Abstract
Objective
. Genes likely play a substantial role in the etiology of attention-deficit hyperactivity disorder (ADHD). However, the genetic architecture of the disorder is unknown, and prior genome-wide association studies have not identified a genome-wide significant association. We have conducted a third, independent multi-site GWAS of DSM-IV-TR ADHD.
Method
. Families were ascertained at Massachusetts General Hospital (MGH, N=309 trios), Washington University at St Louis (WASH-U, N=272 trios), and University of California at Los Angeles (UCLA, N=156 trios). Genotyping was conducted with the Illumina Human1M or Human1M-Duo BeadChip platforms. After applying quality control filters, association with ADHD was tested with 835,136 SNPs in 735 DSM-IV ADHD trios from 732 families.
Results
. Our smallest p-value (6.7E-07) did not reach the threshold for genome-wide statistical significance (5.0E-08) but one of the 20 most significant associations was located in a candidate gene of interest for ADHD, (SLC9A9, rs9810857, p=6.4E-6). We also conducted gene-based tests of candidate genes identified in the literature and found additional evidence of association with SLC9A9.
Conclusion
. We and our colleagues in the Psychiatric GWAS Consortium are working to pool together GWAS samples to establish the large data sets needed to follow-up on these results and to identify genes for ADHD and other disorders.
Keywords: ADHD, Genome-Wide Association Study, SLC9A9
Introduction
Family, twin and adoption studies show that genes play a substantial role in the etiology of attention-deficit hyperactivity disorder (ADHD) 1 but the genetic architecture of the disorder is unknown. Meta-analysis of seven ADHD linkage scans identified several genomic regions with suggestive evidence of linkage but only one region (16q22–q24) was statistically significant genome-wide. 2 An important and widely applied tool in psychiatric genetics is the genome-wide association study (GWAS).
Genome-wide association studies of ADHD have been completed in two European samples: a family study of 958 affected offspring trios (i.e. DNA from an affected offspring and both parents) from the International Multi-center ADHD Genetics (IMAGE) study 3 and a case-control study of 343 adult ADHD cases (304 controls). 4 Neither study identified a genome-wide significant association, 4, 5 but secondary analysis of ADHD symptom scores in the IMAGE study 6 suggested association with an overlapping gene of interest from Lesch et al 4 (CDH13) located on chromosome 16q24.2–q24.3 under the genome-wide significant chromosomal region identified in meta analysis. 2 We have conducted an independent multi-site GWAS of DSM-IV-TR ADHD in 735 affected-offspring trios using the Illumina Human1M and Human1M-Duo BeadChip platforms.
Method
Subjects
Families were ascertained at Massachusetts General Hospital (MGH, N=309 trios), Washington University at St Louis (WASH-U, N=272 trios), and University of California at Los Angeles (UCLA, N=156 trios). Children were 6–17 years of age at initial assessment and met criteria for DSM-IV-TR attention-deficit hyperactivity disorder. All study procedures were reviewed and approved by the subcommittee for human subjects of each respective institution. All subjects’ parents or guardians signed written informed consent forms and children older than 7 years of age signed written assent forms.
Diagnostic Criteria and Assessment
DSM-IV-TR criteria for ADHD require at least six of nine symptoms of inattention and/or hyperactivity impulsivity to be endorsed. Symptoms associated with significant impairment in different settings by 7 years of age. As detailed in the following sections subjects were not initially identified according to DSM-IV-TR criteria but only those subjects meeting full diagnostic criteria (of any DSM-IV-TR ADHD subtype) were enrolled in this genome-wide association study.
MGH (N=309)
Families were recruited for genetic studies of pediatric psychopathology at the Clinical and Research Program in Pediatric Psychopharmacology and Adult ADHD at Massachusetts General Hospital. Screening and recruitment for some subjects (N= 121) occurred prior to the publication of DSM-IV. Initial affection status for those subjects was based on DSM-IIIR criteria but lifetime DSM-IV-TR criteria was asked at follow up, and only those subjects endorsing a life-time DSM-IV-TR diagnosis of ADHD were enrolled. Remaining subjects were screened and assessed according to DSM-IV-TR criteria (N=188). Psychiatric assessments were made with K-SADSE (Epidemiologic Version). We conducted direct interviews with subjects older than 12 years of age and indirect interviews with their mothers (i.e., mothers complete the structured interview about their offspring) for all subjects and combined data from direct and indirect interviews by considering a diagnostic criterion positive if it was endorsed in either interview. A committee of board-certified child and adult psychiatrists or psychologists who were blind to the subject’s ADHD status, referral source and all other data resolved diagnostic uncertainties. Diagnoses presented for review were considered positive only when the committee determined that full DSM-IV-TR diagnostic criteria were met to a clinically meaningful degree. Potential probands were excluded if they had major sensorimotor handicaps (deafness, blindness), psychosis/schizophrenia, autism, inadequate command of the English language, or a Full Scale IQ less than 80.
WASH-U (N=272)
Participating families were selected from a population-representative sample identified through birth records of the state of Missouri, for a genetic epidemiological study of the prevalence and heritability of ADHD. The original sample included 812 complete male and female twin pairs and six individual twins ages 7 to 19 years at the time of interview identified from the Missouri Family Registry from 1996 to 2002. As detailed in previous reports, 7, 8 families were invited into the study if at least one child exhibited three or more inattentive symptoms on a brief screening interview. Parents reported on their children and themselves, and the youths on themselves, using the Missouri Assessment of Genetics Interview for Children (MAGIC). 9 The MAGIC is a semi-structured psychiatric interview allowing DSM-III, DSM-IV, and ICD-10 diagnoses, and exhibited excellent reliability and 1-year stability of diagnoses for both parent report and child self-report of the major DSM-IV diagnostic categories pertinent to youths. Adolescents and parents were interviewed in person or by telephone about half the time. All interviews with children 12 years and younger were in person. DSM-IV diagnoses of ADHD were based upon parental reports (most of the time, maternal). Families were excluded if a parent/guardian reported mental retardation or if the parent/guardian and twins could not speak English. 7
UCLA (N=156)
Study subjects were drawn from 540 children and adolescents ages 5 to 18 years (mean 10.6, SD 3.2) and 519 of their parents ascertained from 370 families with ADHD-affected sibling pairs. Detailed descriptions of recruitment methods, screening, and subject assessment have been previously described. 10 Briefly, lifetime psychiatric diagnoses were based on semi-structured diagnostic interviews conducted by master’s level clinical psychologists or highly trained interviewers with extensive experience and reliability training in psychiatric diagnoses. Children and adolescents were assessed using the Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime version (K-SADS-PL). Adult parents were assessed using the Schedule for Affective Disorders and Schizophrenia-Lifetime version (SADS-LA-IV), supplemented with the K-SADS Behavioral Disorders module for diagnosis of ADHD and disruptive behavior disorders. Direct interviews were supplemented with parent and teacher versions of the Swanson, Nolan, and Pelham, version IV (SNAP-IV) rating scale, as well as a parent-completed CBCL and Teacher Report Form. Parents also completed current ratings of self and spouse behavior with the ADHD Rating Scale IV. Best estimate diagnoses were assigned using all of the available clinical information according to strict DSM-IV criteria and reviewed by senior clinicians (J.J.M., J.T.M.). Inter-rater reliabilities were computed with a mean weighted kappa of 0.84 across all diagnoses with a greater than 5% occurrence in the sample. Subjects were excluded from participation if they were positive for any of the following: neurological disorder, head injury resulting in concussion, lifetime diagnoses of schizophrenia or autism, or estimated Full Scale IQ < 70. Subjects on stimulant medication were asked to discontinue use for 24 hours prior to their visit.
Genotyping
DNA was extracted from blood at each participating institution and Genizon BioSciences Inc. conducted genotyping with funding from Pfizer Inc. Genomic DNA samples from the MGH and WASH-U were genotyped using the Illumina Human1M BeadChip (N=1,057,265 SNPs) while the UCLA samples were genotyped using the Illumina Human 1M-Duo array (N=1,151,846 SNPs). Genotyping calls were generated after clustering all available data within platform at Genizon and then merged into a single file of 1,172,613 SNPs. To generate a data set of markers common to all sites, we removed SNPs that were either not included on both arrays (N=128,718 SNPs) or failed preliminary quality-control (QC) procedures conducted at Genizon (99% call rate for all samples and for all SNPs, gender check, Mendelian errors) on both the 1M and 1M-Duo arrays (N=9,500 SNPs), the 1M array only (N=39,753 SNPs) or the 1M-Duo array only (N=11,201 SNPs). Once the data from the multiple sites and different Illumina arrays were merged, there were 983,441 SNPs genotyped across the complete sample of 737 trios.
The results of further SNP and sample exclusions are presented in Table 1. As in the primary report from the IMAGE GWAS, 5 the majority of additional marker exclusions were made on the basis of call rate (i.e. the proportion of samples successfully genotyped for that SNP) conditional on minor allele frequency (MAF; i.e. the frequency of the least common allele for the SNP) (Figure S1, available online). Because rarer markers have a greater possibility of misclassification that would be more likely to bias tests of association, 11–13 we included SNPs with 0.01 ≤ MAF < 0.05 and call rate >99%; 0.05 ≤ MAF <0.1 and call rate >97%, MAF ≥ 0.1 and call rate >95%. Any SNPs found to be out of Hardy-Weinberg Equilibrium (p<1.0E-6) in founders were excluded from further consideration. We checked for sample duplication by examining identity-by-state for all pairs of individuals and found none. Probably owing to the extensive filtering of SNPs by Genizon and removed in the first step of data management, there were few additional marker or sample exclusions based on Mendelian errors, gender discrepancies or inbreeding (Table 1). The distribution of missingness, Mendel Errors and heterozygosity was similar across all samples in the filtered samples (Figure S2, available online).
Table 1.
Sample and Marker Quality Control
| Quality Control Metric | N Excluded |
|---|---|
| Non-Overlapping SNPs | 128,718 |
| No Founder Genotypes | 60,454 |
| Call Rate; MAF | 142,386 |
| Mendel Errors >2 | 11 |
| Hardy-Weinberg (p<0.10–6) | 5,908 |
|
| |
| SNPs Remaining | 835,136 |
|
| |
| Sex Discrepancy | 1 |
| F Inbreeding Coef. >0.125 | 1 |
| Family Mendel errors >2% | 0 |
|
| |
| Trios Remaining | 735 |
Note: MAF = minor allele frequency; SNP = single nucleotide polymorphism;
Association Analyses
We used PLINK 14 to conduct transmission disequilibrium tests. To control for multiple comparisons we adopted the conservative recommendation of Dudbridge et al and Pe’er et al 15, 16 and considered p-values less than 5.0E-08 to be statistically significant genome-wide. We also examined 43 candidate genes (N=3,603 SNPs) of interest based upon those examined in Neale et al 5 or based upon the recent literature: ADRA1A, ADRA1B, ADRA2A, ADRA2C, ADRB2, ADRBK2, ARRB1, BDNF, CDH13, CHRNA4, COMT, CSNK1E, DBH, DDC, DRD1, DRD2, DRD3, DRD4, FADS1, FADS2, HES1, HTR1B, HTR1E, HTR2A, HTR2C, HTR3B, MAOA, MAOB, NFIL3, NR4A2, PER1, PER2, SLC18A2, SLC6A1, SLC6A2, SLC6A3, SLC6A4, SLC9A9, SNAP25, STX1A, SYT1, TPH1, and TPH2. The analytic unit of interest for these tests was the gene, and we utilized the set-based tests provided in PLINK. 14 The set-based tests estimate the significance of each SNP and then calculate the average chi-squared statistic for the most significant SNPs per gene. Permutation tests were used to obtain empirical significance levels of the gene-based test, while accounting for the number of tests conducted within gene and the lack of independence of SNPs within gene. Sets could be comprised of up to 5 SNPs and gene-wide statistical significance was estimated with 50,000 permutations.
Results
After applying quality control filters, association with ADHD was tested with 835,136 SNPs in 735 DSM-IV-TR ADHD trios from 732 families; clinical demographics of the sample are presented in Table 2. Subjects recruited at WASH-U were older (p<0.001) and more likely to be diagnosed with predominantly inattentive ADHD (p<0.001) compared with both the MGH and the UCLA samples. Subjects recruited at UCLA were less likely to be Caucasian compared with both the MGH and WASH-U samples (p<0.001). Thus, pooled sample was predominantly Caucasian, male, 12 years of age on average with roughly equal prevalence of combined-type and predominantly inattentive-type ADHD (Table 2).
Table 2.
Sample Composition
| Total (N=735) | MGH (N=308) | WASH-U (N=272) | UCLA (N=155) | |
|---|---|---|---|---|
| Age | 12.3±4.0 | 11.5±4.1 | 13.7±3.8 | 11.5±3.5 |
| Sex (% Male) | 472 (64%) | 197 (64%) | 169 (62%) | 106 (68%) |
| Race (% Caucasian) | 676 (92%) | 299 (97%) | 254 (93%) | 123 (79%) |
| ADHD Subtype | ||||
| Combined-Type | 349 (48%) | 203 (67%) | 64 (24%) | 82 (55%) |
| Inattentive-Type | 327 (45%) | 75 (24%) | 190 (70%) | 62 (42%) |
| Hyperactive/Impulsive-Type | 46 (6%) | 24 (8%) | 17 (6%) | 5 (3%) |
Note: ADHD = Attention-Deficit/Hyperactivity Disorder; MGH = Massachusetts General Hospital; WASH-U = Washington University at St Louis; UCLA = University of California, Los Angeles.
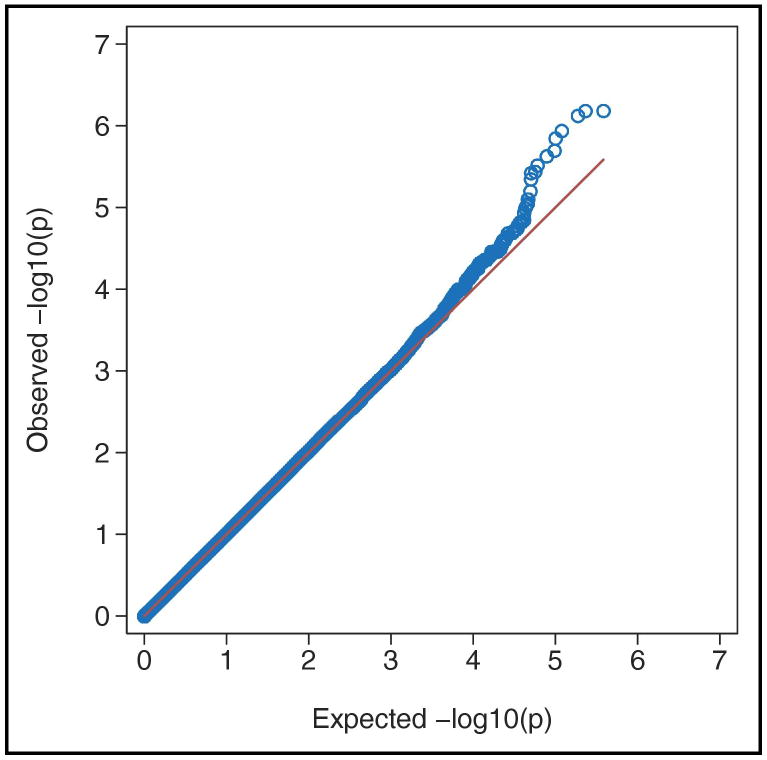
The QQ-plot of observed versus expected p-values presented in Figure 1 demonstrates that there was no appreciable inflation of the test statistic in the analyzed sample, as the mean chi-square was very close to 1.0 (1.005) and lambda was 1.0. Our smallest p-value (6.7E-07) did not reach the threshold for genome-wide statistical significance (i.e., 5.0E-08). The 20 most significant results are presented in Table 3. These top hits contain one SNP in an ADHD candidate gene (SLC9A9) with an associated significance level (6.4E-6) comparable to that observed in other studies.
Figure 1. Quantile-Quantile Plot of Association Results.
Observed results of association results are plotted for 835,136 single nucleotide polymorphisms (SNPs) against the expected distribution under the null hypothesis of no association.
Table 3.
Top 20 Associations from Genome-Wide Association Results
| CHR | Gene Symbol | SNP | Position | allele | MAF | HWE p-value | T : U | OR | p-value |
|---|---|---|---|---|---|---|---|---|---|
| 21 | C21orf34 | rs2823819 | 16750162 | C | 0.176 | 0.241 | 257 : 156 | 1.65 | 6.70E-07 |
| 16 | EMP2 | rs11074889 | 10540666 | A | 0.156 | 0.620 | 239 : 142 | 1.68 | 6.71E-07 |
| 17 | CCDC46 | rs8074751 | 61073004 | C | 0.388 | 0.510 | 278 : 405 | 0.69 | 1.18E-06 |
| 4 | BMPR1B | rs1859156 | 96053057 | T | 0.206 | 0.264 | 192 : 297 | 0.65 | 2.05E-06 |
| 15 | ATPBD4 | rs4923705 | 34080897 | C | 0.267 | 0.046 | 327 : 217 | 1.51 | 2.40E-06 |
| 6 | LOC389365 | rs438259 | 8975335 | A | 0.278 | 0.019 | 332 : 223 | 1.49 | 3.71E-06 |
| 2 | UGT1A9 | rs2602381 | 234249063 | A | 0.477 | 0.402 | 416 : 293 | 1.42 | 3.85E-06 |
| 3 | SLC9A9 | rs9810857 | 144465589 | T | 0.491 | 0.465 | 420 : 299 | 1.41 | 6.41E-06 |
| 4 | ELOVL6 | rs10011926 | 111246376 | A | 0.217 | 0.818 | 201 : 301 | 0.67 | 8.07E-06 |
| 7 | LOC643308 | rs10487524 | 144472464 | T | 0.146 | 0.917 | 226 : 141 | 1.60 | 9.12E-06 |
| 11 | TMEM16E | rs10833716 | 22164303 | C | 0.178 | 0.532 | 166 : 257 | 0.65 | 9.66E-06 |
| 9 | LOC392382 | rs10120476 | 110039137 | T | 0.010 | 1.000 | 27 : 3 | 9.00 | 1.18E-05 |
| 17 | CCDC46 | rs2107654 | 61063535 | T | 0.492 | 0.192 | 296 : 411 | 0.72 | 1.52E-05 |
| 5 | MCTP1 | rs11953346 | 94167203 | C | 0.364 | 0.004 | 259 : 367 | 0.71 | 1.59E-05 |
| 8 | LOC645809 | rs4132831 | 136024990 | A | 0.367 | 0.736 | 283 : 395 | 0.72 | 1.70E-05 |
| 5 | DUSP1 | rs7702178 | 172129603 | G | 0.175 | 0.588 | 255 : 167 | 1.53 | 1.84E-05 |
| 3 | RBMS3 | rs17023218 | 29317721 | A | 0.217 | 0.591 | 199 : 294 | 0.68 | 1.88E-05 |
| 13 | LECT1 | rs6561686 | 52187095 | C | 0.048 | 0.075 | 44 : 94 | 0.47 | 2.08E-05 |
| 6 | LOC729167 | rs9350410 | 22140989 | G | 0.362 | 0.866 | 393 : 283 | 1.39 | 2.33E-05 |
| 6 | LOC728614 | rs760609 | 114814793 | A | 0.425 | 0.558 | 423 : 309 | 1.37 | 2.51E-05 |
Note: CHR = Chromosome; HWE p-value = Hardy-Weinberg Equilibrium p-value; MAF: minor allele frequency; OR = odds ratio; SNP = single nucleotide polymorphism; T:U = ratio of transmitted to untransmitted alleles.
We also conducted gene-based tests of association with previously examined candidate genes for ADHD (Table S1, available online). Several genes (ADRBK2, CHRNA4, COMT, DRD2, DRD4, HTR1E, HTR3B, MAOA, MAOB, PER1, PER2, SLC18A2, SLC6A2, SLC6A3, SLC6A4, TPH1, TPH2) contained no SNPs that were statistically significant at p<0.05. Of the remaining candidate genes examined, 19 had at least one significant association at p<0.05 (ADRA1A, ADRA1B, ADRA2A, ADRA2C, ADRB2, BDNF, DBH, DDC, DRD1, DRD3, HES1, HTR1B, HTR2A, HTR2C, NR4A2, SLC6A1, SNAP25, STX1A, SYT1), but these associations were not significant after adjusting for the number of tests conducted within each gene. Association results for genes with set-based tests that were significant genome-wide are presented in Table 4.
Table 4.
Significant Gene-Based Set-Tests of Association with Selected Candidates
| CHR | Gene | chi-square | p-value | SNP | Position | A1 | MAF | HWE p-value | T : U | OR | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | SLC9A9 | 15.0 | 0.0017 | rs9810857 | 144465589 | T | 0.4905 | 0.4647 | 420 : 299 | 1.41 | 6.41E-06 |
| rs9757063 | 144465620 | C | 0.374 | 1 | 395 : 293 | 1.35 | 1.01E-04 | ||||
| rs6801526 | 144418562 | A | 0.05892 | 0.635 | 104 : 56 | 1.86 | 1.48E-04 | ||||
| rs9857832 | 144463570 | A | 0.2686 | 0.8422 | 331 : 241 | 1.37 | 1.68E-04 | ||||
| rs6781977 | 144407697 | G | 0.3661 | 0.9105 | 384 : 297 | 1.29 | 8.57E-04 | ||||
| 22 | CSNK1E | 10.7 | 0.0038 | rs17753394 | 37008333 | G | 0.1689 | 0.4568 | 161 : 243 | 0.66 | 4.51E-05 |
| rs9622773 | 37008692 | T | 0.1703 | 0.5182 | 164 : 244 | 0.67 | 7.48E-05 | ||||
| rs2075983 | 37020644 | T | 0.251 | 0.4448 | 236 : 304 | 0.78 | 3.43E-03 | ||||
| rs135757 | 37033599 | T | 0.2543 | 0.9451 | 242 : 309 | 0.78 | 4.31E-03 | ||||
| rs1534891 | 37025045 | T | 0.1394 | 0.5139 | 160 : 200 | 0.80 | 3.50E-02 | ||||
| 11 | FADS1 | 6.1 | 0.019 | rs174556 | 61337211 | T | 0.2779 | 0.1934 | 276 : 337 | 0.82 | 1.38E-02 |
| 11 | ARRB1 | 8.4 | 0.024 | rs508435 | 74702464 | A | 0.4237 | 0.3634 | 392 : 306 | 1.28 | 1.13E-03 |
| rs667791 | 74677076 | C | 0.3971 | 0.4139 | 305 : 387 | 0.79 | 1.83E-03 | ||||
| rs11236388 | 74691440 | T | 0.07595 | 0.0938 | 79 : 120 | 0.66 | 3.66E-03 | ||||
| rs472112 | 74699149 | C | 0.4431 | 0.1011 | 383 : 310 | 1.24 | 5.55E-03 | ||||
| rs657561 | 74698027 | T | 0.3164 | 0.5465 | 283 : 342 | 0.83 | 1.83E-02 | ||||
| 9 | NFIL3 | 7.8 | 0.041 | rs1412470 | 93253823 | C | 0.2953 | 0.316 | 264 : 334 | 0.79 | 4.20E-03 |
| rs4744055 | 93288903 | G | 0.3508 | 0.8187 | 293 : 366 | 0.80 | 4.46E-03 | ||||
| rs4743835 | 93286276 | T | 0.3236 | 0.6333 | 283 : 354 | 0.80 | 4.91E-03 | ||||
| rs4743837 | 93286593 | T | 0.1789 | 0.5341 | 190 : 248 | 0.77 | 5.58E-03 | ||||
| rs4744045 | 93250033 | A | 0.2948 | 0.2861 | 264 : 331 | 0.80 | 6.02E-03 | ||||
| 11 | FADS2 | 7.1 | 0.047 | rs174627 | 61394042 | T | 0.14 | 0.7459 | 150 : 201 | 0.75 | 6.49E-03 |
| rs968567 | 61352140 | A | 0.1567 | 0.4892 | 172 : 224 | 0.77 | 8.97E-03 |
Note: CHR = Chromosome; HWE p-value = Hardy-Weinberg Equilibrium p-value; MAF = minor allele frequency; OR = odds ratio; SNP = single nucleotide polymorphism; T:U = ratio of transmitted to untransmitted alleles.
A recent meta-analysis of candidate gene association studies of childhood ADHD estimated the pooled association with 38 polymorphisms (29 SNPs) in 18 genes. 17 Twenty of the 29 SNPs were genotyped for the current study and only one was significant at p<0.05 in single SNP tests (rs6280 in DRD3, p=0.018). Neither of the significant SNPs from the meta-analysis and included in our study were statistically significant (rs27072 p=0.7; rs3746544 p=0.9), and none of the genes (SLC6A3, DRD4, DRD5, SLC6A4, HTR1B, CHRNA4 and SNAP25) implicated by statistically significant association with markers not available in our sample were statistically significant in gene-based set-tests (Table S1, available online).
Discussion
This study is the second independent genome-wide association study of children with ADHD and the first conducted in children from the United States. We failed to identify any statistically significant genome-wide findings, but found some additional evidence for a role of SLC9A9 in the etiology of ADHD. SLC9A9 codes a hydrogen/sodium exchanger located in the membranes of subcellular structures and is expressed in the brain and the heart and skeletal muscle, placenta, kidney and liver. 18 SLC9A9 was first identified at a break point in a family in which ADHD symptoms were correlated with a chromosome 3 inversion. 18 It was also associated with ADHD in the IMAGE study of 51 candidate genes for ADHD 3 and in subsequent quantitative analyses of ADHD symptoms and age at onset. 6, 19 In the current study SLC9A9 was the most significant candidate gene examined (p=0.0017 gene-wide) and the only candidate with a SNP (rs9810857-T, p=6.4E-6) among the top 20 significant associations in the genome-wide scan of 835,136 SNPs.
Our study provides little support for association with additional candidate genes for ADHD previously implicated via meta-analysis; 17, 20 only 6 of the 42 candidates examined demonstrated nominal gene-wide statistical significance, and we failed to replicate any of the positive associations reported with individual SNPs examined and subjected to meta-analysis. This is not surprising considering the number of trios examined and that any associated genes are likely to exert a small or modest increase in risk (i.e. odds ratios <1.5). The sample size required to detect such an association (OR=1.5, p=5E-08, MAF=0.25) is at least 1,188 trios. However, many tens of thousands of subjects will likely be need to uncover true risk loci of smaller effect. For example, GWAS with samples of 29,136, 34,433 and 71,225 subjects were needed to identify replicated genetic loci for blood pressure. 21, 22
Sample heterogeneity could also be driving the lack of strong positive findings in ADHD. For example, different assessment procedures among the three samples (different instruments and different ways of combining information sources) could have increased the phenotypic heterogeneity of cases. We have also demonstrated that persistent ADHD is a more familial form of the disorder and that ADHD symptoms typically persist in 30–60% of childhood cases. 23,24 Focusing on childhood samples may introduce additional noise by including a large subgroup of cases who will remit from ADHD and may have a less “genetic” etiology. Additional genome-wide studies focused on adult ADHD are needed to follow-up on the promising results suggested in Lesch et al. 4 It is also possible that studies of derived phenotypes or of gene-environment interaction might yield evidence for genome-wide significance, although such strategies have not proven effective for the IMAGE GWAS. 6, 19, 25, 26
Alternatively, it may be that the accumulation of rare variants confers the most risk for ADHD rather than variation in common SNPs studied here. The initial evidence suggesting SLC9A9 as a potential candidate for ADHD came from a pericentric inversion of chromosome 3 identified in a single family, for example. 18 Elia et al 27 found no over representation of copy number deletions or insertions in ADHD youth but found that inherited copy number variations were located in genes with prior evidence of involvement with neuropsychiatric conditions related to ADHD.
Like the extant literature of genome-wide association studies of ADHD, the current study represents an intermediate step towards understanding genetic influences on the disorder. 28 If it is true that ADHD is influenced by many common gene variants with small individual effects, then much larger genome-wide association studies need to be conducted. We and our colleagues in the Psychiatric GWAS Consortium 29 PGC2009, are working to pool together GWAS samples for ADHD and other disorders in the belief that pooled analyses of very large data sets will be needed to identify genes for ADHD and its associated impairments.
Supplementary Material
Table S1. Gene-Based Set-Test Association Results for Candidate Genes
Figure S1: Single Nucleotide Polymorphism (SNP) Filtering by Minor Allele Frequency (MAF) Conditional on Call Rate (CR) Included SNPs
Figure S2: Site-Specific Distribution of Quality Controls Metrics
MGH: Massachusetts General Hospital; WASH-U; Washington University at St Louis; UCLA; University of California, Los Angeles
Acknowledgments
Genotyping was supported with funding from Pfizer Inc. Subject ascertainment and assessment was supported buy the following sources: NIH Grants R13MH059126, R01MH62873, U01MH085518 and R01MH081803 to S.V. Faraone; NINDS grant NS054124 S. Loo; NIMH grant MH01966 to J. McGough; NIDA grant K24 DA016264 to T. Wilens; NIMH grant MH63706 to S Smalley; and NIMH grants K08MH001503 and R01MH066237 to J. Wozniak.
Footnotes
Supplemental material cited in this article is available online.
This article represents one of several articles published in the xxx issue of the Journal of the American Academy of Child and Adolescent Psychiatry that explores the intersection of genetics and mental health disorders in children and adolescents. The editors invite the reader to investigate the additional articles on this burgeoning area of developmental psychopathology.
Disclosure:
Dr. Mick receives research support from Ortho-McNeil Janssen Scientific Affairs, Pfizer, Shire Pharmaceuticals, and has served on the advisory board for Shire Pharmaceuticals.
Dr. Biederman receives research support from Alza, AstraZeneca, Bristol Myers Squibb, Eli Lilly and Co., Janssen Pharmaceuticals Inc., McNeil, Merck, Organon, Otsuka, Shire, the National Institute of Mental Health (NIMH), and the National Institute of Child Health and Human Development (NICHD). He has served on the speakers’ bureau for Fundacion Areces, Medice Pharmaceuticals, and the Spanish Child Psychiatry Association In previous years, Dr. Biederman received research support, consultation fees, or speaker’s fees for/from Abbott, AstraZeneca, Celltech, Cephalon, Eli Lilly and Co., Esai, Forest, Glaxo, Gliatech, Janssen, McNeil, NARSAD, NIDA, New River, Novartis, Noven, Neurosearch, Pfizer, Pharmacia, The Prechter Foundation, Shire, The Stanley Foundation, UCB Pharma, Inc. and Wyeth.
Dr. McCracken has received research support from Bristol-Myers Squibb, Aspect, and Seaside Pharmaceuticals. He has served as a consultant to Novopharm and BioMarin, and has received speaker honoraria from the Tourette Syndrome Association, Veritas, and CME Outfitters
Dr. McGough has served as a consultant to and received research support and consulting honoraria from Eli Lilly & Company, and Shire Pharmaceuticals.
Dr. Wilens has received grant support from Abbott, McNeil, Lilly, the National Institutes of Health (NIH) – the National Institutes on Drug Abuse (NIDA), Merck, and Shire. He has served on the speaker’s bureaus for Eli Lilly, McNeil, Novartis, and Shire. He has served as a consultant for Abbott, Astra-Zeneca, McNeil, Eli Lilly, the National Institutes of Health (NIH) – the National Institutes on Drug Abuse (NIDA), Novartis, Merck, and Shire. Dr. Wilens receives royalties from Guilford Press for his book, Straight Talk About Psychiatric Medications for Kids.
Dr. Wozniak receives royalties from Bantam Books for her book, “Is Your Child Bipolar?” Dr. Wozniak has served on the speakers’ bureau for McNeil, Primedia/Massachusetts General Hospital (MGH), Psychiatry Academy, and Eli Lilly. Dr. Wozniak is on the advisory board and has served as a consultant for Pfizer and Shire. She receives research support from McNeil, Shire, and Eli Lilly.
Dr. Faraone has, in the past year, received consulting fees and has been on advisory boards for Eli Lilly, Ortho-McNeil and Shire Development and has received research support from Eli Lilly, Pfizer, Shire and the National Institutes of Health. In previous years, Dr. Faraone has received consulting fees or has been on Advisory Boards or has been a speaker for Shire, McNeil, Janssen, Novartis, Pfizer and Eli Lilly. In previous years he has received research support from Eli Lilly, Shire, Pfizer and the National Institutes of Health.
Drs. Todorov, Smalley, Hu, Loo, Todd (deceased), Dechairo, Nelson, Reiersen, Neale and Ms. Byrne, and Mr. Guiney report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Dr. Eric Mick, Massachusetts General Hospital and Harvard Medical School, Boston, MA
Dr. Alexandre Todorov, Washington University School of Medicine, St Louis, MO
Dr. Susan Smalley, University of California Los Angeles
Dr. Xiaolan Hu, Molecular Medicine, Pfizer Inc
Dr. Sandra Loo, University of California Los Angeles
Dr. Richard D. Todd, Washington University School of Medicine, St Louis, MO
Dr. Joseph Biederman, Massachusetts General Hospital and Harvard Medical School, Boston, MA
Ms. Deirdre Byrne, Massachusetts General Hospital and Harvard Medical School, Boston, MA
Dr. Bryan Dechairo, Molecular Medicine, Pfizer Inc
Mr. Allan Guiney, Massachusetts General Hospital and Harvard Medical School, Boston, MA
Dr. James McCracken, University of California Los Angeles
Dr. James McGough, University of California Los Angeles
Dr. Stanley F. Nelson, University of California Los Angeles
Dr. Angela M. Reiersen, Washington University School of Medicine, St Louis, MO
Dr. Timothy E. Wilens, Massachusetts General Hospital and Harvard Medical School, Boston, MA
Dr. Janet Wozniak, Massachusetts General Hospital and Harvard Medical School, Boston, MA
Dr. Benjamin M. Neale, Center for Human Genetic Research, Massachusetts General Hospital, Boston MA
Dr. Stephen V. Faraone, SUNY Upstate Medical University
References
- 1.Mick E, Faraone SV. Genetics of attention deficit hyperactivity disorder. Child Adolesc Psychiatr Clin N Am. 2008 Apr;17(2):261–284. doi: 10.1016/j.chc.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Zhou K, Dempfle A, Arcos-Burgos M, et al. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008 Dec 5;147B(8):1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brookes K, Xu X, Chen W, et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 2006 Oct;11(10):934–953. doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- 4.Lesch KP, Timmesfeld N, Renner TJ, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008 Nov;115(11):1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 5.Neale BM, Lasky-Su J, Anney R, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008 Dec 5;147B(8):1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lasky-Su J, Neale BM, Franke B, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008 Dec 5;147B(8):1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 7.Todd RD, Sitdhiraksa N, Reich W, et al. Discrimination of DSM-IV and latent class attention-deficit/hyperactivity disorder subtypes by educational and cognitive performance in a population-based sample of child and adolescent twins. J Am Acad Child Adolesc Psychiatry. 2002 Jul;41(7):820–828. doi: 10.1097/00004583-200207000-00014. [DOI] [PubMed] [Google Scholar]
- 8.Neuman RJ, Sitdhiraksa N, Reich W, et al. Estimation of prevalence of DSM-IV and latent class-defined ADHD subtypes in a population-based sample of child and adolescent twins. Twin Res Hum Genet. 2005 Aug;8(4):392–401. doi: 10.1375/1832427054936646. [DOI] [PubMed] [Google Scholar]
- 9.Todd RD, Joyner CA, Heath AC, Neuman RJ, Reich W. Reliability and stability of a semistructured DSM-IV interview designed for family studies. J Am Acad Child Adolesc Psychiatry. 2003 Dec;42(12):1460–1468. doi: 10.1097/00004583-200312000-00013. [DOI] [PubMed] [Google Scholar]
- 10.Smalley SL, McGough JJ, Del’Homme M, et al. Familial clustering of symptoms and disruptive behaviors in multiplex families with attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2000 Sep;39(9):1135–1143. doi: 10.1097/00004583-200009000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Cutler DJ, Zwick ME, Carrasquillo MM, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001 Nov;11(11):1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Hum Hered. 2002;54(1):22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- 13.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005 Feb;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008 Apr;32(3):227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008 May;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 17.Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009 Jun 9; doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- 18.de Silva MG, Elliott K, Dahl HH, et al. Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J Med Genet. 2003 Oct;40(10):733–740. doi: 10.1136/jmg.40.10.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasky-Su J, Anney RJ, Neale BM, et al. Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008 Dec 5;147B(8):1355–1358. doi: 10.1002/ajmg.b.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faraone SV, Perlis RH, Doyle AE, et al. Molecular genetics of attention deficit hyperactivity disorder. Biol Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 21.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009 May 10; doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 May 10; doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faraone SV, Biederman J, Mick E. The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol Med. 2006 Feb;36(2):159–165. doi: 10.1017/S003329170500471X. [DOI] [PubMed] [Google Scholar]
- 24.Faraone SV, Biederman J, Monuteaux MC. Toward guidelines for pedigree selection in genetic studies of attention deficit hyperactivity disorder. Genet Epidemiol. 2000;18(1):1–16. doi: 10.1002/(SICI)1098-2272(200001)18:1<1::AID-GEPI1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 25.Sonuga-Barke EJ, Lasky-Su J, Neale BM, et al. Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet. 2008 Dec 5;147B(8):1359–1368. doi: 10.1002/ajmg.b.30860. [DOI] [PubMed] [Google Scholar]
- 26.Anney RJ, Lasky-Su J, O’Dushlaine C, et al. Conduct disorder and ADHD: Evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008 Oct 24;147B(8):1369–1378. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
- 27.Elia J, Gai X, Xie HM, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2009 Jun 23; doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale BM, Purcell S. The positives, protocols, and perils of genome-wide association. Am J Med Genet B Neuropsychiatr Genet. 2008 Oct 5;147B(7):1288–1294. doi: 10.1002/ajmg.b.30747. [DOI] [PubMed] [Google Scholar]
- 29.A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009 Jan;14(1):10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Gene-Based Set-Test Association Results for Candidate Genes
Figure S1: Single Nucleotide Polymorphism (SNP) Filtering by Minor Allele Frequency (MAF) Conditional on Call Rate (CR) Included SNPs
Figure S2: Site-Specific Distribution of Quality Controls Metrics
MGH: Massachusetts General Hospital; WASH-U; Washington University at St Louis; UCLA; University of California, Los Angeles