Abstract
BACKGROUND
The molecular causes of many hematologic cancers remain unclear. Among these cancers are chronic neutrophilic leukemia (CNL) and atypical (BCR-ABL1–negative) chronic myeloid leukemia (CML), both of which are diagnosed on the basis of neoplastic expansion of granulocytic cells and exclusion of genetic drivers that are known to occur in other myeloproliferative neoplasms and myeloproliferative– myelodysplastic overlap neoplasms.
METHODS
To identify potential genetic drivers in these disorders, we used an integrated approach of deep sequencing coupled with the screening of primary leukemia cells obtained from patients with CNL or atypical CML against panels of tyrosine kinase–specific small interfering RNAs or small-molecule kinase inhibitors. We validated candidate oncogenes using in vitro transformation assays, and drug sensitivities were validated with the use of assays of primary-cell colonies.
RESULTS
We identified activating mutations in the gene encoding the receptor for colonystimulating factor 3 (CSF3R) in 16 of 27 patients (59%) with CNL or atypical CML. These mutations segregate within two distinct regions of CSF3R and lead to preferential downstream kinase signaling through SRC family–TNK2 or JAK kinases and differential sensitivity to kinase inhibitors. A patient with CNL carrying a JAK-activating CSF3R mutation had marked clinical improvement after the administration of the JAK1/2 inhibitor ruxolitinib.
CONCLUSIONS
Mutations in CSF3R are common in patients with CNL or atypical CML and represent a potentially useful criterion for diagnosing these neoplasms. (Funded by the Leukemia and Lymphoma Society and others.)
Therapy with small-molecule kinase inhibitors has improved the outcomes in patients who have certain types of cancer with kinase-pathway dependence caused by defined genetic abnormalities.1,2 Extrapolation of this model to other cancers requires knowledge of operationally important genetic mutations that result in corresponding activation of kinase pathways. Despite advances in our understanding of the molecular pathobiology of certain types of hematologic cancers, many of these disorders are still diagnosed on the basis of neoplastic cell type and additional exclusionary criteria.
Chronic neutrophilic leukemia (CNL) and atypical chronic myeloid leukemia (CML) are rare hematologic neoplasms that are characterized by leukocytosis and hypercellularity of bone marrow consisting predominantly of granulocytic cells, the absence of the Philadelphia chromosome with translocation t(9;22) (BCR-ABL1), and the absence of rearrangements in genes encoding plateletderived growth factor receptors alpha and beta (PDGFRA/B) and fibroblast growth factor receptor 1 (FGFR1). CNL is diagnosed on the basis of the expansion of neutrophils in both bone marrow and blood (segmented neutrophils and band forms, >80% of white cells) and is classified as a myeloproliferative neoplasm, according to World Health Organization (WHO) diagnostic criteria. (A histopathological sample from a patient with CNL is provided in Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org.)
Atypical CML is characterized by granulocytic dysplasia and an increased number of neutrophil precursors in both the peripheral blood and the bone marrow (typically, ≥10% of white cells) and is therefore classified as one subtype of the WHO category of myelodysplastic–myeloproliferative neoplasms.3,4 Some patients with CNL 5,6 and most patients with atypical CML have nonspecific cytogenetic abnormalities 7 or (infrequently) the JAK2 V617F mutation,8,9 findings that reveal the clonal nature of these diseases. The genetic basis for both CNL and atypical CML remains unknown, although certain subtypes of myeloproliferative neoplasms have been operationally defined according to the molecular abnormalities (e.g., BCR-ABL1 in CML) or are characterized by a high frequency of specific genetic abnormalities (e.g., JAK2 V617F in polycythemia vera, essential thrombocythemia, and primary myelofibrosis 8,10–13 and KIT D816V in systemic mastocytosis 14,15).
CSF3R is the receptor for colony-stimulating factor 3 and is thought to play a prominent role in the growth and differentiation of granulocytes.16,17 CSF3R mutations have been described in patients with severe congenital neutropenia, which can evolve into acute myeloid leukemia (AML).18–20 It was recently reported that in a patient with congenital neutropenia, a secondary CSF3R mutation developed at the time of transformation to AML.21 These nonsense or frameshift mutations, which have been described previously, truncate the cytoplasmic tail of CSF3R, impair its internalization, and alter its interactions with proteins such as SHP-1/2 and SOCS family members. 22–24 These structural and functional alterations are thought to perturb the capacity of CSF3R to regulate granulocyte differentiation and to increase granulocytic proliferative capacity. 25–27 CSF3R signals through the JAK– STAT pathway, the nonreceptor tyrosine kinase SYK, 28,29 and the SRC family kinase LYN. CSF3R signaling through LYN was recently shown to be mediated by the phosphatase SHP-2 and the adapter protein GAB2.28–31 With the exception of isolated case reports, 32 mutations in CSF3R have not been reported in patients with cases of de novo leukemia.
METHODS
STUDY DESIGN
All clinical samples were obtained after written and oral informed consent was provided by the patients. The study was approved by the institutional review boards at the University of Texas Southwestern Medical Center, University of Colorado, Stanford University, Washington University in St. Louis, or Oregon Health and Science University (OHSU). All studies in mice were performed according to a protocol approved by an OHSU committee on institutional animal care and use. No commercial support was provided for this study. Ruxolitinib was obtained through health care insurance for treatment of the index patient. An expanded description of the methods is provided in the Supplementary Appendix.
DEEP SEQUENCING AND SCREENING OF PRIMARY CELLS
We hypothesized that patients with CNL or atypical CML may harbor oncogenes that would lead to sensitivity to small-molecule kinase inhibitors. To test this hypothesis, we used a functional-genomics approach in evaluating primary cells from 27 patients with CNL or atypical CML, as well as specimens from patients with a variety of other hematologic cancers. We performed deep sequencing with coverage of coding regions of 1862 genes representing all kinases, phosphatases, non-kinase growth factor or cytokine receptors, and selected adapter genes (Tables S1 and S2 in the Supplementary Appendix). Wherever possible, we also screened these primary leukemia cells against panels of tyrosine kinase– specific small interfering RNAs (siRNAs)33,34 or small-molecule kinase inhibitors. 35 We previously validated this approach on specimens with proofof-principle molecular lesions (e.g., BCR-ABL1, FLT3-ITD, JAK2 V617F, and KRAS G13D), and we also used this strategy to identify previously unknown molecular targets in leukemia specimens (e.g., two base-pair “GG” insertions at position 1886 of the myeloproliferative leukemia virus oncogene [MPL1886InsGG] and ROR1). 33–35
RESULTS
FREQUENCY OF CSF3R MUTATIONS
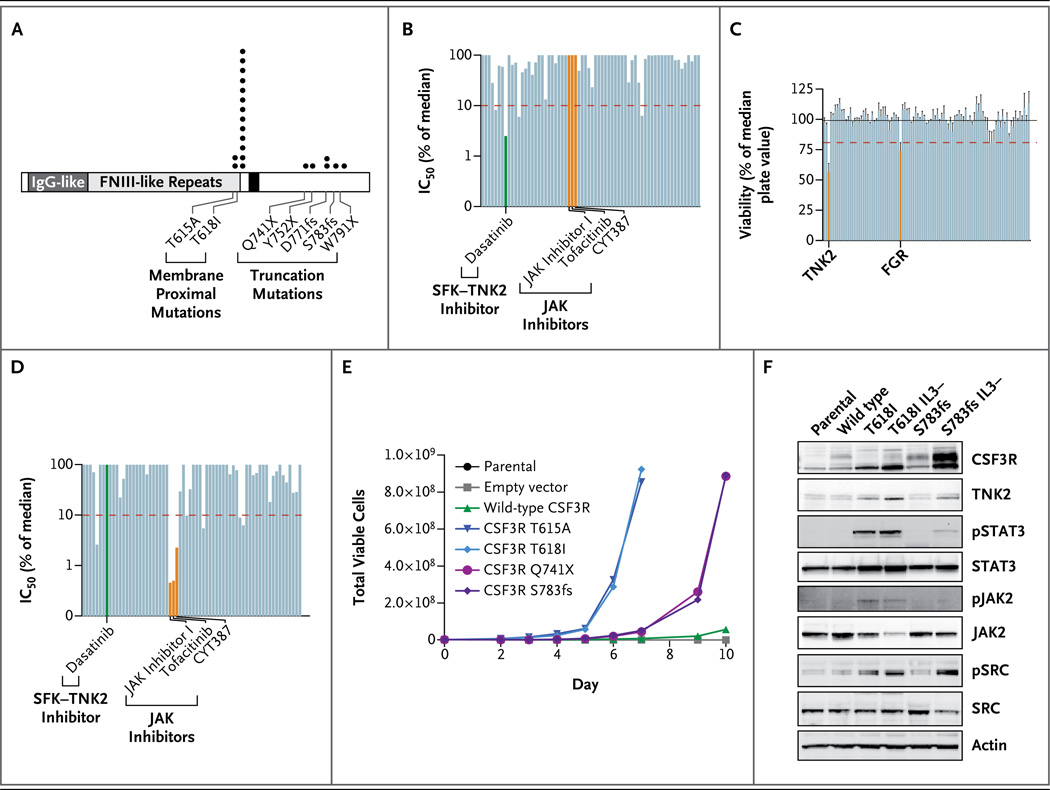
We found enrichment of mutations in CSF3R in 16 of 27 patients (59%) with CNL or atypical CML (Table 1 and Fig. 1A, and Table S3 in the Supplementary Appendix). Sequence variants that were identified included membrane proximal mutations (T615A and T618I) and a number of different frameshift or nonsense mutations that truncate the cytoplasmic tail of CSF3R (D771fs, S783fs, Y752X, and W791X). Similar mutations that truncate the CSF3R cytoplasmic domain have been described in patients with congenital neutropenia that progresses to AML after long-term treatment with granulocyte colony-stimulating factor (G-CSF). 18–20 Representative deep-sequencing data and validation on Sanger sequencing for patients with mutant CSF3R are shown in Figures S2 and S3 in the Supplementary Appendix. Five patients (Patients 3 through 7) had both membrane proximal and truncation mutations (Table S3 in the Supplementary Appendix), and we confirmed that these compound mutations can occur on the same CSF3R allele with no requisite order for sequential acquisition of mutations (Table S4 in the Supplementary Appendix).
Table 1.
Summary of CSF3R Mutational Status in the Study Samples, According to the Type of Hematologic Cancer.*
| Diagnosis |
CSF3R Mutation |
Estimate of Variant Frequency |
|---|---|---|
|
no. of samples/ total no. |
% | |
| Chronic neutrophilic leukemia or atypical chronic myeloid leukemia |
16/27 | 59 |
| Acute myeloid leukemia | 3/292 | 1 |
| T-cell acute lymphoblastic leukemia | 0/8 | 0 |
| Early T-cell precursor T-cell acute lymphoblastic leukemia |
1/3 | NA |
| B-cell acute lymphoblastic leukemia | 0/41 | 0 |
Data are based on deep sequencing and Sanger-sequencing validation of samples obtained from 27 patients with chronic neutrophilic leukemia or atypical chronic myeloid leukemia and from patients with the other listed hematologic cancers. NA denotes not available because of the small number of samples.
Figure 1. Sensitivity to Kinase Inhibition in Leukemia Specimens with Transforming Mutations in CSF3R.
Panel A shows the location and recurrence of CSF3R mutations found in samples from 16 of 27 patients with chronic neutrophilic leukemia (CNL) or atypical chronic myeloid leukemia (CML), along with samples from patients with other types of leukemia. The mutation locations and number of observations are indicated by black circles. The Q741X mutation was found in a sample obtained from a patient with acute myeloid leukemia (AML), and one of the T618I mutations was found in a sample from a patient with early T-cell precursor T-cell acute lymphoblastic leukemia (ETP-T-ALL). Five patients with CNL or atypical CML had both membrane proximal and truncation mutations. (For details, see Table S3 in the Supplementary Appendix.) Two additional CSF3R mutations (Q739X and T618I, which are not shown) have been reported in AML specimens sequenced by the Cancer Genome Atlas.21 CSF3R coordinates are numbered according to the conventions of the Ensembl genome browser, a numbering system that differs from historical CSF3R numbering owing to the inclusion of the 23-amino-acid signal peptide, despite the absence of this signal peptide from the mature protein. Panel B shows the sensitivity of white cells from Patient 3, who had CNL and a CSF3R S783fs mutation (Table S3 in the Supplementary Appendix), to a panel of 66 small-molecule kinase inhibitors. The 50% inhibitory concentration (IC50) of each drug is plotted as a percentage of the median IC 50 for each drug from 150 samples obtained from patients with leukemia.35 A specimen was considered to be hypersensitive to an inhibitor if the IC50 was less than 10% of the median IC50 for that inhibitor for the entire cohort (as indicated by the dashed red line). This specimen was hypersensitive to dasatinib (green) and insensitive to JAK kinase inhibitors (orange). SFK denotes SRC-family kinase, and TNK2 tyrosine kinase nonreceptor 2. Panel C shows the sensitivity of white cells from Patient 3 to small interfering RNAs (siRNAs) directed against all known tyrosine kinases, as described previously.33,34 Silencing of TNK2 and an SRC family kinase, FGR, resulted in a substantial decrease in cell viability. All cell-viability values after silencing with each individual siRNA have been normalized to the median value of the entire panel. The bars on the graph represent the mean normalized cell viability from triplicate data points for each individual siRNA. The T bars represent standard errors. The black horizontal line indicates the mean of all values across the entire siRNA panel, and the red dashed line indicates a threshold of significance, which is calculated as the mean minus 2 SD for all values. In addition to carrying the CSF3R S783fs mutation, Patient 3 had a minority of clones with a CSF3R S783fs–T615A compound mutation, but this small percentage of cells did not have an effect on sensitivity to inhibitors in short-term assays. Panel D shows the sensitivity of white cells from a patient with ETP-T-ALL and a CSF3R T618I mutation to the same panel of 66 small-molecule kinase inhibitors that was used to test cells from Patient 3, as described in Panel B. These cells were insensitive to dasatinib (green) and sensitive to JAK kinase inhibitors (orange). Panel E shows interleukin-3–dependent Ba/F3 cells that were infected with murine retrovirus expressing wild-type CSF3R, membrane proximal mutations, or truncation mutations. Uninfected parental Ba/ F3 cells and empty-vector infected Ba/F3 cells were used as controls. Over a 10-day period, both classes of CSF3R mutations were capable of transforming Ba/F3 cells to interleukin-3–independent growth, and the membrane proximal mutations (T615A and T618I) transformed cells in this assay substantially faster than the truncation mutants (Q741X and S783fs). Panel F shows Ba/F3 cells expressing CSF3R T618I or S783fs mutations before or after interleukin-3– independent transformation (IL3- indicates transformed cells). Cell lysates were subjected to immunoblot analysis for CSF3R, TNK2, phospho-STAT3 (pSTAT3), total STAT3, phospho-JAK2 (pJAK2), total JAK2, phospho-SRC (pSRC), total SRC, and actin. Parental Ba/F3 cells or Ba/F3 cells expressing wild-type CSF3R were included as controls.
We identified a CSF3R mutation in 1 of 92 patients with AML, and 2 of 200 patients with AML in the Cancer Genome Atlas AML data set had a CSF3R mutation, 21 indicating that the incidence of such mutations in AML is low (1%) (Table 1). We identified a CSF3R membrane proximal mutation (T618I) in 1 of 3 patients with early T-cell precursor T-cell acute lymphoblastic leukemia (ETP-T-ALL) (Table 1, and Fig. S4 in the Supplementary Appendix). We found no additional CSF3R mutations in 8 patients with T-cell ALL or 41 patients with B-cell ALL (Table 1). Finally, we sequenced samples from 3 patients with reactive neutrophilia, and none had CSF3R mutations. Taken together, these data suggest that mutations in CSF3R are a defining molecular abnormality of CNL and atypical CML, and testing for CSF3R mutations could aid in the diagnosis of these diseases.
DEPENDENCE ON SRC FAMILY–TNK2 OR JAK KINASES
We next sought to determine whether specimens harboring mutant CSF3R show in vitro sensitivity to small-molecule inhibitors of kinases or siRNA directed against kinases that become dysregulated downstream of mutant CSF3R. Analysis of cells from Patient 3, who had CNL with the CSF3R S783fs mutation (Table S3 and Fig. S2 in the Supplementary Appendix), revealed dramatic sensitivity to the multikinase inhibitor dasatinib (Sprycel, Bristol-Myers Squibb) but no sensitivity to inhibitors of JAK family kinases (Fig. 1B). Further interrogation with our panel of tyrosine kinase– specific siRNAs revealed sensitivity to silencing of tyrosine kinase nonreceptor 2 (TNK2) and an SRC family kinase, FGR, both of which are potently inhibited by dasatinib 36 (Fig. 1C). We also performed drug-sensitivity profiling on samples from two patients with the CSF3R T618I mutation (one with CNL and one with ETP-T-ALL). In contrast to the drug-sensitivity pattern in the patients with truncation mutations, both samples showed sensitivity to inhibitors that target JAK family kinases (including ruxolitinib [Jakafi, Incyte]) but resistance to dasatinib (Fig. 1D, and Fig. S3D and S3E in the Supplementary Appendix). Taken together, the functional genomic data on the samples from these three patients suggest that there are two different classes of CSF3R mutations: truncation mutations, which result in dysregulation of SRC family–TNK2 kinases, and membrane proximal mutations, which result in dysregulation of JAK family kinases. The data also suggest that truncation mutations confer sensitivity to dasatinib but not to JAK kinase inhibitors, where-as the reverse is true for membrane proximal mutant cells.
DISTINCT SIGNALING-PATHWAY DYSREGULATION
To test the relative transforming capacity of truncation mutations, as compared with membrane proximal mutations, in CSF3R, we performed an assay to measure cytokine-independent growth using the interleukin-3–dependent Ba/F3 cell line (Fig. 1E). Both classes of CSF3R mutations were capable of inducing transformation of Ba/F3 cells to interleukin-3–independent growth, and the membrane proximal mutations (T615A and T618I) transformed cells in this assay substantially faster than did the truncation mutations (Q741X and S783fs).
Once we confirmed the transformation capacity of the CSF3R mutations, we investigated the differential signaling and drug sensitivity suggested by our functional screening of samples of CSF3R mutant leukemia. Ba/F3 cells expressing the T618I or S783fs mutation before or after interleukin-3–independent transformation, along with cells expressing wild-type CSF3R or parental control cells, were starved of the interleukin-3 growth factor, and cell lysates were analyzed by means of immunoblotting. The cells with the S783fs mutation showed higher expression of CSF3R than did the cells with the wild-type allele (Fig. 1F), and this difference was magnified after long-term culture in the absence of interleukin-3, a finding consistent with results of previous studies showing disruption of receptor internalization in the context of truncation mutations.22–24,37 After withdrawal of interleukin-3, Ba/F3 cells with CSF3R mutations expressed high levels of endogenous TNK2 and increased phosphorylation of SRC family kinases, providing validation of the initial TNK2 and FGR siRNA sensitivities observed in samples obtained from a patient with a CSF3R truncation mutation (Fig. 1C) and suggesting that TNK2 is a previously unrecognized downstream mediator of CSF3R signaling.
To further investigate the potential signaling differences between the two classes of CSF3R mutations, we performed immunoblot analysis for JAK–STAT phosphorylation. The T618I mutant induced high levels of STAT3 and JAK2 phosphorylation, in sharp contrast to the lower levels induced by the S783fs mutant (Fig. 1F). Taken together, these data indicate that the two classes of CSF3R mutations have different transformation potential and downstream signaling consequences.
USE OF TYROSINE KINASE INHIBITORS
To further test the sensitivities of CSF3R truncation and membrane proximal mutations to inhibitors of SRC family–TNK2 or JAK kinases, we transduced bone marrow cells from mice with CSF3R S783fs, CSF3R T618I, or an empty vector control and plated them in a colony-formation assay. The empty-vector control cells, which expressed endogenous levels of wild-type CSF3R, required 10 ng of exogenous G-CSF per milliliter to form colonies; in contrast, CSF3R S783fs mutant cells required 0.4 ng of G-CSF per milliliter to elicit colony formation, and the T618I mutant grew in the absence of any added G-CSF.
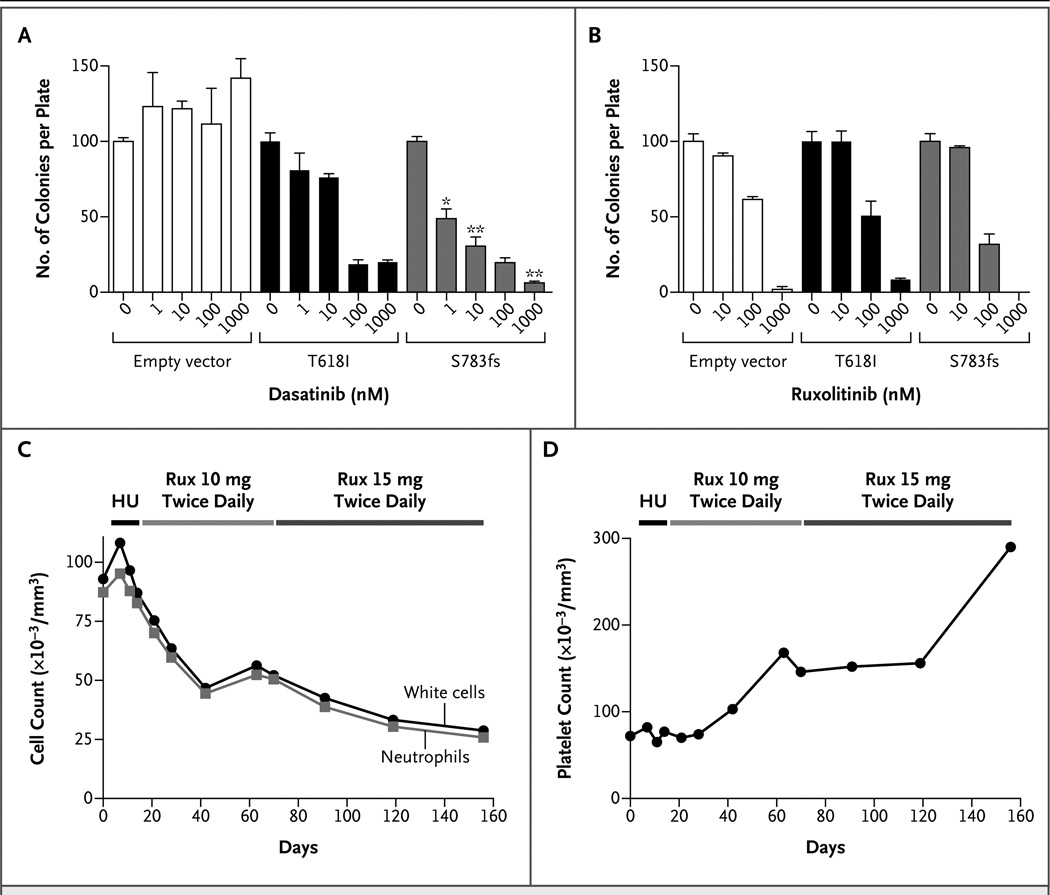
Treatment with dasatinib had a dramatic effect on S783fs-driven colony formation, with a 50% inhibitory concentration (IC50) of approximately 1 nM (Fig. 2A). The T618I mutant was relatively insensitive to dasatinib (IC50, approximately 100 nM), an observation that is consistent with the results for primary cells from patients, and the empty-vector control cells were completely insensitive to dasatinib. All cells showed similar sensitivity to the JAK kinase inhibitor ruxolitinib, with an IC50 of approximately 100 nM, which is equivalent to the sensitivity of cells with a defined JAK dependency 38 (Fig. 2B).
Figure 2. Use of Tyrosine Kinase Inhibitors to Treat Dysregulated Signaling Induced by CSF3R Mutations.
Panel A shows the effect of dasatinib on colony formation in bone marrow cells from mice that were infected with mutant CSF3R-containing retroviruses or an empty vector; the control cells expressed endogenous wild-type CSF3R. Cells were grown in methylcellulose containing the minimal amount of granulocyte colony-stimulating factor (G-CSF) necessary to form colonies (10 ng per milliliter for the empty vector, 0.4 ng per milliliter for the S783fs mutation, and no G-CSF for the T618I mutation). Cells were plated with increasing concentrations of dasatinib (0, 1, 10, 100, and 1000 nM). The experiment was performed in triplicate with the number of colonies normalized to those in the untreated controls. Values represent the mean percent colonies; the T bars indicate standard errors. A single asterisk indicates P<0.07, and a double asterisk indicates P<0.005 for the comparison between the T618I mutation and the S783fs mutation at equivalent doses of dasatinib. Panel B shows the results of a similar colony-formation assay, in which the cells were plated with ruxolitinib (0, 10, 100, or 1000 nM). Panel C shows the results for Patient 9, who had CNL and a CSF3R T618I mutation and in whom earlier testing indicated sensitivity to ruxolitinib (Rux) in vitro (Fig. S3C and S3E in the Supplementary Appendix). This patient was treated with 500 mg of hydroxyurea (HU) daily starting on day 13. Hydroxyurea was stopped on day 21 and oral ruxolitinib (at a dose of 10 mg twice daily) was administered. On day 70, the dose of ruxolitinib was increased to 15 mg twice daily. The numbers of white cells and neutrophils (absolute neutrophil count) are shown. Panel D shows normalized platelet counts while Patient 9 was undergoing the treatment regimen shown in Panel C.
The ruxolitinib sensitivity of the empty-vector and S783fs-mutant cells must be understood in the context of the requirement of exogenous G-CSF for the stimulation of colony growth, in which the exogenous G-CSF preferentially stimulates JAK–STAT signaling. Primary cells from patients with a CSF3R truncation mutation showed sensitivity to dasatinib but not to JAK kinase inhibition when cultured in the absence of exogenous G-CSF (Fig. 1B and 1C). In contrast, colony formation of the T618I cells showed sensitivity to ruxolitinib despite the fact that exogenous G-CSF was not required for colony outgrowth, a finding consistent with the sensitivity of primary cells from patients with CSF3R membrane proximal mutations to JAK kinase inhibition (Fig. 1D). Taken together, these data show that CSF3R truncation mutants studied in vitro are sensitive to SRC family–TNK2 inhibitors, and membrane proximal mutants are sensitive to JAK kinase inhibitors.
CLINICAL EFFICACY OF RUXOLITINIB IN A PATIENT WITH CSF3R T618I
Primary cells from Patient 9, who had CNL with a CSF3R T618I mutation (Table S3 and Fig. S3C in the Supplementary Appendix), showed in vitro hypersensitivity to ruxolitinib (IC50, 127 nM) (Fig. S3E in the Supplementary Appendix). Treatment of this patient with oral ruxolitinib (at a dose of 10 mg twice daily) resulted in a marked decrease in the total number of white cells and the absolute neutrophil count (Fig. 2C). Increasing the dose of ruxolitinib to 15 mg twice daily led to a further decrease in both the white-cell count and the absolute neutrophil count. This treatment also resulted in normalization of the platelet count (Fig. 2D).
DISCUSSION
Rapid improvements in sequencing technology have resulted in a wealth of cancer-genome data, but understanding which genomic aberrations can be targeted as sites for potential treatment remains challenging. By integrating functional and genomic analyses of primary leukemia specimens, we identified CSF3R mutations as drivers of leukemia and also identified tyrosine kinase inhibitors that effectively target downstream CSF3R-signaling pathways. We found mutations in CSF3R in 59% of patients with CNL or atypical CML — myeloid neoplasms for which no diseasespecific genetic markers have been identified to date. The high frequency of activating mutations in CSF3R in these leukemias, which are characterized by high numbers of neutrophils, is consistent with its function as the receptor for the growth factor that promotes neutrophil differentiation and proliferation.16,17
The CSF3R mutations represent a biologically unifying feature of CNL and atypical CML and define a new molecular subset of hematologic cancers. The incorporation of CSF3R mutational status into current diagnostic criteria for CNL and atypical CML may help refine the molecular classification of myeloproliferative neoplasms and myeloproliferative–myelodysplastic overlap neoplasms. Although CNL and atypical CML are currently defined as separate neoplasms by the WHO, distinguishing between the two diagnoses can sometimes be challenging for clinicians and hematopathologists. The categorization relies partly on arbitrary thresholds for the total white-cell count (e.g., ≥25,000 per cubic millimeter for CNL and ≥13,000 per cubic millimeter for atypical CML), the percentage of total white cells that are immature granulocytes (<10% for CNL and ≥10% for atypical CML), and the presence or absence of dysgranulopoiesis (absent in CNL and characteristic of atypical CML).
Similar to the identification of the JAK2 V617F mutation across a spectrum of related myeloproliferative neoplasms (e.g., polycythemia vera, essential thrombocythemia, and primary myelofibrosis), the phenotype of CSF3R mutation–positive neoplasms may be modified by additional unknown molecular abnormalities or host genetic factors, such as mutations in the gene encoding SET-binding protein 1 (SETBP1). 39 In addition, assessment of CSF3R mutational status may be useful for the evaluation of diseases characterized by neutrophilia in which the clinical basis is not readily apparent.
CSF3R has been shown to signal through downstream SRC family and JAK-kinase pathways, 28,29 and we have identified a novel CSF3R downstream substrate, TNK2. These downstream kinase pathways make CSF3R mutations an attractive marker for tyrosine kinase inhibitors. The two types of CSF3R mutations may have differential susceptibility to classes of tyrosine kinase inhibitors, with CSF3R truncation mutations showing activation of SRC family–TNK2 kinase signaling and sensitivity to dasatinib, and CSF3R membrane proximal mutations showing preferential activation of the JAK signaling pathway (Fig. 3). Our observation that a patient with a membrane proximal mutation had an excellent clinical response to the JAK inhibitor ruxolitinib, resulting in a marked decrease in the numbers of white cells and neutrophils and an increased platelet count (Fig. 2C and 2D), constitutes a proof of concept. Although anecdotal, this observation provides an impetus for further investigation of tyrosine kinase inhibitors for the treatment of patients with neutrophilic leukemia who have CSF3R mutations.
Figure 3. Model for Activation and Signaling of CSF3R Mutations.
Truncation mutations in CSF3R (the receptor for G-CSF) result in increased expression levels. Downstream signaling mediators — SRC family kinases (SFKs) and TNK2 — are preferentially activated by these truncation mutations. Consequently, leukemic cells harboring the mutations are highly sensitive to dasatinib. Truncation mutations in CSF3R may also show sensitivity to JAK kinase inhibitors in the context of JAK kinase stimulation downstream of high ligand concentrations. In contrast, membrane proximal mutations in CSF3R show completely ligand-independent function. In this capacity, the dominant mode of signaling appears to operate through the JAK-STAT pathway. Thus, patients with membrane proximal mutations may be candidates for treatment with JAK kinase inhibitors, such as the JAK1/2 inhibitor ruxolitinib.
Although CSF3R truncation mutations have been shown to lead to constitutive overexpression of the receptor and ligand hypersensitivity, 22–24,37 the mechanism of action of the membrane proximal mutation does not appear to involve similar receptor overexpression, since the membrane proximal mutants do not show analogous overexpression in the Ba/F3 model (Fig. 1F). Our data show that T618I is capable of inducing colony formation in the absence of G-CSF ligand, which suggests constitutive activation of the receptor. Data from a recent study 21 identified the same mutation in a patient with congenital neutropenia and sequential acquisition of CSF3R mutations as the disease evolved toward AML. In our study, several patients with CNL or atypical CML had both truncation and membrane proximal mutations, and the signaling of these compound mutations and their sensitivities to tyrosine kinase inhibition also warrant characterization in future studies.
Complex genetic alterations are common in a multitude of tumor types. CSF3R truncation mutations accelerate tumor formation in the presence of other genetic modifiers but alone are incapable of causing AML.40 Although CSF3R mutations have been reported in patients with congenital neutropenia that progressed to AML, the prevalence of CSF3R mutations in de novo AML is low (approximately 1%).21 It is possible that this low frequency is due to the required contribution from other genetic alterations for transformation to AML.
In conclusion, the presence of CSF3R mutations identified a distinct diagnostic subgroup of more than 50% of patients with CNL or atypical CML in our study. The oncogenic CSF3R mutations are molecular markers of sensitivity to inhibitors of SRC family–TNK2 and JAK kinases and may provide a new avenue for therapy.
Supplementary Material
Acknowledgments
Supported in part by grants from the Leukemia and Lymphoma Society, the Training Program in Molecular Hematology (5T32HL007781–20, to Dr. Maxson), the Charles and Ann Johnson Foundation (to Dr. Gotlib), the National Cancer Institute (1-K99-CA151670–01A1, to Dr. Agarwal), the National Center for Advancing Translational Sciences (5UL1RR024140) and the National Cancer Institute (5P30CA069533–13) (both to Mr. Bottomly and Drs. Wilmot and McWeeney), the National Heart, Lung, and Blood Institute (K08 HL106576, to Dr. Oh), and the St. Baldricks Foundation (to Dr. Chang), and by grants from the V Foundation for Cancer Research, Gabrielle’s Angel Foundation for Cancer Research, the William Lawrence and Blanche Hughes Fund, and the National Cancer Institute (4 R00CA151457–03) (all to Dr. Tyner). Dr. Druker is an investigator of the Howard Hughes Medical Institute, and Dr. Deininger is a Scholar in Clinical Research of the Leukemia and Lymphoma Society.
We thank Bob Searles and the Massively Parallel Sequencing Shared Resource at Oregon Health and Science University (OHSU) for sequence capture and deep sequencing of DNA samples, Dorian LaTocha and the OHSU Flow Cytometry Core for sorting of Ba/F3 cells, and Parveen Abidi and Larry Okumoto of the Stanford Hematology Division Tissue Bank.
APPENDIX
The authors’ affiliations are as follows: the Division of Hematology and Medical Oncology (J.E.M., A.G.F., A.A., C.A.E., C.E.T., B.J.D.), Knight Cancer Institute (J.E.M., A.G.F., A.A., C.A.E., D.B., B.W., S.K.M., C.E.T., B.H.C., M.M.L., B.J.D., J.W.T.), Oregon Clinical and Translational Research Institute (D.B., B.W., S.K.M.), Division of Bioinformatics and Computational Biology (S.K.M.), Division of Pediatric Hematology and Oncology, Department of Pediatrics (B.H.C.), Department of Anatomic Pathology (M.M.L.), and Department of Cell and Developmental Biology (J.W.T.), Oregon Health and Science University, and Howard Hughes Medical Institute (C.A.E., C.E.T., B.J.D.) — both in Portland; Stanford Cancer Institute, Stanford University School of Medicine, Stanford, CA (J.G.); the Division of Hematology, Oncology, and Bone Marrow Transplantation, University of Colorado School of Medicine, Aurora (D.A.P.); Hematology/ Oncology, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas (J.B.P., R.H.C.); St. Luke’s Oncology and Hematology, St. Luke’s Regional Cancer Center, Duluth, MN (B.G.); the Department of Medicine, Hematology Division, Washington University School of Medicine, Washington University in St. Louis, St. Louis (S.T.O.); and Huntsman Cancer Institute, University of Utah, Salt Lake City (M.W.D.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 3.Bain BJ, Brunning RD, Vardiman JW, Thiele J. Chronic neutrophilic leukaemia. In: Swerdlow SH, Campo E, Lee Harris N, et al., editors. WHO classification of tumors of haematopoietic and lymphoid tissues. 4. Lyon, France: IARC Press; 2008. pp. 38–39. [Google Scholar]
- 4.Vardiman JW, Bennett JM, Bain BJ, Brunning RD, Thiele J. Atypical chronic myeloid leukaemia, BCR-ABL1 negative. In: Swerdlow SH, Campo E, Lee Harris N, et al., editors. WHO classification of tumors of haematopoietic and lymphoid tissues. 4. Lyon, France: IARC Press; 2008. pp. 80–81. [Google Scholar]
- 5.Froberg MK, Brunning RD, Dorion P, Litz CE, Torlakovic E. Demonstration of clonality in neutrophils using FISH in a case of chronic neutrophilic leukemia. Leukemia. 1998;12:623–626. doi: 10.1038/sj.leu.2400938. [DOI] [PubMed] [Google Scholar]
- 6.Matano S, Nakamura S, Kobayashi K, Yoshida T, Matsuda T, Sugimoto T. Deletion of the long arm of chromosome 20 in a patient with chronic neutrophilic leukemia: cytogenetic findings in chronic neutrophilic leukemia. Am J Hematol. 1997;54:72–75. doi: 10.1002/(sici)1096-8652(199701)54:1<72::aid-ajh11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Hernández JM, del Cañizo MC, Cuneo A, et al. Clinical, hematological and cytogenetic characteristics of atypical chronic myeloid leukemia. Ann Oncol. 2000;11:441–444. doi: 10.1023/a:1008393002748. [DOI] [PubMed] [Google Scholar]
- 8.Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [Erratum, Lancet 2005;366:122.] [DOI] [PubMed] [Google Scholar]
- 9.Steensma DP, Dewald GW, Lasho TL, et al. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106:1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James C, Ugo V, Le Couédic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 11.Jones AV, Kreil S, Zoi K, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 12.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 13.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 14.Longley BJ, Tyrrell L, Lu SZ, et al. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive masto- cytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet. 1996;12:312–314. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- 15.Nagata H, Worobec AS, Oh CK, et al. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci U S A. 1995;92:10560–10564. doi: 10.1073/pnas.92.23.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beekman R, Touw IP. G-CSF and its receptor in myeloid malignancy. Blood. 2010;115:5131–5136. doi: 10.1182/blood-2010-01-234120. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 18.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating–factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 19.Dong F, Hoefsloot LH, Schelen AM, et al. Identification of a nonsense mutation in the granulocyte-colony-stimulating factor receptor in severe congenital neutropenia. Proc Natl Acad Sci U S A. 1994;91:4480–4484. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 21.Beekman R, Valkhof MG, Sanders MA, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119:5071–5077. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- 22.Dong F, Qiu Y, Yi T, Touw IP, Larner AC. The carboxyl terminus of the granulocyte colony-stimulating factor receptor, truncated in patients with severe congenital neutropenia/acute myeloid leukemia, is required for SH2-containing phosphatase-1 suppression of Stat activation. J Immunol. 2001;167:6447–6452. doi: 10.4049/jimmunol.167.11.6447. [DOI] [PubMed] [Google Scholar]
- 23.van de Geijn GJ, Gits J, Aarts LH, Heijmans-Antonissen C, Touw IP. G-CSF receptor truncations found in SCN/AML relieve SOCS3-controlled inhibition of STAT5 but leave suppression of STAT3 intact. Blood. 2004;104:667–674. doi: 10.1182/blood-2003-08-2913. [DOI] [PubMed] [Google Scholar]
- 24.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/ acute myeloid leukemia. Blood. 1999;93:447–458. [PubMed] [Google Scholar]
- 25.Hermans MH, Ward AC, Antonissen C, Karis A, Löwenberg B, Touw IP. Perturbed granulopoiesis in mice with a targeted mutation in the granulocyte colony-stimulating factor receptor gene associated with severe chronic neutropenia. Blood. 1998;92:32–39. [PubMed] [Google Scholar]
- 26.Hunter MG, Avalos BR. Granulocyte colony-stimulating factor receptor mutations in severe congenital neutropenia transforming to acute myelogenous leukemia confer resistance to apoptosis and enhance cell survival. Blood. 2000;95:2132–2137. [PubMed] [Google Scholar]
- 27.Mitsui T, Watanabe S, Taniguchi Y, et al. Impaired neutrophil maturation in truncated murine G-CSF receptor-trans-genic mice. Blood. 2003;101:2990–2995. doi: 10.1182/blood.V101.8.2990. [DOI] [PubMed] [Google Scholar]
- 28.Corey SJ, Dombrosky-Ferlan PM, Zuo S, et al. Requirement of Src kinase Lyn for induction of DNA synthesis by granulo-cyte colony-stimulating factor. J Biol Chem. 1998;273:3230–3235. doi: 10.1074/jbc.273.6.3230. [DOI] [PubMed] [Google Scholar]
- 29.Corey SJ, Burkhardt AL, Bolen JB, Geahlen RL, Tkatch LS, Tweardy DJ. Granulocyte colony-stimulating factor receptor signaling involves the formation of a three-component complex with Lyn and Syk protein-tyrosine kinases. Proc Natl Acad Sci U S A. 1994;91:4683–4687. doi: 10.1073/pnas.91.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futami M, Zhu QS, Whichard ZL, et al. G-CSF receptor activation of the Src kinase Lyn is mediated by Gab2 recruitment of the Shp2 phosphatase. Blood. 2011;118:1077–1086. doi: 10.1182/blood-2009-12-261636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu QS, Robinson LJ, Roginskaya V, Corey SJ. G-CSF-induced tyrosine phosphorylation of Gab2 is Lyn kinase dependent and associated with enhanced Akt and differentiative, not proliferative, responses. Blood. 2004;103:3305–3312. doi: 10.1182/blood-2003-06-1861. [DOI] [PubMed] [Google Scholar]
- 32.Plo I, Zhang Y, Le Couédic JP, et al. An activating mutation in the CSF3R gene induces a hereditary chronic neutrophilia. J Exp Med. 2009;206:1701–1707. doi: 10.1084/jem.20090693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bicocca VT, Chang BH, Masouleh BK, et al. Crosstalk between ROR1 and the pre-B-cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyner JW, Deininger MW, Loriaux MM, et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc Natl Acad Sci U S A. 2009;106:8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyner JW, Yang WF, Bankhead AIII, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–296. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis MI, Hunt JP, Herrgard S, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 37.Hunter MG, Avalos BR. Deletion of a critical internalization domain in the G-CSFR in acute myelogenous leukemia preceded by severe congenital neutropenia. Blood. 1999;93:440–446. [PubMed] [Google Scholar]
- 38.Quintás-Cardama A, Vaddi K, Liu P, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010;115:3109–3117. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piazza R, Valletta S, Winkelmann N, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunter G, Woloszynek JR, Link DC. A truncation mutant of CSF3R cooperates with PML-RARalpha to induce acute myeloid leukemia in mice. Exp Hematol. 2011;39:1136–1143. doi: 10.1016/j.exphem.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.