Abstract
The accessory plasmid pAtC58 of the common laboratory strain of Agrobacterium tumefaciens confers numerous catabolic functions and has been proposed to play a role in virulence. Genomic sequencing of evolved laboratory strains of A. tumefaciens revealed the presence of multiple deletion events in the At plasmid, with reductions in plasmid size ranging from 25% to 30% (115–194 kb). Flanking both ends of the sites of these deletions is a short-nucleotide repeat sequence that is in a single copy in the deleted plasmids, characteristic of a phage- or transposon-mediated deletion event. This repeat sequence is widespread throughout the C58 genome, but concentrated on the At plasmid, suggesting its frequency to be nonrandom. In this study, we assess the prevalence of the larger of these deletions in multiple C58 derivatives and characterize its functional significance. We find that in addition to elevating virulence gene expression, this deletion is associated with a significantly reduced carriage cost to the cell. These observations are a clear demonstration of the dynamic nature of the bacterial genome and suggest a mechanism for genetic plasticity of these costly but otherwise stable plasmids. Additionally, this phenomenon could be the basis for some of the dramatic recombination events so ubiquitous within and among megaplasmids.
Keywords: adaptation, phage, deletion, genomic rearrangements, genetic plasticity, virulence
Introduction
Rhizobiaceae is a family of bacteria that includes many of the nitrogen-fixing rhizobia as well as plant pathogenic species of Agrobacterium. The mutualistic or parasitic relationship of these bacteria with their host plants is often dependent on genes carried on their megaplasmids (Hynes et al. 1986; Goodner et al. 1999; Schwartz 2009). These large and frequently conjugative plasmids can comprise up to 10–20% of their host bacteria’s genetic material and are thus important determinants for the ecology of these microbes. Taxonomic classification within this family is often muddied because of the variation and distinct evolutionary histories of these plasmids relative to the chromosomes of the same or similar species (Sawada et al. 2003; Velazquez et al. 2010). This variation is due in part to the horizontal transfer of these plasmids between distantly related bacteria, as well as high rates of recombination that are associated with these replicons (Demezas et al. 1995; Castillo-Ramirez et al. 2009; Meyer 2009; Orozco-Mosqueda et al. 2009).
Several examples, particularly in the Rhizobiaceae family, demonstrate that a substantial portion of the existing genetic diversity is plasmid determined, where isolates of the same or closely related species share the same genomic core with plasmid-concentrated variation (Schofield et al. 1987; Gonzalez et al. 2003; Cervantes et al. 2011; Perez-Segura et al. 2013). One study compared isolates of two Rhizobium species and found that accessory plasmids were related by syntenic blocks of sequence, suggestive of high levels of recombination and genomic rearrangements within and between plasmids (Crossman et al. 2008). Similarly, a genome comparison between the two species A. tumefaciens C58 and Agrobacterium sp. H13-3 (formerly Rhizobium lupini) revealed synteny between both the linear and circular chromosomes, but evidence for a great deal of shuffling between the two plasmids pAtC58 and pAspH13-3 (Wibberg et al. 2011). One possible explanation for these plasmid-specific genomic rearrangements is that, as conjugative elements, plasmids are able to interact with genes of many different species. In so doing, selection may favor the acquisition or loss of certain genes depending on the host. Additionally, repeat sequences are extremely prevalent in rhizobial genomes and it has long been considered that genetic restructuring is predominantly mediated by transposable elements. For example, a study of genetic diversity associated with the symbiotic plasmids of Sinorhizobium meliloti (pSymA and pSymB) show a much higher concentration of transposable elements on its plasmids relative to the chromosomes (Giuntini et al. 2005). Interestingly, there is also significant sequence similarity between specific regions of pAtC58 and pSymA. For example, the genes mediating conjugal transfer of these plasmids, in addition to many other rhizobial megaplasmids, are homologous to one another (Perez-Mendoza et al. 2005; Ding and Hynes 2009). In addition, there appears to be a great deal of plasmid-specific genetic variation, characterized in part, by large deletions occurring throughout the population.
The At plasmid of the plant pathogen and model organism Agrobacterium tumefaciens C58 is a member of the repABC family, defined in the two published C58 genomes to be a 0.54 Mb circular replicon (Goodner et al. 2001; Wood et al. 2001). Sequence similarities exist between pAtC58 and many other important megaplasmids in the Rhizobiaceae family, such as pSym, p42, and pRL, and it has been considered to play a potential accessory role in pathogenesis and virulence of the strain. Although less thoroughly studied relative to its co-resident plasmid, the Ti, or tumor-inducing plasmid pTiC58, pAtC58 is undoubtedly also very important for the ecology of the species. The At plasmid is self-conjugal and encodes genes that confer the ability to catabolize nutrients such as γ-butyrolactones (GBLs) and deoxy-fructosyl glutamine (DFG) that are abundant in the soil and rhizosphere (Baek et al. 2005; Chai et al. 2007). In addition to this, this plasmid is extremely difficult to cure, likely due to the presence of toxin–antitoxin systems on the plasmid (Morton ER, Platt TG, Fuqua C, and Bever JD, submitted).
In this article, we report on the genetic plasticity of pAtC58, specifically the identification of a large deletion, which effectively removes over one-third of the sequenced pAtC58 plasmid. We find this lesion to be widespread, but specific to one C58 laboratory strain lineage. Importantly, this deletion significantly reduces the high cost of pAtC58. The deletion site is flanked by a short repeat sequence notably enriched on the At plasmid and is strikingly associated with a second unique pAtC58 deletion event that developed over the course of a mutation accumulation experiment. This second deletion is distinct in its location from the larger and more pervasive deletion event, but is also flanked by these short repeat sequences.
Materials and Methods
Strains, Plasmids, and Growth Conditions
All strains and plasmids used in this study are described in table 1. Antibiotics and reagents were purchased from VWR (Radnor, PA), Sigma-Aldrich (St. Louis, MO), Fisher Scientific (Pittsburgh, PA), and New England Biolabs (Ipswich, MA). We obtained oligonucleotide primers from Integrated DNA Technologies (Coralville, IA) and used Omega Biotek E.Z.N.A miniprep and gel extraction kits (Norcross, GA) for isolation of nucleic acids. Plasmids were transferred into Agrobacterium strains via either conjugation or electroporation using standard approaches (Morton and Fuqua 2012a). Unless otherwise stated, A. tumefaciens strains were grown in AT minimal media supplemented with 0.5% glucose (wt/vol) and 15 mM (NH4)2SO4 (ATGN) (Tempé et al. 1977) and incubated at 28 °C in a rotary aerator or on 1.5% agar plates. Induction broth for activating Ti plasmid virulence gene expression contained acetosyringone (200 μM) and was prepared using the recipe described in Morton and Fuqua (2012b). Antibiotics for A. tumefaciens (or Escherichia coli) were used at the following concentrations: 2,500 (25) μg/ml streptomycin (Sm), 250 (100) μg/ml spectinomycin (Sp), and 300 (100) μg/ml kanamycin (Km). Oligonucleotide sequences developed for this study are provided in supplementary table S2, Supplementary Material online.
Table 1.
Strains and Plasmids Used in this Study
| Species | Strain | Characteristics | Plasmids | Reference |
|---|---|---|---|---|
| Agrobacterium tumefaciens | C58-CU | Wild-type C58 (isolated from cherry gall; Geneva, NY) | pTiC58, pAtC58-CU | Gift from Steven Beer (Cornell University) |
| A. tumefaciens | C58-UI | Wild-type C58 laboratory strain | pTiC58, pAtC58 (truncated) | Gift from Steve Farrand (University of Illinois) |
| A. tumefaciens | ERM76 | Isogenic to C58-CU; pTiC58 transconjugant | pTiC58 | Morton ER, Platt TG, Fuqua C, and Bever JD, submitted |
| A. tumefaciens | ERM81 | Isogenic to C58-CU | pTiC58, pAtC58-CU | Morton ER, Platt TG, Fuqua C, and Bever JD, submitted |
| A. tumefaciens | ERM77 | C58-CU ΔtetRAΩaada (SpR; SmR) | pTiC58, pAtC58-CU | Morton ER, Platt TG, Fuqua C, and Bever JD, submitted |
| A. tumefaciens | ERM80 | Isogenic to C58-CU; carrying pAtC58 from C58 S.F. | pTiC58, pAtC58 | This study |
| A. tumefaciens | ERM101 | ERM80 ΔtetRAΩaada (SpR; SmR) | pTiC58, pAtC58-CU | This study |
| A. tumefaciens | NT1 | pTiC58-cured derivative of C58 | pAtC58 | Watson et al. (1975) |
| A. tumefaciens | NTL4 | pTiC58-cured derivative of C58; ΔtetRA | pAtC58 | Luo et al. (2001) |
| A. tumefaciens | A348 | A136 transformed with pTiA6 | pAtC58, pTiA6 | Garfinkel et al. (1981) |
| A. tumefaciens | A136 | C58 heat-cured of pTiC58 | pAtC58 | Watson et al. (1975) |
| A. tumefaciens | CIRS | pTiC58-cured derivative of C58 | pAtC58 | Ellis et al. (1982) |
| A. tumefaciens | AB150 | Isogenic to UIA5 carrying pAtC58 from A136 | pAtC58 | Nair et al. (2003) |
| A. tumefaciens | AB151 | Isogenic to AB150; electroporated with pTiC58 | pTiC58, pAtC58 | Nair et al. (2003) |
| A. tumefaciens | AB152 | Isogenic to UIA5; electroporated with pTiC58 | pTiC58 | Nair et al. (2003) |
| A. tumefaciens | AB153 | Isogenic to AB150; electroporated with pTiC58 | pTiC58, pAtC58 | Nair et al. (2003) |
| Escherichia coli | S17-1 λpir | pSM243cd derivative with PvirB-lacZ | pSW209 | Gift from Steve Winans (Cornell University) |
Whole-genome sequencing of A. tumefaciens strains was performed with total genomic DNA used to generate a paired-end library following the modified protocol of Lazinski and Camilli (http://tucf-genomics.tufts.edu/documents/htseq_protocol_for_illumina_paired.pdf, last accessed July 8, 2013). Approximately 20 µg sheared genomic DNA was blunt-ended using the NEB Quick Blunting Kit (New England Biolabs). Following conversion to blunt ends, the large fragment of DNA Polymerase I (Klenow fragment) was used to add one deoxyadenosine to the 3′-ends of the DNA preparation. This DNA preparation was ligated with an adapter mix consisting of primers OLJ131 and OLJ137 using the NEB Quick Ligation Kit (New England Biolabs). Finally, library amplification was performed by polymerase chain reaction (PCR) with primers OLJ139 and OLJ140. Sequencing was performed on an Illumina HiSeq 2000 at the Tufts University Core Facility.
Curing the At Plasmid
The At plasmid was cured from C58 using a combination of approaches. First, the At plasmid was marked by inserting an ampicillin resistance gene (blaC) into the intergenic region of Atu5161 and Atu5162, by an allelic replacement cloning strategy (Morton and Fuqua 2012a). Similar to the method described by Uraji et al. (2002), a curing plasmid (pEM123), was constructed by cloning the entire replication region, 4,170 bp including the promoter, repABC, and the incβ region of pAtC58 into pNPTS138 (Spratt et al. 1986) using BamHI and SpeI restriction sites (see fig. S1, Supplementary Material online, for plasmid map and description). This vector contains a counterselectable sacB gene conferring sucrose sensitivity in addition to a kanamycin-resistance gene. This curing plasmid was conjugated into C58 by an E. coli S17-1 λpir donor. Plating on ATGN supplemented with kanamycin selected for A. tumefaciens pEM123 transconjugants. The two plasmids, pEM123 and pAtC58, use the same replication and partitioning machinery, and are therefore incompatible (Uraji et al. 2002; Cevallos et al. 2008). As a consequence, transconjugants that contain both plasmids give rise to cells that lack either the At or the curing plasmid. Growth and several days of passaging in the presence of kanamycin was carried out to select for only those cells that carry pEM123 and had lost pAtC58. Because this method alone was not sufficient to promote loss of the plasmid, cells were subsequently exposed to heat shock (42 °C) for 60 s. Isolates from this population were screened for loss of the At plasmid by ampicillin-sensitivity and negative PCR results using multiple regions distributed across the At plasmid. Subsequently, these presumptive pAtC58- derivatives were grown in the presence of 5% sucrose and absence of kanamycin to select for cells that had lost the curing vector via segregation. These sucrose-resistant colonies were patched onto ATGN supplemented with kanamycin to screen for antibiotic-sensitive clones. Sucrose-resistant and kanamycin-sensitive clones were screened for the absence of repABC.
Diagnosis of At Plasmid Deletion
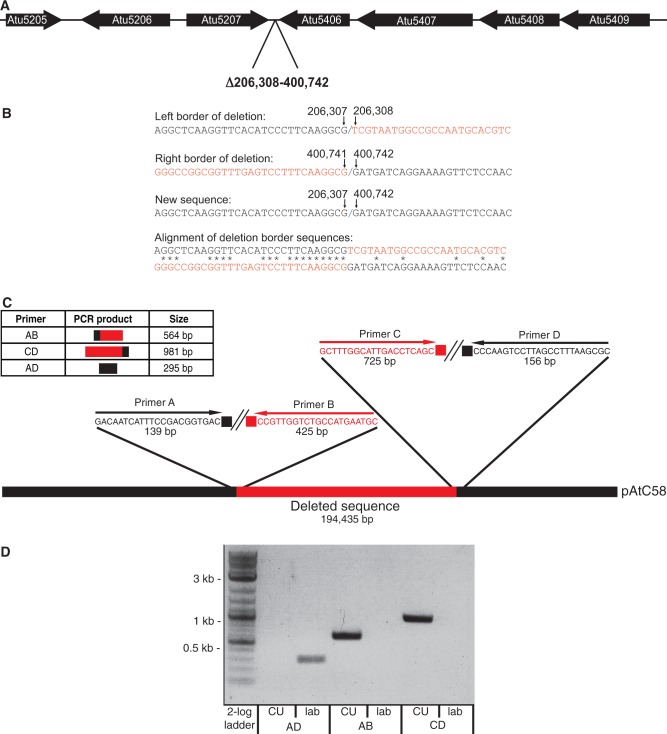
To determine the presence or absence of the described deletion, four primers (supplementary table S2, Supplementary Material online) were designed to amplify either 1) across the entire deletion or 2) across its borders from just external of the area deleted in the truncated pAtC58 to within this area. Figure 1 shows a schematic of the diagnostic primer design. Primers A and D, flanking the deletion, will amplify a 295-bp product for strains harboring the truncated At plasmid. For strains with the full-length form of the plasmid, the distance between primers A and D is too long (∼200 kb) and will thus yield a negative PCR result (fig. 1). For positive confirmation of strains with the full-length plasmid, primer pairs AB and CD will yield 564 and 981 bp products, respectively. The truncated plasmid is missing sequence that is homologous to the internal primers B and C so these reactions should not result in any amplified product. Subsequent sequencing of the purified PCR product(s) provided additional confirmation either for the presence or the absence of the deletion.
Fig. 1.—
Schematic of Agrobacterium tumefaciens laboratory strain pAtC58 194 kb deletion. (A) Gene map for the genes directly flanking the deletion. (B) Sequence at both borders of the deletion, the new sequence formed after the deletion at the junction site, as well as an alignment of the flanking ends. Red letters denote sequence included in the deletion. Black letters denote sequence that is still present in the truncated form of the plasmid. (C) Diagnostic primer design around both flanking ends of the deletion. Sequence included in the deletion is represented in red, and that which remains on the full-length form of the plasmid is in black. The chart shows each of the primer pairs and the corresponding expected product and product size. Pairs AB and CD only amplify from the full-length plasmid and the pair AD only yields the shown product for the truncated form of the plasmid. (D) Electrophoresis gel showing the PCR products resulting from amplification with diagnostic primers AD, AB, and CD described in the text.
Generation of Gene Ontologies for Full-Length and Truncated At Plasmids
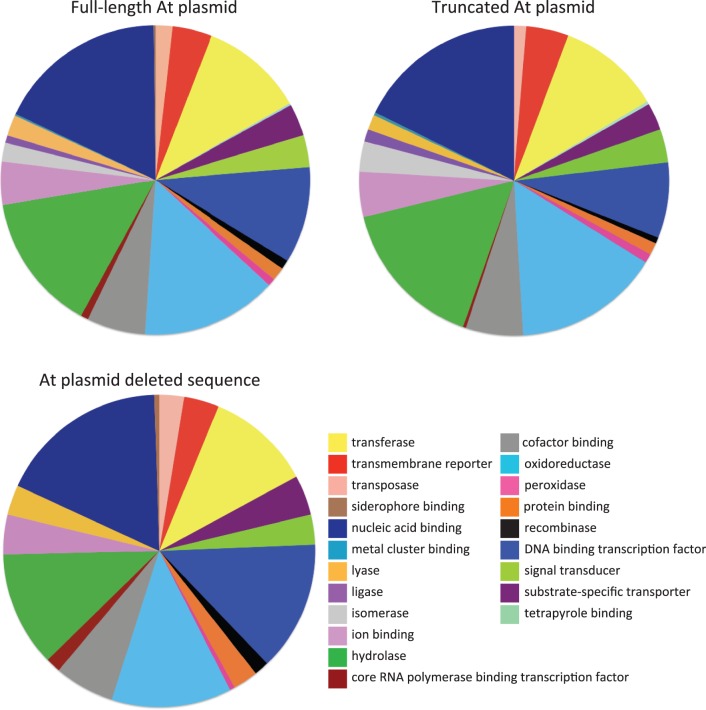
GO annotations were generated for the published sequences of the full-length and truncated At plasmids of A. tumefaciens C58, as well as the deleted sequence using Blast2Go Software (Conesa et al. 2005). The initial BLASTX search was performed against the NCBI database of nonredundant sequences using a minimum expectation value of 1 × 10−3 and a high scoring segment pair cut-off of 33. Annotations were generated using the pre-E-value Hit Filter of 1 × 10−6, an annotation cut-off of 55, and a GO weight of 5. Annotations were then merged with GO terms that were assigned based on their protein functional domains by performing analysis with InterProScan (Mulder et al. 2005). Graphs were generated based on function and Node Score with a Score alpha of 0.6. The sequences were unfiltered and the combined graphs shown in figure 3 represent distribution of genes GO level 3.
Fig. 3.—
Ontology of the full-length At plasmid, the truncated laboratory strain At plasmid (ΔAtu5208-Atu5405) and the deleted sequence. Pie charts show the distribution of Gene Ontology terms (Level 3) of molecular function for the deletion, the full-length and truncated At plasmids. This distribution is based on annotated genes only.
Measurement of Gene Induction
Strains ERM76, ERM80, and ERM81 (table 1) were transformed with the reporter plasmid, pSW209Ω (S.C. Winans, Cornell University) which carries a PvirB::lacZ fusion (PvirB from pTiA6). Cells were grown approximately 24 h in Induction Broth (pH 5.6 and 200 µM acetosyringone) (Winans et al. 1988). When cultures reached mid-log phase of growth, they were assayed for β-galactosidase activity as described (Morton and Fuqua 2012b). Activity is presented in terms of Miller Units; a quantitative measure of specific activity that accounts for gene-expression mediated β-galactosidase activity, normalized to growth. The equation for calculating Miller Units (MU) is as follows:
where A420 represents absorbance of o-nitrophenol, OD600 represents optical density of the culture, t is the time of the reaction, and V is the culture volume. Each treatment was carried out in triplicate.
Measuring the Cost of the At Plasmid Truncation
To determine the fitness costs associated with the truncated portion of the At plasmid, isogenic strains harboring either the full-length or truncated At plasmids were competed in carbon-limiting ATGN media (C1) that was supplemented with 1 mM glucose (compared with 28 mM in standard ATGN). All competitions were inoculated with approximately 6 × 106 cells at a 1:1 ratio. A spectinomycin/streptomycin (SmR/SpR) cassette was integrated in the tetRA locus of each strain by allelic replacement (Morton and Fuqua 2012a). To control for the costs associated with the aadA marker (conferring resistance to Sp and Sm antibiotics) the strain carrying the cassette was reversed for half of the competitions. At the beginning of each competition, strains were used to inoculate 2 ml of ATGN media and incubated at 28 °C for 24 h until reaching exponential mid-log phase. Individual strains were subsequently washed in 1× AT buffer three times to remove excess glucose from the starting media. The cultures were each normalized to an optical density at 600 nm (OD600) of approximately 0.2, at which point they were mixed 1:1 into 2 ml carbon-limiting media to a final OD600 of 0.01. The competition cultures were then incubated at 28 °C for 24 h, after which all mixed cultures were subcultured 1:100 into 2 ml of C1 media and incubated as before. This passaging was repeated the same way through the seventh passage. At the first, second, and seventh passages, a dilution series of each mixed culture were plated onto ATGN and ATGN plus Sm and Sp so that the frequencies of each strain could be estimated. Replicates for this experiment were run on four separate occasions. Relative fitness was estimated as the ratio of the number of doublings by each strain during the course the competition experiment as described (Lenski 1988).
This experiment was analyzed with a mixed effect model using the mixed procedure of SAS software. The mixed model included the experiment and the marker orientation as random effects. This analysis allowed us to remove variance associated with the marker and differences between experimental runs and thereby isolate the effect of the 0.2 Mb of At plasmid DNA by comparing least square means of the different competitions.
Generation and Sequence Analysis of Mutation Accumulation Lines
Agrobacterium tumefaciens C58-CU was grown to mid-log phase in Tryptone Yeast (TY) liquid media before plating for isolated colonies onto TY agar plates. From these plates, 75 colonies were randomly chosen and struck for isolated colonies onto TY agar plates (one MA line per plate), representing the second passage from the first bottleneck. For all subsequent passages, a single colony was chosen for streaking onto fresh TY agar plates. To limit isolation bias, the colony closest to the cross-secting line of each Petri plate (designated by a plate-sized template) was chosen for passaging. Following 125 passages (∼3,350 generations), a colony from each MA line was selected for inoculation into 2 ml TY liquid media and subsequently frozen at −80 °C in 25% glycerol. Cells from frozen permanents of 10 randomly chosen MA lines were inoculated into 2 ml TY liquid media. The DNA of mid-log phase cultures was subsequently isolated using Wizard Genomic DNA purification kit (Promega).
101-bp paired-end Illumina (Illumina Hi-Seq platform) sequencing was applied such that each A. tumefaciens MA line was sequenced to a coverage depth of approximately 100× with an average library fragment size (distance between paired end reads) of approximately 175 bp. The paired-end reads for each line were individually mapped against the A. tumefaciens C58 reference genome (assembly and annotation available from the National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov, last accessed July 8, 2013) using two separate alignment algorithms: BWA (Li and Durbin 2009) and NOVOALIGN (available at www.novocraft.com, last accessed July 8, 2013). The resulting pileup files were converted to SAM format using SAMTOOLS (Li et al. 2009). Using in-house perl scripts, the alignment information was further parsed to generate forward and reverse mapping information at each site, resulting in a configuration of eight numbers for each line (A, a, C, c, G, g, T, t), corresponding to the number of reads mapped at each genomic position in the reference sequence. To identify small- and medium-sized deletion and insertion events (>11 bp), we further implemented the pattern growth alignment algorithm PINDEL (Ye et al. 2009) and the high-resolution structural variation mapper BREAKDANCER (Chen et al. 2009).
Results
Identification and Description of the pAtC58 Deletion
The genome of our laboratory strain of A. tumefaciens C58 was resequenced by Illumina and compared with the published C58 genome (Wood et al. 2001; Goodner et al. 2001). With the expectation to find numerous genetic changes after years of passaging and maintenance in the laboratory, we identified only a small number of base substitutions and small nucleotide polymorphisms between our laboratory lineage and the published C58 genomes. However, in addition to this, we discovered that the At plasmid has incurred a large approximately 0.19 Mb deletion relative to the previously sequenced strain. The truncated At plasmid is approximately 0.35 Mb with deletion of all genes between Atu5207 and Atu5408 (positions 206,308–400,472). Figure 1A and B depict the deletion and shows the resulting new sequence where Atu5207 and Atu5408 are convergent with one another. Using primers that are flanking the deletion we were able to amplify across the deletion junction, validating the data of the genome sequence (fig. 1C and D), whereas primers that should prime out of the missing segment into flanking sequences on each end failed to amplify. It is important to point out that this was a clean deletion and there are no signs of any genetic shuffling in the remaining portion of the plasmid.
A Short Repeat Sequence Flanks pAtC58 Deletions
Interestingly, flanking both ends of the missing pAtC58 segment in the published genome sequence is a short 13-bp (TTCNTTCAGGCG) repeat. In the truncated form of the plasmid, the two ends have recombined to leave one copy of the repeat sequence, a hallmark of a phage- or transposon-type excision event. We find that variations on this sequence occur quite frequently throughout the At plasmid (fig. 2; supplementary table S1, Supplementary Material online). A 9-bp repeat within the larger 13 bp (TTCAGGCG) appears 13 times on the At plasmid. Supplementary figure S2, Supplementary Material online, shows an alignment of this sequence and it is apparent that the 13 bp sequence is also quite common, particularly in regions proximal to the deletion. Supplementary table S1, Supplementary Material online, shows the gene annotation (Atu) numbers marking the locations of all 9-bp repeats including 4 within the deletion itself. One can see that the repeat sequence is rather evenly dispersed around the plasmid with a few exceptions (fig. 2). At Atu5007, there is one repeat within the predicted coding sequence, and one that is just upstream of it (downstream of Atu5006). In addition to this, the repeat sequence can be found in the two adjacent genes, Atu5435 and Atu5436. There is no clear signature for where this repeat can be found based on the predicted functions of these genes.
Fig. 2.—
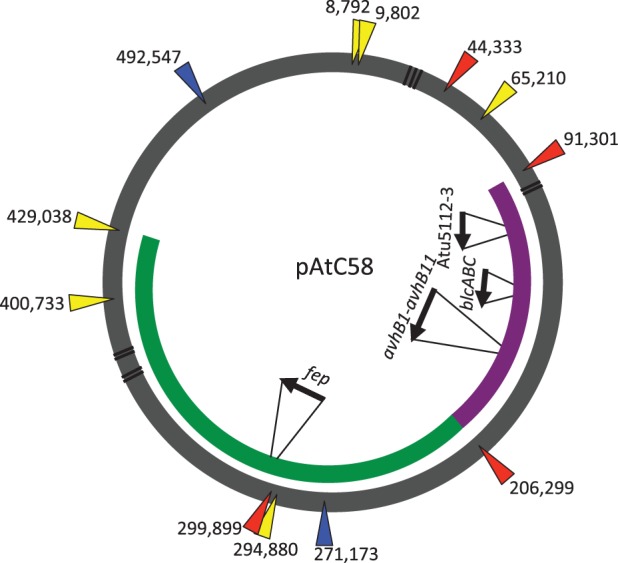
Distribution and corresponding starting base number for each 9-bp repeat sequence located throughout pAtC58, where the green band shows the region of sequence that is deleted in the laboratory At plasmid and the purple band indicates the region of sequence deleted in the mutation accumulation line. Yellow, blue, and red arrows correspond to locations of the 9, 11, or 13-bp repeat, respectively. Black lines indicate positions of putative transposase or transposase fragments and gray triangles and arrows denote recognized genes or operons of importance.
In quantifying the frequency of this repeat sequence on each of the four replicons in C58, we find that they are notably overrepresented on the At plasmid relative to the other 3 replicons (the Ti plasmid, the linear chromosome, and the circular chromosome). Table 3 shows the number of occurrences of the 9-, 11-, and 13-bp repeat sequences, normalized to the size of each replicon. The difference becomes quite apparent for the 11-bp sequence that appears six times throughout the 0.54 Mb At plasmid, only once on the Ti plasmid, and only five times on both chromosomes combined (5.8 Mb of DNA), representing a 5-fold to 100-fold difference in frequency. For the 13-bp sequence which is flanking the deletion with the exception of one variable position (and thus not included in the tally in table 3) it is completely restricted to the At plasmid, and there it exists at four locations which are spaced around the plasmid. It is interesting to note that neither of these 11 and 13 bp sequences exist on the related pSym plasmids of S. meliloti.
Table 3.
Frequency of 9-, 11-, and 13-bp Repeat Sequence in All Four Replicons of Agrobacterium tumefaciens C58
| Replicon |
||||
|---|---|---|---|---|
| At Plasmid (0.54 Mb) | Ti Plasmid (0.21 Mb) | Linear Chromosome (2.08 Mb) | Circular Chromosome (2.84 Mb) | |
| TTCAAGGCG | 13 (0.024) | 2 (0.0093) | 33 (0.0159) | 39 (0.0137) |
| CCTTCAAGGCG | 6 (0.0111) | 1 (0.0047) | 2 (0.001) | 3 (0.001) |
| TCCCTTCAAGGCG | 4 (0.0074) | 0 (<0.0047) | 0 (<0.0005) | 0 (<0.0004) |
Values in parentheses indicate frequency per kilobase.
Gene Ontology of Full-Length and Truncated At Plasmids
GO analysis of the full-length and truncated At plasmids reveals that both plasmids have the same relative proportion of genes associated with each functional category (fig. 3). Thus, although a large fraction of the plasmid was lost, the basic functional distribution remained the same. This is not surprising, provided that the truncation is due to a single deletion event but it does demonstrate that genes of distinct functional categories are relatively evenly dispersed around the plasmid, some of which are likely to be redundant. The truncated At plasmid is approximately 64% of the size of the full-length plasmid, and the relative number of genes that fall into each GO category is comparable with this ratio, only deviating for smaller categories. The only exception to this is for genes with predicted isomerase activity, of which there are 10 on both the full-length and truncated forms of the At plasmid (none are present on the deleted segment). Interestingly, there are 9 genes that were identified as having transposase activity, only 5 of which remain on the truncated form of the plasmid. A cluster of genes homologous to the fep iron siderophore uptake system (Atu5311-5316) were removed from the At plasmid with the deletion. The importance of iron in the regulation of cellular processes within A. tumefaciens has been clearly established (Hibbing and Fuqua 2011). However, between strains harboring the full-length versus truncated At plasmids we found no difference in growth during iron limitation. It is important to point out that most of the more thoroughly characterized genes on pAtC58, including those associated with conjugation (avhB) and γ-butyrolactone catabolism (bcl), are all still present on the truncated At plasmid. The plasmid was experimentally determined to be fully conjugative and is predicted to harbor many of the same catabolic properties.
Tracing the Origins of the pAtC58 Plasmid Deletion
The large deletion (∼0.2 Mb) is found on a major replicon of an important pathogen and model organism and its prevalence has important consequences for any labs studying the biology of A. tumefaciens. C58 is one of the more widely used strains. Using primers designed to amplify 1) across the deletion for the truncated plasmid or 2) across both of the deletion borders for the full-length plasmid (fig. 1A and B), we characterized the At plasmids of 12 frequently used A. tumefaciens C58 derivatives (table 2). Based on this method of diagnostics, we found that the deletion was found throughout several C58 laboratory strains (acquired from colleagues at Williams College, C58-WC and the University of Illinois, Urbana-Champaign, C58-UI), but not present in cells grown from an archival C58 stock (C58-CU) obtained through the Department of Plant Pathology at Cornell University in Ithaca, NY. The original C58 isolate was obtained from a cherry tree gall in Geneva, NY, in 1958, and this Cornell stock is thought to represent the closest stock to this isolate (Hamilton and Fall 1971).
Table 2.
Characterization of the At Plasmid of Common Laboratory Agrobacterium tumefaciens Strains
| Strain | Across Deletiona,b | From within 5′-End of Deletiona,b | From within 3′-End of Deletiona,b |
|---|---|---|---|
| C58 (Indiana University) | + | − | − |
| C58 (University of Illinois) | + | − | − |
| C58 (Williams College) | + | − | − |
| C58 (University of Washington) | − | + | + |
| C58-CU | − | + | + |
| NT1 | − | + | + |
| NTL4 | − | + | + |
| C1RS | − | + | + |
| A348 | − | + | + |
| A136 | − | + | + |
| AB150 | − | + | + |
| AB151 | − | + | + |
| AB153 | **c | + | + |
aTable summarizes the results of diagnostic PCR using primers to amplify either across the ΔAtu5208-Atu5405 deletion (column 2) or from within the deletion (columns 3 and 4).
bAmplification across the deletion, but not from within is indicative of the truncated form of the plasmid. Amplification from within the deleted sequence, but not across indicates that the full-length At plasmid is present in that strain.
cAsterisks (**) indicate that there were distinct band of unique size.
All of the C58 derivatives with the deletion are reported to originate from the C58-UI stock from the University of Illinois, Urbana-Champaign (collection of S.K. Farrand). The C58-UI stock is itself derived from laboratory stocks at the University of Washington, Seattle, in the laboratory of E.W. Nester (S.K. Farrand, personal communication). Individual cultures from all of the available UW stocks were tested using the diagnostic PCR and none were found to have incurred the deletion (table 2).
A collection of several other purported C58 derivatives in our laboratory stocks (tables 1 and 2) were all found not to have incurred this deletion event. The C58-derived AB strains were generated by Nair et al. (2003) to be isogenic with one another so that the At and Ti plasmids were introduced independently into the plasmidless C58 derivative UIA5, to generate a pAtC58 plasmid-harboring derivative (AB150), one that carries only pTiC58 (AB152) and a Ti + At + derivative (AB153). AB151 is another strain that carries both the At and Ti plasmids and was generated the same way as AB153. All of these strains, except for AB153 generated the amplification patterns predicted for the full length pAtC58 plasmid (table 2). AB153 is unique in that it has a distinct At plasmid deletion profile such that primers designed to amplify across the deletion not only yield a band that is ∼3 kb in size (larger than the 295-bp product characteristic of the C58 strains which carry the deletion), but also yield the amplicons predicted for the full length plasmid. This suggests that a duplication event has occurred in the AB153 strain resulting in priming from multiple sites along the plasmid.
Common pAtC58 Truncation Correlates with Increased vir Expression
Previous studies have demonstrated that the presence of the At plasmid corresponds to an increase in tumor-size in Ti plasmid carrying strains (Nair et al. 2003). Based on this observed interaction between the At plasmid and the level of disease induced by the Ti plasmid, we were interested in determining whether the At plasmid deletion had any impact on associated phenotypes. In this experiment, we introduced a PvirB-lacZ transcriptional fusion expression construct, pSW209Ω (Wang et al. 2000) into three C58-derived strains: C58-CU (the original isolate harboring the full-length At plasmid), our own C58 lab strain (carrying the truncated At plasmid), and C58 pAt- (a plasmidless derivative to which the Ti plasmid was reintroduced by conjugation). We found that under inducing conditions, the presence of the full-length At plasmid is associated with an approximately 50% decrease in the expression of the virB virulence operon relative to the derivative with the full length At plasmid (fig. 4). This suggests that some factor(s) that directly or indirectly negatively regulates expression of these virulence genes is encoded between Atu5207 and Atu5408. We did not find that the presence of the truncated pAtC58, relative to the full-length plasmid, had any impact on tumor size or number based on standard assays in which potato slices were inoculated with these strains (data not shown). However, the correlation between the level of vir gene expression and tumor size and frequency in this assay have not been thoroughly tested, and thus we are hesitant to speculate as to whether this decreased vir expression might significantly impact disease incidence and severity.
Fig. 4.—
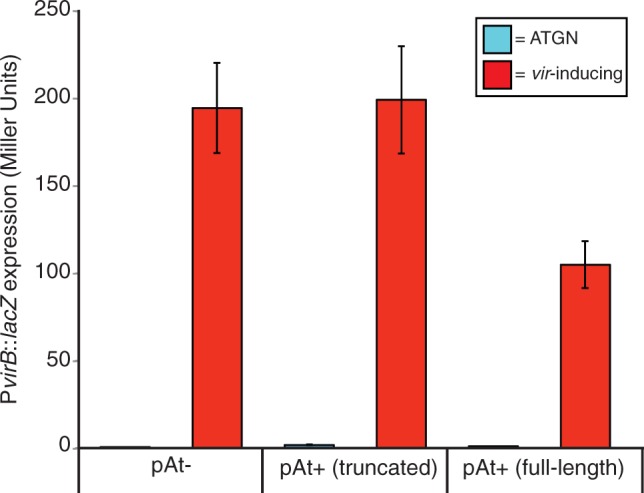
Expression of PvirB-lacZ in Agrobacterium tumefaciens C58 pAt-, pAt+ truncated (ΔAtu5208-Atu540), and pAt+ full-length. Cells were grown in liquid for 24 h in either ATGN minimal media (pH 7.0, 50 mM phosphate) (blue bars) or virulence-induction broth (pH 5.6, 50 µM phosphate, 200 µM acetosyringone) (red bars). Error bars represent standard deviation between biological replicates (n = 3). P < 0.0001, unpaired student t test.
Decreased Plasmid Carriage Cost due to pAtC58 Truncation
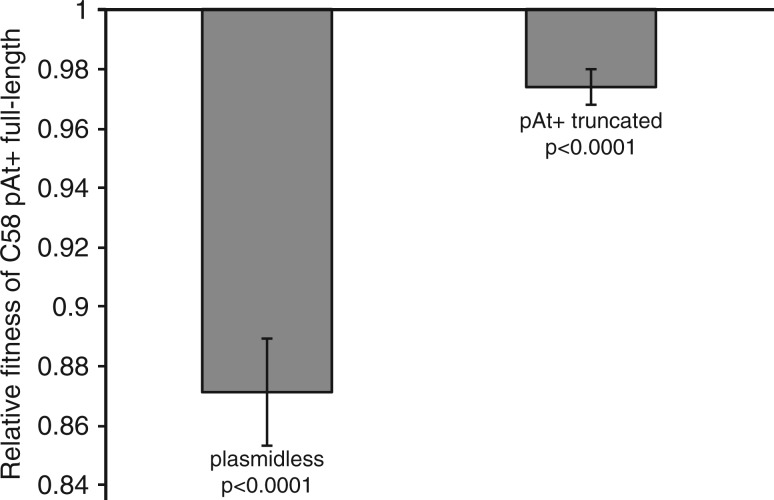
To determine the carriage cost of the truncated portion pAtC58 plasmid, a competition experiment was carried out between the original A. tumefaciens C58-CU isolate and our laboratory C58 strain harboring the truncated form of the plasmid. Strains were mixed at a 50/50 starting ratio and passaged 7 times for a total of approximately 46 generations. At the end of the experiment the relative fitness of each strain was determined (fig. 5). Competitions between a C58 strain harboring the full-length At plasmid and a plasmid-cured derivative (independent analysis) revealed a high cost indicated by a relative fitness decrease of 0.129 (P < 0.0001). The results of the competition also showed a significantly higher relative fitness of the strain harboring the truncated plasmid relative to the full-length one (diff = 0.103; P < 0.0001). This increase in fitness of the common laboratory strain indicates that there is a significant cost associated with the deleted genetic material, and suggests that the loss of this portion of the plasmid was adaptive under laboratory conditions. This suggests that approximately 74.57% of the cost associated with the full-length At plasmid is linked with the deleted genetic material, despite the fact that many genes, including those associated with conjugation and replication are still present on the truncated form of the plasmid.
Fig. 5.—
Relative fitness based on pairwise competitions between Agrobacterium tumefaciens C58 pAt+ full-length cells versus 1) a plasmidless derivative and 2) the pAt+ truncated laboratory strain (ΔAtu5208-Atu5405). In each experiment, both strains in the competition pair were normalized to a 50/50 starting ratio and competed in carbon-limiting media for seven passages and a total of 46 generations. Each bar represents data from two separate experiments, for which the statistics were calculated independently as described. Data summarizing the results of competition between the C58 pAt+ (full-length) and C58 plasmidless strains include four independent experimental runs with four replicates for each competition pair. For strains harboring either the full-length or the truncated form of the At plasmid, data include results of three experimental runs with eight replicates for each competition pair.
Distinct Large Deletion Flanked by Repeat Sequence
A large deletion flanked by the identified repeat sequence is not an isolated event. As a consequence of a separate experiment that was designed to determine the basal mutation rates of multiple organisms, including A. tumefaciens C58-CU, we discovered a second deletion event that was qualitatively similar to the first. Sequencing of 10 mutation accumulation lines (after ∼3,350 generations and 125 bottlenecks) revealed that the At plasmid in one line had undergone a different, approximately 0.115 Mb deletion (fig. 2). One border of this second deletion was shared with that of the laboratory strain (between Atu5207 and Atu5408) whereas the other end was located at the border of an another one of the repeat sequences between Atu5087 and Atu5088. Although smaller, this second deletion removed the At plasmid conjugation genes, the blcABC operon, in addition to a putative toxin-antitoxin pair (Atu5112 and Atu5113).
Discussion
Similar to many rhizobial plasmids, the At plasmid is comprised of a mosaic of sequences that show synteny to both plasmid and chromosomal genes of multiple species of soil bacteria ranging from other α-proteobacteria to γ-proteobacteria such as Azotobacter and Pseudomonas spp. This type of large megaplasmid deletion has been observed in other plasmids, such as the symbiotic megaplasmid of the nitrogen-fixer, R. phaeseoli. In this case, the deletion event was one of several frequently occurring genomic rearrangements of the nodulation and nitrogen fixation genes (∼120 kb), generated by recombination between reiterated nif operons (Romero et al. 1991).
Repeat sequences are potential markers for lysogenic phage or transposon-mediated recombination events. There are many examples of IS elements, integrative and conjugative elements (ICEs), and pathogenicity/symbiosis islands that can generate short duplications upon insertion, which are resolved after excision. This duplication event is similar to what is found flanking either end of the deleted pAtC58 region that we report here. These repeat sequences are prevalent throughout microbial genomes and have been found flanking large deletions in human mitochondrial DNA (Iida et al. 1985; Schon et al. 1989). Interestingly, in a comparison of two different C58 genomes, originally sequenced in 2001 by (Goodner et al. 2001; Wood et al. 2001), the presence of multiple replicon specific classes of repeats were identified (Slater et al. 2013). These inverted repeats (CIR elements) are distinct from those described in this study, but may also be important for genome evolution in the genus. The observation that the pAtC58-enriched 11- and 13-bp repeats cannot be found anywhere on the closely related pSym plasmids suggests that 1) their prevalence is not random, and 2) if their presence is a signature of some transposable element, it is specific to this A. tumefaciens strain. This has strong implications regarding the genetic plasticity of the At plasmid and demonstrates a clear mechanism driving these rather dramatic changes.
It is interesting to note that there are a total of nine transposable elements that were identified using GO Software. These include Atu5025, Atu5027, Atu5028, Atu5092, Atu5094, Atu5352, Atu5353, Atu5370, and Atu5371. None of these putative transposases or transposase fragments correspond with the locations of the 9-bp repeat sequence that was found flanking either deletion.
The high cost associated with the deleted portion of the laboratory strain At plasmid suggests that passage in the lab may have selected for fixation of this deletion event. The full-length At plasmid incurs a large cost to host cells, and is extremely difficult to cure (Morton ER, Platt TG, Fuqua C, and Bever JD, submitted). There are several putative toxin–antitoxin systems on the plasmid, and one of these antitoxins has been identified from a genome-wide screen of essential genes (Curtis PD, Brun YV, personal communication). Both the toxin (Atu5112) and antitoxin (Atu5113) of this system, homologous to HipA and HipB, are present on the truncated form of the plasmid. The difficulty in curing the At plasmid is likely due, at least in part, to the presence of this and similar systems. Interestingly, this gene pair was removed in the mutation accumulation line, suggesting that a previous mutation might have rendered the toxin in that line inactive. Under laboratory conditions, high carriage costs of the At plasmid would select for its loss. However, if plasmid-free segregants or cured derivatives die immediately (due to activity of these toxin–antitoxin systems), they would never become established in the population. It is possible that deletions such as those described here, are not uncommon events, and due to the strong growth advantage conferred by this genetic change they would be able to rapidly outcompete the remaining population.
It is impossible at this juncture to know what type of event triggered the deletions we have discovered in pAtC58, but given the nature of the deletions they may be phage-mediated. It is clear that all related C58 lineages we have tested with origins from the University of Illinois, Urbana-Champaign, have the ΔAtu5208-Atu5407 deletion. The original C58 isolate used to establish this collection was reportedly obtained from the strain collection of E.W. Nester at the University of Washington, Seattle (Farrand SK, personal communication). This is curious because the two separate isolates whose genomes were sequenced were also obtained from this strain collection. Our PCR analysis of as many of the UW stocks as were available has not revealed the same deletion we have identified in the C58-UI lab lineages (although one stock appeared to have a different deletion or rearrangement in this same region). This suggests that the deletion was incurred after acquisition of this C58 lineage from the Nester laboratory. The observation that the AB153 strain generated separately during isogenic strain construction also has a rearrangement in this region, of a different size, adds to the impression that this may be a very recombinationally active region of the pAtC58 plasmid.
Considering the high frequency of genetic changes that occur in bacteria and their clonal mode of reproduction, the process of basic laboratory maintenance can easily generate severe bottlenecks of significant consequence. For example, given the reduced cost of the truncated pAtC58 in the laboratory strain, it is certainly possible that something as simple as isolation of the largest colony on a Petri plate during cultivation could easily have led to fixation of this deletion among certain lineages. Based on GO analysis, there is no immediately obvious source for the higher cost of the full-length At plasmid. However, little is known about the regulation of many of the genes on the plasmid and it is possible that the energetic costs associated with their expression are quite high. Additionally, there are a considerable number of ABC transporters, the expression of which may be energetically costly. It is also possible that At plasmid regulatory factors interact with chromosomal genes to impact the burden of the plasmid on the host cell. Outside of the lab, in the soil and rhizosphere, the benefits associated with these deleted genes are likely to be sufficiently significant to select for their retention.
In addition to relieving a significant portion of the cost of the At plasmid, the deletion common to the majority of C58-derived laboratory stains, has a positive impact on virulence gene expression. Interest in A. tumefaciens has often been due to its role as a plant pathogen and ability to transfer foreign DNA into plant cells (Chilton et al. 1977). A quantitative correlation between subtle differences in virB expression and effective T-DNA transfer and integration has not been established, but it is possible that at some point in history there was selection for this increase in virB expression. These results are contradictory to observations that cells harboring the At plasmid form larger tumors (Nair et al. 2003). It is important to point out that those experiments were performed with the AB151-AB153 strains that exhibit laboratory growth characteristics that dramatically differ from other C58-derived strains, as well as one another (Morton ER, unpublished results).
Further studies are required to determine the precise mechanism driving these deletion events. Patchy homology among other rhizobial megaplasmids suggests that this phenomenon could be widespread and the source of rapid genetic variation between plasmids and bacteria. For large plasmids such as pAtC58 that incur a high cost to their host cells (Morton ER, Platt TG, Fuqua C, and Bever JD, submitted), this genomic instability could be adaptive, by conferring a certain degree of genetic plasticity.
Supplementary Material
Supplementary figures S1 and S2 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Greg Velicer, Jay Lennon, and Brynn Heckel for helpful feedback with the manuscript; Aaron Buechlein for help with plasmid annotation; and Tom Platt, Rachel Hissong, and Katie Savage for organization and maintenance of the mutation accumulation lines. They also thank several laboratories who supplied their agrobacterial stocks for analysis, most notably the Nester and Farrand labs. They thank Michael Lynch, Pat Foster, Way Sung, Haixu Tang, and Steve Finkel for design and financial support of the mutation accumulation (MA) experiment, sequencing, and subsequent analysis. The MA project is supported through National Institutes of Health grant (F32GM103164) to W.S. and US Department of Defense grant ONRBAA10-002. This work on the pAtC58 deletions was supported by the National Institutes of Health grant (GM092660) to J.D.B. and C.F.
Literature Cited
- Baek CH, et al. Genes for utilization of deoxyfructosyl glutamine (DFG), an amadori compound, are widely dispersed in the family Rhizobiaceae. FEMS Microbiol Ecol. 2005;53:221–233. doi: 10.1016/j.femsec.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Castillo-Ramirez S, Vazquez-Castellanos JF, Gonzalez V, Cevallos MA. Horizontal gene transfer and diverse functional constrains within a common replication-partitioning system in alphaproteobacteria: the repABC operon. BMC Genomics. 2009;10:536. doi: 10.1186/1471-2164-10-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes L, et al. The conjugative plasmid of a bean-nodulating Sinorhizobium fredii strain is assembled from sequences of two Rhizobium plasmids and the chromosome of a Sinorhizobium strain. BMC Microbiol. 2011;11:149. doi: 10.1186/1471-2180-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevallos MA, Cervantes-Rivera R, Gutierrez-Rios RM. The repABC plasmid family. Plasmid. 2008;60:19–37. doi: 10.1016/j.plasmid.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Chai YR, Tsai CS, Cho HB, Winans SC. Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J Bacteriol. 2007;189:3674–3679. doi: 10.1128/JB.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton MD, et al. Stable incorporation of plasmid DNA into higher plant cells: molecular basis of crown gall tumorigenesis. Cell. 1977;11:263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Crossman LC, et al. A common genomic framework for a diverse assembly of plasmids in the symbiotic nitrogen fixing bacteria. PLoS One. 2008;3:e2567. doi: 10.1371/journal.pone.0002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demezas DH, Reardon TB, Strain SR, Watson JM, Gibson AH. Diversity and genetic-structure of a natural population of Rhizobium leguminosarum Bv trifolii isolated from Trifolium subterraneum L. Mol Ecol. 1995;4:209–220. [Google Scholar]
- Ellis JG, Kerr A, Petit A, Tempe J. Conjugal transfer of nopaline and agropine Ti plasmids: the role of agrocinopines. Mol Gen Genet. 1982;186:269–274. [Google Scholar]
- Ding H, Hynes MF. Plasmid transfer systems in the rhizobia. Can J Microbiol. 2009;55:917–927. doi: 10.1139/w09-056. [DOI] [PubMed] [Google Scholar]
- Garfinkel DJ, et al. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- Giuntini E, et al. Large-scale genetic variation of the symbiosis-required megaplasmid pSymA revealed by comparative genomic analysis of Sinorhizobium meliloti natural strains. BMC Genomics. 2005;6:158. doi: 10.1186/1471-2164-6-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V, et al. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 2003;4:158. doi: 10.1186/gb-2003-4-6-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodner B, et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- Goodner BW, et al. Combined genetic and physical map of the complex genome of Agrobacterium tumefaciens. J Bacteriol. 1999;181:5160–5166. doi: 10.1128/jb.181.17.5160-5166.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton RH, Fall MZ. The loss of tumor initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia. 1971;27:229–230. doi: 10.1007/BF02145913. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C. Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J Bacteriol. 2011;193:3461–3472. doi: 10.1128/JB.00317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes MF, et al. The two megaplasmids of Rhizobium meliloti are involved in the effective nodulation of alfalfa. Mol Gen Genet. 1986;202:356–362. [Google Scholar]
- Lenski RE. Experimental studies of pleiotropy and epistasis in Escherichia coli. 1. Variation in competitive fitness among mutants resistant to virus T4. Evolution. 1988;42:425–432. doi: 10.1111/j.1558-5646.1988.tb04149.x. [DOI] [PubMed] [Google Scholar]
- Iida S, Hiestandnauer R, Arber W. Transposable element IS1 intrinsically generates target duplications of variable length. Proc Natl Acad Sci U S A. 1985;82:839–843. doi: 10.1073/pnas.82.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Clemente TE, Farrand SK. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol Plant Microbe Interact. 2001;14:98–103. doi: 10.1094/MPMI.2001.14.1.98. [DOI] [PubMed] [Google Scholar]
- Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton ER, Fuqua C. Genetic manipulation of Agrobacterium. Curr Protoc Microbiol. 2012a doi: 10.1002/9780471729259.mc03d02s25. Chapter 3:Unit3D.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton ER, Fuqua C. Phenotypic analyses of Agrobacterium. Curr Protoc Microbiol. 2012b doi: 10.1002/9780471729259.mc03d03s25. Chapter 3:Unit3D.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, et al. Bioinformatics tools for large-scale functional classification of proteins—the InterPro database. NY: Nova Science Publishers; 2005. [Google Scholar]
- Nair GR, Liu Z, Binns AN. Reexamining the role of the accessory plasmid pAtC58 in the virulence of Agrobacterium tumefaciens strain C58. Plant Physiol. 2003;133:989–999. doi: 10.1104/pp.103.030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Mosqueda MD, Altamirano-Hernandez J, Farias-Rodriguez R, Valencia-Cantero E, Santoyo G. Homologous recombination and dynamics of rhizobial genomes. Res Microbiol. 2009;160:733–741. doi: 10.1016/j.resmic.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Perez-Mendoza D, et al. Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids. J Bacteriol. 2005;187:7341–7350. doi: 10.1128/JB.187.21.7341-7350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Segura G, Perez-Oseguera A, Cevallos MA. The repAC replication system of the Rhizobium leguminosarum pRL7 plasmid is functional: implications regarding the origin and evolution of repABC plasmids. Plasmid. 2013;69:49–57. doi: 10.1016/j.plasmid.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Romero D, et al. Amplification and deletion of a Nod-Nif region in the symbiotic plasmid of Rhizobium phaseoli. J Bacteriol. 1991;173:2435–2441. doi: 10.1128/jb.173.8.2435-2441.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H, Kuykendall LD, Young JM. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol. 2003;49:155–179. doi: 10.2323/jgam.49.155. [DOI] [PubMed] [Google Scholar]
- Schofield PR, Gibson AH, Dudman WF, Watson JM. Evidence for genetic exchange and recombination of rhizobium plasmids in a soil population. Appl Environ Microbiol. 1987;53:2942–2947. doi: 10.1128/aem.53.12.2942-2947.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, et al. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- Schwartz E. Microbial megaplasmids. Berlin, Heidelberg, Germany: Springer-Verlag; 2009. [Google Scholar]
- Slater S, et al. Reconciliation of sequence data and updated annotation of the genome of Agrobacterium tumefaciens C58, and distribution of a linear chromosome in the genus Agrobacterium. Appl Environ Microbiol. 2013;79:1414–1417. doi: 10.1128/AEM.03192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG, Hedge PJ, Heesen ST, Edelman A, Broomesmith JK. Kanamycin-resistant vectors that are analogs of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- Tempé J, Petit A, Holsters M, Van Montagu M, Schell J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci U S A. 1977;74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraji M, Suzuki K, Yoshida K. A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet Syst. 2002;77:1–9. doi: 10.1266/ggs.77.1. [DOI] [PubMed] [Google Scholar]
- Velazquez E, et al. Analysis of core genes supports the reclassification of strains Agrobacterium radiobacter K84 and Agrobacterium tumefaciens AKE10 into the species Rhizobium rhizogenes. Syst Appl Microbiol. 2010;33:247–251. doi: 10.1016/j.syapm.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Wang YL, Mukhopadhyay A, Howitz VR, Binns AN, Lynn DG. Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene. 2000;242:105–114. doi: 10.1016/s0378-1119(99)00541-7. [DOI] [PubMed] [Google Scholar]
- Watson B, Currier TC, Gordon MP, Chilton MD, Nester EW. Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 1975;123:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibberg D, et al. Complete genome sequencing of Agrobacterium sp H13-3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J Biotechnol. 2011;155:50–62. doi: 10.1016/j.jbiotec.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Winans SC, Kerstetter RA, Nester EW. Transcriptional regulation of the virA gene and virG gene of Agrobacterium tumefaciens. J Bacteriol. 1988;170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DW, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning ZM. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.