Abstract
Adhesion-G protein-coupled receptors (GPCRs) are a poorly studied subgroup of the GPCRs, which have diverse biological roles and are major targets for therapeutic intervention. Among them, the Brain Angiogenesis Inhibitor (BAI) family has been linked to several psychiatric disorders, but despite their very high neuronal expression, the function of these receptors in the central nervous system has barely been analyzed. Our results, obtained using expression knockdown and overexpression experiments, reveal that the BAI3 receptor controls dendritic arborization growth and branching in cultured neurons. This role is confirmed in Purkinje cells in vivo using specific expression of a deficient BAI3 protein in transgenic mice, as well as lentivirus driven knockdown of BAI3 expression. Regulation of dendrite morphogenesis by BAI3 involves activation of the RhoGTPase Rac1 and the binding to a functional ELMO1, a critical Rac1 regulator. Thus, activation of the BAI3 signaling pathway could lead to direct reorganization of the actin cytoskeleton through RhoGTPase signaling in neurons. Given the direct link between RhoGTPase/actin signaling pathways, neuronal morphogenesis and psychiatric disorders, our mechanistic data show the importance of further studying the role of the BAI adhesion-GPCRs to understand the pathophysiology of such brain diseases.
Keywords: adhesion-GPCR, dendrite, morphogenesis, ELMO1, BAI3, Purkinje cell
Introduction
The increased complexity of behaviors that appeared during evolution has been correlated with an increased molecular complexity of upstream signaling membrane proteins.1 Genetic studies have linked mutations and copy number variations in genes coding for these upstream signaling pathways with many psychiatric and neurodevelopmental diseases, such as autism and schizophrenia. Deciphering these signaling pathways and how they regulate the formation of a functional neuronal network in mammals will thus help understand their contribution to brain diseases.
The Brain Angiogenesis Inhibitor (BAI) family is part of the poorly understood family of adhesion-G protein-coupled receptors (GPCRs).2 Adhesion-GPCRs are unique in that they contain a very long extracellular domain with multiple modules potentially conferring adhesive and recognition properties. The few studies of those receptors have shown their roles in physiology, including in the central nervous system, and pathology. For example, CELSR3 deficiency leads to abnormal neuronal migration, defects in tract development and reduced dendritic development.1, 3, 4, 5 Mutations in the adhesion-GPCR GPR56 lead to deficits in neuronal migration in patients with bilateral frontoparietal polymicrogyria.6 The BAI proteins are highly expressed in the brain and have been identified at post-synaptic densities in the forebrain7 and in the cerebellum.8 These proteins have several structural features that suggest their potential involvement in the development of functional neuronal networks: their extracellular domain contains several thrombospondin type 1 repeats domains that could provide adhesive and recognition properties. A PDZ-binding domain in their C-terminus could enable their association with synaptic scaffolding proteins such as PSD95. Sequence analysis shows that this subfamily of adhesion-GPCRs has homologs only in vertebrates, and that these homologs are extremely well conserved (Supplementary Figure 1). The BAI proteins may thus have important functions in controlling the development of complex cognitive abilities that are specific to vertebrates. This is highlighted by the fact that BAI proteins could contribute to behaviors defective in psychiatric disorders: single nucleotide polymorphisms and copy number variations in the BAI3 gene have been associated with schizophrenia,9, 10, 11 bipolar disorder12 and addiction,13 and the Bai2 knockout mouse has an anti-depressant phenotype.14
The regulation of dendrite morphogenesis in neurons is key to the formation of functional neuronal networks and is deficient in several neurodevelopmental disorders, such as autism, Fragile X syndrome or schizophrenia.15, 16, 17, 18 This process involves stabilization of dynamic filopodia through regulation of the actin cytoskeleton,19 in particular by the modulation of RhoGTPases.20, 21 Direct interference with the activity of RhoGTPases, such as RAC1, or their guanylate exchange factor activators, such as Tiam1, betaPIX, kalirin and the ELMO1/DOCK180 complex, leads to defects in dendrite morphogenesis.22, 23, 24 However, which upstream pathways coordinate RhoGTPases activation by integrating extracellular cues during dendrite morphogenesis is not well understood. The BAI1 receptor regulates phagocytosis through the modulation of the ELMO1/DOCK180/RAC1 signaling pathway.25 BAI1 interacts with ELMO1 through a motif conserved in BAI2 and BAI3, suggesting that the control of the small GTPase RAC1 through the ELMO1/DOCK180 module is a general feature of the BAI receptors and might be important for their role in the central nervous system. Here we show that the BAI3 protein controls dendritic arborization growth and complexity in neurons, partially through its interaction with ELMO1.
Materials and methods
BAI3 constructs, knockdown and transgenic mice
The BAI3-wild-type (WT) construct was cloned into the pEGFP-C2 vector from mouse cDNA clone no. BC099951. The Quikchange Site-Directed Mutagenesis kit (Agilent technologies, Santa Clara, CA, USA) was used to change the RKR sequence to AAA (residues 1431–1433) for the BAI3-WT-A construct. The BAI3-FLT construct codes for the entire BAI3 protein with an insertion of green fluorescent protein (GFP) after amino acid 1349. In BAI3-EMT, the cytoplasmic tail is replaced by GFP after amino acid 1174. The BAI3-SCT construct is a fusion between GFP and the cytoplasmic tail of BAI3 starting at amino acid 1166. The cDNA coding for BAI3-EMT was subcloned in the BamHI site of the L7/pcp2 promoter.26 A HindIII fragment was then purified for microinjection in the male pronucleus of C57BL/6N oocytes (Institut Clinique de la Souris, Strasbourg, France). The small hairpin RNA (shRNA) sequence for BAI3 was: 5′-ggtgaagggagtcatttat-3′, and was subcloned under the H1 promoter in either pSUPER vector for transfection in cultured hippocampal neurons or in a lentiviral vector that also drives GFP expression.27
Results
The adhesion-GPCR BAI3 modulates dendrite morphogenesis in neurons
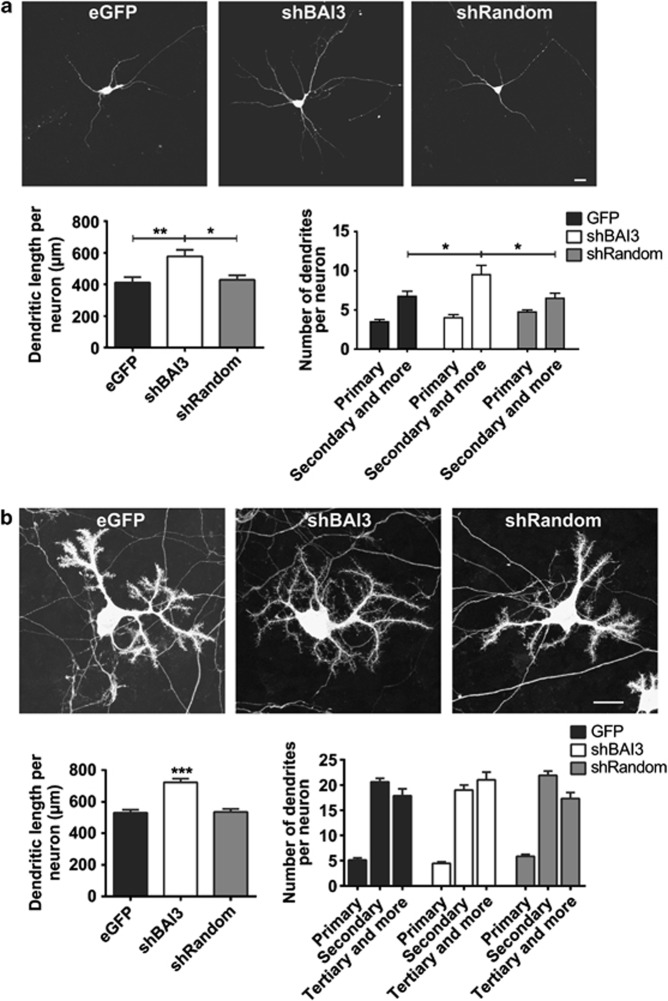
The BAI3 receptor was found to localize to actin-rich cell protrusions, such as filopodia and lamelipodia in HEK-293H cells, and dendrites and filopodia in cultured DIV5 hippocampal neurons (Supplementary Figure 2). Moreover, quantitative reverse transcription PCR (qRT-PCR) analysis shows expression of the endogenous BAI3 in developing hippocampal neurons in culture (Supplementary Figure 3). Given these data and the fact that BAI1 regulates RAC1, a major modulator of actin function, and dendrite and spine morphogenesis, we hypothesized that the BAI3 receptor has a role in the regulation of the actin cytoskeleton and dendrite morphogenesis in neurons. We first used a RNA interference strategy to knockdown the expression of the BAI3 protein in cultured hippocampal neurons, a classical model for the study of signaling pathways controlling dendrite morphogenesis (Supplementary Figure 3). Our quantitative analysis showed a significant increase in total dendrite length per neuron after BAI3 knockdown compared with control conditions (Figure 1a). We also observed a tendency for an increased total number of dendrites per neuron following BAI3 knockdown due to a significant increase in the number of dendrites of order 2 and more. As BAI3 is highly expressed in cerebellar Purkinje cells in vivo, a neuronal type of exquisite complexity in terms of dendritic arborization, we tested BAI3′s role in this neuronal type by transducing cerebellar mixed cultures with a lentivirus driving the expression of a shRNA directed against BAI3 or the corresponding controls. Knockdown of BAI3 also increased dendrite length in Purkinje cells significantly (Figure 1b). Hence, the role of the BAI3 protein in dendrite morphogenesis is a general feature of this adhesion-GPCR that can be found in multiple neuronal types.
Figure 1.
Knockdown of BAI3 promotes growth and branching of dendrites in several neuronal populations. (a) DIV5 hippocampal neurons were fixed two days after transfection with mCherry, and either a vector driving the expression of eGFP or a small hairpin RNA against BAI3 (shBAI3) or a control small hairpin RNA (shRandom). N=35–40 neurons per condition, four independent experiments. (b) DIV7 cerebellar cultures infected at DIV4 with a lentivirus driving either GFP alone, shBAI3 or shRandom were immunostained for calbindin, a Purkinje cell-specific marker. N=40–50 neurons per condition, three independent experiments. Scale bars: 20 μm. ‘*' denotes P<0.05, ‘**' denotes P<0.01, ‘***' denotes P<0.001.
BAI3 interacts with ELMO1, a regulator of RAC1 activity
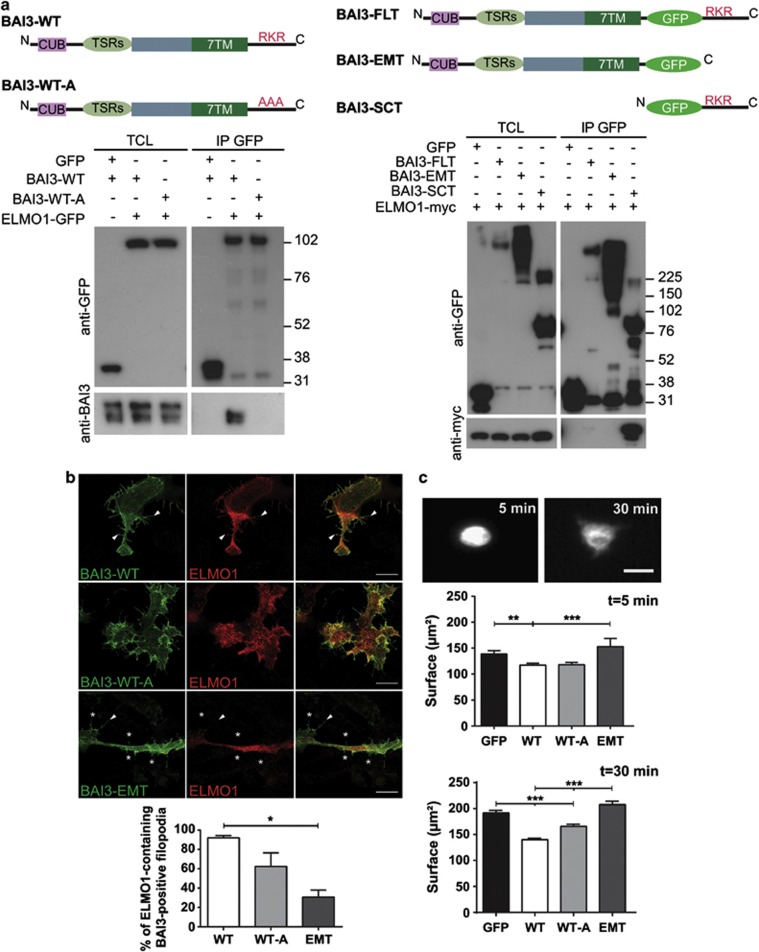
Next we were interested in determining the signaling pathway used by the BAI3 receptor to control dendrite morphogenesis. The BAI1 receptor interacts with the N-terminal part of ELMO1, through an RKR motif present in its cytoplasmic tail25 and conserved throughout the BAI family. To test whether BAI3 also binds ELMO1 through the same motif, we performed coimmunoprecipitation experiments using extracts of HEK-293H cells cotransfected with several tagged mutants of BAI3 and ELMO1 (Figure 2a). Using an anti-GFP antibody, we were able to affinity purify the wild-type form of BAI3 (BAI3-WT) from cells expressing ELMO1-GFP, but not from cells expressing soluble GFP, showing the specific interaction of BAI3 with ELMO1 (Figure 2a). This result was further confirmed by performing the reverse experiment in which we affinity purified different GFP-tagged forms of the BAI3 receptor and checked for the copurification of myc-tagged ELMO1. ELMO1-myc was coimmunopurified with the cytoplasmic tail of BAI3 (BAI3-SCT, soluble cytoplasmic tagged), but not with the mutant BAI3 receptor lacking the whole cytoplasmic domain (BAI3-EMT, extracellular membrane tagged). The interaction was reduced when using the full-length BAI3 with a GFP inserted in its cytoplasmic tail close to the RKR motif (BAI3-FLT, full length tagged), and totally abolished by mutagenesis of the RKR motif in the BAI3 protein (BAI3-WT-A). These results were further confirmed by immunofluorescence analysis and quantification, which showed that the colocalization of BAI3 with ELMO1 in actin-rich filopodia of transfected HEK-293H cells was dependent on the presence of the cytoplasmic tail of BAI3, and more particularly of the RKR motif (Figure 2b). Overall, these results show that ELMO1 interacts with the RKR motif located in the cytoplasmic tail of the BAI3 receptor.
Figure 2.
BAI3 regulates cell morphogenesis, partially through its interaction with ELMO1, a critical regulator of Rac1 signaling. (a) Top: schematic representations of the BAI3 constructs. Bottom affinity-purified proteins from transfected HEK-293H cells were detected by immunoblot analysis with anti-GFP, anti-BAI3 antibodies and anti-myc antibodies. ELMO1 was either tagged with GFP (expected molecular weight 112 kDa, left) or with myc (expected molecular weight 85 kDa, right). Control experiments were performed in parallel on HEK-293H cells expressing a soluble GFP instead of the GFP-tagged constructs. TCL: total cell lysate. IP GFP: samples affinity purified using an anti-GFP antibody. (b) Top: immunostaining of HEK-293H cells cotransfected with ELMO1-myc (detected with an anti-myc antibody) and either BAI3-WT, BAI3-WT-A (detected with an antibody against the N-terminus of the receptor) or BAI3-EMT (detected with an antibody against GFP). Arrows denote filopodia colabelled for BAI3 and ELMO1; asterisks denote filopodia missing ELMO1. Scale bar: 10 μm. Bottom: quantification of BAI3/ELMO1 colocalization in filopodia. Mean±s.e.m., n=40 cells per condition, 4 independent experiments. (c) Top: example of HEK293 cell shapes after 5 or 30 min of spreading on fibronectin. Scale bar: 20 μm. Middle and bottom: quantification of the cell surface area at 5 or 30 min of spreading. Mean±s.e.m., n=600–900 cells per condition, six independent experiments. ‘*' denotes P<0.05, ‘**' denotes P<0.01, ‘***' denotes P<0.001.
The BAI3 protein regulates cell morphogenesis, partly through binding of ELMO1
ELMO1 is part of the RAC1 guanylate exchange factor ELMO1/DOCK18028 and BAI1 has been shown to regulate RAC1 activity through its binding to the ELMO1/DOCK180 module.25 The modulation of dendrite morphogenesis mediated by BAI3 could thus be a result of the regulation of Rac1 activity. We tested this hypothesis using a classical in vitro assay, the cell-spreading assay,28 which consists in measuring the spreading of transfected HEK-293H cells at different time points after plating on fibronectin (Figure 2c). BAI3-expressing cells showed a significant reduction in their spreading both at 30 min (BAI3-WT: 140±3 μm2; GFP: 192±5 μm2, respectively) and at 5 min (BAI3-WT: 118±3 μm2; GFP: 139±6 μm2) when compared with control GFP-expressing cells. This effect on cell spreading was totally absent when the cytoplasmic tail of BAI3 was deleted (BAI3-EMT), and partially abolished when the RKR motif was mutated (BAI3-WT-A). Thus overexpression of the BAI3 receptor inhibits cell spreading through its cytoplasmic tail, partially through ELMO1 binding, suggesting that BAI3 signaling could indeed regulate dendrite morphogenesis through RAC1 modulation.
The BAI3/ELMO1 interaction is involved in the regulation of dendrite morphogenesis
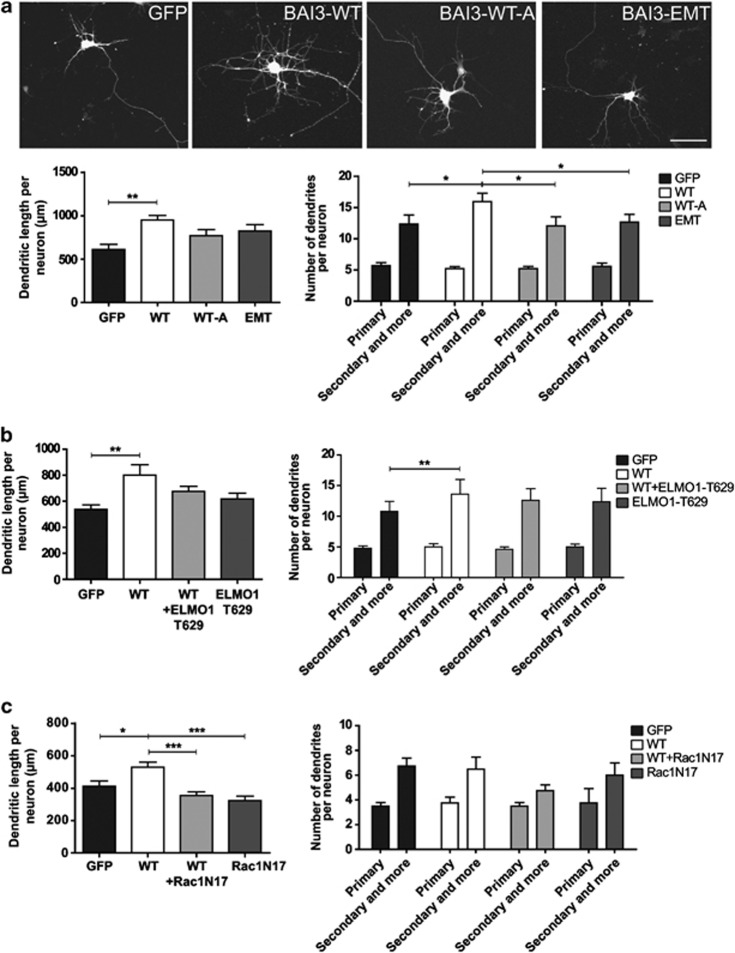
Our data suggested the implication of a new BAI3/ELMO1 signaling pathway controlling neuronal morphogenesis. We first confirmed that the BAI3 receptor could colocalize with ELMO1-myc in DIV5 hippocampal neurons, in particular in developing dendrites (Supplementary Figure 2). We then transfected hippocampal neurons with either a construct coding for BAI3 WT or BAI3 mutant constructs deficient for ELMO1 interaction (BAI3-WT-A, BAI3-EMT, see Figure 2). As shown in Figure 3, overexpression of the BAI3 protein results in a 55% increase in the total dendritic length per neuron when compared with GFP-expressing neurons, as well as in an increase in the number of branches of order 2 and more per neuron (Figure 3a). These effects were partially abolished when mutant BAI3 constructs unable to bind ELMO1 were used (see Figure 3a) or when BAI3-WT was cotransfected with a truncated ELMO1 unable to bind DOCK180 (see Figure 3b). Finally cotransfecting a dominant-negative RAC1 with BAI3-WT totally prevented BAI3′s induced promotion of dendritogenesis (see Figure 3c). Taken together, our data show that the BAI3 protein regulates dendrite morphogenesis by regulating RAC1 activity, partially through binding to the ELMO1/DOCK180 complex. They also suggest another, yet to be found, BAI3 signaling pathway associated with domains other than its C-terminus.
Figure 3.
Overexpression of BAI3 promotes dendritic arborization growth and complexity through interaction with ELMO1 and Rac1 activity. (a) Top: confocal images of representative neurons transfected with the indicated constructs at DIV3 and fixed at DIV5. Scale bar: 50 μm. Bottom: the total dendrite length and the number of dendrites per neuron were quantified on DIV5 hippocampal neurons transfected with mCherry and either a GFP control construct, wild-type BAI3 (WT) or BAI3 mutants (WT-A, EMT). Mean±s.e.m., n=30 neurons per construct, four independent experiments. (b) Cotransfection of BAI3-WT and a mutant ELMO1 protein unable to bind DOCK180 (ELMO1T629) partially abolishes BAI3's promotion of dendrite morphogenesis in DIV5 hippocampal neurons. Mean±s.e.m., n=55 neurons per construct, five independent experiments. (c) Cotransfection of BAI3-WT and a dominant-negative Rac1 (Rac1N17) totally prevents BAI3's effect on dendrite morphogenesis in DIV5 hippocampal neurons. Mean±s.e.m., n=40 neurons per construct, four independent experiments. ‘*' denotes P<0.05, ‘**' denotes P<0.01, ‘***' denotes P<0.001.
The BAI3 protein regulates dendrite morphogenesis in vivo
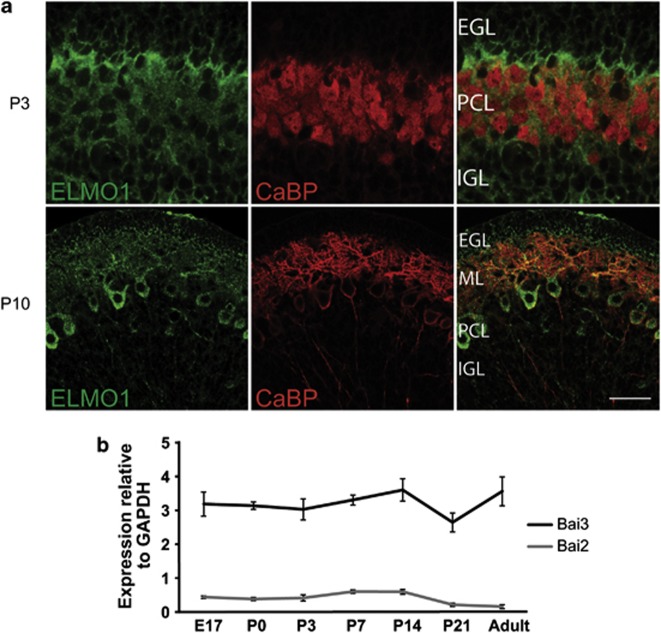
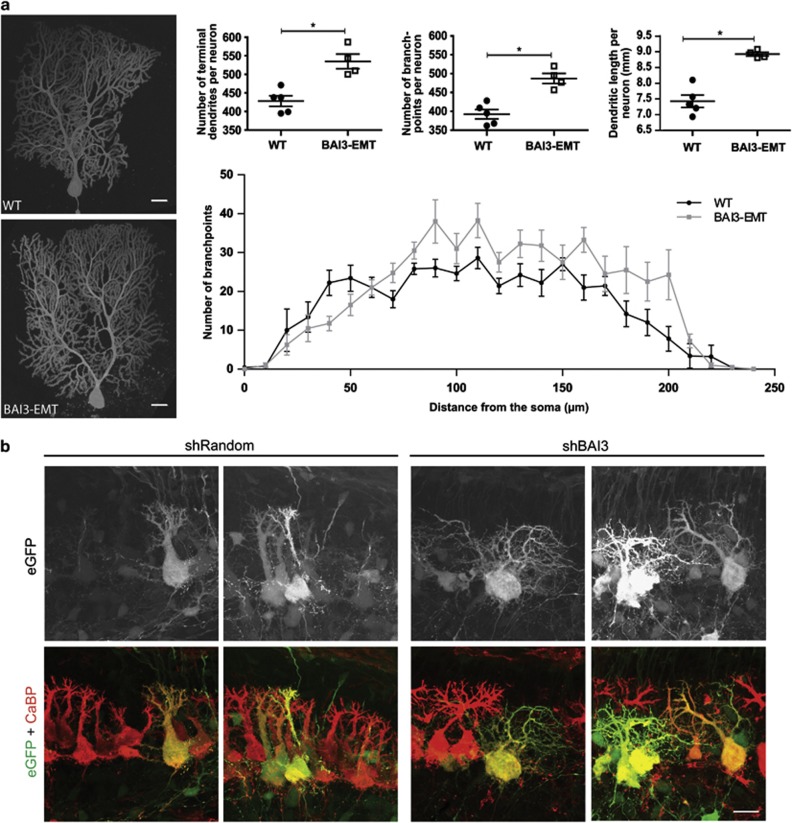
Katoh et al.29 have shown by in situ hybridization that ELMO1 is expressed in multiple neuronal populations in the mouse brain, including Purkinje cells. Double immunolabeling of mouse cerebellar sections using an antibody against the ELMO1 protein and an antibody against calbindin, a Purkinje cell marker (Figure 4a), showed the presence of ELMO1 in the growing tips of Purkinje cell dendrites at postnatal day 3. By P10, it was filling the whole Purkinje cell dendritic arborization. This pattern, together with the expression of BAI3 in Purkinje cells8, 30 and in the cerebellum during development (Figure 4b), is in agreement with a potential role of the BAI3/ELMO1 signaling pathway in the morphogenesis of Purkinje cells, whose elaborate dendritic arborization is the result of extensive reorganization between P0 and P15 in the mouse cerebellum. To test this role in vivo, we generated transgenic mice expressing the BAI3-EMT mutant protein, which lacks the entire cytoplasmic domain and the ability to regulate neuronal morphogenesis (see Figures 2 and 3). In addition, the BAI3-EMT construct reduces by 44% the effect of the BAI3-WT receptor on dendritogenesis in hippocampal neurons (Supplementary Figure 4), and can thus act partially as a dominant-negative form. We used the Pcp2 promoter to specifically drive the expression of BAI3-EMT in Purkinje cells in the cerebellum.26 This specific expression was confirmed by immunoblot and was detected as early as P3 by GFP immunohistochemistry (Supplementary Figure 5). It was estimated by qRT-PCR to be equivalent to 60% of the endogenous Bai3 gene. Calbindin immunostaining of cerebellar sections and morphometric measurements did not reveal any gross reorganization as a consequence of the expression of the mutant construct (Supplementary Figure 5). However, quantitative analysis of single-labeled Purkinje cells from Pcp2/BAI3-EMT mice revealed important changes in their dendritic morphology: increased dendritic length, number of junctions and terminal dendrites when compared with wild-type Purkinje cells (Figure 5a). Scholl analysis shows that the complexity of Purkinje cell dendritic arborization is particularly increased in distal parts of the cells relative to the soma. Finally, we analyzed the effects of BAI3 knockdown in vivo by injecting the lentiviral vectors in the cerebellar cortex of P6 pups (Figure 5b), and imaging after 4 days of infection. Defects in dendritic arborization were clearly visible as dendrites were longer, thinner and misoriented in Purkinje cells transduced with the shRNA against BAI3 compared with the control shRNA. Alltogether, these results show a major role for BAI3 in regulating Purkinje cell dendritic arbor formation in vivo.
Figure 4.
The BAI3 receptor and ELMO1 are expressed in the developing cerebellum. (a) Cerebellar sections from wild-type mice were immunolabelled for the endogenous ELMO1 protein and for calbindin (CaBP), a marker of Purkinje cells, at P3 and P10. EGL: external granular layer, ML: molecular layer, PCL: Purkinje cell layer, IGL: internal granular layer. Scale bar: 50 μm. (b) qRT-PCR shows a high expression of the Bai3 gene during postnatal development in the mouse cerebellum relative to the Gapdh gene.
Figure 5.
The BAI3 protein controls neuronal development and dendrite morphogenesis in vivo. (a) Left: images of representative reconstructed Purkinje cells in 1—month-old Pcp2/BAI3-EMT transgenic mice and age-matched wild-type (WT) mice. Right: quantification of total dendritic length, number of junctions and terminal branches per Purkinje cell as well as Scholl analysis. Mean±s.e.m., n=4–5 Purkinje cells per genotype. Student's t-test followed by Mann–Whitney, ‘*' denotes P<0.05. (b) Purkinje cells transduced at P6 with lentivirus particles driving either shBAI3 or control shRandom (eGFP positive) were analyzed at P10 using calbindin immunostaining (CaBP). Massive defects in dendritic arborization were observed after BAI3 knockdown when compared with non transduced adjacent cells or cells transduced with the control virus. Scale bars: 20 μm.
Discussion
The brain is composed of thousands of different types of neurons that differ drastically in their morphology, in particular in the shape and complexity of their dendritic arborization. This morphology underlies functional differences between neurons, in particular how they integrate signals coming from different inputs. Its proper development is thus essential for the normal function of the central nervous system, and deficits in neuronal morphogenesis have been correlated to psychiatric disorders such as schizophrenia. We have now provided evidence, both in vitro and in vivo, for a new signaling pathway regulating dendrite morphogenesis involving the BAI receptor BAI3, a member of the poorly studied family of adhesion-GPCRs, and the protein ELMO1, an important regulator of the RAC1 RhoGTPase.
The regulation of dendrite morphogenesis involves integration of extracellular signals and intrinsic molecular programs in order to control the growth and branching of the actin cytoskeleton. The BAI receptors constitute a new regulator of this process that can sense extracellular signals and signal in the cell through their interaction with effectors such as ELMO1. Another family of adhesion-GPCRs, the CELSR proteins, has been shown to have a role in dendrite morphogenesis through regulation of intracellular calcium signaling. Knockdown of CELSR2 in organotypic cultures in pyramidal neurons and Purkinje cells induces a simplification of their dendritic arborization,31 whereas CESLR3 has an opposite role.3 This function is conserved as the Drosophila homolog, Flamingo, is also involved in neuronal morphogenesis and more particularly in regulating dendritic field through repulsion.32 Our results show that control of neuronal morphogenesis could be a property of many adhesion-GPCRs in vivo. Given the diversity of domains found in the extracellular part of adhesion-GPCRs, an attractive hypothesis is that each type of adhesion-GPCR might regulate the morphology of particular neuronal populations and thus contributes to the diversity of shape, and of function, in the vertebrate central nervous system.
What is the signaling pathway of BAI3 during neuronal morphogenesis? Our results show that its interaction with the protein ELMO1 is partially involved in this process. Previous results have shown that BAI1 can regulate RAC1 and phagocytosis through an interaction with ELMO1.25 Regulation of RhoGTPases is essential for driving changes in the actin cytoskeleton and cell morphogenesis during development. Moreover modifying RAC1 activity in neurons is known to induce changes in dendrite morphogenesis22 and interferes with BAI3's function as shown by our results. Hence BAI3's interaction with ELMO1 constitutes a direct pathway linking extracellular cues and intracellular modification of the actin cytoskeleton during neuronal development. It will be of interest to analyze the role of other potential intracellular partners of BAI3. In particular, IRSp53/BAIAP2 was originally identified as a partner for BAI1,33 and has since been shown to regulate actin morphogenesis through binding of small RhoGTPAses and the WAVE complex.34 Small G protein binding, although yet to be demonstrated for BAI proteins, could also have an important role through binding of the third intracytoplasmic loop. Extracellularly, binding of the secreted protein C1QL1 has recently been shown to promote synapse elimination in vitro, but no effect was demonstrated on dendrite morphogenesis.35 These results were obtained in mature hippocampal neurons and, taken together with our data, suggest that the function of BAI proteins in the regulation of dendrite morphogenesis is critical at early stages of neuronal development. Alternatively, other unknown ligands of BAI3 might be critical for this function.
Several candidate genes linked with neurodevelopmental disorders are proteins regulating neuronal morphogenesis.36 For example, mutations in SHANK3 associated with autism induce defects in spine morphogenesis and in actin polymerization.37 Neuregulin1′s mutation at valine 321, previously linked to schizophrenia, has been shown to prevent neuregulin's control of dendritic arborization growth and complexity.36 Given the evidence associating BAI genes with psychiatric disorders,9, 13, 14, 10, 12 our data reveals a new pathway involved in the etiology of these brain diseases through regulation of dendrite morphogenesis, and highlights the potential of the BAI signaling pathway for therapeutic intervention.
Acknowledgments
We would like to thank Dr Sophie Vriz and Dr Jean-Louis Bessereau for critical reading of the manuscript. The ELMO1-GFP and ELMO1T629-GFP constructs were a kind gift of Dr Lorraine Santy (PennState, USA). We would like to thank R Schwartzman and S Bolte at the IFR83 imaging facility (UPMC, Paris, France), and J Teillon at the CIRB imaging facility (Collège de France, Paris, France). We would like to thank J Bakouche and V Gautheron for helping us with cloning and mouse colonies. This work was supported by the CNRS (PEPS, PICS), ATIP-AVENIR, Association Française du Syndrome de Rett, UPMC (Emergence), Ecole des Neurosciences de Paris. VL was supported by the ED3C (UPMC) and the Ministère de la Recherche et de l′Enseignement Supérieur, AU by UPMC and AVENIR, SMS by NERF and LABEX MEMOLIFE, and MT by FRM, PI by ANR-09-MNPS-038 and ANR-2010-JCJC-1403-1.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Emes RD, Pocklington AJ, Anderson CNG, Bayes A, Collins MO, Vickers CA, et al. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat Neurosci. 2008;11:799–806. doi: 10.1038/nn.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnadóttir TK, Fredriksson R, Schiöth HB. The adhesion GPCRs: a unique family of G protein-coupled receptors with important roles in both central and peripheral tissues. Cell Mol Life Sci. 2007;64:2104–2119. doi: 10.1007/s00018-007-7067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Kawaguchi S-Y, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, et al. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci. 2007;10:963–969. doi: 10.1038/nn1933. [DOI] [PubMed] [Google Scholar]
- Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Zhou L, Bar I, Achouri Y, Campbell K, De Backer O, Hebert JM, et al. Early forebrain wiring: genetic dissection using conditional Celsr3 mutant mice. Science. 2008;320:946–949. doi: 10.1126/science.1155244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Jeong S-J, Jin Z, Strokes N, Li S, Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci USA. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CNG, Blackstock WP, et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97 (Suppl 1:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol. 2009;7:e83. doi: 10.1371/journal.pbio.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRosse P, Lencz T, Burdick KE, Siris SG, Kane JM, Malhotra AK. The genetics of symptom-based phenotypes: toward a molecular classification of schizophrenia. Schizophr Bull. 2008;34:1047–1053. doi: 10.1093/schbul/sbn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-M, Chao Y-L, Huang A-L, Cheng M-C, Chen Y-J, Lee K-F, et al. Identification and characterization of three inherited genomic copy number variations associated with familial schizophrenia. Schizophr Res. 2012;139:229–236. doi: 10.1016/j.schres.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Lips ES, Cornelisse LN, Toonen RF, Min JL, Hultman CM, Holmans PA, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry. 2011;17:996–1006. doi: 10.1038/mp.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MJ, Nievergelt CM, Kelsoe JR, Welsh DK. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS ONE. 2012;7:e32091. doi: 10.1371/journal.pone.0032091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q-R, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many ‘cell adhesion' genes. Am J Med Genet. 2006;141B:918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- Okajima D, Kudo G, Yokota H. Antidepressant-like behavior in brain-specific angiogenesis inhibitor 2-deficient mice. J Physiol Sci. 2010;61:47–54. doi: 10.1007/s12576-010-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill ME, Jones KA, Rafalovich I, Xie Z, Barros CS, Müller U, et al. Control of interneuron dendritic growth through NRG1/erbB4-mediated kalirin-7 disinhibition. Mol Psychiatry. 2011;17:99–107. doi: 10.1038/mp.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lomassese S, Nissant A, Mota T, Néant-Féry M, Oostra BA, Greer CA, et al. Fragile X mental retardation protein regulates new neuron differentiation in the adult olfactory bulb. J Neurosci. 2011;31:2205–2215. doi: 10.1523/JNEUROSCI.5514-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalus P, Müller TJ, Zuschratter W, Senitz D. The dendritic architecture of prefrontal pyramidal neurons in schizophrenic patients. Neuroreport. 2000;11:3621–3625. doi: 10.1097/00001756-200011090-00044. [DOI] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol. 1996;91:117–119. doi: 10.1007/s004010050401. [DOI] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Cerri C, Fabbri A, Vannini E, Spolidoro M, Costa M, Maffei L, et al. Activation of Rho GTPases triggers structural remodeling and functional plasticity in the adult rat visual cortex. J Neurosci. 2011;31:15163–15172. doi: 10.1523/JNEUROSCI.2617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L, Cline HT. Rho GTPases and activity-dependent dendrite development. Curr Opin Neurobiol. 2004;14:297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-Y, Oh MH, Bernard LP, Macara IG, Zhang H. The RhoG/ELMO1/Dock180 signaling module is required for spine morphogenesis in hippocampal neurons. J Biol Chem. 2011;286:37615–37624. doi: 10.1074/jbc.M111.268029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont A-C, Elliott MR, Lu M, Haney LB, Ma Z, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- Smeyne RJ, Chu T, Lewin A, Bian F, S Crisman S, Kunsch C, et al. Local control of granule cell generation by cerebellar Purkinje cells. Mol Cell Neurosci. 1995;6:230–251. doi: 10.1006/mcne.1995.1019. [DOI] [PubMed] [Google Scholar]
- Avci HX, Lebrun C, Wehrlé R, Doulazmi M, Chatonnet F, Morel M-P, et al. Thyroid hormone triggers the developmental loss of axonal regenerative capacity via thyroid hormone receptor α1 and kruppel-like factor 9 in Purkinje cells. Proc Natl Acad Sci USA. 2012;109:14206–14211. doi: 10.1073/pnas.1119853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–464. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- Katoh H, Fujimoto S, Ishida C, Ishikawa Y, Negishi M. Differential distribution of ELMO1 and ELMO2 mRNAs in the developing mouse brain. Brain Res. 2006;1073-1074:103–108. doi: 10.1016/j.brainres.2005.12.085. [DOI] [PubMed] [Google Scholar]
- Kee HJ, Ahn KY, Choi KC, Won Song J, Heo T, Jung S, et al. Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett. 2004;569:307–316. doi: 10.1016/j.febslet.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kengaku M, Hirano T, Takeichi M, Uemura T. Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Dev Cell. 2004;7:205–216. doi: 10.1016/j.devcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Gao F-B, Kohwi M, Brenman JE, Jan LY, Jan YN. Control of dendritic field formation in Drosophila. Neuron. 2000;28:91–101. doi: 10.1016/s0896-6273(00)00088-x. [DOI] [PubMed] [Google Scholar]
- Oda K, Shiratsuchi T, Nishimori H, Inazawa J, Yoshikawa H, Taketani Y, et al. Identification of BAIAP2 (BAI-associated protein 2), a novel human homologue of hamster IRSp53, whose SH3 domain interacts with the cytoplasmic domain of BAI1. Cytogenet Cell Genet. 1999;84:75–82. doi: 10.1159/000015219. [DOI] [PubMed] [Google Scholar]
- Abou-Kheir W, Isaac B, Yamaguchi H, Cox D. Membrane targeting of WAVE2 is not sufficient for WAVE2-dependent actin polymerization: a role for IRSp53 in mediating the interaction between Rac and WAVE2. J Cell Sci. 2008;121:379–390. doi: 10.1242/jcs.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliger MF, Martinelli DC, Sudhof TC. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci USA. 2011;108:2534–2539. doi: 10.1073/pnas.1019577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hancock ML, Role LW, Talmage DA. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. J Neurosci. 2010;30:9199–9208. doi: 10.1523/JNEUROSCI.0605-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.