Summary
In vitro follicle growth has emerged as a technology that can provide new information about folliculogenesis and serve as part of a suite of methods currently under development to assist women whose fertility is threatened by cancer treatments. Though it has been shown that in vitro-grown follicles secrete peptide and steroid hormones, much of the follicular transcriptome remains unknown. Thus, microarray analysis was performed to characterize the transcriptome and secretome of in vitro-grown follicles. One prominently regulated gene product was cartilage oligomeric matrix protein (Comp): its mRNA was upregulated during the final 4 days of culture (P < 0.05) and COMP protein could be detected in medium from individual follicles. COMP expression localized to mural granulosa cells of large antral follicles both in vitro and in vivo, with maximal expression immediately preceding ovulation in cycling and chorionic gonadotropin-primed female mice. COMP was co-expressed with two known markers of follicle maturation, inhibin βA and gremlin, and was expressed only in TUNEL-negative follicles. In addition to other gene products identified in the microarray, COMP has potential utility as a marker of follicle maturation.
Introduction
The cyclical selection and maturation of ovarian follicles is fundamental to female fertility. Factors from the anterior pituitary and the ovary stimulate immature follicles to develop to the preovulatory stage, which involves both somatic cell proliferation and oocyte growth and maturation. Stimulated by the luteinizing hormone (LH) surge, Graafian follicles release mature egg(s) in coordination with somatic compartment luteinization. Thus, a highly coordinated in vivo mechanism times follicular maturation leading to the release of mature eggs within an ideal hormonal milieu for implantation.
The ovary provides the optimal environment for follicle growth; mimicking these conditions in vitro holds great potential for clinical applications. Currently, fertility preservation for women undergoing gonadotoxic therapies involves hormone stimulation protocols followed by oocyte or embryo banking (Jeruss and Woodruff, 2009; Hirshfeld-Cytron et al., 2011; von Wolff et al., 2011; Rodriguez-Wallberg and Oktay, 2012). If successful, cryopreservation of oocytes or embryos provides the potential for fertility restoration post-treatment. Such protocols, however, may encounter delays due to disease treatment or may be contraindicated in children/adolescents or women with certain types of hormone-sensitive malignancies. In vitro follicle growth has emerged as a potential reproductive solution for these patients. Clinical in vitro follicle growth would involve isolation of ovarian tissue without significant delay in disease treatment, cryopreservation of immature follicles, and in vitro growth when the patient or surrogate is ready to carry a pregnancy (Smitz et al., 2010). To date, several in vitro follicle growth systems have been developed for rodents (Spears et al., 1994; Cortvrindt et al., 1996; Eppig and O'Brien, 1996; Smitz and Cortvrindt, 1999; O'Brien et al., 2003; Xu et al., 2006a,b), large mammals (Newton et al., 1999; Gutierrez et al., 2000; Telfer et al., 2000; Wu et al., 2001; Picton et al., 2003; Thomas et al., 2007), and humans (Roy and Treacy, 1993; Abir et al., 1997, 1999, 2001, 2006; Hovatta et al., 1997; Wright et al., 1999; Scott et al., 2004; Telfer et al., 2008; Amorim et al., 2009). The three-dimensional (3D), alginate hydrogel-basedin vitro follicle growth system maintains follicle architecture and critical cell–cell interactions between the somatic cells and oocyte throughout maturation. Maintenance of follicle architecture is essential for growing follicles from large mammalian species, including the dog, rhesus monkey, baboon, and human (Xu et al., 2009a,b, 2011b; Songsasen et al., 2011).
One of the next steps in the emerging in vitro follicle growth field is to gain a better understanding of the follicular transcriptome throughout growth. Follicles with similar starting size can have significant heterogeneity in growth rate and gamete quality, which likely recapitulates in vivo physiology (Xu et al., 2010, 2011a). Therefore, in vitro follicle growth will provide the greatest clinical utility when follicles can be individually monitored for maturity and quality. Estrogen and inhibin levels are commonly used metrics of follicle growth (Xu et al., 2009a, 2010; Dunning et al., 2011), but the expression of these proteins throughout folliculogenesis (multi-layered secondary to preovulatory) limits their use as specific markers of terminal follicle maturity. Additional secreted proteins may be identified through characterization of the ovarian follicular secretome, and will provide important information regarding in vivo and in vitro follicle development.
To identify secreted factors, we performed a genomewide expression analysis of in vitro-grown follicles from the secondary through preovulatory stages. Cartilage oligomeric matrix protein (Comp) was the most significantly upregulated transcript during in vitro follicle growth and was comparable between in vitro- and in vivo-grown follicles. Our data show that Comp is one of several secreted factors that could add important information about the status of the follicle or its enclosed oocyte in vitro and in vivo.
Results
Genome-Wide Expression Analysis of In Vitro-Grown Follicles
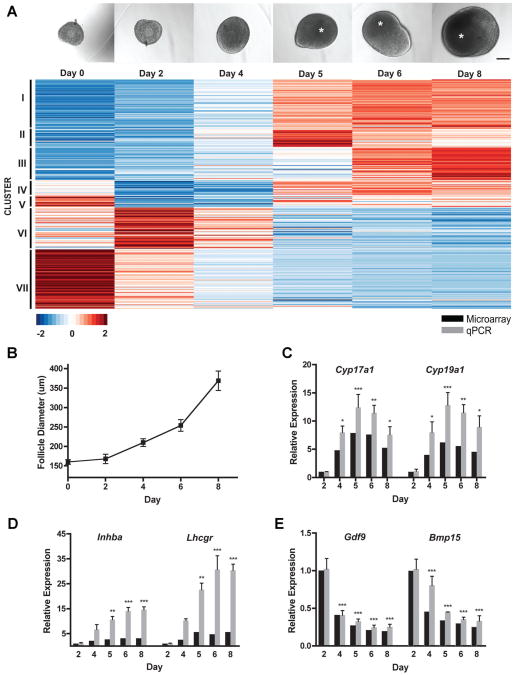
In our 3D hydrogel system, alginate preserves follicular architecture with the oocyte completely surrounded by granulosa cells (Pangas et al., 2003). When grown in this system over an 8-day culture period (Fig. 1A), multi-layer, secondary mouse follicles increased in diameter from 160±6 μm to 369±25 μm (Fig. 1B). A fluid-filled antral cavity formed between days 4 and 6, and expanded throughout follicle growth. We subsequently performed a microarray that compared gene expression in follicles cultured for variable durations (0, 2, 4, 5, 6, and 8 days) to better understand transcriptional changes during follicular growth. To identify differentially expressed genes over time, two independent statistical approaches were employed using different software packages. First, the Linear Models for Microarray Data (limma) package analysis identified statistically significant changes in gene expression between each time point (Smyth, 2004). A second independent analysis was performed using the timecourse package (Tai and Speed, 2006). Each analysis identified 212 genes that significantly changed in follicles over the 8-day culture period, with 91 genes identified by both methods (Supplemental Fig. 1, Supplemental Table 1) and a total of 333 genes by the union of methods. A supervised clustering analysis showed that genes could be classified in seven different clusters based on dynamic transcriptional expression, including follicles before encapsulation (“day 0”) (Fig. 1A). The first three clusters were highly upregulated after day 4, while the remaining clusters were highly downregulated after antrum formation. This set of differentially expressed transcripts was enriched in genes encoding ligands and enzymes (z-scores of 6 and 4, respectively), and the top gene ontologies included cellular responses to endogenous and exogenous stimuli and reproduction and the reproductive system (P <10−14). The microarray results were validated by quantitative real-time reverse transcriptase-PCR (qPCR) analysis using well-characterized genes within the follicle (Cyp17a1, Cyp19a1, Inhba, Lhcgr, Gdf9, Bmp15; Fig. 1C–E).
Figure 1.
Microarray analysis of in vitro-grown follicles. A: Representative gross morphology of an individual follicle encapsulated in alginate at 0, 2, 4, 5, 6, and 8 days in vitro. Asterisks denote antral cavities. Scale bar, 100 μm. Heat-map representation of results from genome-wide expression analysis of in vitro-grown follicles. K-means clustering of the scaled average intensity of the significant genes indicated seven clusters. B: Average follicle diameters from days 0 to 8 (n = 423 individual follicles). Data are expressed as mean ± standard error mean. C–E: Microarray validation. Microarray and qPCR results of Cyp17a1, Cyp19a1 (C), Inhba, Lhcgr (D), Gdf9, and Bmp15 (E) expression in cultured follicles compared to expression on day 2. For microarray P ≤ 0.01, n = 2–3. For qPCR, n = 3; *P < 0.05, **P < 0.01, ***P< 0.001, according to one-way ANOVA followed by Bonferroni's Multiple Comparison Test.
Expression of Secreted Proteins During In Vitro Follicle Growth
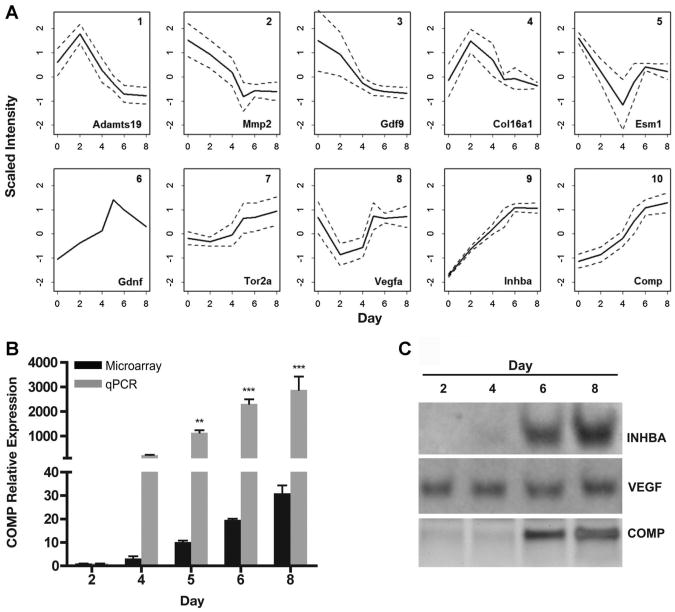
Potential noninvasive markers of follicle growth were investigated by focusing on the temporal expression patterns of genes encoding secreted proteins. To that end, the 63 genes encoding secreted proteins (Supplemental Table 2) were further classified into 10 groups based on their dynamic patterns using self-organizing maps (SOMs) (Fig. 2A). The gene encoding a secreted protein that had the greatest differential expression over the culture period was Comp (P=5.07 × 10−6, Supplemental Tables 1 and 2), which was classified in group 10 (Fig. 2A). Group 10 included proteins that initially had low levels of expression, and increased most dramatically during the latter 4 days of culture. In addition, expression patterns for groups 8 and 9 increased in the latter half of the culture period and contained genes encoding secreted proteins known to be associated with follicle maturity, including vascular endothelial growth factor A (Vegfa) and inhibin βA(Inhba) (Woodruff et al., 1988; Celik-Ozenci et al., 2003; Greenaway et al., 2004; Xu et al., 2010). As expected, both Vegfa (group 8, Fig. 2A) and Inhba (group 9, Fig. 2A) were expressed at low levels at the beginning of culture, and increased steadily throughout the culture period (days 2–8). Validation of Comp expression by qPCR showed an increase in mRNA levels to an almost 3,000-fold change by day 8 of culture compared to day 2 (Fig. 2B). The upregulation of these genes at the protein level was verified in conditioned medium that was collected from individual follicles. VEGFA was detected on day 2 and was present through day 8 and INHBA was detected in the medium on days 6 and 8 (Fig. 2C). COMP was detected in culture media beginning on day 6, and increased on day 8 (Fig. 2C).
Figure 2.
COMP is a secreted protein that increases significantly during in vitro follicle growth and can be detected in culture medium. A: Scaled trends of the intensity using self-organized maps (SOMs) for the genes encoding secreted proteins during ovarian follicle maturation. Solid lines indicate the average, scaled mean intensity of the group and dotted lines represent 1 standard deviation (note that group 6 contains only Gdnf). Listed is a representative gene from each group (bottom right). See Supplemental Table 2 for the complete list of genes encoding secreted proteins. B: Gene expression profiles comparing Comp derived from the microarray analysis (black) and qPCR validation (gray). *P <0.05, **P <0.01, ***P < 0.001 relative to expression at culture day 2 (n=3 independent experiments, one-way ANOVA followed by Bonferroni's Multiple Comparison Test). C: Western blot detection of VEGFA, INHBA, and COMP in conditioned media from individual follicles cultured in vitro. Images are representative of three independent experiments.
COMP Expression During In Vitro and In Vivo Follicular Growth
Next, the localization of COMP expression within follicles grown both in vitro and in vivo was characterized. Following 8 days of in vitro growth, preovulatory follicles were fixed and processed for immunohistochemistry with a COMP-specific antibody. COMP expression was restricted to mural granulosa cells within the follicle periphery (Fig. 3A). To assess COMP expression in vivo, an immunohistochemical analysis was applied to ovaries from adult female mice throughout the estrous cycle (Fig. 3B). COMP expression was specific to large antral follicles during the afternoon of proestrus, immediately preceding ovulation. Additionally, COMP localized to granulosa cells and was present in the antral cavity. COMP was not detected in smaller follicles or in corpora lutea. The ovarian surface epithelium was COMP-positive at all stages of the estrous cycle, however (Fig. 3B). To examine expression in larger mammalian species, we also examined histological sections of bovine ovaries. While small antral follicles did not stain for COMP, granulosa cells exhibited robust expression in larger antral follicles (Supplemental Fig. 2). Thus, COMP was expressed in mural granulosa cells of large antral follicles in vitro and in preovulatory follicles immediately preceding the LH surge in vivo.
Figure 3.
Ovarian COMP expression in vitro and in vivo. A: Image of a representative follicle cultured for 8 days and immunostained for COMP protein (i), counterstained with DAPI (ii), and photomerged (iii). Scale bar, 100 μm. Confocal microscopy image of mural granulosa cells within the follicle showing COMP localized to cytoplasmic and extracellular compartments (iv). Scale bar, 15 μm. B: COMP immunofluorescence of murine ovarian sections at different stages of the estrous cycle. Ovarian sections taken at diestrus, proestrus at 10:00 am, proestrus at 4:00 pm, estrus, and metestrus. Arrowheads signify COMP-positive staining within the ovarian surface epithelium. Scale bar, 200 μm. Images are representative of 2–3 independent experiments.
Gonadotropin Regulation of COMP
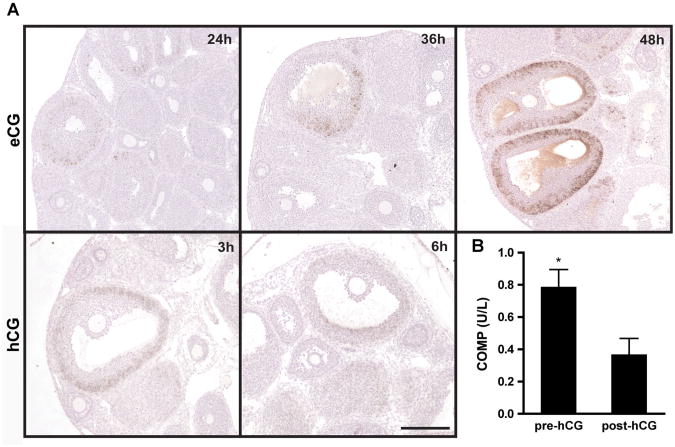
Since follicular COMP expression was restricted in cycling mice to proestrus, we investigated the effects of gonadotropin stimulation on COMP expression. We induced follicle growth in prepubertal mice with equine chorionic gonadotropin (eCG). Ovaries at 12hr after eCG injection did not stain positively for COMP (data not shown). By 24hr post-injection, however, COMP protein was detected in granulosa cells of growing follicles, and levels progressively increased 36 and 48hr post-injection (Fig. 4A). COMP localized to mural granulosa cells and expression patterns were consistent with those observed in vitro and in cycling animals during proestrus (Fig. 3B). At 48 hr after eCG stimulation, mice were injected with hCG to induce ovulation. By3 hr post-hCG injection, a decline in the intensity of COMP expression was observed (Fig. 4A). At14 and 20 hr post-injection (i.e., post-ovulation, which occurs approximately 12hr after hCG injection), COMP protein was no longer detectable within the cells of the corpora lutea (data not shown). To test the potential role of LH/human chorionic gonadotropin (hCG) in downregulating COMP expression, primary granulosa cells were isolated from mice 48hr following eCG injection and treated with hCG. As determined by ELISA (enzyme-linked immunosorbent assay), COMP protein decreased by more than 50% post-hCG administration (P < 0.05) (Fig. 4B). Thus, throughout folliculogenesis COMP is stimulated and inhibited by gonadotropins.
Figure 4.
Gonadotropin regulation of COMP. A: Histological ovarian sections stained for COMP from prepubertal mice treated with 5IU eCG 12, 24, and 36hr post-injection (upper panel). Mice were injected with hCG 48hrpost-eCG, and slides were stained for COMP 3 and 6hr post-hCG injection (lower panel). Scale bar, 200 μm. Images are representative of three independent experiments. B: ELISA results for COMP in culture medium from primary granulosa cells before and after 1.5 IU hCG treatment *P < 0.05 (n=3 independent experiments).
Co-Expression of Antral Follicle Markers
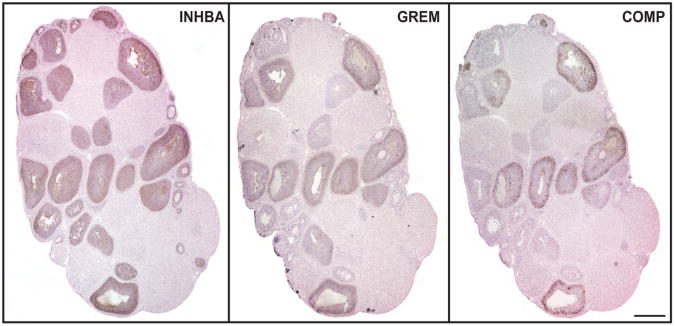
The subpopulation of follicles that expressed COMP was subsequently analyzed by immunohistochemistry for other known markers of follicle development. In adjacent sections of fixed ovaries from eCG-stimulated mice, we identified populations of follicles using two established markers of antral follicles (inhibin βA and gremlin) and COMP (Fig. 5). As expected, inhibin βA was expressed in follicles ranging from multi-layered secondary to large Graafian follicles (Woodruff et al., 1988). Further, gremlin was present in a subset of inhibin βA-positive follicles, namely antral stage and later (Pangas et al., 2004). Importantly, COMP was restricted to an even smaller subset of large Graafian follicles that were inhibin βA- and gremlin-positive. Thus, COMP expression was highly restricted to a subpopulation of follicles expressing known markers of maturity.
Figure 5.
COMP is a selective marker of terminally mature follicles. Histological ovarian sections stained for inhibin βA, gremlin, and COMP from prepubertal mice treated with 5 IU eCG 36 hr post-injection. Scale bar, 400 μm. Images are representative of three independent experiments.
COMP Expression and Apoptosis in Antral Follicles
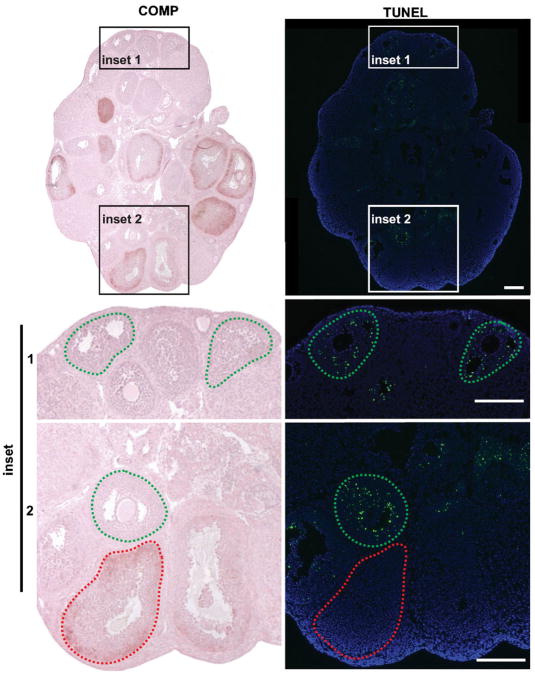
Since we found COMP expression was restricted to a subset of mature follicles, we further characterized this follicular subpopulation. COMP has been reported to have an anti-apoptotic role in chondrocytes (Hecht et al., 2004; Gagarina et al., 2008), thus we performed a TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) assay to probe its possible function in the ovary. From adjacent histological sections of ovaries from mice 36 hr post-eCG injection, follicles that did not contain COMP were TUNEL-positive, whereas antral follicles that expressed COMP were TUNEL-negative (Fig. 6). Thus, the subpopulation of follicles that express COMP was not apoptotic.
Figure 6.
Follicles that express COMP are TUNEL-negative. Ovarian section from a prepubertal mouse 48 hr after injection with eCG immunohistochemically stained for COMP. Adjacent ovarian sections stained for TUNEL. Follicles delineated in green and red dashed lines highlight COMP-positive and TUNEL-positive staining, respectively. Scale bars, 200 μm. Images are representative of three independent experiments.
Discussion
Folliculogenesis is precisely coordinated to support follicle selection and oocyte maturation, leading to the release of a developmentally competent egg. Transcriptional changes throughout in vitro follicular maturation were analyzed in order to better understand follicle development and to identify new markers of maturation. The ovarian follicle secretome was identified computationally, and we elected to analyze the most highly regulated genes. Known secreted factors, such as inhibin βA, VEGF, and gremlin, were studied together with a new marker of terminal follicle development, COMP. COMP was identified as a selective marker of follicle maturity and increased throughout follicle growth both in vitro and in vivo.
In the alginate in vitro follicle growth system, secondary follicles are cultured for 8 days and treated with hCG to induce terminal maturation and attain cumulus oocyte complexes. This time period is associated with several critical transformations, namely development of the antral cavity and acquisition of oocyte meiotic competence (Pan et al., 2005). We identified a number of significant changes in the genes expressed at these time points, which could be classified into seven groups by k-means clustering. In addition, we further classified genes encoding secreted proteins into 10 groups based on their dynamic patterns using SOMs. We observed a high correlation between groups 2, 8, 9, and 10 (r2> 0.95), which could indicate that proteins in group 9 regulate the delayed expression in groups 2, 8, and 10. Furthermore, the expression of proteins in group 10 increased from low to high during culture, suggesting that these factors may be associated with terminal follicle maturation. Network analysis of these key transitions and signaling processes is the subject of future studies. The secreted proteins that were identified in maturing follicles included known endocrine factors, such as inhibin βA and the steroidogenic enzymes.
In addition to known hormone profiles of growing follicles, newly identified secreted factors could be used as part of a screening paradigm for in vitro follicle growth. One of the genes identified in group 10, Comp, was the most significantly upregulated transcript throughout in vitro growth. It was selected as a candidate secreted biomarker of terminal follicle maturity because Comp mRNA increased during the latter 4 days of culture and its protein could be detected in medium from individual follicles. COMP expression localized to mural granulosa cells of large antral follicles in vivo and in vitro, and we observed maximal expression immediately preceding ovulation in cycling and eCG-primed mice. In addition, we found that hCG decreased COMP expression in primary granulosa cells, suggesting that gonadotropins stimulate and inhibit COMP throughout folliculogenesis. Perhaps most interestingly, COMP expression was restricted to the largest preovulatory follicles and to a subset of both inhibin βA- and gremlin-positive follicles. These data suggest that there are developmental subsets of follicles delineated by expression of inhibin βA, gremlin, and COMP, provides new opportunities to discover the hierarchy and timing of secreted factors measured in maturing single follicles in vitro.
While we showed that antral follicles express abundant COMP, its precise biological function has yet to be elucidated. Comp knockout mice displayed no apparent phenotype, suggesting that COMP acts via redundant pathways (Svensson et al., 2002). In cartilage, COMP was identified as a potent anti-apoptotic agent (Gagarina et al., 2008), and it may play a similar role in the ovary. We found that antral follicles that did not express COMP were apoptotic and, conversely, COMP-positive follicles were not apoptotic. The majority of work characterizing COMP (also known as thrombospond in 5) has been performed in cartilage, tendon, ligament, and bone (Morgelin etal., 1992; DiCesare et al., 1995; Shen et al., 1995; Motaung et al., 2011). To date, reports of COMP in the ovary have described its downregulation in inhibin-null (whole ovaries) and Smad 1/5 conditional knock out (granulosa cell tumors) mice (Pangas et al., 2008; Nagaraja et al., 2010). Given our observation that COMP is primarily expressed in preovulatory follicles, the decline in Comp in these knock out mice is likely due to defective folliculogenesis and supports its specific expression in large antral follicles. While this report is the first temporal and spatial characterization of COMP in the ovary, much remains to be investigated about other possible reproductive functions. Further studies will investigate its utility in large animal in vitro follicle growth, where long culture periods (more than 30 days) and loss of oocyte visibility provides the impetus to discover biomarkers like COMP. Toward this end, we showed that COMP is expressed in large antral bovine follicles. COMP can thus be tested in the context of bovine, primate, and other large mammalian species to complement hormone measurements.
In summary, we identified COMP as a marker of terminally mature follicles selected to ovulate in vivo, and Comp was the most significantly upregulated transcript during in vitro follicle growth. COMP may function as an antiapoptotic factor in preovulatory follicles, and its expression may be used as a positive marker of follicle development in culture. Biomarkers that can distinguish follicular stages and thereby optimize the time for selection of oocytes to be matured could significantly enhance in vitro follicle growth technologies.
Materials and Methods
Animals
CD1 mice were housed in a temperature- and lightcontrolled environment (14 hr light: 10 hr dark), and provided with food and water ad libitum. Animals were fed Teklad Global irradiated 2919 low-phytoestrogen chow. To minimize differences in nutrient availability, eight females were housed per dam (pups were sacrificed at birth, if necessary). Animals were treated in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the established IACUC protocol at Northwestern University.
To stimulate follicular growth, prepubertal, 18-day-old female mice were injected intraperitoneally with 5 IU of eCG (Calbiochem, La Jolla, CA) as described previously (Wilson and Zarrow, 1962). Mice injected with eCG (and their saline-injected control counterparts) were sacrificed at 12, 24, 36, and 48 hr post-injection and ovaries were explanted. At 48 hr post-eCG injection, a subset of mice was injected with 5IU of hCG, and the ovaries removed at 3, 6, 14, and 20hr post-injection. Collected ovaries were immediately fixed in Modified Davidson's Fixative (Electron Microscopy Sciences, Hatfield, PA) for 16–18hr at 4°C, then dehydrated and embedded in paraffin. The ovaries were sectioned at 5 μm and adhered to glass slides.
Postnatal day 30 CD1 female mice were housed 1–2 per cage and vaginal cytology was used for the determination of the estrous cycle. Smears were taken daily between 9:30 am and 10:00 am, were imaged on an inverted stereomicroscope (Leica Microsystems, Buffalo Grove, IL), and stages of the estrous cycle were classified according to the vaginal cell morphology (Nelson et al., 1982). Mice that underwent three consecutive 4- or 5-day cycles were sacrificed on the morning of metestrus, diestrus, proestrus, or estrus. Mice were also sacrificed at 4:00 pm on proestrus to capture an additional time point prior to ovulation, which occurs during proestrus between 3:00 am and 4:00 am. Ovaries were removed, fixed, and processed for histology as described above. To ensure that the observed vaginal cytology matched the proper cycle stage, blood was drawn at the time of sacrifice by cardiac puncture at each defined time point.
Follicle Isolation, Encapsulation, and Culture
Multi-layered secondary follicles (150–180 μm in diameter) were mechanically isolated from ovaries of 16-day-old mice and individually encapsulated in 0.5% alginate (FMC BioPolymers, Philadelphia, PA), as previously described (Kreeger et al., 2006; Xu et al., 2006a). Alginate-encapsulated follicles were placed in individual wells of a 96-well plate containing 100 μl of growth medium (alpha minimum essential medium (alphaMEM) supplemented with 10mIU/ml recombinant FSH (Organon, Roseland, NJ), 3mg/ml bovine serum albumin (MP Biomedicals, Irvine, CA), 1 mg/ml bovine fetuin (Sigma, St. Louis, MO), 5 μg/ml insulin, 5 μg/ml transferrin, and 5ng/ml selenium) and cultured for 2, 4, 5, 6, or 8 days in a 5% CO2:21% O2 atmosphere. Day 0 samples correspond to follicles that were isolated and immediately frozen. Half of the culture medium was exchanged every 2 days, and conditioned medium stored at −80°C. At each time point, follicles were removed from growth medium, pooled (20–40 follicles per group, n = 3 independent experiments), and transferred into 1 ml of Liebovitz L-15 medium containing 10U/ml alginate lyase (Sigma) for 20min at 37°C to remove them from alginate. Follicles were aspirated, transferred into microcentrifuge tubes, flash frozen in liquid nitrogen, and stored at −80°C until subsequent RNA isolation was performed.
Microarray Assay and Analysis
RNA was purified from follicles using the Qiagen RNeasy Micro Kit according to the manufacturer's protocol (Qiagen, Valencia, CA). RNAquality and quantity was assessed both by NanoDrop (Thermo Scientific, Wilmington, DE) and BioAnalyzer 2100 Expert (Agilent Technologies, Santa Clara, CA). RNA samples were submitted to the Northwestern University Genomics Core at the Center for Genetic Medicine for genome-wide RNA expression analysis using the MouseRef-8 v2.0 Expression BeadChip Kit (Illumina, San Diego, CA) that targets over 18, 140 unique genes. Briefly, mRNA samples were in vitro-labeled using the TargetAmp 1-Round Aminoallyl-aRNA Kit (Epicentre, Madison, WI) and randomly hybridized to BeadChips. Raw signal intensities of each probe were obtained using BeadStudio (Illumina). The subsequent statistical analyses were done in R (2008). Inadequate Illumina microarray probes, due to misannotations or intronic coverage, were removed from analysis using the Mouse WG V2.0 R0 file within ReMOAT (http://remoat.sysbiol.cam.ac.uk) (Team, 2008; Barbosa-Morais et al., 2010). Adequately annotated probes were considered to be above background if at least two out of the three replicates measured were above background at P ≤ 0.01. The data were transformed using the variance stabilization transformation method (Lin etal., 2008) and normalized by robust spline normalization (Du etal., 2008). Differentially expressed genes overtime were assessed using limma (Smyth, 2004) and timecourse (Tai and Speed, 2006) packages within Bioconductor (http://www.bioconductor.org). Genes were considered differentially expressed in limma if they exhibited a fold-change ≥ 1.4 per day and a false discovery rate corrected P ≤ 0.02. For timecourse analysis, data were ranked by Hotelling statistics. In this instance, Hotelling statistics indicated the variation in temporal expression profiles of the studied genes; large Hotelling numbers indicated very dynamic genes. Due to the fact that there is no direct translation of the Hotelling statistics into a comparable P-value for limma, a hard cut-off of 212 genes was used for the timecourse. Raw microarray data were deposited in the Gene Expression Omnibus (GEO) database (accession number GSE42795; http://www.ncbi.nlm.nih.gov/geo/).
Clustering Analysis and Gene Ontology
Before clustering, the data were scaled according to its mean and standard deviation. Clustering analysis of the differentially expressed genes over time was done using k-means. The number of clusters was chosen based on the minimal number of partitions that minimized standard error within each cluster. The final results were plotted using heatmap from the gplots package, in which colors were selected from RColorBrewer (http://colorbrewer2.org/). Genes that encode for secreted proteins were clustered based on a SOM using a Gaussian neighboring function and rectangular topology. The number of SOMs was selected based on the minimum number that did not generate new profiles. The described packages were attained through CRAN (http://cran.r-project.org/). Genes that encoded for secreted proteins were determined based on the gene ontologies obtained from the Mouse Genomic Database (MGD) using the extracellular region ontology identifier GO:0005576 (Eppig et al., 2012).
Gene Expression Analysis by Real-Time PCR (qPCR)
Full-length cDNA was synthesized from 20 ng mRNA for each sample using the AccuScript High Fidelity 1st Strand cDNA Synthesis Kit (Agilent Technologies). Real-time PCR was performed on the ABI PRISM® 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) using both the Taqman® Universal PCR Master Mix (Roche, Indianapolis, IN) and Taqman® Gene Expression Assays (Applied Biosystems), according to the manufacturer's specifications. Each sample was performed in technical duplicate and biological triplicate, and all results were normalized to Rpl18. Analysis of relative gene expression was done using the comparative Ct (2−ΔΔCT) method (Livak and Schmittgen, 2001).
Immunoblot Analysis
Conditioned medium collected at 2, 4, 6, and 8 days after the start of in vitro culture was loaded on 4–12% SDS–PAGE gels under reducing conditions. Proteins of interest were detected using the following antibodies: rabbit anti-COMP (Abcam, Cambridge, MA), mouse anti-VEGF (Abcam), rabbit anti-inhibin βA (kindly provided by Dr. W. Vale, The Salk Institute), followed by a horseradish peroxidase-conjugated secondary antibody (Zymed, San Francisco, CA). The secondary antibody was detected using Amersham ECL Plus (GE Healthcare, Piscataway, NJ). Signal visualization was performed using a FluorChem HD2 ChemiImager (Alpha Innotech, San Leandro, CA).
Primary Granulosa Cell Culture and COMP Assay
Granulosa cells were collected as previously published (Epstein-Almog and Orly, 1985). Briefly, postnatal day 21 mouse ovaries were collected 44–48 hr after priming with 5 IU eCG. Granulosa cells were released by puncturing large antral follicles with insulin-gauge needles in DMEM/F12 medium supplemented with 3mg/ml bovine serum albumin. A single-cell suspension was acquired by gently pipetting up and down several times. Granulosa cells were cultured for 48 hr in growth medium (DMEM/F12 with 10 μg/ml insulin, 5 nM sodium selenite, 5 μg/ml transferring, and 5% fetal bovine serum). COMP was measured in culture medium using an Animal COMP ELISA kit (MD Bioproducts, St. Paul, MN) according to the manufacturer's specifications. The intra-assay variation was 9.4% and the sensitivity was <0.2 U/L.
Immunohistochemistry and TUNEL Staining
Histological sections were deparaffinized with xylenes and rehydrated. Antigen retrieval was performed using application of 0.01 M sodium citrate (pH 6.0), then endogenous peroxidase activity quenched with 3% hydrogen peroxide. Immunohistochemical staining of the sections was done using the Vectastain Elite Standard ABC Kit (Vector Laboratories, Burlingame, CA). Tissue sections were first blocked with the Avidin/Biotin Block Kit (Vector Laboratories) for 15min, then blocked with 3% bovine serum albumin in Tris-buffered saline supplemented with 10% serum for 1 hr. Proteins of interest were detected using the following antibodies: rabbit anti-COMP (Abcam), rabbit anti-inhibin βA (kindly provided by Dr. W. Vale, The Salk Institute), and goat anti-Gremlin (R&D Systems, Minneapolis, MN). Detection and localization of antibody binding was observed through the use of biotinylated immunoglobulin G secondary antibodies and the avidin-biotinylated horseradish peroxidase complex (Vector Laboratories), which reacted and stained with 3,3′-diaminobenzidine (DAB, Vector Laboratories). Sections were counterstained with hematoxylin, dehydrated, then cover-slipped and mounted with Cytoseal (Electron Microscopy Sciences).
To detect apoptotic cells in situ, the DeadEnd™ Fluorometric TUNEL System was employed as per the manufacturer's suggested protocol (Promega, Madison, WI). Briefly, tissue sections were rehydrated and biotinylated nucleotide incorporated into fragmented DNA using the Terminal Deoxynucleotidyl Transferase Recombinant enzyme. To detect the fragmented DNA, horseradish peroxidase-labeled streptavidin was applied to the sections then stained with a FITC-conjugated antibody and slides were mounted with Vectashield + DAPI (Vector Laboratories).
In cases of confocal imaging, sections were imaged using a TCS SP5 confocal microscope (Leica Microsystems), and maximum 3D projections rendered using LAS Image Analysis software (Leica Microsystems).
Statistics
All experiments were independently performed at least three times, unless otherwise noted. Statistical analyses of the microarray results were performed as described above. For comparisons between groups, a one-way ANOVA followed by Bonferroni post hoc test was performed (Prism4; Graph Pad Software).
Supplementary Material
Acknowledgments
We thank Dr. Lei Lei, Sarah Kiesewetter, Jennifer Jozefik, Erin Jackson, and Dragan Mackovic for their invaluable technical support. Additionally, we are grateful to Dr. Nadereh Jafari, Director of the Genomics Core Facility (Center for Genetic Medicine, Northwestern University), and Lizbeth Gutierrez, Director of the Ovarian Histology Core (Center of Reproductive Sciences, Northwestern University). We would also like to thank Dr. Francesca Duncan for her critical review in the preparation of this manuscript. This work has been funded by NIH U54 HD041857 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. R.M.S. was supported by NIH F30AG040916-01 from the National Institute on Aging.
Grant sponsor: Eunice Kennedy Shriver National Institute of Child Health & Human Development; Grant number: NIH U54 HD041857; Grant sponsor: National Institute on Aging; Grant number: NIH F30 AG040916-01
Abbreviations
- COMP
cartilage oligomeric matrix protein (or thrombospon-din 5)
- e/hCG
equine/human chorionic gonadotropin
- INHBA
inhibin βA
- LH
luteinizing hormone
- qPCR
quantitative real-time reverse-transcriptase polymerase chain reaction
- VEGFA
vascular endothelial growth factor A
Footnotes
Disclosure statement: The authors have nothing to disclose.
Additional supporting information may be found in the online version of this article.
References
- Abir R, Franks S, Mobberley MA, Moore PA, Margara RA, Winston RM. Mechanical isolation and in vitro growth of preantral and small antral human follicles. Fertil Steril. 1997;68:682–688. doi: 10.1016/s0015-0282(97)00264-1. [DOI] [PubMed] [Google Scholar]
- Abir R, Roizman P, Fisch B, Nitke S, Okon E, Orvieto R, Ben Rafael Z. Pilot study of isolated early human follicles cultured in collagen gels for 24hours. Hum Reprod. 1999;14:1299–1301. doi: 10.1093/humrep/14.5.1299. [DOI] [PubMed] [Google Scholar]
- Abir R, Fisch B, Nitke S, Okon E, Raz A, Ben Rafael Z. Morphological study of fully and partially isolated early human follicles. Fertil Steril. 2001;75:141–146. doi: 10.1016/s0015-0282(00)01668-x. [DOI] [PubMed] [Google Scholar]
- Abir R, Nitke S, Ben-Haroush A, Fisch B. In vitro maturation of human primordial ovarian follicles: Clinical significance, progress in mammals, and methods for growth evaluation. Histol Histopathol. 2006;21:887–898. doi: 10.14670/HH-21.887. [DOI] [PubMed] [Google Scholar]
- Amorim CA, Van Langendonckt A, David A, Dolmans MM, Donnez J. Survival of human pre-antral follicles after cryopreservation of ovarian tissue, follicular isolation and in vitro culture in a calcium alginate matrix. Hum Reprod. 2009;24:92–99. doi: 10.1093/humrep/den343. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, Darot JF, Ritchie ME, Lynch AG, Tavare S. A re-annotation pipeline for Illumina BeadArrays: Improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik-Ozenci C, Akkoyunlu G, Kayisli UA, Arici A, Demir R. Localization of vascular endothelial growth factor in the zona pellucida of developing ovarian follicles in the rat: A possible role in destiny of follicles. Histochem Cell Biol. 2003;120:383–390. doi: 10.1007/s00418-003-0586-4. [DOI] [PubMed] [Google Scholar]
- Cortvrindt R, Smitz J, Van Steirteghem AC. In-vitro maturation, fertilization and embryo development of immature oocytes from early preantral follicles from prepuberal mice in a simplified culture system. Hum Reprod. 1996;11:2656–2666. doi: 10.1093/oxfordjournals.humrep.a019188. [DOI] [PubMed] [Google Scholar]
- DiCesare PE, Morgelin M, Carlson CS, Pasumarti S, Paulsson M. Cartilage oligomeric matrix protein: Isolation and characterization from human articular cartilage. J Orthop Res. 1995;13:422–428. doi: 10.1002/jor.1100130316. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dunning KR, Akison LK, Russell DL, Norman RJ, Robker RL. Increased beta-oxidation and improved oocyte developmental competence in response to l-carnitine during ovarian in vitro follicle development in mice. Biol Reprod. 2011;85:548–555. doi: 10.1095/biolreprod.110.090415. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, O'Brien MJ. Development in vitro of mouse oocytes from primordial follicles. Biol Reprod. 1996;54:197–207. doi: 10.1095/biolreprod54.1.197. [DOI] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): Comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein-Almog R, Orly J. Inhibition of hormone-induced steroidogenesis during cell proliferation in serum-free cultures of rat granulosa cells. Endocrinology. 1985;116:2103–2112. doi: 10.1210/endo-116-5-2103. [DOI] [PubMed] [Google Scholar]
- Gagarina V, Carlberg AL, Pereira-Mouries L, Hall DJ. Cartilage oligomeric matrix protein protects cells against death by elevating members of the IAP family of survival proteins. J Biol Chem. 2008;283:648–659. doi: 10.1074/jbc.M704035200. [DOI] [PubMed] [Google Scholar]
- Greenaway J, Connor K, Pedersen HG, Coomber BL, LaMarre J, Petrik J. Vascular endothelial growth factor and its receptor, Flk-1/KDR, are cytoprotective in the extravascular compartment of the ovarian follicle. Endocrinology. 2004;145:2896–2905. doi: 10.1210/en.2003-1620. [DOI] [PubMed] [Google Scholar]
- Gutierrez CG, Ralph JH, Telfer EE, Wilmut I, Webb R. Growth and antrum formation of bovine preantral follicles in long-term culture in vitro. Biol Reprod. 2000;62:1322–1328. doi: 10.1095/biolreprod62.5.1322. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Makitie O, Hayes E, Haynes R, Susic M, Montufar-Solis D, Duke PJ, Cole WG. Chondrocyte cell death and intracellular distribution of COMP and type IX collagen in the pseudoachondroplasia growth plate. J Orthop Res. 2004;22:759–767. doi: 10.1016/j.orthres.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Hirshfeld-Cytron J, Gracia C, Woodruff TK. Nonmalignant diseases and treatments associated with primary ovarian failure: An expanded role for fertility preservation. J Womens Health (Larchmt) 2011;20:1467–1477. doi: 10.1089/jwh.2010.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatta O, Silye R, Abir R, Krausz T, Winston RM. Extracellular matrix improves survival of both stored and fresh human primordial and primary ovarian follicles in long-term culture. Hum Reprod. 1997;12:1032–1036. doi: 10.1093/humrep/12.5.1032. [DOI] [PubMed] [Google Scholar]
- Jeruss JS, Woodruff TK. Preservation of fertility in patients with cancer. N Engl J Med. 2009;360:902–911. doi: 10.1056/NEJMra0801454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreeger PK, Deck JW, Woodruff TK, Shea LD. The in vitro regulation of ovarian follicle development using alginateextracellular matrix gels. Biomaterials. 2006;27:714–723. doi: 10.1016/j.biomaterials.2005.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA. Model-based variancestabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Morgelin M, Heinegard D, Engel J, Paulsson M. Electron microscopy of native cartilage oligomeric matrix protein purified from the Swarm rat chondrosarcoma reveals a five-armed structure. J Biol Chem. 1992;267:6137–6141. [PubMed] [Google Scholar]
- Motaung SC, DiCesare PE, Reddi AH. Differential response of cartilage oligomeric matrix protein (COMP) to morphogens of bone morphogenetic protein/transforming growth factor-beta family in the surface, middle and deep zones of articular cartilage. J Tissue Eng Regen Med. 2011;5:e87–e96. doi: 10.1002/term.358. [DOI] [PubMed] [Google Scholar]
- Nagaraja AK, Middlebrook BS, Rajanahally S, Myers M, Li Q, Matzuk MM, Pangas SA. Defective gonadotropin-dependent ovarian folliculogenesis and granulosa cell gene expression in inhibin-deficient mice. Endocrinology. 2010;151:4994–5006. doi: 10.1210/en.2010-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JF, Felicio LS, Randall PK, Sims C, Finch CE. A longitudinal study of estrous cyclicity in aging C57BL/6J mice: I. Cycle frequency, length and vaginal cytology. Biol Reprod. 1982;27:327–339. doi: 10.1095/biolreprod27.2.327. [DOI] [PubMed] [Google Scholar]
- Newton H, Picton H, Gosden RG. In vitro growth of oocytegranulosa cell complexes isolated from cryopreserved ovine tissue. J Reprod Fertil. 1999;115:141–150. doi: 10.1530/jrf.0.1150141. [DOI] [PubMed] [Google Scholar]
- O'Brien MJ, Pendola JK, Eppig JJ. A revised protocol for in vitro development of mouse oocytes from primordial follicles dramatically improves their developmental competence. Biol Reprod. 2003;68:1682–1686. doi: 10.1095/biolreprod.102.013029. [DOI] [PubMed] [Google Scholar]
- Pan H, O'Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Saudye H, Shea LD, Woodruff TK. Novel approach for the three-dimensional culture of granulosa celloocyte complexes. Tissue Eng. 2003;9:1013–1021. doi: 10.1089/107632703322495655. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Jorgez CJ, Matzuk MM. Growth differentiation factor 9 regulates expression of the bone morphogenetic protein antagonist gremlin. J Biol Chem. 2004;279:32281–32286. doi: 10.1074/jbc.M403212200. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Li X, Umans L, Zwijsen A, Huylebroeck D, Gutierrez C, Wang D, Martin JF, Jamin SP, Behringer RR, Robertson EJ, Matzuk MM. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol. 2008;28:248–257. doi: 10.1128/MCB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton HM, Danfour MA, Harris SE, Chambers EL, Huntriss J. Growth and maturation of oocytes in vitro. Reprod Suppl. 2003;61:445–462. [PubMed] [Google Scholar]
- Rodriguez-Wallberg KA, Oktay K. Options on fertility preservation in female cancer patients. Cancer Treat Rev. 2012;38:354–361. doi: 10.1016/j.ctrv.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Roy SK, Treacy BJ. Isolation and long-term culture of human preantral follicles. Fertil Steril. 1993;59:783–790. [PubMed] [Google Scholar]
- Scott JE, Carlsson IB, Bavister BD, Hovatta O. Human ovarian tissue cultures: Extracellular matrix composition, coating density and tissue dimensions. Reprod Biomed Online. 2004;9:287–293. doi: 10.1016/s1472-6483(10)62143-8. [DOI] [PubMed] [Google Scholar]
- Shen Z, Heinegard D, Sommarin Y. Distribution and expression of cartilage oligomeric matrix protein and bone sialoprotein show marked changes during rat femoral head development. Matrix Biol. 1995;14:773–781. doi: 10.1016/s0945-053x(05)80020-4. [DOI] [PubMed] [Google Scholar]
- Smitz J, Cortvrindt R. Oocyte in-vitro maturation and follicle culture: Current clinical achievements and future directions. Hum Reprod. 1999;14:145–161. doi: 10.1093/humrep/14.suppl_1.145. [DOI] [PubMed] [Google Scholar]
- Smitz J, Dolmans MM, Donnez J, Fortune JE, Hovatta O, Jewgenow K, Picton HM, Plancha C, Shea LD, Stouffer RL, Telfer EE, Woodruff TK, Zelinski MB. Current achievements and future research directions in ovarian tissue culture, in vitro follicle development and transplantation: Implications for fertility preservation. Hum Reprod Update. 2010;16:395–414. doi: 10.1093/humupd/dmp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Songsasen N, Woodruff TK, Wildt DE. In vitro growth and steroidogenesis of dog follicles are influenced by the physical and hormonal microenvironment. Reproduction. 2011;142:113–122. doi: 10.1530/REP-10-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spears N, Boland NI, Murray AA, Gosden RG. Mouse oocytes derived from in vitro grown primary ovarian follicles are fertile. Hum Reprod. 1994;9:527–532. doi: 10.1093/oxfordjournals.humrep.a138539. [DOI] [PubMed] [Google Scholar]
- Svensson L, Aszodi A, Heinegard D, Hunziker EB, Reinholt FP, Fassler R, Oldberg A. Cartilage oligomeric matrix protein deficient mice have normal skeletal development. Mol Cell Biol. 2002;22:4366–4371. doi: 10.1128/MCB.22.12.4366-4371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai YC, Speed TP. A multivariate empirical Bayes statistic for replicated microarray time course data. Ann Stat. 2006;34:2387–2412. [Google Scholar]
- Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Telfer EE, Binnie JP, McCaffery FH, Campbell BK. In vitro development of oocytes from porcine and bovine primary follicles. Mol Cell Endocrinol. 2000;163:117–123. doi: 10.1016/s0303-7207(00)00216-1. [DOI] [PubMed] [Google Scholar]
- Telfer EE, McLaughlin M, Ding C, Thong KJ. A two-step serum-free culture system supports development of human oocytes from primordial follicles in the presence of activin. Hum Reprod. 2008;23:1151–1158. doi: 10.1093/humrep/den070. [DOI] [PubMed] [Google Scholar]
- Thomas FH, Campbell BK, Armstrong DG, Telfer EE. Effects of IGF-I bioavailability on bovine preantral follicular development in vitro. Reproduction. 2007;133:1121–1128. doi: 10.1530/REP-06-0382. [DOI] [PubMed] [Google Scholar]
- von Wolff M, Montag M, Dittrich R, Denschlag D, Nawroth F, Lawrenz B. Fertility preservation in women—A practical guide to preservation techniques and therapeutic strategies in breast cancer, Hodgkin's lymphoma and borderline ovarian tumours by the fertility preservation network FertiPROTEKT. Arch Gynecol Obstet. 2011;284:427–435. doi: 10.1007/s00404-011-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ED, Zarrow MX. Comparison of superovulation in the immature mouse and rat. J Reprod Fertil. 1962;3:148–158. doi: 10.1530/jrf.0.0030148. [DOI] [PubMed] [Google Scholar]
- Woodruff TK, D'Agostino J, Schwartz NB, Mayo KE. Dynamic changes in inhibin messenger RNAs in rat ovarian follicles during the reproductive cycle. Science. 1988;239:1296–1299. doi: 10.1126/science.3125611. [DOI] [PubMed] [Google Scholar]
- Wright CS, Hovatta O, Margara R, Trew G, Winston RM, Franks S, Hardy K. Effects of follicle-stimulating hormone and serum substitution on the in-vitro growth of human ovarian follicles. Hum Reprod. 1999;14:1555–1562. doi: 10.1093/humrep/14.6.1555. [DOI] [PubMed] [Google Scholar]
- Wu J, Emery BR, Carrell DT. In vitro growth, maturation, fertilization, and embryonic development of oocytes from porcine preantral follicles. Biol Reprod. 2001;64:375–381. doi: 10.1095/biolreprod64.1.375. [DOI] [PubMed] [Google Scholar]
- Xu M, Kreeger PK, Shea LD, Woodruff TK. Tissueengineered follicles produce live, fertile offspring. Tissue Eng. 2006a;12:2739–2746. doi: 10.1089/ten.2006.12.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, West E, Shea LD, Woodruff TK. Identification of a stage-specific permissive in vitro culture environment for follicle growth and oocyte development. Biol Reprod. 2006b;75:916–923. doi: 10.1095/biolreprod.106.054833. [DOI] [PubMed] [Google Scholar]
- Xu M, Barrett SL, West-Farrell E, Kondapalli LA, Kiesewetter SE, Shea LD, Woodruff TK. In vitro grown human ovarian follicles from cancer patients support oocyte growth. Hum Reprod. 2009a;24:2531–2540. doi: 10.1093/humrep/dep228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, West-Farrell ER, Stouffer RL, Shea LD, Woodruff TK, Zelinski MB. Encapsulated three-dimensional culture supports development of nonhuman primate secondary follicles. Biol Reprod. 2009b;81:587–594. doi: 10.1095/biolreprod.108.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bernuci MP, Lawson MS, Yeoman RR, Fisher TE, Zelinski MB, Stouffer RL. Survival, growth, and maturation of secondary follicles from prepubertal, young, and older adult rhesus monkeys during encapsulated three-dimensional culture: Effects of gonadotropins and insulin. Reproduction. 2010;140:685–697. doi: 10.1530/REP-10-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lawson MS, Yeoman RR, Pau KY, Barrett SL, Zelinski MB, Stouffer RL. Secondary follicle growth and oocyte maturation during encapsulated three-dimensional culture in rhesus monkeys: Effects of gonadotrophins, oxygen and fetuin. Hum Reprod. 2011a;26:1061–1072. doi: 10.1093/humrep/der049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Fazleabas AT, Shikanov A, Jackson E, Barrett SL, Hirshfeld-Cytron J, Kiesewetter SE, Shea LD, Woodruff TK. Invitro oocyte maturation and preantral follicle culture from the lutealphase baboon ovary produce mature oocytes. Biol Reprod. 2011b;84:689–697. doi: 10.1095/biolreprod.110.088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.