Abstract
Divergence of protein sequences and gene expression patterns are two fundamental mechanisms that generate organismal diversity. Here, we have used genome and transcriptome data from eight mammals and one bird to study the positive correlation of these two processes throughout mammalian evolution. We demonstrate that the correlation is stable over time and most pronounced in neural tissues, which indicates that it is the result of strong negative selection. The correlation is not driven by genes with specific functions and may instead best be viewed as an evolutionary default state, which can nevertheless be evaded by certain gene types. In particular, genes with developmental and neural functions are skewed toward changes in gene expression, consistent with selection against pleiotropic effects associated with changes in protein sequences. Surprisingly, we find that the correlation between expression divergence and protein divergence is not explained by between-gene variation in expression level, tissue specificity, protein connectivity, or other investigated gene characteristics, suggesting that it arises independently of these gene traits. The selective constraints on protein sequences and gene expression patterns also fluctuate in a coordinate manner across phylogenetic branches: We find that gene-specific changes in the rate of protein evolution in a specific mammalian lineage tend to be accompanied by similar changes in the rate of expression evolution. Taken together, our findings highlight many new aspects of the correlation between protein divergence and expression divergence, and attest to its role as a fundamental property of mammalian genome evolution.
Keywords: gene expression evolution, protein evolution, primates, amniotes, correlation analysis
Introduction
Phenotypic evolution depends on mutations that alter protein sequences and mutations that affect gene regulation, but their relative contributions remain to be settled. One line of evidence suggests that the two types of mutations play different roles during evolution, such that genes involved in physiological traits are biased toward changes in protein sequences, whereas genes involved in morphological traits evolve primarily in terms of gene expression (Wray 2007; Haygood et al. 2010; Liao et al. 2010). According to this view, protein divergence and expression divergence can, at least to a certain extent, be considered decoupled processes. In contrast, other studies have reported a positive correlation between protein divergence and expression divergence in pairwise comparisons of mammals (Jordan et al. 2005; Khaitovich et al. 2005; Liao and Zhang 2006a) and several other species (Nuzhdin et al. 2004; Lemos et al. 2005; Sartor et al. 2006; Hunt et al. 2013), as well as among recent gene duplicates in humans (Makova and Li 2003). These results instead suggest that protein divergence and expression divergence are two highly related phenomena, which affect individual genes in similar ways. How can these seemingly opposing views of the roles of protein divergence and expression divergence during evolution be reconciled?
To add further uncertainty, the mechanism underlying the correlation between protein divergence and expression divergence remains poorly understood. One possibility is that the correlation is linked to a specific gene characteristic. As an example, highly expressed genes tend to have slow-evolving protein sequences (Subramanian and Kumar 2004; Drummond and Wilke 2008) and less divergent gene expression patterns (Liao and Zhang 2006b; Gout et al. 2010), meaning that the correlation between protein divergence and expression divergence could be a result of between-gene variation in expression levels. That said, Lemos et al. (2005) found that, in Drosophila, the strength of the correlation was not affected when they took expression level into account. They also excluded protein length and the number of protein–protein interactions as responsible factors and speculated that the correlation was instead due to more general selective constraints that affect gene expression and protein sequences in similar ways (Lemos et al. 2005). It is, however, possible that the true effect of one of these factors was hidden by measurement noise (Drummond et al. 2006; Kim and Yi 2007) or that the evaluation of additional factors could yield different results. For example, Khaitovich et al. (2005) found that, in humans and chimpanzees, the correlation became weaker after correction for expression breadth and the tissues in which genes were expressed. This result is somewhat difficult to interpret, because the explained variance in their original model was very low, but a role for tissue specificity in establishing the correlation would be consistent with the fact that genes experience different selective constraints depending on their tissue expression profile (Duret and Mouchiroud 2000; Khaitovich et al. 2005; Gu and Su 2007; Brawand et al. 2011) and might also provide an explanation for why the correlation is absent in yeast (Tirosh and Barkai 2008).
In this study, we use gene expression and sequence data from eight mammals and one bird to explore the correlation between protein divergence and expression divergence in detail. Our results help clarify the respective roles of these two processes during evolution and add new layers to the current understanding of mammalian genome evolution.
Materials and Methods
Pairwise Expression Divergence
Normalized gene expression values from six organs (brain, cerebellum, heart, kidney, liver, and testis) and nine species (human, chimpanzee, gorilla, orangutan, rhesus macaque, mouse, gray short-tailed opossum, platypus, and nondomesticated chicken) were taken from Brawand et al. (2011). These expression measurements were based on RNA sequencing of adult individuals, typically one male and one female per species. No data were available for orangutan testis. The normalization procedure applied to these data involved ranking genes in terms of their expression level, choosing the 1,000 genes with the most stable expression ranks and then scaling the data, so that the median expression level of these genes would be the same across species and tissues (Brawand et al. 2011). We further took the natural logarithm of all expression values to ensure that an n-fold change in expression would be treated equally, regardless of whether it affected a lowly or a highly expressed gene. Expression estimates below 10−6 were replaced by this value before log transformation.
We analyzed two gene sets: protein-coding genes with 1:1 orthologs in the primate species (the primate data set) or in all the studied species (the amniote data set), based on the assignments by Brawand et al. (2011). In cases where the transcribed regions of two genes overlapped in at least one species, both genes were removed from all subsequent analyses. For all species pairs, we calculated the expression divergence for each gene either as the Euclidean distance between the species means for the different tissues (i.e., by considering a six-dimensional Euclidean space where each dimension corresponds to one tissue) or as 1 − r, where r is the Pearson correlation coefficient between the two tissue expression profiles.
Sequence Divergence
For each gene, for which we had gene expression data, we downloaded its longest coding sequence from the Ensembl database, version 57 (Flicek et al. 2011). Information on the base calling quality in the chimpanzee, orangutan, macaque, opossum, platypus, and chicken genome assemblies was available from the UCSC Genome Browser Database (Fujita et al. 2011), and we used this to mask all bases with a quality score below 40. We aligned the protein-coding sequences using the codon option in PRANK (Löytynoja and Goldman 2008), which has been shown to outperform other alignment algorithms (Fletcher and Yang 2010), and removed all codons that corresponded to a gap in at least one species. Only genes for which at least 150 high-quality bases aligned across all species were used for further analysis. Following this filtering step, the amniote data set contained 3,749 nonoverlapping genes (see Pairwise Expression Divergence), whereas the primate data set contained 10,227. We estimated the number of nonsynonymous substitutions per nonsynonymous site (dN) and synonymous substitutions per synonymous site (dS) in these alignments using the codeml program in the PAML package (Yang 1997), both for pairwise comparisons and for each branch in the species tree (free-ratios model).
Multispecies Expression Divergence
We estimated ancestral gene expression levels in each tissue using AncML (Schluter et al. 1997; Holloway et al. 2007) and the following branch lengths (in million years): ((((((((human:6, chimpanzee:6):1, gorilla:7):7, orangutan:14):11, macaque:25):64, mouse:89):91, opossum:180):20, platypus:200):110, chicken:310) (Brawand et al. 2011). For each branch, we calculated expression divergence for individual tissues and jointly for all tissues (see Pairwise Expression Divergence), and summed the values from all external and internal branches. It should be noted that this method is based on a Brownian motion model and therefore does not take negative selection into account. We found that a more complex approach, in which we fitted an Ornstein–Uhlenbeck model to the data, using the geiger R package (Harmon et al. 2008) and then transformed the tree branch lengths accordingly before running AncML, gave highly similar estimates (for total expression divergence across all species, the Spearman correlation coefficient was 0.99 for the primate data set and 0.98 for the amniote data set). To track how the addition of more species affected our results, we began by summing up the divergence along the branches connecting only two species (human and chimpanzee) and then added one species at a time.
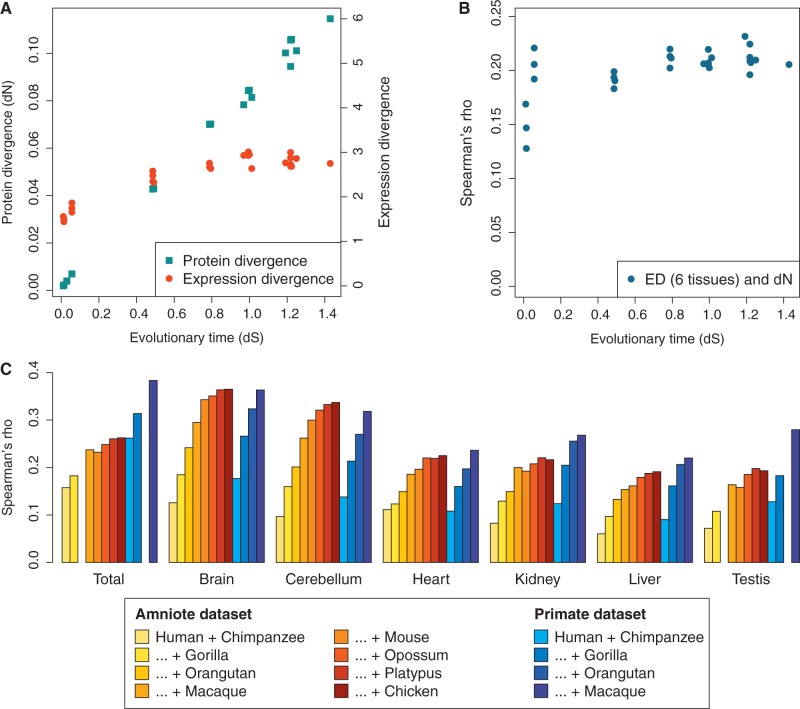
The estimation of ancestral gene expression levels is not trivial, especially for traits that are heavily affected by negative selection and for which the ancestral values might therefore frequently fall outside the range observed in extant species. An approach that relies on the estimation of ancestral states can nevertheless be justified, provided that the gain in power from combining data from multiple species outweighs the shortcomings of the model. In our analysis, it is improbable that biases in the estimation of expression divergence would lead to a biologically irrelevant inflation of the correlation between expression divergence and protein divergence, which is measured independently. The fact that the observed correlation between expression divergence and protein divergence increases with each added species (fig. 1C) therefore confirms the usefulness of our method.
Fig. 1.—
Evolutionary and tissue-specific dynamics of the correlation between protein divergence and expression divergence. (A) Genome-wide patterns of protein and expression divergence in pairwise species comparisons. Estimates of dN and dS were based on a concatenation of all gene alignments, whereas expression divergence was calculated as the median expression divergence for individual genes across six tissues. All values refer to the amniote data set. Because we did not have access to data from orangutan testis, comparisons with this species were excluded. The corresponding analysis using all species and five tissues is presented in supplementary table S1, Supplementary Material online. (B) Spearman correlation coefficients for dN and total expression divergence across six tissues in pairwise species comparisons. Each datapoint represents 3,749 genes from the amniote data set. (C) Spearman correlation coefficients for total and tissue-specific ED against dN when estimates were combined for multiple species. Results are shown for both the amniote (N = 3,749) and primate (N = 10,227) data sets, with the leftmost bar representing the correlation coefficient for human and chimpanzee, the next showing human, chimpanzee and gorilla, and so on. No data were available for orangutan testis.
Enrichment of Gene Ontology Terms
We identified overrepresented gene ontology (GO) terms (Gene Ontology Consortium 2000), using the GOrilla tool (Eden et al. 2009), which is designed to find enrichments at the top of a ranked gene list. All analyses were based on human annotations. We corrected the P values for 115,357 multiple tests using the Benjamini–Hochberg method (Benjamini and Hochberg 1995), with a false discovery rate (FDR) of 0.1% as our cutoff value.
Correlation Analysis
All analyses were performed in R 2.12.2 (R Development Core Team 2011). Partial Spearman correlations with correction for single or multiple factors were calculated with the ppcor package. To summarize gene expression across species, we used the estimated expression levels for the most basal node of the tree. These values are interchangeable with the average across species (Spearman correlation coefficients above 0.99 for all six tissues). The expression levels were averaged across tissues.
Tissue specificity was calculated using the tissue specificity index, τ (Yanai et al. 2005), which is 0 for genes that are uniformly expressed and 1 for genes that are exclusively expressed in a single tissue. To ensure that n-fold expression changes were treated equally, we did not log-transform the expression levels. To calculate neural bias, we divided the total expression in neural tissues (brain and cerebellum) by the total expression in all six tissues.
We downloaded information on gene family size (number of paralogs), length of the coding sequence, and GC content of the transcribed sequence from Ensembl version 57 (Flicek et al. 2011). Data on connectivity, developmental onset of gene expression, phyletic age, and essentiality were taken from the OGEE database, build 304 (Chen et al. 2012). All downloaded data referred to humans, unless otherwise specified in the text.
Branch-Wise Analysis
We calculated branch-specific protein divergence and expression divergence as described above for each branch in the amniote species tree, while leaving out chimpanzee, gorilla, and orangutan to avoid short branch lengths. For each branch, we ranked genes according to the two types of divergence and replaced the divergence estimates with these ranks, to make direct comparisons between branches possible. We then calculated the Spearman correlation coefficient for expression divergence ranks and protein divergence ranks for each gene. The analysis was repeated for expression levels in the six studied species and dN/dS values for the terminal branches leading to those species.
Results
The Correlation between Protein Divergence and Expression Divergence Is Evolutionarily Stable
We based our analysis of protein divergence and expression divergence on data from nine species (Lander et al. 2001; Mouse Genome Sequencing Consortium 2002; International Chicken Genome Sequencing Consortium 2004; Gibbs et al. 2007; Mikkelsen et al. 2007; Warren et al. 2008; Brawand et al. 2011; Locke et al. 2011; Scally et al. 2012). These comprised six placental mammals (human, chimpanzee, gorilla, orangutan, rhesus macaque, and mouse), one marsupial (gray short-tailed opossum), one monotreme (platypus), and one bird (nondomesticated chicken). We further focused on two gene sets: protein-coding genes with 1:1 orthologs in all the studied species (the amniote data set) or in the five primate species (the primate data set). For each gene and each species pair, we calculated protein divergence as the rate of nonsynonymous substitutions (dN) and expression divergence as the Euclidean distance between log-transformed expression values (see Materials and Methods). Gene expression data were available for brain (cerebral cortex or whole brain without cerebellum), cerebellum, heart, kidney, liver, and testis (Brawand et al. 2011), which allowed us to determine the degree of expression divergence for each individual tissue, as well as the total expression divergence across all six tissues.
The dissimilarity between protein and gene expression evolution was clear in our data (fig. 1A). Protein divergence increased steadily with the evolutionary time that separated two species, whereas the increase in expression divergence quickly tapered off. We previously observed this saturation effect using a different divergence measure (Brawand et al. 2011), and the same trend was also demonstrated in fruit flies (Bedford and Hartl 2009). We might expect that the saturation of expression divergence would make it difficult to distinguish fast-evolving and slow-evolving genes, which in turn would lead to a decay of the correlation with protein divergence. This was, however, not the case, given that we obtained comparable correlation coefficients when we compared primates with each other and when we compared mammals with birds (fig. 1B). The pattern was also present when we used an alternative method, based on the Pearson correlation coefficient, to estimate expression divergence (supplementary table S1, Supplementary Material online), in spite of a previous microarray-based study, where no significant correlation was found for dN and this measure of expression divergence (Liao and Zhang 2006a). The Pearson correlation coefficient method is complementary to the Euclidean distance, because it focuses on changes in tissue expression profiles rather than expression values but has been shown to be unreliable under some circumstances (Pereira et al. 2009). A further advantage of the Euclidean distance method is that it is more flexible, in that it is possible to either combine data from all tissues into a single divergence value or to consider each tissue separately. We therefore used this method for our further analyses.
In summary, our results suggest that expression divergence saturates at different levels for different genes and that the constraints that determine the maximum level of expression divergence show substantial overlap with the constraints that affect protein evolution.
Strong Negative Selection Acts to Preserve the Correlation in Neural Tissues
The evolutionary conservation of the correlation between expression divergence and protein divergence suggests that it is the result of long-term selection. Two mechanisms could be responsible: On the one hand, strong negative selection might place similar constraints on both protein sequences and gene expression patterns. On the other hand, strong selection might primarily affect either protein divergence or expression divergence, whereas less constrained genes would be free to change in both regards, thereby driving the correlation. How can these alternatives be distinguished from one another? Conveniently, it is known that the selection on gene expression varies across tissues (Khaitovich et al. 2005; Brawand et al. 2011). We therefore assessed the impact of selection strength by calculating the correlation between protein divergence and tissue-specific expression divergence. Given that the correlation was maintained over evolutionary time, we decided to combine data from multiple species to minimize the contribution of noise to our divergence estimates (see Materials and Methods). The value of this approach is evident from figure 1C, which shows that the observed correlation strength increased with each added species. When all species were included, Spearman’s rho for protein divergence and total expression divergence had reached 0.27 (P < 10−15) for the amniote data set and 0.38 (P < 10−15) for the primate data set. In the latter case, the inclusion of the final species brought about a considerable increase in correlation strength, indicating that the correlation coefficient might have to be revised further upward as transcriptome data become available for additional species.
Strikingly, we identified strong correlations even for individual tissues (fig. 1C), in particular for brain and cerebellum. For the amniote data set, these correlations even exceeded the correlation we observed when all tissues were analyzed together. As neural tissues are associated with particularly strong negative selection on gene expression (Khaitovich et al. 2005; Brawand et al. 2011), these results show that intense negative selection contributes to, rather than detracts from, the overall correlation between protein divergence and expression divergence.
Enrichment for Functional Categories among ED-Biased and dN-Biased Genes
Previous studies reported Pearson correlation coefficients of 0.03 for human and chimpanzee (Khaitovich et al. 2005) and 0.19 for human and mouse (Liao and Zhang 2006a). The correlations revealed by our analysis were therefore substantially stronger (fig. 1C), presumably due the superior sensitivity of RNA sequencing compared with microarrays (Wang et al. 2009). We also found a qualitative difference compared with these earlier results: In our data, the correlation was stronger for closely related species, that is, the primate data set. When we further split the primate data set into genes that also occurred in the amniote data set and those that did not, we found a correlation coefficient of 0.26 for the shared genes (P < 10−15), consistent with our results for the amniote data set, whereas the coefficient reached 0.43 (P < 10−15) for genes specific to the primate data set. Compared with the full primate data set, genes that overlapped with the amniote data set were more frequently associated with biological processes linked to development, with the highest ranking GO term being “anatomical structure development” (P = 0.006 following Benjamini–Hochberg correction for multiple tests). This caused us to speculate that genes belonging to certain functional categories might make differential contributions to the overall correlation.
To further investigate the potential link between correlation strength and gene function, we ranked all genes in the primate data set based on their degree of divergence and ordered the resulting gene list in three ways: according to which genes had the highest expression divergence rank relative to their protein divergence rank (ED-biased genes), a higher relative protein divergence rank (dN-biased genes), or the smallest difference between the two ranks (nonbiased genes) (fig. 2A). At an FDR of 0.1%, there was no enrichment for GO terms referring to biological process or molecular function among the most nonbiased genes, and only a single significant term from the cellular component category: “extracellular region.” The ED-biased genes, on the other hand, showed significant enrichments of 452 GO terms after correction for multiple testing, whereas dN-biased genes were enriched for 78 terms (supplementary table S2, Supplementary Material online). Together, these results suggest that the correlation between protein divergence and expression divergence is a global phenomenon that spans a broad range of gene categories, whereas deviations from the overall patterns tend to be associated with specific biological functions.
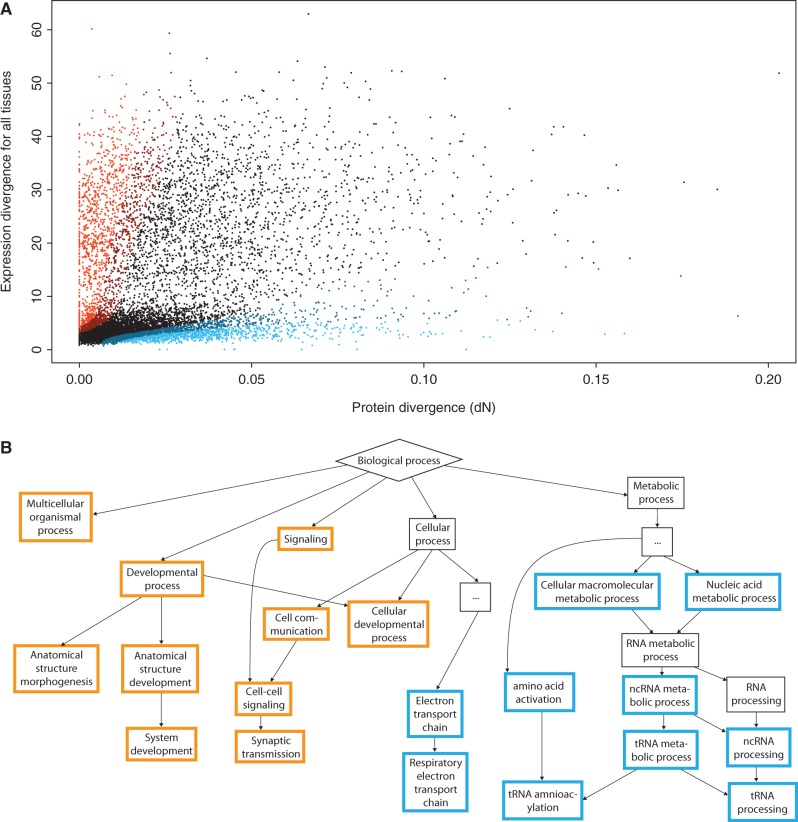
Fig. 2.—
Enrichment of functional categories among ED-biased and dN-biased genes. (A) Expression divergence (six tissues) and dN values for the primate data set (N = 10,227). Increasing bias toward expression divergence is indicated in shades of orange and increasing bias toward protein divergence in blue. (B) Relationships among the 10 most overrepresented GO terms for ED-biased (in orange) and dN-biased (in blue) genes. Some intermediate terms have been omitted for clarity.
We observed clear functional differences between genes that primarily changed their expression pattern or their protein-coding sequences. Among the dN-biased genes, the enriched GO terms were primarily associated with the electron transport chain and tRNA processing, whereas ED-biased genes showed enrichment for processes related to cell communication and the regulation of development (fig. 2B). Perhaps surprisingly, these differences did not seem to chiefly stem from differences in gene expression levels between genes of different functions: For the full data set, there was a negative correlation between the degree of ED bias and the average expression level across tissues (rho = −0.33, P < 10−15, Spearman correlation), but when we repeated the analysis using only genes with above-median expression, the correlation between ED bias and expression level disappeared (rho = −0.01, P = 0.47), whereas we still observed enrichments of the same broad functional categories among the ED-biased and dN-biased genes (supplementary table S2, Supplementary Material online).
For the full data set, many of the GO terms associated with the most ED-biased genes were connected to the nervous system, for example, sensory perception (corrected P value < 10−10), axon guidance (P < 10−9), learning or memory (P < 10−8), and several others. Because we based our analysis on the total ED across six tissues, of which two were neural, we speculated that this might be due to sampling effects. However, the preponderance of neural GO terms persisted when we tested brain and cerebellum individually, and many terms were also found for heart, kidney, liver, and testis (supplementary table S2, Supplementary Material online). This was not surprising considering that ED was correlated across tissues (e.g., for brain and heart: rho = 0.55, P < 10−15), possibly due to similar constraints on gene expression across tissues or a spillover effect, where negative or positive selection on gene expression in a specific tissue also affects divergence in other tissues due to shared regulatory elements. Although the overall correlation between protein divergence and expression divergence is strongest in brain and cerebellum (fig. 1C), it therefore seems that the evolution of genes with specific functions in the primate nervous system is skewed toward changes that affect gene expression.
The Correlation Persists After Correction for 11 Factors
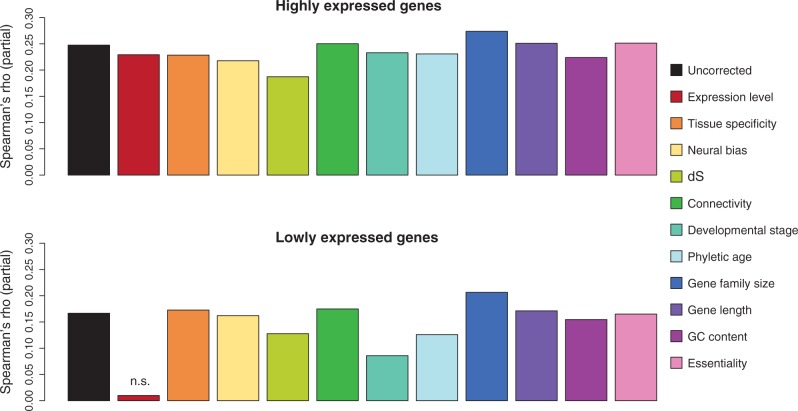
Is the correlation between expression divergence and protein divergence linked to specific gene characteristics or does it reflect more general selective constraints? To investigate this, we performed a partial Spearman correlation analysis, where we corrected the correlation between protein divergence and expression divergence for each of the following factors in turn: average expression level across tissues, tissue specificity, expression bias toward neural tissues, local mutation rate (dS), protein connectivity, developmental stage at which the gene is first expressed, phyletic age, gene family size, GC content of the genomic locus, and essentiality (see Materials and Methods). As our test case, we used estimates of protein divergence and total expression divergence from the multispecies analysis of the primate data set (supplementary table S3, Supplementary Material online). For lowly expressed genes, the difficulty of distinguishing true expression divergence from experimental noise could potentially introduce an expression-dependent bias in our calculations, and we therefore performed separate analyses for highly and lowly expressed genes.
Notably, the correlation between protein divergence and expression divergence was present in both gene sets (fig. 3). If one or more of the investigated factors was crucial for establishing the correlation, we would therefore expect to see a reduction in correlation strength for both highly and lowly genes after correction for the relevant factor. We found that, for lowly expressed genes, the correction for expression level abolished the correlation (fig. 3), possibly due to the bias discussed earlier. A similar, but less pronounced, effect was also seen for some other factors, such as developmental stage, that correlated with expression level and for which the same bias would therefore be present (fig. 4). Importantly, we did not detect the same pattern among the highly expressed genes, where none of the 11 tested factors appeared to have made a substantial contribution to the observed correlation. To some extent, the correlation might have been reinforced by variation in the local mutation rate, consistent with the notion that expression divergence is primarily due to cis-regulatory mutations (Wilson et al. 2008). However, because synonymous mutations are not completely neutral (Chamary et al. 2006), it is also conceivable that dS is subject to the same varying selection pressures that affect protein divergence and expression divergence, that is, the relationship is not causal. In conclusion, we did not find convincing evidence that any of the 11 investigated factors were sufficient to drive the correlation between expression divergence and protein divergence.
Fig. 3.—
Contributions of specific gene characteristics to the correlation between expression divergence and protein divergence. Spearman correlation coefficients for expression divergence (6 tissues) and dN following correction for individual factors, based on the primate data set. Genes with missing data for at least one factor were omitted, leaving a total of 6,228 genes. The data set was further split into highly expressed (above-median expression) and lowly expressed (below-median expression) genes. The black bar represents the uncorrected correlation and the other bars represent partial Spearman correlations, controlling for, from left to right: Total expression across six tissues, tissue specificity (τ), neural bias, dS, protein connectivity, developmental stage at which expression is first observed in humans, phyletic age, gene family size, gene length, GC content of the transcribed region, and essentiality. All correlation coefficients, except after correction for expression level in lowly expressed genes, were significantly different from 0 (P < 0.05) following Benjamini–Hochberg correction for multiple testing.
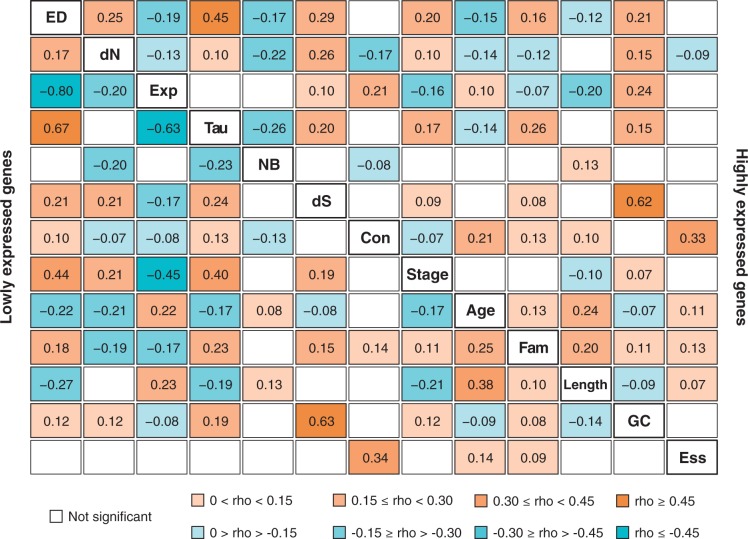
Fig. 4.—
Correlations between gene characteristics. Spearman correlation coefficients for pairwise comparisons of expression divergence (ED), protein divergence (dN), expression level (Exp), tissue specificity (Tau), neural bias (NB), the synonymous substitution rate (dS), protein connectivity (Con), developmental stage at which expression is first observed (Stage), phyletic age (Age), gene family size (Fam), gene length (Length), GC content of the transcribed region, and essentiality (Ess) for genes in the primate data set. Separate calculations were performed for highly expressed (top right) and lowly expressed (bottom left) genes. All shown correlation coefficients were significantly different from 0 (P < 0.05) following Benjamini–Hochberg correction.
Although we were unable to identify a single responsible factor, we speculated that a combination of the investigated factors might drive the correlation in highly expressed genes. We therefore repeated the analysis simultaneously correcting for all 11 factors, but the correlation remained (rho = 0.16, P < 10−15, partial Spearman correlation). As further validation, we constructed a linear model with expression divergence as the dependent variable, and the 11 tested factors and dN as explanatory variables; dN remained highly significant in this analysis (P < 10−15). When we instead excluded dN from the model and calculated the correlation between the residuals and dN, we obtained a Pearson correlation coefficient of 0.14 (P < 10−14). Even when taken together, the 11 factors thus cannot provide a full explanation for the observed correlation.
Naturally, we cannot formally exclude that the impact of one or more of the investigated factors has been underestimated due to noise. Nevertheless, we note that each of the 11 factors showed a significant correlation with at least one other, independently measured, factor (fig. 4), indicating that the estimates are biologically informative. It is also worth noting that the correlation persists in spite of several other differences between the two gene sets. For example, expression level and gene length showed opposite correlation patterns in highly and lowly expressed genes, as was previously shown (Carmel and Koonin 2009). We also observed the same for expression level versus protein connectivity, dS and GC content (fig. 4). Given the many interconnections shown in figure 4, we also consider it unlikely that the analysis of further factors would drastically change our results, because these factors would presumably be correlated with at least one factor in the present data set and would therefore to some extent already have been tested indirectly. Our results therefore suggest that the correlation between expression divergence and protein divergence is not directly linked to between-gene variation in one or more specific gene characteristics but instead is the result of more general selective constraints on gene function.
With this in mind, it is nevertheless surprising that we did not observe a reduced correlation after correction for gene essentiality, given that this parameter should serve as a proxy for the selective constraints that affect a given gene. However, gene essentiality is difficult to measure, especially for humans, and this might have precluded us from appreciating its true impact. We therefore repeated the analysis for a subset of 972 genes, where the human annotation as essential or nonessential was further supported by data from mouse (Chen et al. 2012). Because we only had data for a reduced number of genes, we did not split the set according to expression level. The Spearman correlation coefficient for expression divergence and protein divergence was 0.28 (P < 10−15) for these genes, but after correction for essentiality, it decreased to 0.24 (P < 10−14). Although the decrease was modest, it was nevertheless significant: We repeated the analysis for 100,000 permutations of the essentiality data and in no case did we observe a similar or more extreme decrease. This analysis therefore provides some additional support to the hypothesis that the correlation between expression divergence and protein divergence is driven by general selective constraints, but it is too early to determine the full extent to which these constraints can be captured by measuring gene essentiality.
Concerted Changes of Gene Expression and Protein Sequences during the Evolution of Individual Genes
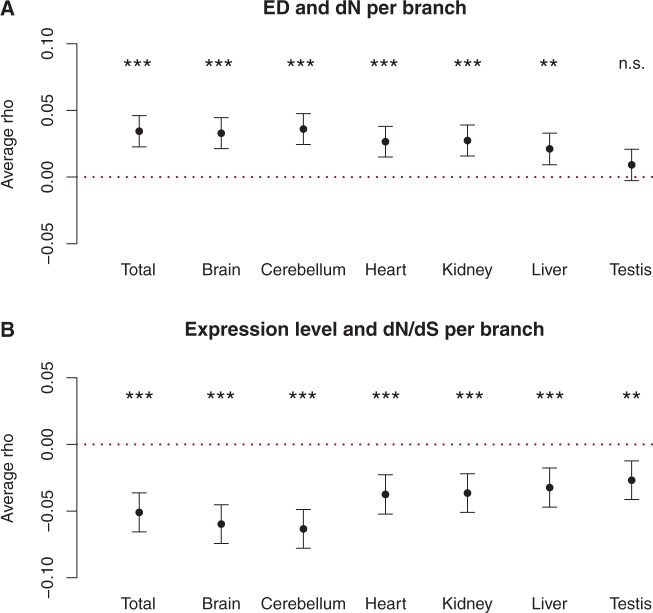
In addition to our analyses of the genome-wide correlation between expression divergence and protein divergence, we asked a complementary question: For any given gene, are periods of rapid protein evolution also associated with rapid expression evolution? To address this, we correlated ranked branch-specific estimates of protein divergence and expression divergence for each gene in the amniote data set (see Materials and Methods). If there is no covariation between the rates of protein and expression evolution, the average correlation coefficient across genes should be 0. In our data, the average correlation coefficient was significantly positive in all cases, except for testis, showing that protein divergence and expression divergence are indeed positively correlated during the evolution of individual genes (fig. 5A). Consistent with figure 1C, the correlation is most pronounced for brain and cerebellum. As an aside, the same approach can also be used to investigate other genomic relationships, such as the impact of expression level on the rate of protein evolution (Subramanian and Kumar 2004; Drummond and Wilke 2008; Gout et al. 2010). We compared expression levels in each species with dN/dS along the corresponding terminal branches and found that, on average, they were negatively correlated (fig. 5B), which suggests that changes in expression level tend to occur in close association with adjustments of the constraint on the protein-coding sequence.
Fig. 5.—
Correlated rates of expression divergence and protein divergence during the evolution of individual genes. (A) Mean value with 95% confidence interval for 3,749 gene-specific Spearman correlation coefficients obtained by correlating ranked branch-specific gene expression divergence and dN values across all branches of the six-species tree (included species: human, macaque, mouse, opossum, platypus, and chicken). The analysis was performed for the combination of all tissues and for each tissue individually. Stars indicate whether the observed coefficients were significantly different from zero (red dotted line), with the following P-value cutoffs after Benjamini–Hochberg correction for multiple testing: Not significant (NS) > 0.05 > * > 0.01 > ** > 0.001 > *** > 0.0001. (B) The analogous analysis for correlations between gene expression levels in the six extant species and dN/dS values for the terminal branches leading to the same species.
The average correlation between branch-specific protein divergence and expression divergence was small, but this is not surprising, because many genes will presumably experience similar selective pressures in all lineages. For these genes, we would not expect to observe correlated patterns of divergence and they would therefore not contribute to the overall signal. In addition, the estimates of expression divergence are likely to be noisy, due to the difficulties of estimating ancestral expression levels (see Materials and Methods). That we still observe a clear effect therefore demonstrates that there exists a robust positive correlation between protein divergence and expression divergence, not only when we compare different genes with each other but also during the evolution of individual genes.
Discussion
We have analyzed the evolutionary relationship between the two principal sources of phenotypic variation between species: expression divergence and protein divergence. Our analyses demonstrate that the positive correlation between these two processes is a general theme of mammalian genome evolution, both in the longer and shorter term: We observe a genome-wide correlation that is stable over evolutionary time, as well as a correlation across phylogenetic branches when we compare orthologs of individual genes. In both cases, the effect is strongest in neural tissues, which implies that the correlation is maintained by strong negative selection. These selective constraints do not appear to be directly linked to the 11 gene characteristics evaluated in this study, with the possible exception of gene essentiality. In particular, we find that the correlation is not explained by tissue specificity or neural bias, which are the two factors in our study that are directly linked to multicellularity. Thus, our results do not support the hypothesis that the correlation arises in multicellular organisms, as a consequence of tissue-dependent selection pressures (Khaitovich et al. 2005; Gu and Su 2007; Tirosh and Barkai 2008). The absence of the correlation between expression divergence and protein divergence in yeast therefore remains unexplained. It is worth noting that yeast, unlike mammals (fig. 4), also lacks a correlation between expression divergence and dS (Tirosh and Barkai 2008).
Although we have attempted to perform an exhaustive analysis, it is of course possible that we have overlooked one or more factors that might be responsible for the correlation between expression divergence and protein divergence. Considering the many associations that exist between different genomic features (fig. 4), we would nevertheless expect to see the indirect effects of such a factor. For example, we have shown that features of developmental gene expression are reflected in data from adult individuals, as shown by the correlations between different aspects of adult gene expression and the developmental stage at which gene expression is first observed (fig. 4). Another possibility is that our results are influenced by the fact that we have measured expression divergence at the mRNA, rather than the protein, level. Given the imperfect correspondence between mRNA and protein expression levels (Vogel and Marcotte 2012), the correlation between protein expression divergence and sequence divergence might therefore be different from that which we report here. Conceivably, this discrepancy might also prevent us from detecting the causal factor underlying the correlation. That said, we are not aware of a mechanism through which post-transcriptional events could obscure the connection to specific gene characteristics, while still allowing us to observe a clear correlation between mRNA expression divergence and sequence divergence. Our results therefore provide substantive evidence in favor of the hypothesis proposed by Lemos et al. (2005), namely that expression divergence and protein divergence are shaped by similar selective constraints but that these constraints are not linked to any specific gene characteristic.
Our analysis also shows how the positive correlation between expression divergence and protein divergence can be reconciled with the unequal contributions seen for morphological and physiological genes (Wray 2007; Haygood et al. 2010; Liao et al. 2010). As seen in figure 2, certain gene categories are more likely to “escape” from the correlation and show disparate values of protein divergence and expression divergence. In particular, we find that genes with disproportionally high expression divergence tend to have developmental and neural functions. This is consistent with what is known about positive selection: Haygood et al. (2010) previously found that human genes with roles in neurogenesis are more likely to show signs of positive selection in their cis-regulatory sequences, in agreement with the hypothesis that adaptive changes in developmental programs are primarily due to changes in gene regulation because these have fewer pleiotropic effects (Wray 2007). However, given that the patterns we observe appear to be primarily due to negative selection and are maintained over long time frames, our results suggest that the bias toward expression divergence among genes involved in development and neural functions is not restricted to genes undergoing adaptive evolution.
Although “escaper” genes tend to belong to particular functional categories, we do not see any enrichment for genes where expression divergence and protein divergence are in proportion to each other. This suggests that the correlation between expression divergence and protein divergence is a global phenomenon, rather than being driven by a few gene types. Together with our observations on the stability and pervasiveness of the correlation, this underlines that the correlated patterns of expression divergence and protein divergence represent a fundamental property of mammalian genome evolution. We therefore suggest that the correlation might best be viewed as an evolutionary “default” state but that specific functional requirements can cause the balance to shift.
Supplementary Material
Supplementary tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Frédéric Schütz for statistical advice, Anamaria Necşulea, Julien Meunier, and Katerina Guschanski for helpful discussion, and the anonymous reviewers for comments that helped improve the manuscript. This research was supported by grants from the European Research Council (Starting Independent Researcher Grant: 242597, SexGenTransEvolution) and the Swiss National Science Foundation (grant 31003A_130287) to H.K. and an EMBO long-term fellowship to M.W.
Literature Cited
- Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci U S A. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Carmel L, Koonin EV. A universal nonmonotonic relationship between gene compactness and expression levels in multicellular eukaryotes. Genome Biol Evol. 2009;1:382–390. doi: 10.1093/gbe/evp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Chen WH, Minguez P, Lercher MJ, Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40:D901–D906. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Eden E, et al. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gout JF, Kahn D, Duret L. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010;6:e1000944. doi: 10.1371/journal.pgen.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Su Z. Tissue-driven hypothesis of genomic evolution and sequence-expression correlations. Proc Natl Acad Sci U S A. 2007;104:2779–2784. doi: 10.1073/pnas.0610797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon LJ, et al. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Haygood R, Babbitt CC, Fedrigo O, Wray GA. Contrasts between adaptive coding and noncoding changes during human evolution. Proc Natl Acad Sci U S A. 2010;107:7853–7857. doi: 10.1073/pnas.0911249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway AK, et al. Adaptive gene expression divergence inferred from population genomics. PLoS Genet. 2007;3:2007–2013. doi: 10.1371/journal.pgen.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Ometto L, Keller L, Goodisman MA. Evolution at two levels in fire ants: the relationship between patterns of gene expression and protein sequence evolution. Mol Biol Evol. 2013;30:263–271. doi: 10.1093/molbev/mss234. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Marino-Ramirez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005;345:119–126. doi: 10.1016/j.gene.2004.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Kim SH, Yi SV. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica. 2007;131:151–156. doi: 10.1007/s10709-006-9125-2. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Liao BY, Weng MP, Zhang J. Contrasting genetic paths to morphological and physiological evolution. Proc Natl Acad Sci U S A. 2010;107:7353–7358. doi: 10.1073/pnas.0910339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol. 2006a;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006b;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- Makova KD, Li WH. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 2003;13:1638–1645. doi: 10.1101/gr.1133803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Pereira V, Waxman D, Eyre-Walker A. A problem with the correlation coefficient as a measure of gene expression divergence. Genetics. 2009;183:1597–1600. doi: 10.1534/genetics.109.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2011. [Google Scholar]
- Sartor MA, et al. A new method to remove hybridization bias for interspecies comparison of global gene expression profiles 'uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res. 2006;34:185–200. doi: 10.1093/nar/gkj413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AO, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Evolution of gene sequence and gene expression are not correlated in yeast. Trends Genet. 2008;24:109–113. doi: 10.1016/j.tig.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, et al. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.