction in cells with singlets, rescuing cohesion to nearly wild type levels. By contrast, depletion of SA2 led to only a 1.3-fold reduction in cells with singlets (Fig. 1K,L). Surprisingly, depletion of the ring subunit Scc1 had only a minimal effect (1.3-fold; similar to SA2), indicating that persistent telomere cohesion in tankyrase-1-depleted cells is mediated largely by SA1, with only minimal contribution by SA2 and the cohesin ring.
A limited requirement for cohesin ring subunits in telomere cohesion
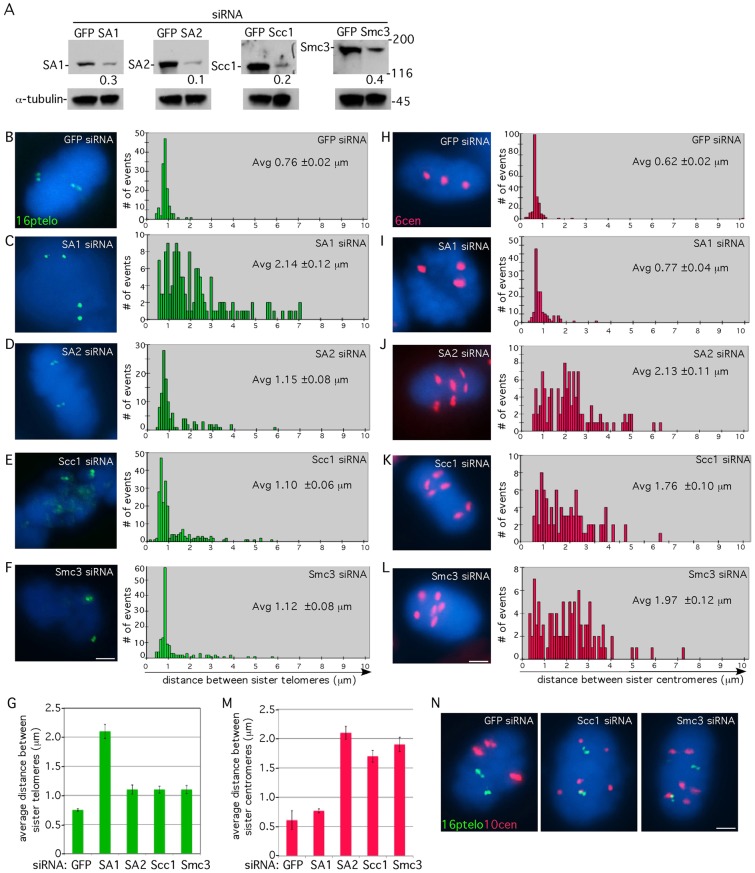
We next asked if the cohesin ring was required to maintain cohesion at telomeres irrespective of tankyrase 1. Following treatment with siRNA for each cohesin subunit, cells were analyzed by immunoblot (Fig. 2A) and by FISH with a 16ptelo probe following mitotic shake-off (Fig. 2B–F,G). In HeLaI.2.11 cells treated with GFP siRNA sister telomeres appeared as closely associated doublets separated by an average distance of 0.76 µm (Fig. 2B). Depletion of SA1 led to a dramatic increase (average distance 2.14 µm) in the distance between most sister telomeres, whereas in SA2-depleted cells most sister telomeres remained close, although a subset showed increased distance [Fig. 2C,D, and shown previously (Canudas and Smith, 2009)]. In cells depleted of the ring subunits Scc1 or Smc3, most sister telomeres remained closely associated as in control cells, although a subset showed increased distance (Fig. 2E,F). Together these data show that in cells depleted of SA2 or the ring (but not SA1), the majority of sister telomeres remain closely associated, suggesting that SA2 and the ring contributed only minimally to telomere cohesion.
Fig. 2.
Telomere cohesion is maintained in mitosis in cohesin-ring-depleted cells. (A) Immunoblot analysis of extracts from HeLaI.2.11 cells transfected with siRNA to GFP, SA1, SA2, Scc1 or Smc3 for 48 hours and probed with the indicated antibodies. Protein levels relative to α-tubulin and normalized to the GFP siRNA control are indicated below the blots. (B–F) Telomere and (H–L) centromere FISH analysis of HeLaI.2.11 cells isolated by mitotic shake-off following 48 hours transfection with GFP (B,H), SA1 (C,I), SA2 (D,J), Scc1 (E,K) or Smc3 (F,L) siRNA with a 16ptelo (green) or 6cen (red) probe. The cen locus is trisomic. DNA was stained with DAPI (blue). Scale bars: 5 µm. Histograms showing the distance between FISH signals (n = 98–225) are on the right with the average (Avg) distance (± s.e.m.) indicated. (G,M) Graphical representation of the average distance (± s.e.m.) between sister telomeres (G) or centromeres (M). (N) Combined telomere and centromere FISH analysis. HeLaI.2.11 cells were transfected with GFP, Scc1 or Smc3 siRNA for 48 hours and probed with 16ptelo (green) and 10cen (red). The cen locus is trisomic. DNA was stained with DAPI (blue). Scale bar: 5 µm.
To validate that cohesin ring function was impaired in the Scc1- and Smc3-depleted cells, we measured centromere cohesion using FISH with a chromosome-specific centromere probe 6cen (Fig. 2H–L,M). In HeLaI.2.11 cells treated with GFP siRNA centromeres were cohered, appearing as closely associated doublets with an average diameter of 0.62 µm (Fig. 2H). Depletion of SA1 led to only a minor effect (1.2-fold increase) in the distance between sister centromeres, whereas in SA2-depleted cells the majority of sister centromeres showed an increased distance [Fig. 2I,J, and shown previously (Canudas and Smith, 2009)]. In cells depleted of the ring subunits Scc1 or Smc3, the distance between most sister centromeres was increased dramatically (Fig. 2K,L). Together these data show that in cells depleted of SA2 or the ring, the majority of sister centromeres have lost cohesion (Fig. 2M), whereas the majority of sister telomeres retain their close association (Fig. 2G). Indeed, double FISH analysis of Scc1- or Smc3-depleted cells showed that within the same cell telomere cohesion remained intact despite a dramatic loss in centromere cohesion (Fig. 2N).
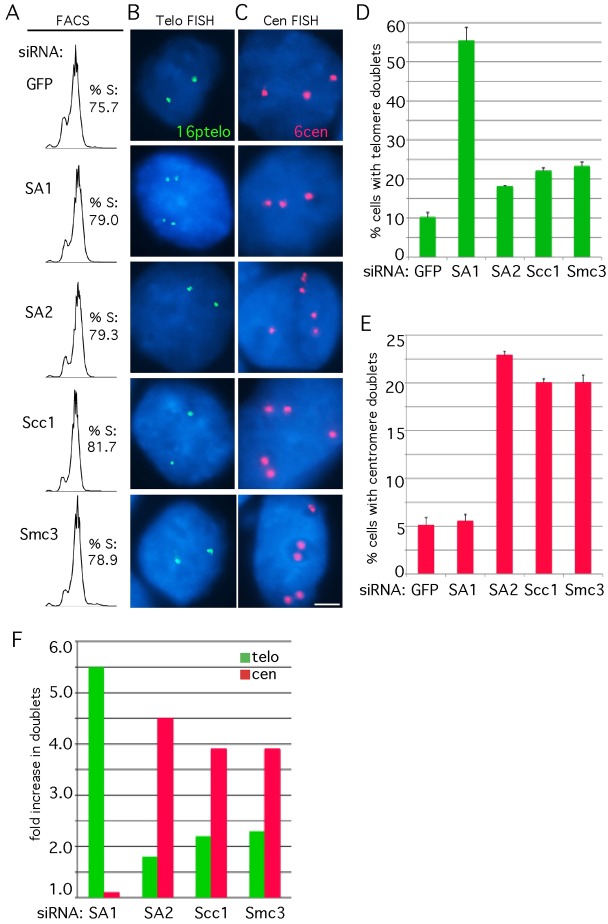
We were surprised that depletion of the ring had only a limited effect on telomere cohesion measured at mitosis. To probe the issue further we queried the impact of ring depletion on establishment of cohesion at telomeres in S phase. siRNA-treated HeLa.I.2.11 cells were synchronized with a double thymidine block, harvested 4 hours after release (mid S phase), and processed for FACS analysis (Fig. 3A) and chromosome-specific telomere (Fig. 3B,D) or centromere (Fig. 3C,E) FISH. FACS analysis indicated that the siRNA treated cells were in S phase (Fig. 3A). In cells treated with GFP siRNA the majority of telomeres and centromeres appeared as singlets, consistent with the view that in mid-S-phase chromosomes are either unreplicated or replicated, but still cohered. Cells depleted of SA1 showed a dramatic (5.5-fold) increase in telomere doublets, whereas SA2-depleted cells showed only a 1.8-fold increase in telomere doublets [Fig. 3B,D, and shown previously (Canudas and Smith, 2009)]. In cells depleted of the ring subunits Scc1 or Smc3, most telomeres remained as singlets, although there was a (2-fold) increase in doublets (Fig. 3B,D,F). Analysis of centromere cohesion under the same conditions revealed the opposite; depletion of SA1 had no effect, whereas depletion of SA2, Scc1 or Smc3 resulted in at least a 4-fold increase in doublets (Fig. 3C,E,F). Together these data support the notion that SA1 drives telomere cohesion, whereas, SA2 along with the cohesin ring drives centromere cohesion.
Fig. 3.
Telomere cohesion is established in S phase in cohesin-ring-depleted cells. HeLa.I.2.11 cells were synchronized by a double thymidine block, treated with siRNA against GFP, SA1, SA2, Scc1 or Smc3 and analyzed by (A) FACS, (B) telomere FISH with a telomere 16ptelo (green), or (C) centromere FISH with a 6cen (red) probe 4 hours after release from the second thymidine block. DNA was stained with DAPI (blue). Scale bar: 5 µm. (D,E) Graphical representation of the frequency of telomere (from B; n = 514 cells or more each) and centromere (from C; n = 441 cells or more each) doublets in S phase, respectively. Values are means ± s.e.m., derived from two independent experiments. (F) Graphical representation of the fold increase in telomere (from D) and centromere (from E) doublets relative to the GFP siRNA control.
SA1 is enriched at telomeres
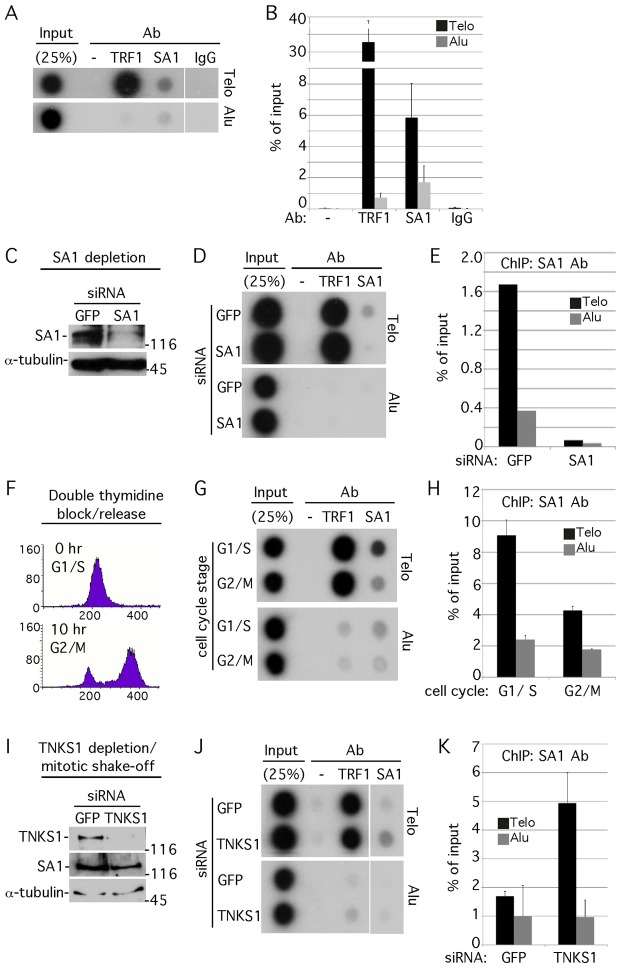
The studies described above raised the possibility that SA1 may be acting in a unique or independent way at telomeres. We thus asked if we could detect accumulation of SA1 at human telomeres. The genomic distribution of SA1 was measured previously using ChIP-ChIP on the non-repetitive ENCODE regions representing 1% of the human genome (Rubio et al., 2008; Wendt et al., 2008). However, these studies did not address the association with telomeres. To address this question, we performed a specialized telomeric ChIP assay that uses a dot blot approach to hybridize the immunoprecipitated DNA with a telomere repeat probe (Telo). Alu repeats are used as a negative control. ChIP analysis of HeLaI.2.11 cells using anti-TRF1 antibody revealed a robust telomere-specific signal (Fig. 4A,B), as expected for a telomeric-DNA-binding protein. ChIP with anti-SA1 antibody revealed a weaker, but specific (and reproducible) telomeric association (Fig. 4A,B). To validate that the increased SA1 signal observed at telomeres was due to SA1 protein, ChIP was performed on SA1-depleted cells. Immunoblot analysis of cells treated with GFP or SA1 siRNA showed efficient knockdown of SA1 (Fig. 4C) and a corresponding knockdown of the SA1-ChIP signal (Fig. 4D,E).
Fig. 4.
SA1 associates with telomeres in vivo. (A,B) Telomeric ChIP analysis of TRF1 and SA1. (A) Autoradiograph showing telomeric DNA ChIP analysis of HeLaI.2.11 cells using the indicated antibodies. Dot blots with the immunoprecipitated DNA were analyzed by Southern blotting with 32P-labelled telomeric (Telo) or Alu repeat (Alu) probes. (B) Graphical representation of the percentage of immunoprecipitated Telo or Alu DNA relative to total input DNA (as in A), derived from four independent experiments. Values are means ± s.d. (C–E) Ablation of the SA1 signal at telomeres by SA1 siRNA. HelaI.2.11 cells were transfected with siRNA to GFP or SA1 for 48 hours and analyzed by (C) immunoblotting with antibodies against SA1 or α-tubulin and (D) telomeric ChIP with antibodies against TRF1 or SA1. (E) Graphical representation of the percentage of SA1-immunoprecipitated Telo or Alu DNA relative to total input DNA (as in D). (F–H) SA1 is reduced at telomeres in mitosis. HeLaI.2.11 cells were synchronized by a double thymidine block, released, and collected by trypsinization at 0 hours (G1/S) or 10 hours (G2/M) and analyzed by (F) FACS (y-axis, cell numbers; x-axis, relative DNA content based on propidium iodide staining) and (G) telomeric ChIP with antibodies against TRF1 or SA1. (H) Graphical representation of the percentage of SA1-immunoprecipitated Telo or Alu DNA relative to total input DNA (as in G). Values are means ± s.e.m., derived from two independent experiments. (I–K) SA1 is increased at telomeres in TNKS1-depleted mitotic cells. HelaI.2.11 cells were transfected with siRNA to GFP or TNKS1 for 48 hours, isolated by mitotic shake-off, and analyzed by (I) immunoblotting with antibodies against TNKS1, SA1 or α-tubulin and (J) telomeric ChIP with antibodies against TRF1 or SA1. (K) Graphical representation of the percentage of SA1-immunoprecipitated Telo or Alu DNA relative to total input DNA (as in J). Values are means ± s.e.m., derived from two independent experiments.
Telomere cohesion is released at mitosis in a tankyrase 1 dependent manner (described above; Fig. 1A–D). Accordingly, we asked if SA1 was released from telomeres at mitosis. HeLaI.2.11 cells were synchronized by a double thymidine block, released, and collected at 0 and 10 hours. FACS analysis indicated that the cells were synchronized in G1/S (0 hours) and G2/M (10 hours; Fig. 4F). ChIP analysis of the cell cycle staged extracts indicated a greater than 2-fold reduction of SA1 at telomeres in mitosis (Fig. 4G,H). To determine if removal of SA1 relied on tankyrase 1, HeLaI.2.11 cells were treated with GFP or TNKS1 siRNA and mitotic cells were isolated by mitotic shake-off. Immunoblot analysis showed efficient knockdown of TNKS1 (Fig. 4I). ChIP analysis revealed a threefold increase in SA1 at telomeres in TNKS1-depleted mitotic cells (Fig. 4J,K). Together the data are consistent with a role for SA1 in telomere cohesion; SA1 is enriched at telomeres, is reduced at telomeres in mitosis when cohesion is removed, and is increased at telomeres in TNKS1-depleted mitotic cells when cohesion persists.
SA1 overexpression promotes sister telomere cohesion
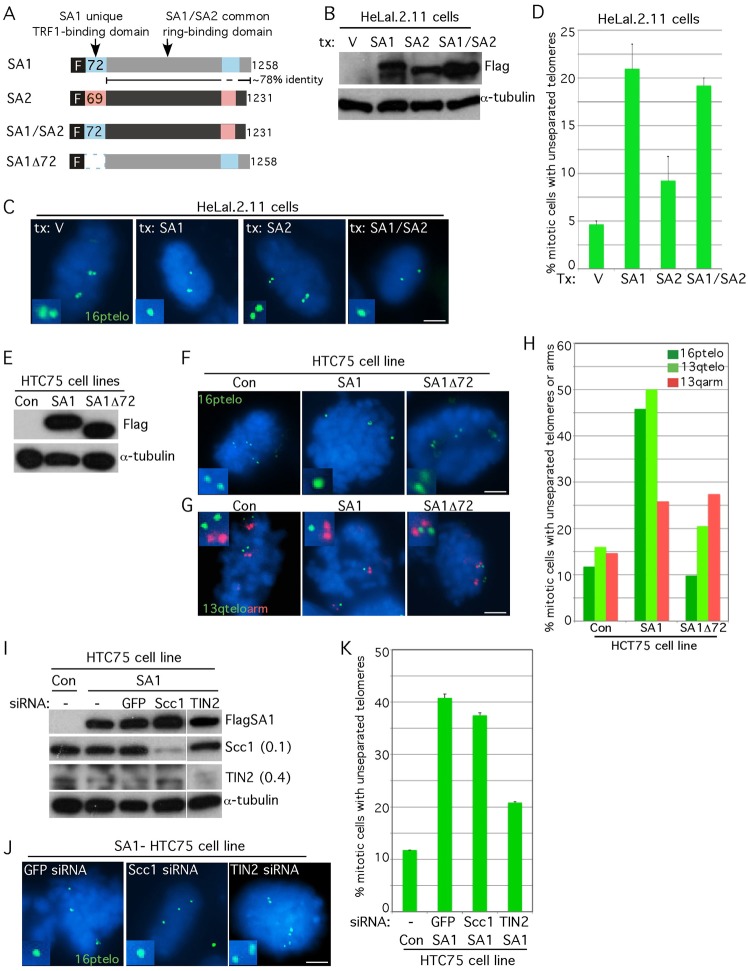
Our studies thus far indicated that SA1 was enriched at telomeres where it was required (to a greater extent than SA2 and the cohesin ring) to establish and maintain sister telomere cohesion. SA1 and SA2 are highly conserved along their length, where they bind to the cohesin ring through association with Scc1, but are distinguished by unique sequences in their N- and C-termini (Fig. 5A). To dissect the telomeric specificity of SA1 versus SA2, HeLaI.2.11 cells were transiently transfected with epitope-tagged alleles, FlagSA1 or FlagSA2. Transfected cells were analyzed by immunoblot (Fig. 5B) and isolated by mitotic shake-off for FISH analysis with a 16ptelo probe. As shown in Fig. 5C,D, overexpression of SA1 induced a fivefold increase in persistent telomere cohesion at mitosis, whereas overexpression of SA2 had a less than twofold effect.
Fig. 5.
SA1 induces persistent sister chromatid cohesion at telomeres. (A) Schematic representation of FLAG-tagged SA1, SA2 and mutant alleles SA1/SA2 and SA1Δ72. (B–D) Overexpression of SA1 but not SA2, induced persistent telomere cohesion. HeLaI.2.11 cells were transfected with vector, SA1, SA2 or SA1/SA2 for 20 hours and analyzed by (B) immunoblot and (C) telomere FISH using a 16ptelo probe (green) following mitotic shake-off. (D) Graphical representation of the frequency of mitotic cells with unseparated telomeres. Values are means ± s.e.m., derived from two independent experiments (n = 68 cells or more each). (E–H) SA1 induced persistent cohesion specifically at telomeres, dependent on its N-terminal domain. Stable HTC75 cell lines overexpressing SA1 or SA1Δ72 or control cells (Con) were analyzed by (E) immunoblot, (F) telomere FISH using a 16ptelo probe or (G) by double FISH with a 13qtelo (green) and 13qarm (red) probe following mitotic shake-off. (H) Graphical representation of the frequency of mitotic cells with unseparated telomeres or arms (n = 102 cells or more each). (I–K) SA1 induced persistent telomere cohesion independent of Scc1, but dependent on TIN2. Stable SA1-HTC75 or control cells were treated without (−) or with siRNA against GFP, Scc1 or TIN2 and analyzed by (I) immunoblot (protein levels relative to α-tubulin and normalized to the GFP siRNA control are indicated next to the blots) and (J) telomere FISH using a 16ptelo probe (green) following mitotic shake-off. (K) Graphical representation of the frequency of mitotic cells with unseparated telomeres. Values are means ± s.e.m., derived from two independent experiments (n = 29 cells or more each). In C, F, G and J DNA was stained with DAPI (blue). Scale bars: 5 µm.
Our previous observation that the 72-amino-acid domain of SA1 (but not the corresponding 69-amino-acid domain in SA2) bound to the shelterin subunit TRF1 (Canudas et al., 2007), suggested that this domain could be important for telomere cohesion. Hence, we generated a chimeric protein replacing the N-terminal domain of SA2 with that of SA1 (SA1/SA2; Fig. 5A). FISH analysis of HeLaI.2.11 cells transiently transfected with an epitope-tagged allele of the chimera (FlagSA1/SA2) induced persistent telomere cohesion similar to SA1 (Fig. 5C,D), consistent with the notion that the unique N-terminal domain of SA1 imparts telomere specificity to SA1-mediated cohesion.
To further dissect the telomere specificity of SA1 in cohesion, HTC75 cell lines stably overexpressing FlagSA1 or an N-terminally deleted allele of SA1 (FlagSA1Δ72) were generated and analyzed by immunoblot (Fig. 5E) and isolated by mitotic shake-off for FISH analysis with a 16ptelo probe. As shown in Fig. 5F,H, the SA1-HTC75 cell line, but not the SA1Δ72-HTC75 line, showed persistent sister telomere cohesion in mitosis, consistent with a role for the 72-amino-acid N-terminal domain of SA1 in telomere cohesion. To determine if the effect of SA1 on cohesion was specific to telomeres, double FISH was performed with probes against the telomere and arm of the same chromosome. As shown in Fig. 5G,H, FISH analysis with the 13qtelo probe (like the 16ptelo probe) showed persistent telomere cohesion dependent on the N-terminal domain of SA1. By contrast, FISH analysis with an arm probe from the same chromosome (13qarm) showed only a slight increase in cohesion and this was independent of the N-terminal domain of SA1.
Finally, to determine the protein requirements for the persistent sister telomere cohesion induced by overexpression of SA1, the SA1-HTC75 cell line was treated with siRNA against GFP control, the cohesin ring subunit Scc1 or the shelterin subunit TIN2 and analyzed by immunoblot (Fig. 5I) and isolated by mitotic shake-off for FISH analysis with a 16ptelo probe. As shown in Fig. 5J,K, depletion of Scc1 did not rescue the persistent cohesion induced by SA1. By contrast, depletion of TIN2, shown previously to be required for sister telomere cohesion (Canudas et al., 2007; Canudas and Smith, 2009), partially rescued the SA1-induced persistent cohesion at telomeres. Together these data indicate that SA1 induces persistent cohesion (specifically at telomeres) that is dependent on its unique N-terminal TRF1-binding domain and is independent of the cohesin ring.
SA1 binds directly to DNA through a N-terminal AT-hook motif
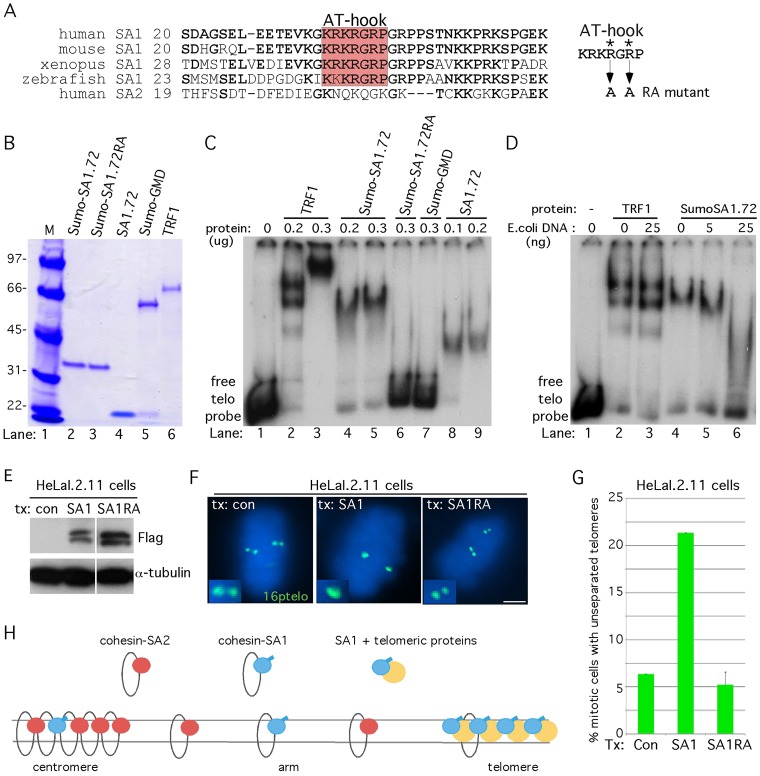
The studies described above indicate a crucial role for the unique N-terminal domain of SA1 in mediating telomere cohesion. Inspection of the amino acid sequence revealed the presence of an AT-hook motif at amino acid position 34 [previously identified in a screen for all detectable AT-hooks in the protein sequence database (Aravind and Landsman, 1998)]. The AT-hook motif, first described in the high mobility group proteins (HMGA1/2), binds to AT-rich DNA sequences in the minor groove of DNA in a non-sequence-specific manner (Reeves, 2001). The AT-hook motif in SA1 (KRKRGRP) is highly conserved, but is not found in SA2 (Fig. 6A). To determine if the AT-hook containing N-terminal domain of SA1 binds to DNA, we generated recombinant proteins (Fig. 6B) and performed gel shift assays with a 32P-labeled duplex (TTAGGG)12 probe and poly(dG-dC) as non-specific competitor (Fig. 6C). As a positive control we used TRF1 (Fig. 6B, lane 6), which binds specifically to telomere repeats (Chong et al., 1995) (shown in Fig. 6C, lanes 2 and 3). For SA1, the 72-amino-acid N-terminal domain was expressed as a Sumo fusion protein and purified from E. coli (Fig. 6B, lane 2). Sumo-SA1.72 (like TRF1) bound to telomeric DNA (Fig. 6C, lanes 4 and 5).
Fig. 6.
SA1 binds to telomeric DNA in vitro and promotes persistent telomere cohesion in vivo through its AT-hook. (A) Alignment of the SA1 domain containing the AT-hook. Amino acids identical to human SA1 are in bold. The AT-hook RA mutation is indicated on the right. (B–D) SA1 binds telomeric DNA. (B) Purified recombinant proteins (500 ng each) were fractionated by SDS-PAGE and visualized by staining with Coomassie Blue. (C,D) Autoradiographs of the telomere repeat binding assay with the indicated recombinant proteins and a 32P-labeled (TTAGGG)12 probe. SA1 binds to telomeric DNA dependent on the AT-hook (C). Addition of E. coli competitor DNA competes for SA1, but not TRF1, binding to telomeric DNA (D). (E–G) SA1 promotes persistent telomere cohesion dependent on its AT-hook. HeLaI.2.11 cells were transfected with a vector control, SA1 and SA1RA for 20 hours and analyzed by (E) immunoblot and (F) telomere FISH using a 16ptelo probe (green) following mitotic shake-off. Scale bar: 5 µm. (G) Graphical representation of the frequency of mitotic cells with unseparated telomeres. Values are means ± s.e.m., derived from two independent experiments (n = 102 cells or more each). (H) Model of how SA1 promotes cohesion independent of the cohesin ring by associating with telomeric DNA and proteins.
To determine if the binding was due to the AT-hook motif, we generated a double point mutation, converting the arginines [predicted to participate in DNA–protein interactions (Huth et al., 1997)] at position 37 and 39 to alanines (Sumo-SA1.72RA, see Fig. 6A). As shown in Fig. 6C, lane 6, Sumo-SA1.72RA did not bind telomeric DNA, indicating a requirement for the AT-hook motif in DNA binding. As additional controls, we show that an unrelated Sumo protein (Sumo-GMD, Fig. 6B, lane 5) (Bisht et al., 2012) did not bind DNA (Fig. 6C, lane 7) and that the SA1-72 protein alone (cleaved and purified away from Sumo, Fig. 6B, lane 4) bound to the (TTAGGG)12 probe (Fig. 6C, lanes 8 and 9), like Sumo-SA1-72.
To determine if SA1 showed the same specificity for telomeric DNA as TRF1, we asked if E. coli DNA could compete with binding of SA1 to telomeric DNA. As shown in Fig. 6D, under conditions where the TRF1-(TTAGGG)12 complex was resistant to competition (Fig. 6D, lanes 2 and 3), the SA1-(TTAGGG)12 complex was competed by E. coli DNA (Fig. 6D, lanes 4 to 6), consistent with the notion that AT-hooks do not bind in a sequence-specific fashion to DNA. Together these data indicate the while SA1 is not a sequence-specific DNA-binding protein like TRF1, it does binds to telomeric (and likely other AT-rich) DNA through its AT-hook motif.
Finally, to determine if the AT-hook was required for telomere cohesion in vivo, we created the same (RA) double point mutation in the full-length FLAG-tagged SA1 allele to generate FlagSA1RA. HeLaI.2.11 cells were transiently transfected with FlagSA1 or FlagSA1RA, analyzed by immunoblot (Fig. 6E), and isolated by mitotic shake-off for FISH analysis with a 16ptelo probe. As shown in Fig. 6F, G, mutation of the AT-hook abrogated the ability of SA1 to induce persistent cohesion at telomeres.
Discussion
Telomeres are unique heterochromatic structures that rely on specialized mechanisms for their replication and protection. Perhaps it is not surprising then that they utilize a novel mechanism for their cohesion. The repetitive, G rich nature of telomeres makes them highly susceptible to replication damage and hence more reliant on homology directed repair with the sister chromatid to restore genome integrity following DNA replication in late S and G2 phases of the cell cycle. Our study indicates that SA1, through its unique AT-hook, drives cohesion at telomeres. We suggest that SA1, along with the shelterin subunits TIN2 and TRF1 and heterochromatin protein HP1γ, promote an intimate association between sister telomeres that allows continual surveillance during and after DNA replication that cannot be achieved by the cohesin ring (see model in Fig. 6H).
Our studies indicate that depletion of the cohesin ring, despite having a robust effect at centromeres, only minimally affects cohesion at telomeres. This is perhaps not surprising since cohesin is highly enriched at centromeres where it plays an essential role holding sister centromeres together against the forces of the mitotic spindle. In our study depletion of the ring subunits (Smc3 and Scc1) phenocopied depletion of SA2 (Figs 2, 3), consistent with the notion that these subunits (SA2, Smc3 and Scc1) act in concert. However, depletion of the ring did not phenocopy depletion of SA1, suggesting that these subunits (SA1, Smc3 and Scc1) do not act in concert. Depletion of SA2, Smc3 or Scc1 had some impact on telomere cohesion, but it was significantly less than the impact of SA1 depletion, suggesting a distinct role for SA1 at telomeres. Indeed, we detected enrichment of SA1 at telomeres by ChIP (Fig. 4), but we did not detect SA2, Scc1 or Smc3 at telomeres (data not shown). While this could be due to differences between antibodies used for SA1 versus the other subunits, given the sparse distribution of cohesin rings along chromosome arms [one every 340 kb (Wendt et al., 2008)], we favor the interpretation that telomeres, which are 20–25 kb in HeLa1.2.11cells, may be devoid of rings. We suggest that the small cohesion defects observed at telomeres upon depletion of SA2 and the ring, are due to loss of cohesion at the level of the whole chromosome, from the neighboring arm or centromere. Together our studies suggest that the robust effect of SA1 depletion or overexpression on telomere cohesion is due to intrinsic properties of the SA1 subunit.
SA1 contains an AT-hook in its N-terminal domain that is highly conserved in SA1 homologs from other species and is not found in SA2 (Fig. 6A). We show that SA1 binds directly to telomeric DNA in vitro (Fig. 6C). However, this binding does not have the specificity of the sequence-specific telomere-repeat-binding protein TRF1, since in the case of SA1, we can compete the binding using E. coli competitor DNA (Fig. 6D), consistent with the idea that AT-hooks bind in a non-sequence-specific manner (Reeves, 2001). We suggest that the AT-hook endows SA1 with the capacity to associate with telomeres where it then binds to TRF1 and other proteins to promote cohesion.
Our finding that SA1 contains a functional AT-hook may shed light on additional roles of SA1 in vivo, revealed recently by work in mice. Knockout of SA1 in mouse resulted in embryonic lethality (Remeseiro et al., 2012a; Remeseiro et al., 2012b). While analysis of SA1-null mouse cells indicated defects in telomere cohesion and telomere replication (Remeseiro et al., 2012a) [consistent with studies in human cells (Canudas and Smith, 2009)], additional observations indicated a role for SA1 in different processes. Analysis of the distribution of SA1-cohesin versus SA2-cohesin in the non-repetitive parts of the mouse genome by ChIP-sequencing indicated striking differences in their genomic distribution; SA1 was enriched at promoters and SA1 null cells showed dramatic changes in gene expression (Remeseiro et al., 2012b). Considering that AT-hook containing proteins have been shown to function in chromatin remodeling and transcription, it is tempting to speculate that SA1 may directly impact transcription through its unique AT-hook.
The initial finding that the Scc3 subunit of cohesin existed as two isoforms in vertebrate cells was surprising. However, given the strong homology between the two (and their association with the ring) it seemed likely that it was simply a gene duplication event that might not lead to dramatic difference in function. However, studies in human and mouse cells show that SA1 is required for sister chromatid cohesion at telomeres and its depletion has dramatic consequences on cell and organismal function despite the presence of SA2. Our work here, identifying a functional AT-hook motif that is unique to SA1, provides a molecular mechanism and sheds slight on the role of SA1 at telomeres and potentially throughout the genome.
Materials and Methods
Plasmids
The SA1 cDNA (kindly provided by Jose Luis Barbero) containing amino acids 1–1258 and the SA2 cDNA containing amino acids 1–1231 were cloned into the vector p3XFLAG-CMV-10 (Sigma) to generate 3XFlagSA1 and 3XFlagSA2. The SA1/SA2 chimera was generated by replacing amino acids 1–69 of FlagSA2 with SA1 amino acids 1–70; the resulting hybrid contained three alanines at the junction site. Sumo-SA1.72 contains amino acids 1–72 of SA1 cloned into the BamHI and XhoI sites of the pET28b-Sumo-6xHis vector (Chen et al., 2008). The SA1RA mutation was created by substituting the arginine (R) residues at position 37 and 39, with alanine (A) residues, by site-directed mutagenesis of Sumo-SA1.72 and 3XFlagSA1 using the oligonucleotide 5′-CAGAGGTCAAAGGAAAAAGAAAAGCGGGTGCTCCTGGCCGGCC-3′. Mutagenesis was performed using the Stratagene QuikChange site-directed mutagenesis kit according to the manufacturer's instructions. For tankyrase 1 constructs, the tankyrase1 cDNA under the CMV promoter, derived from plasmid TT20 (Smith et al., 1998), was inserted into a modified pLKO.1ps vector (a kind gift from Bill Hahn, Dana-Farber Cancer Institute, Boston, MA). The siRNA resistant (Dynek and Smith, 2004) and tankyrase 1 PARP-dead (Cook et al., 2002) plasmids were generated as previously described.
Chromatin lmmunoprecipitation
HeLaI.2.11 cells were processed for ChIP as described previously (Bisht et al., 2012). Following preparation of cell lysates, 25% was saved as input. Immunoprecipitation was performed by addition of the following antibodies: 5 µl of rabbit TRF1 415 (Cook et al., 2002) crude sera, 5 µg goat anti-SA1 BL143G (Bethyl Laboratories, Inc.); or 5 µg goat IgG. Hybridization with a 32P-TTAGGG or Alu probe was performed in Church buffer [0.5 M sodium phosphate buffer (pH 7.2), 1% BSA, 1 mM EDTA, 7% SDS] as described previously (de Lange, 1992). Washed membranes were exposed to Kodak XAR film and ImageJ software was used to quantify the percentage of precipitated DNA relative to the input DNA.
Chromosome-specific FISH
Cells were fixed and processed as described previously (Dynek and Smith, 2004). Briefly, cells were fixed twice in methanol∶acetic acid (3∶1) for 15 minutes, cytospun (Shandon Cytospin) at 2000 rpm for 2 minutes onto slides, rehydrated in 2× SSC at 37°C for 2 minutes, and dehydrated in an ethanol series of 70%, 80% and 95% for 2 minutes each. Cells were denatured at 75°C for 2 minutes and hybridized overnight at 37°C with the following probes from Cytocell: a subtelomeric FITC-conjugated probe (16ptelo); a chromosome-6-specific alpha-satellite TRITC-conjugated centromere probe (6cen); a TRITC-conjugated chromosome 10 centromere probe (10cen); or a dual probe comprising a TRITC-conjugated 13qarm and a FITC-conjugated 13q subtelomere probe. Cells were washed in 0.4× SSC at 72°C for 2 minutes, and in 2× SSC with 0.05% Tween 20 at RT for 30 seconds. DNA was stained 0.2 µg/ml DAPI. The distance between FISH signals was measured using OpenLab software (Perkin Elmer).
Cell synchronization
For FISH analysis across the cell cycle, HelaI.2.11 cells were grown in the presence of 2 mM thymidine for 16 hours, washed three times with PBS, released into fresh medium, and transfected with siRNA. After 10 hours the medium was replaced with medium containing 2 mM thymidine and the cells were incubated for 16 hours, washed three times with PBS, and released into fresh medium. Cells were then harvested by trypsinization at 4, 6, 8 and 10 hours and analyzed by FISH.
For the tankyrase 1 rescue experiment supertelomerase HeLa cells (Cristofari and Lingner, 2006) were synchronized and transfected with siRNA as describe above, but were in a addition, transfected with tankyrase 1 plasmids prior to the second thymidine block. After release for 10 hours, cells were harvested by mitotic shake-off and analyzed by immunoblot and FISH.
For the XAV939 inhibition, HeLaI.2.11 cells were synchronized by a double thymidine block as describe above, released for 10 hours in 0.5% serum with or without 1 mM XAV939, harvested by mitotic shake-off, and analyzed by FISH.
Stable cell lines
The SA1 cDNA (amino acids 1–1258) (Carramolino et al., 1997; Losada et al., 2000) was obtained from The I.M.A.G.E. Consortium. The clone (accession no. AAH64699) contained an internal deletion from amino acids 1150–1186. SA1 and SA1Δ72 (lacking the first 72 amino acids) were cloned into the vector p3XFLAG-CMV-10 (Sigma) to generate 3XFlagSA1 and 3XFlagSA1Δ72, described previously (Canudas et al., 2007), and then inserted into the retroviral vector pLPCX (Clontech). To create stable cell lines, amphotropic retroviruses were generated by transfecting FlagSA1 and FlagSA1Δ72 into phoenix amphotropic cells (ATCC) using calcium phosphate precipitation. HTC75 cells [an HT1080-derived clonal cell line (van Steensel and de Lange, 1997)] were infected and selected in 2 µg/ml puromycin as described (Houghtaling et al., 2004).
Cell extracts
Cells were resuspended in 4 volumes of TNE buffer [10 mM Tris (pH 7.8), 1% Nonidet P-40, 0.15 M NaCl, 1 mM EDTA, and 2.5% protease inhibitor cocktail (PIC) (Sigma)] and incubated for 1 hour on ice. Suspensions were pelleted at 8000 g for 15 minutes. Twenty-five micrograms (determined by Bio-Rad protein assay) of supernatant proteins were fractionated by SDS-PAGE and analyzed by immunoblotting.
Immunoblot analysis
Immunoblots were incubated separately with the following primary antibodies: mouse anti-α-tubulin ascites (1∶10,000; Sigma); goat anti-SA1 BL143G (1 µg/ml; Bethyl Laboratories, Inc.); goat anti-SA2 BL146G (1 µg/ml; Bethyl Laboratories, Inc.); rabbit anti-Scc1 (2 µg/ml; Bethyl Laboratories, Inc.); rabbit anti-Smc3 (1 µg/ml; Abcam); rabbit anti-TIN2 701 (0.5 µg/ml) (Houghtaling et al., 2004); or rabbit anti-tankyrase 1 762 (1.8 µg/ml) (Scherthan et al., 2000), followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (1∶2500; Amersham). Bound antibody was detected by Super Signal West Pico (Thermo Scientific).
siRNA transfection
siRNA transfections were performed with in HeLaI.2.11 cells, a HeLa-derived clonal cell line (van Steensel et al., 1998) or HTC75 cell lines with Oligofectamine (Invitrogen) according to the manufacturer's protocol. The final concentration of siRNA was 100 nM. The following siRNAs (synthesized by Dharmacon Research Inc.) were used: TNKS1 (5′-AACAAUUCACCGUCGUCCUCU-3′) described previously (Dynek and Smith, 2004); SA1.a (5′-GUGAUGCCUUCCUAAAUGA-3′); SA2.a (5′-GUACGGCAAUGUCAAUAUA-3′); and TIN2.a (5′-AACGCCUUUGUAUGGGCCUAA-3′) described previously (Canudas et al., 2007); Scc1 (5′-GGUGAAAAUGGCAUUACGGUU-3′) described previously (Watrin et al., 2006); Smc3 (5′-AUCGAUAAAGAGGAAGUUUUU-3′) described previously (Kueng et al., 2006); and GFP Duplex I. Following 48 hours transfection, cells were isolated by mitotic shake-off and processed for chromosome-specific FISH.
Plasmid transfections
HeLa1.2.11 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Following 20 hours transfection, cells were isolated by mitotic shake-off and processed for chromosome-specific FISH.
Purified recombinant proteins and gel-shift assays
TRF1 was purified from baculovirus as described previously (Bianchi et al., 1997). Recombinant Sumo-SA1.72, Sumo-SA1.72RA, and Sumo-GMD (Bisht et al., 2012) were expressed and purified from E. coli BL21 cells according to standard protocols. Digestion and removal of the SUMO protein tag to generate SA1.72 was performed with SUMO Protease 1 according to the manufacturer's instructions (Life Sensors). Gel-shift assays were performed with a 32P-end-labeled XbaI fragment from plasmid pTH12 (a kind gift from Titia de Lange, The Rockefeller University, New York, NY) containing 12 tandem TTAGGG repeats. Purified recombinant protein (200–300 ng) was incubated for 25 minutes at room temperature in a 20 µl reaction containing 20 mM Hepes-KOH (pH 7.5), 100 mM KCl, 5% glycerol, 1 mM EDTA, 0.1 mM MgCl2, 0.5 mM DTT, 100 ng casein, 5 ng polydG-dC, and 3 ng labeled probe. Competition experiments were performed by adding 5–25 ng E. coli DNA. Samples were fractionated on 4% polyacrylamide native gels run in 0.1% TBE (Tris-Borate-EDTA) at 150 V for 55 minutes in the cold. Prior to loading samples, gels were prerun for 30 minutes at 80 V in the cold. Gels were dried onto Whatman DE81 paper and autoradiograph.
FACS analysis
siRNA transfected, trypsinized cells were washed twice with PBS containing 2 mM EDTA, fixed in cold 70% ethanol and stained with propidium iodide (50 µg/ml) and analyzed using a Becton-Dickenson FACSAN and FlowJo 8.8.6 software.
Image acquisition
Images were acquired using a microscope (Axioplan 2; Carl Zeiss, Inc.) with a Plan Apochrome 63× NA 1.4 oil immersion lens (Carl Zeiss, Inc.) and a digital camera (C4742-95; Hamamatsu Photonics). Images were acquired and processed using Openlab software (Perkin Elmer).
Acknowledgments
We thank members of the Smith lab and Tom Meier for comments on the manuscript and helpful discussion.
Footnotes
Author contributions
K.K.B. and Z.D. designed and performed experiments and interpreted data. S.S. designed experiments, interpreted data, and wrote the manuscript.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health [grant number R01CA116352 to S.S.]. Deposited in PMC for release after 12 month.
References
- Anderson D. E., Losada A., Erickson H. P., Hirano T. (2002). Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156, 419–424 10.1083/jcb.200111002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Landsman D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26, 4413–4421 10.1093/nar/26.19.4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Smith S., Chong L., Elias P., de Lange T. (1997). TRF1 is a dimer and bends telomeric DNA. EMBO J. 16, 1785–1794 10.1093/emboj/16.7.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht K. K., Dudognon C., Chang W. G., Sokol E. S., Ramirez A., Smith S. (2012). GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity. Mol. Cell. Biol. 32, 3044–3053 10.1128/MCB.00258-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S., Smith S. (2009). Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 187, 165–173 10.1083/jcb.200903096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S., Houghtaling B. R., Kim J. Y., Dynek J. N., Chang W. G., Smith S. (2007). Protein requirements for sister telomere association in human cells. EMBO J. 26, 4867–4878 10.1038/sj.emboj.7601903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S., Houghtaling B. R., Bhanot M., Sasa G., Savage S. A., Bertuch A. A., Smith S. (2011). A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 25, 1807–1819 10.1101/gad.17325211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carramolino L., Lee B. C., Zaballos A., Peled A., Barthelemy I., Shav-Tal Y., Prieto I., Carmi P., Gothelf Y., González de Buitrago G. et al. (1997). SA-1, a nuclear protein encoded by one member of a novel gene family: molecular cloning and detection in hemopoietic organs. Gene 195, 151–159 10.1016/S0378-1119(97)00121-2 [DOI] [PubMed] [Google Scholar]
- Chang C. R., Wu C. S., Hom Y., Gartenberg M. R. (2005). Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19, 3031–3042 10.1101/gad.1356305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yang Y., van Overbeek M., Donigian J. R., Baciu P., de Lange T., Lei M. (2008). A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319, 1092–1096 10.1126/science.1151804 [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. (1995). A human telomeric protein. Science 270, 1663–1667 10.1126/science.270.5242.1663 [DOI] [PubMed] [Google Scholar]
- Cook B. D., Dynek J. N., Chang W., Shostak G., Smith S. (2002). Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22, 332–342 10.1128/MCB.22.1.332-342.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G., Lingner J. (2006). Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25, 565–574 10.1038/sj.emboj.7600952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. (1992). Human telomeres are attached to the nuclear matrix. EMBO J. 11, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- Dynek J. N., Smith S. (2004). Resolution of sister telomere association is required for progression through mitosis. Science 304, 97–100 10.1126/science.1094754 [DOI] [PubMed] [Google Scholar]
- Gilson E., Géli V. (2007). How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 10.1038/nrm2259 [DOI] [PubMed] [Google Scholar]
- Gruber S., Haering C. H., Nasmyth K. (2003). Chromosomal cohesin forms a ring. Cell 112, 765–777 10.1016/S0092-8674(03)00162-4 [DOI] [PubMed] [Google Scholar]
- Haering C. H., Löwe J., Hochwagen A., Nasmyth K. (2002). Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 10.1016/S1097-2765(02)00515-4 [DOI] [PubMed] [Google Scholar]
- Holzmann J., Fuchs J., Pichler P., Peters J. M., Mechtler K. (2011). Lesson from the stoichiometry determination of the cohesin complex: a short protease mediated elution increases the recovery from cross-linked antibody-conjugated beads. J. Proteome Res. 10, 780–789 10.1021/pr100927x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling B. R., Cuttonaro L., Chang W., Smith S. (2004). A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631 10.1016/j.cub.2004.08.052 [DOI] [PubMed] [Google Scholar]
- Huang S. M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S. et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- Huth J. R., Bewley C. A., Nissen M. S., Evans J. N., Reeves R., Gronenborn A. M., Clore G. M. (1997). The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4, 657–665 10.1038/nsb0897-657 [DOI] [PubMed] [Google Scholar]
- Kueng S., Hegemann B., Peters B. H., Lipp J. J., Schleiffer A., Mechtler K., Peters J. M. (2006). Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967 10.1016/j.cell.2006.09.040 [DOI] [PubMed] [Google Scholar]
- Losada A., Yokochi T., Kobayashi R., Hirano T. (2000). Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150, 405–416 10.1083/jcb.150.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir R., Yalon-Hacohen M., Segev Y., Schultz A., Skorecki K. L., Selig S. (2002). Replication and/or separation of some human telomeres is delayed beyond S-phase in pre-senescent cells. Chromosoma 111, 147–155 10.1007/s00412-002-0199-z [DOI] [PubMed] [Google Scholar]
- Palm W., de Lange T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T. et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Reeves R. (2001). Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277, 63–81 10.1016/S0378-1119(01)00689-8 [DOI] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Carretero M., Martínez P., Drosopoulos W. C., Cañamero M., Schildkraut C. L., Blasco M. A., Losada A. (2012a). Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31, 2076–2089 10.1038/emboj.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Gómez-López G., Pisano D. G., Losada A. (2012b). A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 31, 2090–2102 10.1038/emboj.2012.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio E. D., Reiss D. J., Welcsh P. L., Disteche C. M., Filippova G. N., Baliga N. S., Aebersold R., Ranish J. A., Krumm A. (2008). CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA 105, 8309–8314 10.1073/pnas.0801273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Jerratsch M., Li B., Smith S., Hultén M., Lock T., de Lange T. (2000). Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol. Biol. Cell 11, 4189–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., de Lange T. (2000). Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299–1302 10.1016/S0960-9822(00)00752-1 [DOI] [PubMed] [Google Scholar]
- Smith S., Giriat I., Schmitt A., de Lange T. (1998). Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 10.1126/science.282.5393.1484 [DOI] [PubMed] [Google Scholar]
- Stedman W., Kang H., Lin S., Kissil J. L., Bartolomei M. S., Lieberman P. M. (2008). Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27, 654–666 10.1038/emboj.2008.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. A., Chaiken M. F., Wang F., Price C. M. (2012). Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 730, 12–19 10.1016/j.mrfmmm.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström L., Lindroos H. B., Shirahige K., Sjögren C. (2004). Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 10.1016/j.molcel.2004.11.026 [DOI] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Gieffers C., Peters B. H., Peters J. M. (2000). Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151, 749–762 10.1083/jcb.151.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. (2000). Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2, 492–499 10.1038/35019529 [DOI] [PubMed] [Google Scholar]
- Ünal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J. E., Koshland D. (2004). DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- van Steensel B., de Lange T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743 10.1038/385740a0 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 10.1016/S0092-8674(00)80932-0 [DOI] [PubMed] [Google Scholar]
- Wallace J. A., Felsenfeld G. (2007). We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17, 400–407 10.1016/j.gde.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E., Schleiffer A., Tanaka K., Eisenhaber F., Nasmyth K., Peters J. M. (2006). Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 16, 863–874 10.1016/j.cub.2006.03.049 [DOI] [PubMed] [Google Scholar]
- Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T. et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- Yalon M., Gal S., Segev Y., Selig S., Skorecki K. L. (2004). Sister chromatid separation at human telomeric regions. J. Cell Sci. 117, 1961–1970 10.1242/jcs.01032 [DOI] [PubMed] [Google Scholar]
- Zhang N., Kuznetsov S. G., Sharan S. K., Li K., Rao P. H., Pati D. (2008). A handcuff model for the cohesin complex. J. Cell Biol. 183, 1019–1031 10.1083/jcb.200801157 [DOI] [PMC free article] [PubMed] [Google Scholar]