Abstract
The promise of targeting epigenetic abnormalities for cancer therapy has not been realized for solid tumours, although increasing evidence is demonstrating its worth in haematological malignancies. In fact, true clinical efficacy in haematopoietic-related neoplasms has only become evident at low doses of epigenetic-targeting drugs (namely, inhibitors of histone deacetylase and DNA methyltransferases). Describing data from preclinical studies and early clinical trial results, we hypothesize that in using low-dose epigenetic-modulating agents, tumour cells can be reprogrammed, which overrides any immediate cytotoxic and off-target effect observed at high dose. We suggest that such optimization of drug dosing and scheduling of currently available agents could give these agents a prominent place in cancer management—when used alone or in combination with other therapies. If so, optimal use of these known agents might also pave the way for the introduction of other agents that target the epigenome.
Introduction
One of the most of exciting areas of biology over the past decade has been the expanding understanding of how epigenetic control influences the patterns of gene expression in cells.1–3 This knowledge has immediate translational implications for targeting epigenetic abnormalities in cancer for therapeutic purposes.
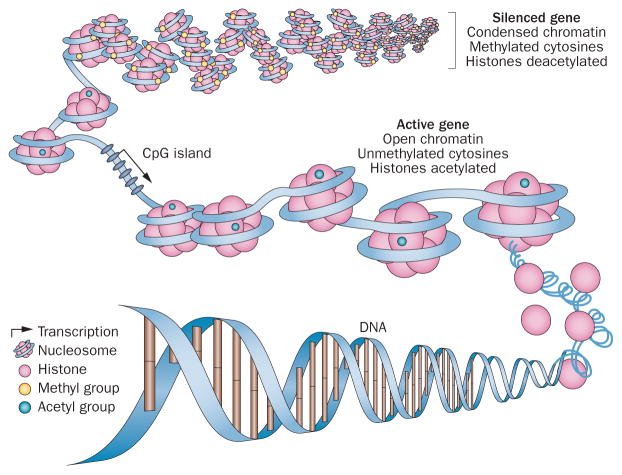
In principle, ‘epigenetics’ refers to the somatically heritable differences in gene expression not attributable to intrinsic alterations in the primary sequence of DNA.4 In this paradigm, the DNA serves as a ‘hard drive’ of genetic information that requires the ‘software’ package of epigenetic control to determine its full transcriptional output.1,2 In a single individual, cells of different types all have the same genome, but have multiple complex and different ‘epigenomes’ that define their respective phenotypes.1–3 Epigenetic control occurs throughout the genome—as well as in regulatory elements distant from the genes they control—and can mediate interactions between chromosomes.3 Core elements of the epigenetic regulation of gene expression include how DNA is packaged or wrapped around nucleosomes, and how those nucleosomes are positioned throughout the genome (Figure 1).5–7 Key regulatory features include how chromatin and nucleosomes are modified by a complex series of enzymes and their subsequent interaction with proteins that recognize these modifications.5–8 Cancers of all types, and even different cell populations within a single patient’s cancer, have different genomes resulting from the many mutations that alter the DNA hard drive. In addition, extensive and biologically significant changes in the epigenetic software package are increasingly recognized; these changes constitute the ‘cancer epigenome’.1,2 Rapid advances in technology are enabling the ever more-robust interrogation of this complex control of gene expression and the findings are informing drug-development strategies.3
Figure 1.
Epigenetic regulation of gene expression. Gene expression is controlled in the promoter regions by a combination of DNA methylation and chromatin configuration. In normal cells, gene expression is silenced by condensing chromatin, methylating (at cytosine) DNA and deactylating histones. By contrast, active genes are those with open nucleosome spacing around the transcription start site, are unmethylated and associated with acetylated histones. CpG islands that are rich in cytosine and guanine—and are typically unmethylated to promote gene expression—can be epigenetically silenced by hypermethylation in cancer. Adapted from Figueiredo, L. M. et al. Nat. Rev. Microbiol. 7, 504–513 (2009).
Accordingly, the cancer cell can be summed up to represent a complex interplay between genetic and epigenetic abnormalities that, from beginning to end, drive the evolution of each patient’s malignancy. One exciting development is the recognition that virtually all tumours harbour mutations in genes that encode proteins that control the epigenome.9–20 As we start to understand the precise relationships between these mutations and the consequences for cancer cell phenotypes, these observations will increasingly guide therapeutic strategies for epigenetic-modulating agents. The current theory is that the mutated genes ‘set up’ downstream abnormalities in the epigenome.21–23 The downstream genes affected by these epigenetic abnormalities are generally wild-type for the underlying DNA sequence. Thus, normal function might be restored since, theoretically, reverting the software package of epigenetic abnormalities should be easier than repairing the hard drive that harbours the upstream mutations.
In the past when genome-wide data were lacking, the epigenetic research community focused heavily on regulatory mechanisms biased to gene start sites.20 This focus arose from the initial discoveries that cancer-specific hypermethylation changes were often found in CpG islands (so-called as they are rich in cytosine and guanine) in gene promoters. Studies are now expanding our view of the genome-wide role of epigenetics.1,2 The concept of epigenetic therapy has also focused largely on changes at gene promoters, which will surely shift to (and be integrated with) studies of cancer epigenetic abnormalities across the genome.
Indeed, the rapidly growing field of therapeutic epigenetics is being enhanced by the merging of laboratory and clinical data that are instructing us on how epigenetic modulators best provide meaningful benefit to patients. To date, the best-characterized and only FDA-approved epigenetic modulating antineoplastic agents have targeted abnormalities in DNA methylation and histone acetylation (via histone deacetylase; HDAC) in haematological malignancies.24–32 Emerging data is now instructing the epigenetics research community on how to better use epigenetic drugs, including lowering doses and the rational sequence of treatment approaches with other agents.67 Importantly, these lessons are informing the clinical assessment of epigenetic drugs in patients with solid tumours, with compelling results, which we discuss in this Review. We focus on the growing body of data centred on promoters, which illustrates that targeting epigenetic abnormalities is an exciting concept with the potential to improve the management of patients with cancer. We believe the principles of epigenetic therapy will help guide novel therapeutic concepts, and the rapidly strengthening focus on the entire cancer epigenome will enrich these therapeutic possibilities even further.
Principles of epigenetic therapy
The epigenetic targets
Currently, the most studied and recognized cancer-specific epigenetic changes are alterations in DNA methylation and histone acetylation, which have been extensively reviewed previously.2,20,33,34 Briefly, these modifications include global hypomethylation, regional hypermethylation and chromatin events that are closely tied to DNA methylation changes.
Hypomethylation of tumour genomes is often observed within the body of genes and in regions flanking genes.1,20,34 Meanwhile, cancer-specific regional hypermethylation (of hundreds of genes per tumour) typically involves normally unmethylated, CpG-enriched DNA—the CpG islands residing in and around proximal gene promoters.2,20 This hypermethylation can be associated with decreased gene expression. Indeed, some data indicate that ‘shore’ regions (adjacent to CpG islands) distant to transcriptional start sites can also have an important role in gene expression through their hypermethylation. 35 The increased methylation patterns in promoters provide an alternative mechanism to mutations by which loss-of-function mutations of key tumour suppressor genes can occur, and are now recognized as an early and central event in carcinogenesis.2,20,33 Finally, cancer-specific epigenetic chromatin events also typically involve modifications of histone proteins, including universal decreases in histone acetylation as well as decreases in an active mark of gene transcription (methylation at lysine 4 of histone 3; H3K4) at promoters of DNA hypermethylated genes. These changes result in a closed chromatin configuration and decreased gene expression (Figure 1). Increased H3K4 methylation creates resistance to DNA methylation.124,125
Reversing low histone acetylation is one possible method for epigenetic cancer therapy, by targeted inhibition of histone deacetylase.36–38 However, it is important to consider that, in the interplay between DNA methylation and chromatin abnormalities in cancer, either aberration can be the dominant driver for tumorigenesis. Thus, chromatin modifications can, even in the absence of DNA methylation, be determinants of altered patterns of gene expression.5,20 For example, increased polycomb silencing protein complexes (PcG), which result in increases in the repressive chromatin mark of H3K27 methylation, seem to render many genes vulnerable to abnormal promoter DNA methylation during tumorigenesis.20,39–41 This cascade results in the silencing of genes involved in important protections against neoplasia, including tumour suppressors, regulators of immune escape and apotosis. These chromatin changes, as well as abnormal DNA methylation, comprise a growing focus of strategies to reverse epigenetic abnormalities to provide novel cancer treatments.42–44
Optimizing epigenetic therapy
Against the backdrop of these complexities for epigenetic control of gene expression in cancer, epigenetic therapy can be a unique type of targeted therapy. In effect, the aim is to reverse the abnormal cancer epigenome in a scenario not unlike the cellular reprogramming used when, for example, inducing an embryonic phenotype from a mature cell to prepare induced pluripotent stem cells (iPSCs).45,46 For iPSCs, every aspect of the epigenome of the mature cell must be driven ‘backwards’ to create the stem cells and this requires changing the DNA methylation and chromatin patterns. Interestingly, the DNA demethylating agents and inhibitors of HDACs used as therapeutic agents can act as small molecules to improve the efficiency of preparing iPSCs.47 This fact emphasizes the mechanisms underpinning these drugs in the management of patients with cancer. We hypothesize that, by targeting virtually any component of the abnormal epigenetic processes in cancer, we will almost always affect the multiple altered signalling networks48,49 that drive tumorigenesis to therapeutic advantage (Figure 2).
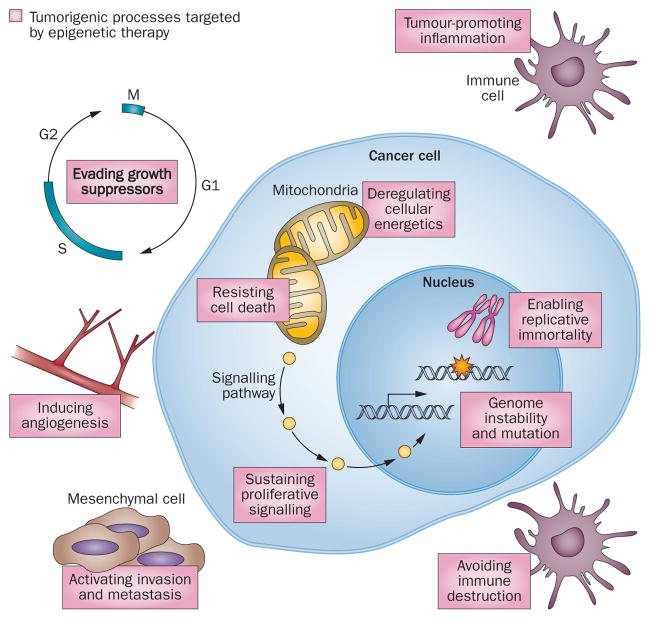
Figure 2.
Epigenetic control is involved in all the hallmarks of tumour initiation and survival. Tumorigenesis, propagation and survival are maintained through a complex interplay of multiple cellular biological processes, all of which are regulated to an extent by epigenetic control of gene expression. Targeting one signalling pathway or biological function can result in compensatory modulation of other, off-target drivers of cell survival. Epigenetic therapy offers the ability to concurrently target, and reverse, multiple aberrant signalling pathways as well as the expected compensatory changes in other pathways.49
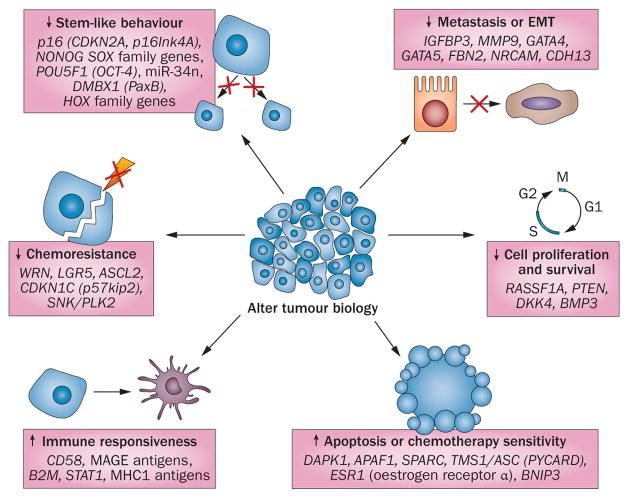
Thus, reversing DNA methylation or chromatin abnormalities in cancer will seldom target one specific gene, which could be construed as a nonspecific mechanism of action of epigenetic drugs. However, in reality this ‘lack’ of specificity might lie at the heart of its value. The challenges are to understand and maximize (for clinical advantage) the biological facets of epigenetic therapy. A key starting point to consider is, although cancer-specific DNA hypermethylation can affect several known tumour suppressor genes—such as CDKN2A (p16), MLH1, APC and other canonical WNT pathway genes2,20—many more genes are simultaneously hypermethylated in each patient’s cancer.1,2,20 Indeed, for every genetic mutation in a given patient’s tumour, hundreds of genes have epigenetic abnormalities within the deviant cell signalling pathways that drive tumorigenesis.32,33 Among the genes scattered throughout the pathways, many important candidate genes or groups of genes exist, the loss of function or expression of which can be critical to the initiation and progression of tumours as well as to their sensitivity and resistance to chemotherapy (Figure 3). Consequently, alterations to these genes can ‘lock in’ abnormal cell self-renewal or diminish the differentiation capacity of cancer cells.50,51 For traditional targeted therapies aimed at a single gene mutation, such as EGFR mutations in lung cancer,52,53 the existence of multiple signalling abnormalities poses a great challenge for the durability of the therapeutic responses. Compensatory intrasignalling and intersignalling pathway events can lead to resistance to the inhibition of any single gene-mediated event to which a cancer cell is addicted.54–58 The broad advantage of epigenetic therapy lies in reversing these changes simultaneously via multiple signalling pathways, which can durably damp down many of the compensatory events that ultimately defeat other targeted approaches (Figure 2). That is, epigenetic agents can function predominantly as mediators of a cell reprogramming effect to inhibit cancer initiation and progression. Furthermore, epigenetic agents are useful when used either alone or in combination to sensitize tumour cells to cytotoxic agents or to slow or reverse resistance to other targeted approaches. We believe these concepts are crucial for how one designs and tests—preclinically and clinically—agents touted as epigenetic therapy.
Figure 3.
Concurrent widespread changes in gene expression with epigenetic therapy. Anticancer efficacy of treatment with epigenetic-modulating agents is ssociated with widespread changes in gene expression that affect multiple biological processes. Gene expression is increased through direct reversal of epigenetic modifications of genomic DNA, whereas, for cancer-promoting genes, gene expression is reduced by the re-expression of their regulatory genes. Abbreviation: EMT, epithelial–mesenchymal transition.
DNA demethylating agents
The efficacy of decitabine and 5-azacitidine (both inhibitors of DNA methyltransferase) in the management of myelodysplasia and acute myelogenous leukaemia (AML) is well known. These drugs were initially designed and tested as cytotoxic chemotherapy in the 1960s59,60 and some 20 years later were discovered to have DNA demethylation activity.61 Their development and subsequent FDA approval—decitabine was approved in 2006 for myelodysplasia and AML, whereas 5-azacitidine was approved in 2004 for myelodysplasia—was achieved by markedly reducing the doses to overcome limiting toxicities, which also likely enhanced their targeted effects on DNA methyltransferases (Table 1). Indeed, the development of decitabine and 5-azacitidine was severely hampered for decades because, as for the vast majority of all anticancer drugs, initial trials used standard designs to escalate drug doses until maximally tolerated doses were reached. These doses proved to be substantially toxic to patients (likely because of off-target epigenetic modulation) and dose de-escalation was needed to achieve both improved tolerability and activity.32 The lessons learned from this evolution should strongly inform laboratory research and clinical trials that examine DNA demethylating agents for the management of advanced lung cancer62 and possibly other solid tumours.
Table 1.
Selected trials of epigenetic therapy
| Regimen | n | Dose | Study design | Clinical results | Survival benefit |
|---|---|---|---|---|---|
| Cutaneous T-cell lymphoma | |||||
| Romidepsin28 | 71 | 14mg/m2 on day 1, 8 and 15 of a 28-day cycle | Single arm, phase II | 34% response rate with 6% complete responses | Median duration 11–15 months in one study |
| Vorinostat123 | 74 | 400mg/day | Phase II (multiple doses) | 24% response rate | Not assessed/median time to progression 30 weeks |
| Myelodysplasia | |||||
| 5-azacitidine versus best supportive care25 | 191 | 75mg/m2 per day subcutaneously for 7 days every 28 days | Randomized phase III with crossover design* | 23% response rate versus 0%; Improved time to leukaemia (21 months versus 13 months); Improved quality of life | After 6 months, median overall survival of 18 months versus 11 months (P=0.03) |
| Decitabine versus best supportive care27 | 170 | 5mg/m2 intravenously over 4 hours every 8 hours for 3 consecutive days every 6 weeks | Randomized phase III | 17% response rate versus 0%; Trend to increased time to leukaemic transformation or death (12 months versus 8 months, P=0.16) | None |
| SGI-110‡ | 78 | Ongoing testing of dosing schedules in phase I study | Phase I/II§ | Heavily pretreated AML group: 1 patient with complete response without platelet recovery and 1 patient with complete response but incomplete blood count recovery; Myelodysplasia group previously treated with azacitidine: 1 patient with marrow complete remission and 1 patient with haematological improvement | Not assessed |
| Ovarian cancer | |||||
| Decitabine + carboplatin88 | 17 | Decitabine: 10mg/m2 intravenously on days 1–5 of a 28-day cycle Carboplatin: AUC 5 intravenously on day 8 of a 28-day cycle |
Phase II | 35% response rate | PFS of 10.2 months with 53% progression-free at 6 months |
| 5-azacitidine + carboplatin87 | 30 | Azacitidine: 75mg/m2 subcutaneously daily for days 1–5 of a 28-day cycle Carboplatin: AUC 5 intravenously on day 2 of a 28-day cycle |
Phase Ib/II | 13.8% response rate | 7.5-month duration of benefit 23-month overall survival 5.6-month PFS |
| Non-small-cell lung cancer | |||||
| 5-azacitidine + entinostat||62 | 45 | 5-Azacitidine: 40mg/m2 on days 1–6 and days 8–10 of a 28-day cycle Entinostat: 7mg on days 3 and 10 of a 28-day cycle |
Phase I/II | 1 patient with complete response 1 patient with partial response 4 out of 19 patients treated with subsequent therapy with partial response |
6.4-month overall survival 20% of patients with ≥1-year survival (range 1–4 years) |
| Entinostat + erlotinib versus unreported placebo112 | 132 | Erlotinib: 150mg daily for 28 days Entinostat: 10mg on days 1 and 15 of a 28-day cycle |
Randomized phase II | 4-month PFS similar (18% versus 20%) | In subset of patients with high E-cadherin levels, improved overall survival compared with placebo (9.4 months versus 5.4 months; HR 0.35, 95% CI 0.13–0.92; P=0.03) |
| Carboplatin + paclitaxel + vorinostat or placebo113 | 94 | Carboplatin: AUC 6 on day 3 of a 3-week cycle Paclitaxel: 200mg/m2 on day 3 of a 3-week cycle Vorinostat: 400mg daily on days 1–14 of a 3-week cycle |
Randomized, blinded phase II | 34% versus 12.5% response rate | No survival benefit, but trend to overall survival benefit in vorinostat arm |
| ER-positive breast cancer | |||||
| Exemestane + entinostat versus placebo114 | 130 | Exemestane: 25mg daily Entinostat: 5mg weekly |
Randomized phase II | Decreased risk of progression by 27% | PFS 4.3 months versus 2.3 months (P=0.06) Overall survival 26.9 months versus 19.8 months (P=0.04) |
Patients randomly assigned to supportive care could cross over to azacitidine treatment if their disease worsened. Worsening was considered an increased FAB subtype (to RAEB or RAEB-T), progression to AML, remaining RBC transfusion-dependent before and during study or progressive bone marrow failure.
Patients with AML.
Trial is ongoing.
Refractory disease.
Abbreviations: AML, acute myeloid leukaemia; AUC, area under the plasma drug concentration-time curve; ER, oestrogen receptor; FAB, French–American–British; HR, hazard ratio; PFS, progression-free survival; RAEB, refractory anaemia with excessive blasts; RAEB-T, RAEB in transformation; RBC, red blood cell.
As gleaned from the studies of lessons of decitabine and 5-azacitidine, 126 low doses of the drugs minimized toxicity while potentially retaining the inhibition of DNA methyltransferases by incorporating into DNA and then covalently binding all three biologically active forms of these enzymes. By this mechanism, the agent irreversibly inhibits DNA methyltransferase catalytic sites for methylation and triggers their degradation.42,63–66 With higher doses of the drugs, off-target effects can be exerted, such as triggering DNA damage to evoke cell-cycle check points, which is immediately cytotoxic.42 When used at low doses in patients with myelodysplasia, pharmacokinetic studies indicated that tumour cells are exposed to nanomolar concentrations of the drug at which cytotoxic effects are not the prevailing mode of action. Indeed, our laboratory studies data these concepts. 67 We showed that one 3-day treatment of cancer cells of multiple histologies using low-dose 5-azacitidine resulted in minimal cell death, but gene expression (lasting weeks) increased cell signalling, including increased expression of immune response and pro-apoptotic genes. Moreover, this single ex vivo treatment followed by implantation of cells in immunosuppressed mice resulted in slowed tumour engraftment and sub-sequent tumour growth through multiple passages over several months. Low nanomolar doses seem to minimize off-target effects and enable cancer cellsto undergo cellular reprogramming associated with a loss of long-term clonogenic or tumorigenic capacity.67 These effects are accompanied by partial genome-wide DNA demethylation and altered gene expression events that potentially reverse signalling events in the multiple pathways (Figure 2) thought to drive tumorigenesis.67
Importantly, these antitumour effects have been observed in cultured and primary cancer cells that constitute tumour stem-like cells with increased self-renewal capacity.67 This result suggests that certain epigenetic therapies could achieve the highly desired goal of blunting these cells, which are resistant to most standard therapies.68 The results can also help explain why many patients with myelodysplasia and AML take several months to respond to low-dose 5-azacitidine–progressive exhaustion of resistant stem-like cells could require an extended period of time for the effects to be evident. As a case in point, Silverman et al.69 reported that a considerable number of patients (48%) with high-risk myelodysplasia who continued on DNA demethylating drugs past their first response had increased magnitudes of response with further treatment.
Our multi-institutional, multidisciplinary Stand-Up to Cancer (SU2C) group has worked to accelerate the laboratory and clinical research of epigenetic therapy for cancer treatment in the most common human cancers. For example, on the basis of these translational and clinical data, we have taken a low-dose 5-azacytidine strategy, combined with the HDAC inhibitor entinostat, to phase II trials for common solid tumours (non-small-cell lung cancer [NSCLC], colorectal and breast cancers). Encouragingly, the regimen, which is well-tolerated and clinically active in patients with myelodysplasia and AML,70 seems to have compelling activity in patients with solid tumours.62 Specifically, results in patients with advanced, heavily pretreated NSCLC are promising and suggest that epigenetic therapy could represent a major change in the management of this deadly cancer.62 Although the epigenetic therapy alone achieved responses in only 3% of patients who were heavily pre-treated (median of three prior therapies), the responses were remarkably durable and resulted in survival from just under 3 years to over 4 years.62 For comparison, in the phase III study showing efficacy of pemetrexed as a second-line therapy in advanced NSCLC (in patients who had only one prior therapy), no patients survived past 22 months.
These encouraging results could be improved upon if patients are appropriately preselected. Such personalization will require analyses of pretreatment and post-treatment tissue samples, which were not available from our first trials, to evaluate possible methylation-specific (and other) biomarkers that could identify patients most likely to benefit from the therapy. Such biopsy studies are essential for all trials moving forward; the approaches for identifying useful biomarkers are constantly being enriched by a large body of research in cancer epigenetics. For example, our small pilot study of candidate genes in NSCLC revealed that the demethylation of a set of four epigenetically silenced genes (p16, CDH13, RASSF1A and APC) could be detected in sequential blood samples of patients and tracked with improved progression-free survival and overall survival. 55 These four genes had previously been reported to be predictors of early recurrence in early stage lung cancer.71 Indeed, many such candidate genes can be identified for lung and other solid tumours from the extensive studies in the field.20,72–74 Moreover, genome-wide surveys of DNA methylation changes in multiple tumour types, such as those in the Cancer Genome Atlas Project (TCGA),72–74 will provide a great number of gene promoter and other genomic sites of DNA methylation abnormalities to consider. For example, the DNA promoter sites encoding microRNAs (miRNAs) can become abnormally methylated, which is associated with loss of expression of the miRNA.75 These changes could be critical with respect to pathway changes that are affected.
The basal methylation status of genes, responses of these loci to the initial cycles of low doses of DNA demethylating drugs, the accompanying gene expression changes and the pathways altered will all be pivotal to guide personalization of any epigenetic treatment paradigm. Such individualization must also consider the integration of DNA methylation patterns with the genetic abnormality of each tumour type approached. This need has been elevated by the previously mentioned frequency of mutations in genes encoding for protein regulators of the epigenome.20,76 DNA methylation patterns specifically accompanying these genetic changes are already being recognized,77 and the genes involved might be important for tailoring biomarker approaches to specific tumour types and their subgroups. Finally, DNA polymorphisms—some of which are inherited—can affect gene regulatory regions and are beginning to be linked to abnormal gene promoter DNA methylation events in cancer.78–80 These polymorphisms probably function by providing a decreased basal transcriptional activity of the gene, making it vulnerable to the evolution of abnormal DNA methylation during the initiation and progression of tumours.78,80,81
Nearly 20% of the heavily pretreated patients with advanced NSCLC enrolled in our initial epigenetic therapy trial lived for ≥1 year (and up to >4 years) after the initial therapy.62 Expected survival at 6 months for patients after three lines of therapy, such as ours, is 48% based on a large, retrospective observational study.82 This survival might be attributable to prolonged response or disease stabilization from the epigenetic therapy. However, we observed an unusually robust response to subsequent cytotoxic therapies, with which the majority of patients were treated.62 Approximately 25% of the patients, which is considerable in the NSCLC setting, achieved Response Evaluation Criteria in Solid Tumors (RECIST) responses to therapies following even short courses (~two cycles) of the epigenetic treatment.62 These responses clinically reinforce the laboratory findings that long-lasting cellular reprogramming occurs as a result of epigenetic targeting. If these clinical benefits are validated in the already planned larger clinical trials (which is currently under consideration by the institutional review board at Johns Hopkins University), the management of NSCLC could be substantially improved.
Other researchers are achieving results in the laboratory and in clinical trials that are consistent with these initial findings. For example, Humenuik et al.83 showed that colorectal cancer cell lines exposed to increasing 5-fluorouracil concentrations generate acquired resistance to downregulated UMP-CMP kinase, which is an important effector of 5-fluorouracil activity. With low-dose decitabine treatment, UMP-CMP kinase levels increased and the resistance to 5-fluorouracil was reversed.83 In another study, combination decitabine and trichostatin A (an HDAC inhibitor) resulted in decreased expression of the multidrug resistance transporter ATP-binding cassette sub-family G member 2 (encoded by ABCG2) as well as markers of enhanced self-renewal populations in ovarian cancer cells, and increased the sensitivity of these cells to cisplatin in vivo.84 In relapsed childhood acute lymphoblastic leukaemia (ALL) and B-lymphoblastic leukeamia, blasts isolated from patients displayed a methylation signature indicating that acquired resistance to standard chemotherapy is associated with epigenetic changes; pretreatment with vorinostat (an HDAC inhibitor) and decitabine restored chemosensitivity in these cells.85 A study based on these findings of decitabine and vorinostat in combination with chemotherapy in relapsed childhood ALL is ongoing and preliminary activity has been reported (P. Brown, personal communication). Other studies have suggested that low-dose decitabine is efficacious in terms of antitumour effects, as well as promoting immune recognition through increasing expression of cancer testis antigen, in a model of pancreatic cancer.86 Such results have been translated to the clinic to meaningful effect, notably in ovarian cancer—multiple groups have reported the use of demethylating agents to restore platinum sensitivity in patients with platinum-resistant ovarian cancer. Fu et al.87 reported a phase I/II study of 5-azacitidine and carboplatin that demonstrated durable responses and stable disease (median duration of therapy 7.5 months) in 46% of patients with platinum-resistant or refractory ovarian cancer. A similar study of decitabine and carboplatin in the same patient population reported a 40% 6-month progression-free survival rate, with one patient having a complete response.88 Larger trials testing demethylating agents to overcome platinum resistance are currently ongoing.
The mechanisms leading to sensitization for subsequent cytotoxic therapies must be established in the laboratory. However, many of the pathways found to be altered in our recent low-dose studies, such as decreased activity of cell-cycle pathways and enhanced apoptosis, are predicted to provide such sensitization.67 In fact, many of the proteins altered—such as FOXM1, polo kinase 1 and Aurora kinase A and B—are being targeted by the pharmaceutical industry for such purposes.67 These studies support the hypothesis that epigenetic therapy functions by reprogramming cancer cells to a more chemosensitive, less stem-cell-like phenotype through the re-expression of various groups of genes in multiple anticancer pathways.
Histone deacetylase inhibitors
HDAC inhibitors are the other major class of epigenetic modulating agents that have been tested and approved for the treatment of cancer. As single agents, approved use is limited to lymphoma—both romidepsin and vorino-stat have FDA approval in cutaneous T-cell lymphoma, whereas romidepsin is also approved for use in patients with relapsed peripheral T-cell lymphoma.28–31 Multiple HDAC inhibitors are in preclinical and clinical trials, the details of which have been discussed elsewhere.36
Targeting HDACs is more complex than targeting DNA methytransferases because this group of proteins has multiple subclasses with mechanisms of action still under contention.89 In fact, proteins other than histones are targets of the lysine deacetylation effects of these drugs. For example, even inhibitors that preferentially target nuclear class I HDACs can alter the transcriptional activity of the tumour suppressor p53 by altering its acetylation status.90 Furthermore, not all HDACs are located in the nucleus,89 which means they probably do not target histone acetylation. For example, class III HDACs are a separate group of NAD-dependent enzymes, among which NAD-dependent protein deacetylase sirtuin-1 (hSIRT1) is best studied. hSIRT1 regulates key histone marks, with important transcriptional control consequences that include H4K16 acetylation. 91,92 This enzyme has increasingly been implicated in the altered gene expression in cancer, and its inhibition might have clinical relevance.93 hSIRT1 inhibition is associated with p53-independent apoptosis in cancer cells after DNA damage, and its inhibition could have therapeutic efficacy when used alone or in combination with other drugs. Uncertainty persists regarding which of the many potential targets of HDAC inhibitors are responsible for their anticancer effects.94 However, multiple studies have demonstrated that HDAC inhibition results in reproducible increases in gene expression of important tumour suppressor genes (including p53, SFRP1, miR-375 and DAPK)95–97 and can slow tumour growth and sensitize cells to other therapies, including cytotoxic agents and signal transduction inhibitors.
Against this background, multiple mechanisms of action of HDAC inhibitors have been observed in preclinical studies, both epigenetic and cytotoxic.37,38,98–100 Similar to results of DNA demethylating agents, these effects are probably dose-dependent.37,38,98–100 For example, HDAC inhibitors being used at high doses in the clinic seem to cause rapid extension of DNA damage and impaired repair of induced DNA breaks.100,101 The consequences for these effects are rapid cell-cycle arrest and cell death typical of cytotoxic agents.37,98–100 Presumably, such rapid effects are not the ultimate goal and do not represent the optimal use of these drugs (comparable to DNA demethylating agents). In fact, effects of these high doses might well explain why they have not achieved high efficacy for the treatment of most common human cancers. That is, these immediate effects preclude the cellular reprogramming that alters the disease biology in a durable, clinically meaningful fashion.
Given these limitations, HDAC inhibitors could still be extremely powerful tools—if used with the right timing, doses and combinations—to provide a robust component of epigenetic therapy with widespread efficacy. As noted earlier, one key advantage of epigenetic therapy is the ability to inhibit subpopulations of cancer cells that drive tumorigenesis and are usually resistant to most therapies. Although the precise definition of these stem-like cells is still controversial, the heterogeneity of cancer cells on a molecular level is well-accepted.102–105 Most importantly, extensive plasticity among cell populations in both normal cells and in tumours is needed to generate these cells, otherwise cells would be unable to change their phenotype.102–105 Although the resistance of these stem-like cells to therapies might, in some instances, be fixed by mutations, solid experimental evidence demonstrates that epigenetic factors might regulate both the plasticity of their formation and the mediation of treatment resistance.68 A widely noted example of this balance stems from a series of experiments by Settleman and colleagues, 68 which suggests that resistance to treatment of multiple cancer cell types, against both targeted therapy agents and traditional chemotherapy drugs, centres on the creation or persistence of drug-tolerant, stem-like cells that are driven by epigenetic control. One protein that was highly upregulated in stem-cell-like, therapy-resistant populations was lysine-specific demethylase 5A (encoded by KDM5A), a histone demethylase that controls the H3K4 mark for active transcription.68 Settleman et al.68 also showed that low doses of clinically relevant HDAC inhibitors, including entinostat, could reversibly, and without initial cytotoxicity, eliminate drug-resistant cells persisting in cultures treated with highly active targeted inhibitors. Furthermore, a related enzyme, lysine-specific demethylase 5B, has been shown to be a key driver for stem-like cells in cultured and primary human melanoma.106 These findings have tremendous relevance for potential strategies to maximally employ HDAC inhibitors in the management of human cancers.
Given this background, whether HDAC inhibitors will provide clinical benefit to patients remains to be seen. As previously mentioned, the HDAC inhibitors romidepsin and vorinostat have both been approved by the FDA for haematological malignancies owing to their ability to produce remarkable, if not considerably durable, responses in approximately 30–40% of patients with T-cell cutaneous lymphomas.28–31 However, the mechanisms underlying the robust initial sensitivity of these tumours have yet to be defined. Obviously, gaining such insight would markedly enrich strategies for using HDAC inhibitors in the clinic. The paucity of definitive cell culture lines for T-cell cutaneous lymphomas seems to limit progress in this arena.
Outside of these successes, HDAC inhibitors used alone have had little success, as yet, in clinical trials for a variety of solid tumour types, including thymic cancer, glioblastoma, renal cell carcinoma, hepatocellular carcinoma, 107–111 ovarian cancer and others.37,38,98 However, the work of Settleman et al.68 suggests that HDAC inhibitors could reverse treatment resistance or sensitize tumours to treatment, which might have some bearing on the growing body of clinical data consistent with this hypothesis. Very prominent among these are studies in patients with NSCLC and breast cancer that suggest combining HDAC inhibitors with established therapies might work. In a randomized phase II trial, the HDAC inhibitor entinostat combined with the EGFR inhibitor erlotinib showed no overall benefit relative to erlotinib alone in patients with recurrent advanced NSCLC. However, in a subset of patients with high baseline E-cadherin levels, a significant overall survival benefit was evident (9.4 months versus 5.4 months, P = 0.03).112 In a second study, vorinostat therapy increased response rates significantly from 12.5% to 34% when combined with carboplatin and paclitaxel in treatment-naive patients with metastatic NSCLC, with a trend towards improved progression-free survival and overall survival as well.113 Finally, in a phase II trial in patients with breast cancer, entinostat significantly increased survival when combined with an aromatase inhibitor.114 At median follow up of 25 months, treatment with exemestane with entinostat versus exemestane alone resulted in an 8.3-month improvement in overall survival (P = 0.04). This latter study is perhaps the most promising of those in solid tumours and warrants additional testing for validation. However, for these studies, the true mechanisms for the results are—crucially—far from clarified and the doses of HDAC inhibitors used highlight specific on-target effects rather than off-target cytotoxic effects.
Future outlooks
We believe that the best hope for future clinical trials with DNA demethylating agents and HDAC inhibitors lies in critically analysing what emerging laboratory and clinical studies have to teach us. Firstly, the data support that these drugs will not work best when doses that produce immediate cytotoxic effects are used; such cytotoxic doses prevent the reprogramming of cancer cells and preclude durable clinical consequences. As with all anticancer therapies, even with lower doses, a therapeutic window must be established in which normal tissue is relatively preserved and tumour tissue is affected. The major challenge for all these epigenetic therapies is determining what that window is, which will require close scrutiny of both acute and chronic toxic effects. Assessing methylation and expression differences in normal versus tumour tissues using in vivo laboratory studies and sequential biopsy analyses, where feasible, in patients participating in well-designed translational studies, should enable the necessary quantitative assessments.
Secondly, these agents are best used in the clinic when adequate time for response is possible because the epigenetic reprogramming takes longer to become apparent than the actions of traditional chemotherapies. Accordingly, the RECIST criteria commonly used to assess clinical responses within 6–8 weeks will often be suboptimal for monitoring clinical trials of epigenetic therapy—patients might need to be continued on therapy if they are clinically stable. This paradigm has been used in the immunology arena whereby responses are expected to take time to develop, and interim disease progression followed by response has been observed.115 Furthermore, trials need not depend solely on short-term criteria for monitoring results. Long-term clinical benefits must be rigorously assessed using careful follow-up measurements of response to subsequent therapies and, of course, overall survival should be maintained as the key end point incorporated into clinical trial designs, as we emphasized in our lung cancer trial.62 Immediate response criteria would have precluded the documentation of the benefits of DNA demethylating agents for patients with myelodysplasia and AML, which only emerged after months of treatment and, in some cases, a worsening of disease before any benefit became apparent.
These caveats also apply to trials of other targeted agents; an excellent example is of antitolerance-disrupting immunotherapy approaches, which required prolonged time courses.115,116 Accordingly, in monitoring epigenetic therapy in patients, acknowledging the likelihood that traditional initial tumour responses will not often be evident is important. Rather, the end points of clinical trials should permit the evaluation of the biological changes to the behaviour of the tumour—end points such as changes in the pace of the disease or response to subsequent therapy as well as overall survival.
With these criteria in mind, we believe current DNA demethylating agents and HDAC inhibitors have a promising future in the treatment of the most common solid tumours. Careful biomarker work within the trials and, preclinical work conducted in parallel, is mandatory to fully evaluate the mechanisms underpinning any observed clinical efficacy. Genome-wide studies of DNA methylation, lysine acetylation studies, gene-expression studies and the assessment of protein targets all must be combined with candidate gene approaches to define all possible predictive biomarkers. Direct examination of tumour cells or tissues should be complemented with serum sample analyses to identify and monitor patients who are most likely to benefit from these agents.
Perhaps the greatest impact of epigenetic therapy for cancer management will involve combination with current chemotherapeutic approaches. Reversing resistance or sensitizing cancers to multiple therapy types, including hormonal therapies, immunomodulatory therapies and standard chemotherapy, are now very promising and rational concepts. However, particular care must be taken with dosing and scheduling if these agents are to be combined with standard agents, for reasons of efficacy and tolerability. For example, dosing must be chosen to minimize toxicity for the epigenetic therapy lead-in and to maintain optimal dosage of the standard agent. Wisely designed clinical trials should be increasingly instituted to realize this promise.
We must also consider the development of new agents for targeting the cancer epigenome. Such agents will include new derivatives of current drugs as well as novel drug design concepts. One example is the design of a prodecitabine drug, SGI-110, which is in phase I/II trials and shows promise in patients with in myelodysplasia and AML.117 Additionally, an oral form of 5-azacitidine, which has obvious compliance and convenience advantages, is now also in clinical trial.118 Finally, assessing other therapeutic targets, such as histone methyltransferases and the BET family of proteins, is an emerging theme.37,38,119–122 The bromodomain inhibitors of the BET family, which are hypothesized to block activation of MYC target genes mediated by BET proteins, are now entering clinical trials. As with DNA methyltransferase and HDAC inhibitors, these new targets invite exploration for use in combination regimens and must be explored preclinically to determine any underlying mechanistic features that can improve efficacy.
Conclusions
The rapidly growing field of therapeutic epigenetics is being enhanced by robust translational approaches that have married laboratory and clinical data to suggest how epigenetic drugs best provide meaningful benefit to patients. These data suggest that epigenetic agents can have considerable clinical effect through reprogramming malignant cells, which could fundamentally change the management of patients with cancer. With thoughtful preclinical and clinical study design, we are poised to fully elevate epigenetic therapy to a prime position in cancer therapy.
Key points.
Evidence of the effects of epigenetic-modulating agents has revealed their dramatic consequences on cellular programming, in particular reversing stem-cell-like behaviour and chemoresistance
Treatment with epigenetic drugs affects multiple cell signalling pathways, including those regulating immune response and evasion, apoptosis, cell survival and DNA-damage repair
If used optimally, the widespread targets of these agents can be their greatest feature—cancer cells abnormally regulate many diverse pathways, which likely results in major therapeutic barriers
Previous research and clinical use of these agents might have been hampered because their effects were assessed too early and the doses used were too high
Lower doses of these agents results in less cytotoxicity to normal tissue, and should provide the added time and exposure needed for the ‘reprogramming’ effect of epigenetic therapy to become evident
Review criteria.
The PubMed and MEDLINE databases were searched using several terms including but not limited to: “epigenetics”, “DNA methylation”, “histone acetylation”, “DNA methyltransferase inhibitors”, “HDAC inhibitors”, “epigenetic gene mutations”, “epigenetics and stem-cells”, “epigenetics and reprogramming”, “chemoresistance” and “chemosensitivity”. Papers were not limited by language or date.
Acknowledgments
A portion of the authors’ work cited in this article was supported by grants from SU2C (Stand Up To Cancer), Waxman Foundation, Hodson Endowment and the NCI (National Cancer Institute) CA043318. The authors thank Kathy Bender for help with manuscript preparation and submission.
Footnotes
Competing interests S. B. Baylin declares an association with MDxHealth. See the article online for full details of the relationship. The other authors declare no competing interests.
Author contributions N. Azad and S. B. Baylin researched the data for the article and wrote the manuscript. All authors contributed to the discussion of the article’s content. C. A. Zahnow and C. M. Rudin edited the manuscript before submission.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ecker JR, et al. Genomics: ENCODE explained. Nature. 2012;489:52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 4.Holliday R. DNA methylation and epigenetic mechanisms. Cell Biophys. 1989;15:15–20. doi: 10.1007/BF02991575. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Caparros M, editor. Epigenetics. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 7.Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 8.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 9.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thol F, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 11.Metzeler KH, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdel-Wahab O, et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25:1200–1202. doi: 10.1038/leu.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzeler KH, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guglielmelli P, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118:5227–5234. doi: 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 16.Tefferi A, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tefferi A, et al. IDH mutations in primary myelofibrosis predict leukemic transformation and shortened survival: clinical evidence for leukemogenic collaboration with JAK2V617F. Leukemia. 2012;26:475–480. doi: 10.1038/leu.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tefferi A, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Carvalho DD, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baylin SB, Jones PA. A decade of exploring the cancer epigenome — biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 22.Berdasco M, Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum Genet. 2013;132:359–383. doi: 10.1007/s00439-013-1271-x. [DOI] [PubMed] [Google Scholar]
- 23.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 24.Kaminskas E, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 25.Silverman LR, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 26.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 27.Kantarjian H, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 28.Piekarz RL, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittaker SJ, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol. 2010;28:4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 30.O’Connor OA, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol. 2006;24:166–173. doi: 10.1200/JCO.2005.01.9679. [DOI] [PubMed] [Google Scholar]
- 31.Duvic M, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issa JP. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: dose, duration, and patient selection. Nat Clin Pract Oncol. 2005;2(Suppl 1):S24–29. doi: 10.1038/ncponc0355. [DOI] [PubMed] [Google Scholar]
- 33.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 34.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 35.Doi A, et al. Differential methylation of tissue-and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 37.Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2:405–413. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 41.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 42.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Issa JP, Kantarjian HM. Introduction: emerging role of epigenetic therapy: focus on decitabine. Semin Hematol. 2005;42(Suppl):S1–S2. doi: 10.1053/j.seminhematol.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 45.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 49.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Morey L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Easwaran H, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 53.Tsao MS, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 54.Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–280. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- 55.Astsaturov I, et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal. 2010;3:ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poulikakos PI, Rosen N. Mutant BRAF melanomas–dependence and resistance. Cancer Cell. 2011;19:11–15. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Solit DB, Rosen N. Resistance to BRAF inhibition in melanomas. N Engl J Med. 2011;364:772–774. doi: 10.1056/NEJMcibr1013704. [DOI] [PubMed] [Google Scholar]
- 58.Soda M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 59.Sorm F, Vesely J. Effect of 5-aza-2′-deoxycytidine against leukemic and hemopoietic tissues in AKR mice. Neoplasma. 1968;15:339–343. [PubMed] [Google Scholar]
- 60.Notari RE, DeYoung JL. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci. 1975;64:1148–1157. doi: 10.1002/jps.2600640704. [DOI] [PubMed] [Google Scholar]
- 61.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 62.Juergens RA, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferguson AT, et al. Role of estrogen receptor gene demethylation and DNA methyltransferase. DNA adduct formation in 5-aza-2’deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem. 1997;272:32260–32266. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- 64.Gabbara S, Bhagwat AS. The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem J. 1995;307:87–92. doi: 10.1042/bj3070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghoshal K, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Tsai HC, et al. Transient low doses of DNAdemethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silverman LR, et al. Continued azacitidine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer. 2011;117:2697–2702. doi: 10.1002/cncr.25774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fandy TE, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114:2764–2773. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brock MV, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 72.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 76.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 79.Chan EC, Lam SY, Fu KH, Kwong YL. Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1, and NQO1 genes in Chinese patients with non-small cell lung cancers: relationship with aberrant promoter methylation of the CDKN2A and RARB genes. Cancer Genet Cytogenet. 2005;162:10–20. doi: 10.1016/j.cancergencyto.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 80.Chan TL, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 81.Ligtenberg MJ, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 82.Girard N, et al. Third-line chemotherapy in advanced non-small cell lung cancer: identifying the candidates for routine practice. J Thorac Oncol. 2009;4:1544–1549. doi: 10.1097/JTO.0b013e3181bbf223. [DOI] [PubMed] [Google Scholar]
- 83.Humeniuk R, et al. Decreased levels of UMP kinase as a mechanism of fluoropyrimidine resistance. Mol Cancer Ther. 2009;8:1037–1044. doi: 10.1158/1535-7163.MCT-08-0716. [DOI] [PubMed] [Google Scholar]
- 84.Meng F, Sun G, Zhong M, Yu Y, Brewer MA. Anticancer efficacy of cisplatin and trichostatin A or 5-aza-2′-deoxycytidine on ovarian cancer. Br J Cancer. 2013;108:579–586. doi: 10.1038/bjc.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhatla T, et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood. 2012;119:5201–5210. doi: 10.1182/blood-2012-01-401687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shakya R, et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res. 2012;73:885–896. doi: 10.1158/0008-5472.CAN-12-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu S, et al. Phase 1b–2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer. 2011;117:1661–1669. doi: 10.1002/cncr.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matei D, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gray SG, Ekström TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 90.Solomon JM, et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pruitt K, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pediconi N, et al. hSirT1-dependent regulation of the PCAF–E2F1–p73 apoptotic pathway in response to DNA damage. Mol Cell Biol. 2009;29:1989–1998. doi: 10.1128/MCB.00552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cooper SJ, et al. Reexpression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol Cancer Ther. 2012;11:2105–2115. doi: 10.1158/1535-7163.MCT-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Isozaki Y, et al. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int J Oncol. 2012;41:985–994. doi: 10.3892/ijo.2012.1537. [DOI] [PubMed] [Google Scholar]
- 97.Shin H, Lee YS, Lee YC. Sodium butyrate-induced DAPK-mediated apoptosis in human gastric cancer cells. Oncol Rep. 2012;27:1111–1115. doi: 10.3892/or.2011.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mund C, Lyko F. Epigenetic cancer therapy: Proof of concept and remaining challenges. Bioessays. 2010;32:949–957. doi: 10.1002/bies.201000061. [DOI] [PubMed] [Google Scholar]
- 99.Verbrugge I, Johnstone RW, Bots M. Promises and challenges of anticancer drugs that target the epigenome. Epigenomics. 2011;3:547–565. doi: 10.2217/epi.11.82. [DOI] [PubMed] [Google Scholar]
- 100.Robert C, Rassool FV. HDAC inhibitors: roles of DNA damage and repair. Adv Cancer Res. 2012;116:87–129. doi: 10.1016/B978-0-12-394387-3.00003-3. [DOI] [PubMed] [Google Scholar]
- 101.Kachhap SK, et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS ONE. 2010;5:e11208. doi: 10.1371/journal.pone.0011208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gilbertson RJ, Graham TA. Cancer: resolving the stem-cell debate. Nature. 2012;488:462–463. doi: 10.1038/nature11480. [DOI] [PubMed] [Google Scholar]
- 103.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 106.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yeo W, et al. Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J Clin Oncol. 2012;30:3361–3367. doi: 10.1200/JCO.2011.41.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hainsworth JD, et al. A phase II trial of panobinostat, a histone deacetylase inhibitor, in the treatment of patients with refractory metastatic renal cell carcinoma. Cancer Invest. 2011;29:451–455. doi: 10.3109/07357907.2011.590568. [DOI] [PubMed] [Google Scholar]
- 109.Giaccone G, et al. Phase II study of belinostat in patients with recurrent or refractory advanced thymic epithelial tumors. J Clin Oncol. 2011;29:2052–2059. doi: 10.1200/JCO.2010.32.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iwamoto FM, et al. A phase I/II trial of the histone deacetylase inhibitor romidepsin for adults with recurrent malignant glioma: North American Brain Tumor Consortium Study 03–03. Neuro Oncol. 2011;13:509–516. doi: 10.1093/neuonc/nor017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mackay HJ, et al. Phase II trial of the histone deacetylase inhibitor belinostat in women with platinum resistant epithelial ovarian cancer and micropapillary (LMP) ovarian tumours. Eur J Cancer. 2010;46:1573–1579. doi: 10.1016/j.ejca.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Witta SE, et al. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J Clin Oncol. 2012;30:2248–2255. doi: 10.1200/JCO.2011.38.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramalingam SS, et al. Carboplatin and paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:56–62. doi: 10.1200/JCO.2009.24.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yardley DA, Ismail-Khan R, Klein P. Results of ENCORE 301, a randomized, phase II, double-blind, placebo-controlled study of exemestane with or without entinostat in postmenopausal women with locally recurrent or metastatic estrogen receptor-positive (ER+) breast cancer progressing on a nonsteroidal aromatase inhibitor (AI) [abstract 268] J Clin Oncol. 2011;29(Suppl 27):268. doi: 10.1200/JCO.2012.43.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Topalian SL, et al. Safety, activity, and immune orrelates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Issa JP, et al. Interim results from a randomized phase 1–2 first-in-human (FIH) study of PK/PD guided escalating doses of SGI-110, a novel subcutaneous (SQ) second generation hypomethylating agent (HMA) in relapsed/refractory MDS and AML [abstract LB-214] Cancer Res. 2012;72(Suppl 1):LB-214. [Google Scholar]
- 118.Garcia-Manero G, et al. Phase I study of oral azacitidine in myelodysplastic syndromes, chronic myelomonocytic leukemia, and acute myeloid leukemia. J Clin Oncol. 2011;29:2521–2527. doi: 10.1200/JCO.2010.34.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chung CW, et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem. 2011;54:3827–3838. doi: 10.1021/jm200108t. [DOI] [PubMed] [Google Scholar]
- 120.Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- 121.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 124.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kantarjian H, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]