Abstract
Cancer stem cells (CSCs) constitute a subpopulation of cancer cells that have the potential for self-renewal, multipotent differentiation, and tumorigenicity. Studies on CSC biology and CSC-targeted therapies depend on CSC isolation and/or enrichment methodologies. Scientists have conducted extensive research in this field since John Dick's group successfully isolated CSCs based on the expression of the CD34 and CD38 surface markers. Progress in CSC research has been greatly facilitated by the enrichment and isolation of these cells. In this review, we summarize the current strategies used in our and other laboratories for CSC isolation and enrichment, including methods based on stem cell surface markers, intracellular enzyme activity, the concentration of reactive oxygen species, the mitochondrial membrane potential, promoter-driven fluorescent protein expression, autofluorescence, suspension/adherent culture, cell division, the identification of side population cells, resistance to cytotoxic compounds or hypoxia, invasiveness/adhesion, immunoselection, and physical property. Although many challenges remain to be overcome, it is reasonable to believe that more reliable, efficient, and convenient methods will be developed in the near future.
Introduction
Although the existence of extreme heterogeneity in primary cancers and immortalized cancer cell lines has long been recognized; the relative contributions of heritable and nonheritable mechanisms, such as stochastic mutation, clonal evolution, and phenotypic plasticity, to this heterogeneity remain controversial [1]. The concept of cancer stem cells (CSCs) was recently proposed to explain tumor heterogeneity. CSCs, a limited subpopulation of tumor-initiating cells (TICs), are defined as cells that retain extensive self-renewal potential through multiple generations and have the ability to recreate the heterogeneity of the original tumor through asymmetric division [2]. Despite the controversy surrounding this theory, the study of CSCs is important for the following reasons. (i) If tumors are a type of stem cell disease and are derived from CSCs, then our previous results for cancer must be reassessed because many substantial and extreme differences may exist between CSCs and other subpopulations of cancer cells. The systematic study of the cellular genetics, biological characteristics, and signal transduction mechanisms of CSCs will help elucidate the mechanisms of carcinogenesis. (ii) The concept of CSCs forces us to evaluate our current understanding of cancer metastasis. CSCs have the ability to detach from the primary tumor and invade the surrounding tissue by undergoing the epithelial-to-mesenchymal transition (EMT); therefore, CSCs might be the cause of tumor dissemination, which is the primary cause of death among cancer patients [3]. (iii) The CSC theory also has profound implications in terms of cancer therapy, and we should re-examine our previous experience in this area. Although chemo- and radiotherapy can kill most of the cells in a tumor, CSCs may be left behind. These cells can regenerate the original tumor due to their enhanced resistance, which makes these cells less susceptible to conventional therapies [4,5].
Thus, strategies to identify CSCs and to efficiently and reliably isolate them from a heterogeneous tumor mass may have fundamental roles in CSC studies, the results of which will have profound implications both for tumor development and for therapeutic outcomes. In this review, we will briefly discuss the progress made in CSC isolation and enrichment during the past 10 years, particularly during the last 4–5 years.
It should be emphasized that putative CSC or CSC-enriched populations obtained using any of these strategies must be tested rigorously by serial xenotransplantation in immunocompromised mice, the gold standard for the identification of CSCs [6]. Self-renewal can be confirmed by this assay, in which prospectively re-isolated CSC populations are placed into secondary recipients. Multipotency is typically demonstrated by the ability of the cells to generate tumor xenografts that reflect the cellular heterogeneity of the original tumor [6,7].
Strategies for Isolating and Enriching CSCs
Surface markers
Cellular surface markers have been used for the isolation of CSCs. In 1994, Dick provided the first evidence of the existence of CSCs derived from acute myeloid leukemia using fluorescence activated cell sorting (FACS) based on CD34 and CD38 (CD34+CD38−) surface marker expression [8,9]. Since then, CSCs have been isolated from many types of solid tumors by FACS and magnetic cell sorting using the following specific surface markers: CD24, CD44, CD133, CD13, CD14, CD15, Stro-1, Cripto-1, CXC chemokine receptor type 4 (CXCR4), Lin, Thy1, stage-specific embryonic antigen-1 (SSEA-1), epithelial cell adhesion molecule (EpCAM), epithelial specific antigen, CD20, ATP-binding cassette (ABC) transporter B5, CD166, A2B5, leucine-rich-repeat-containing G-protein-coupled receptor 5 (LGR5), CD49f, CD90, CD117, stem cell antigen-1 (Sca-1), epidermal growth factor receptor (EGFR), CD271, and CD47 [8–52]. This surface marker-based approach has become the most commonly used method to isolate CSCs from heterogeneous tumor cell populations and has significantly contributed to progress in CSC research. However, many of the surface markers used for sorting have been identified empirically and were identified on normal stem cells (SCs), such as embryonic stem cells (ESCs) and adult stem cells (ASCs). Questions have been raised regarding the specificity and reliability of these markers for the identification of CSCs. For example, CD133 is an important cell surface marker present on neural stem cells (NSCs) and has been the most extensively used marker for the isolation of CSCs from many types of cancers, such as glioblastomas, breast cancer, prostate cancer, colon cancer, pancreatic cancer, liver cancer, Wilms tumor, and neuroblastomas [53]. However, Beier et al. provided the first evidence that CD133+ CSCs maintain only a subset of primary glioblastomas. Kemper et al. found that glioma stem cells (GSCs) identified using antibodies against CD133 have inconsistent tumorigenic potential. The AC133 epitope, but not the CD133 protein, is lost upon CSC differentiation [54]. Some cell lines derived from primary glioblastomas grow adherently in vitro and are driven by CD133− tumor cells that exhibit apparent SC-like properties but distinct molecular profiles and growth characteristics in vitro and in vivo [55]. A subpopulation of A2B5+ glioma cells with the capacity to form tumors exists in some human gliomas that contain very few or no detectable CD133+ cells. These A2B5+ cells are phenotypically distinct from CD133+ cells [19]. In human lung cancer and rat glioma cell lines, both CD133+ and CD133− subpopulations possess clonogenic, self-renewal, and tumorigenic capacities [56,57]. A similar phenomenon has also been found in colon cancer. During the process of metastasis, CD133+ tumor cells may give rise to a more aggressive CD133− subset capable of tumor initiation in mice [58]. CD133 is expressed regardless of the differentiation state of colon cancer cells and may not be restricted to CSCs. The isolated CD133− cells still have the capacity to give rise to tumors in nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice [58]. The same result has also been recently demonstrated for head and neck squamous cell carcinoma [59]. Moreover, several studies have indicated that CD133 is expressed by differentiated epithelial cells in a variety of organs [60]. Park et al. performed immunohistochemical analyses of several stem cell-related markers in breast cancer cells with different stemness and differentiation characteristics. They found that the frequency of tumor cells positive for stem cell-like cell markers and markers of more differentiated cells varied according to the tumor subtype and histologic stage [61]. Furthermore, a recent study found that CD133 expression in colorectal cancer cells is modulated by the microenvironment [62]. In addition to colorectal cancer, there are other cancers in which the expression of CD133 is regulated, such as gliomas and prostate cancer. When GSCs were sorted into CD133+ and CD133− fractions, promoter methylation was observed in the CD133− fraction; this methylation was reversible using demethylation agents and could be reproduced by in vitro methylation. The researchers thought that CD133 might be a marker that is only co-regulated with other more relevant factors that help determine the stem cell characteristics, such as Sp1 and c-myc [63]. In both benign and malignant primary prostate tissues, the regulation of CD133 is independent of DNA methylation but is under the dynamic control of chromatin condensation [64].
Suspension culture
In 1992, Reynolds et al. established a serum-free culture method to obtain neurospheres from brain tissue [65]. This simple and convenient method for NSC enrichment has not only been widely used to study neurogenesis but has also been applied for CSC isolation from brain tumors and many other types of cancers with different combinations of supplemental factors. These tumor spheres express high levels of SC markers and exhibit a great degree of tumorigenicity. Using sphere assays for tumor cells, a number of groups have demonstrated that CSCs efficiently form tumor spheres in a clonogenic manner [66]. These tumor spheres are also chemoresistant and exhibit the upregulation of drug-resistance proteins [67]. However, the use of the sphere assay for CSC isolation has several problems. As shown by Pollard and others [68,69], the efficiency of successfully isolating CSCs from any given tumor is low (from 1% to 30%). The majority of cells in the sphere are differentiated and/or dying progeny [70]. During serial passaging, sphere cells spontaneously differentiate and/or undergo apoptosis [70]. Additionally, it is worth noting that sphere cells in some cancer cell lines are no more tumorigenic in vivo than cells grown on plastic and that cells that generate spheres in culture do not always give rise to tumors after more than 10 months following implantation [66]. Recently, Sudha Krishnamurthy designed an alternative strategy, termed the orosphere assay, for the propagation of head and neck CSCs. An orosphere is defined as a nonadherent colony of cells sorted from a tumor mass and cultured in three-dimensional soft agar or on ultralow attachment plates. This strategy is suitable for the propagation of head and neck cancer cells and can retain the stemness and self-renewal capacity of these cells [71].
Activity of intracellular enzymes
The aldehyde dehydrogenase (ALDH) family is composed of cytosolic isoenzymes that are responsible for oxidizing intracellular aldehydes [72]. Normal SCs, including ESCs and ASCs, have high ALDH activity [73–78]. Increased activity of the Class 1 ALDH family (ALDH1) has been found in different types of CSCs [79–84]. In these studies, researchers found that the ALDH1+ cancer cells isolated from tumor masses using the aldefluor assay and FACS analysis exhibited obvious CSC properties in vitro, including self-renewal, multipotency, and drug-resistance, and these isolated cells expressed SC markers. Additionally, these cells had CSC properties in vivo, including tumorigenicity and the generation of a heterogeneous cancer cell population. These data suggest that ALDH1 could be used as a CSC marker [79–85]. Some researchers have also found that this ALDH1+ CSC pool is regulated by the expression of several growth factor receptors, including epidermal growth factor receptor type 2 (HER2, also known as ErbB), through the PI3-kinase/Akt pathway [86].
The 26S proteasome is a key regulator of many cellular functions, including cell cycle control, DNA repair, cell death, and survival. Cancer cells grown in sphere cultures enriched for CSCs exhibited decreased 26S proteasome activity relative to their respective monolayer cultures. Vlashi et al. engineered human glioma and breast cancer cells to express stably, ZsGreen fused to the carboxyl-terminal degron of ornithine decarboxylase, resulting in a fluorescent fusion protein that accumulates in cells in the absence of 26S proteasome activity. They found that the ZsGreen+ cells had increased sphere-forming capacity and higher expression levels of CSC markers than ZsGreen− cells in vitro. In vivo, ZsGreen+ cells were more tumorigenic than ZsGreen− cells [87]. Spheres from lung cancer cell lines have also been found to exhibit decreased 26S proteasome activity [88].
The Jumonji AT-rich interactive domain 1B (JARID1B), which belongs to the highly conserved family of Jumonji/ARID1 histone demethylases, is capable of removing three methyl groups from histone H3 lysine 4 (H3K4) [89]. Using the H3K4 demethylase JARID1B as a biomarker, Roesch et al. characterized a small subpopulation of slow proliferating melanoma cells and found that the JARID1B+ subpopulation is essential for continuous tumor growth. Compared with melanoma cells that do not express this enzyme [90], JARID1B+ cells cycled more slowly but also generated more progeny and were more tumorigenic [91]. The same result was found for uveal melanoma cells [92]. However, the presence of a subset of cells functionally enriched for tumorigenicity in melanoma is in contrast with the findings obtained by Morrison and coworkers. [93–95].
Intracellular concentration of reactive oxygen species
Reactive oxygen species (ROS) have been implicated in many intracellular physiopathological processes, including cell cycle progression, proliferation, apoptosis, and differentiation. Normal SCs, such as ESCs, induced pluripotent stem cells, NSCs, and hematopoietic stem cells, have been shown to contain lower levels of ROS than their more mature progeny. This difference is essential for maintaining SC function [96–103]. Interestingly, the results obtained by Diehn et al. indicated that as observed for normal SCs, subsets of CSCs in some tumors also contain lower ROS levels and enhanced ROS defenses compared with their differentiated descendants and non-CSCs [104,105]. Following the purification of CD44+CD24−/loLin− CSCs from breast tumors, 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (DCF-DA) staining revealed that the CSC-enriched population contained a considerably lower DCF-DA intensity than the corresponding non-CSC population. A similar phenomenon has also been found in head and neck tumors. These mechanisms may be attributed to the upregulation of the ROS defense system via the overexpression of scavenging molecules and an enhanced capacity for glutathione synthesis and protection against ROS [106]. Although the detailed mechanisms remain unknown, it may be possible to establish a method to isolate CSCs based on the intensity of fluorescent stains for ROS, such as DCF-DA and dihydroethidium. In our laboratory, researchers are attempting to isolate CSCs using this method.
Mitochondrial membrane potential
The mitochondrial and energy/metabolism-related features of ASCs and ESCs have sparked the interest of an increasing number of researchers [107–110]. It is thought that mitochondrial function and integrity may affect SC viability, the proliferative and differentiation potential of these cells, and their lifespan [111]. Heerdt et al. found that the mitochondrial membrane potential (Δψm) is associated with a cell's tumorigenicity. Two isogenic cell lines were subcloned and established from the SW620 colonic carcinoma cell line, and they exhibited significant and stable differences in the intrinsic Δψm [112]. These differences in Δψm are linked to important tumorigenic properties. Compared with cells with a lower Δψm, cells with an intrinsically higher Δψm exhibit significantly higher resistance to apoptotic inducers and hypoxia, increased invasive behavior, and enhanced initiation of angiogenesis [113,114]. Our recent results have indicated that side population (SP) cells and sphere-forming A549 lung cancer cells have a fairly high Δψm (tumor sphere, 90.92±18.2; monolayer subpopulation, 50.53±3.35; P<0.05; SP, 133.48±29.33; non-SP, 111.37±20.7; P<0.05) [105, 115]. In most of the A549 cells with Δψm values in the highest 5% (ΔψmH), CD133 expression was clearly detected. However, the expression of this SC marker was nearly absent in the subpopulation with Δψm values in the lowest 5% (ΔψmL) [115]. Michelakis et al. measured the Δψm in cells from freshly excised glioblastoma multiforme (GBM) tissue, primary cell lines, GBM-SCs isolated from tumors, and differentiated cells derived from GBM-SCs. The highest potential was found in the putative GBM-SCs. Both the primary and GBM-SC-derived secondary GBM cells (15-day differentiation) had Δψm values similar to those of the parent tumors. Simultaneous staining with a CD133 antibody and tetramethylrhodamine methyl ester (TMRM) demonstrated that CD133+ cells had a higher Δψm than the neighboring non-GBM-SCs in vivo [116]. These data suggest that differences in intrinsic Δψm of carcinoma cells are likely to be associated with subtle shifts in the biochemical pathways and/or cell phenotypes that play fundamental roles in determining the probability of tumorigenesis. Compared with their differentiated descendants, CSCs may have a different Δψm. Using approaches, such as flow cytometry, Δψm heterogeneity can be detected based on varying fluorescence intensities (Rhodamine-123 (Rh123), TMRM, and JC-1), and difference in the Δψm can used to isolate and/or enrich CSCs from a tumor population [115].
Promoter-driven fluorescent protein expression
ESCs and multiple types of CSCs share a genetic expression pattern [117]. Some core transcription factors associated with the stemness of ESCs, such as octamer-binding transcription factor 4 (Oct-4), sex determining region Y-box 2 (Sox2), Nanog, and c-kit, are also overexpressed in CSCs. In contrast, the activities of these factors are weak or even absent in the differentiated descendants [118,119]. Thus, one can isolate CSCs from a heterogeneous tumor mass based on the different activities of these stemness factors. Compared with surface marker-based strategies, this approach may provide a more target-specific tool with a wider range of applicability to isolate CSCs due to the critical roles of these transcription factors in controlling cell stemness. However, due to the intracellular expression of these proteins, one cannot isolate CSCs directly using these proteins but must use a reporter system in which a specific gene promoter drives the expression of a reporter protein.
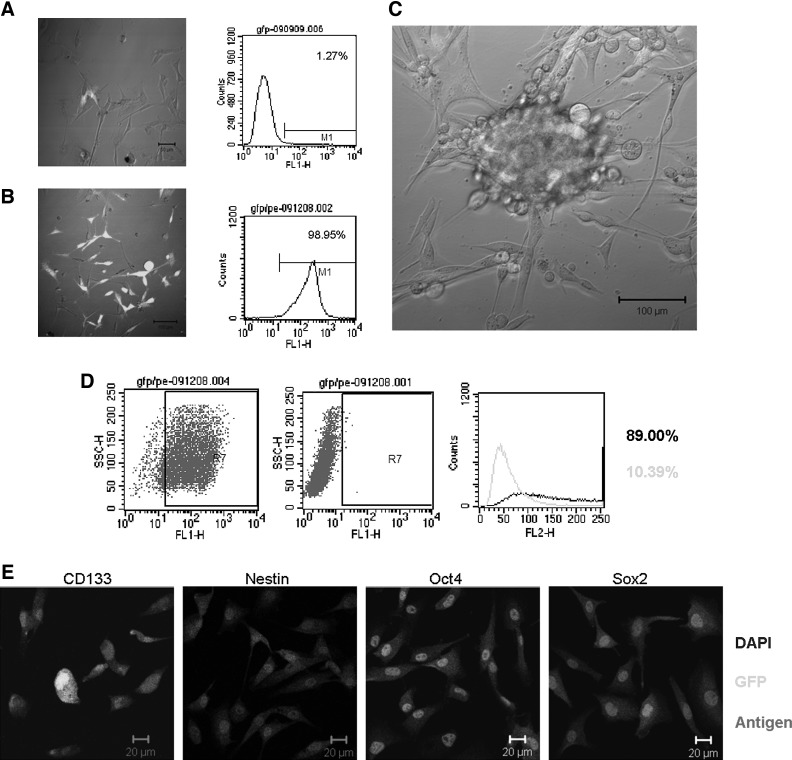
In recent years, reporter systems employing a specific gene promoter driving green fluorescent protein (GFP)/red fluorescent protein (RFP) expression have been widely used to isolate CSCs. These genes include SH2 domain containing 5′-inositol phosphatase-1 (s-SHIP) [120], Oct4 [121], Nanog [122], nucleostemin [123], T-cell factor/lymphoid enhancer factor [124], and Sox2 [125]. Additionally, early transposon (ETn), which is highly transcribed during early mouse embryogenesis in ESCs and in embryonic carcinoma cell lines [126], could potentially be used to isolate CSCs [127]. In our laboratory, we used the Nanog promoter as a reporter system to successfully isolate a small subpopulation of Nanog-positive cells from hepatocellular carcinoma (HCC) tissue. These cells demonstrated an enhanced ability for self-renewal, clonogenicity, and tumor initiation. Moreover, these cells were also multipotent and chemoresistant [128]. Recently, we used the EOS system (based on a lentiviral vector with the ETn promoter and Oct4/Sox2 enhancer) developed by Hotta et al. to isolate CSCs from the glioma cell line U87, obtaining very exciting preliminary results. As shown in Fig. 1, GFP+ U87 cells can form tumor spheres in serum-free stem cell culture medium, and they express several stem cell markers, such as CD133, Sox2, Oct4, nestin, and SSEA-1. Because of the complex gene expression pattern, the selection of the appropriate vector and the number of promoter/enhancer copies should be carefully considered.
FIG. 1.
Isolation of cancer stem cells (CSCs) based on EOS system (based on a lentiviral vector with the ETn promoter and Oct4/Sox2 enhancer, plasmid purchased from Addgene). (A) GFP expression in U87 cells transfected with the EOS system. The image was taken 5 days after transfection. Flow cytometry analysis of GFP expression in U87 cells transfected with the EOS lentiviral vector. (B) Puromycin selection was applied starting 7 days after infection. The image of puromycin-resistant GFP-expressing cells was taken after 2 weeks of puromycin selection. The effect of puromycin selection was analyzed by flow cytometry. (C) Tumor sphere-forming cells exhibited higher GFP expression than adherent nonsphere-forming cells in serum-free neural stem cell (NSC) medium containing DMEM/F12, B27, EGF, and bFGF. (D) Flow cytometry analysis showing the percentage of SSEA-1+ cells among the GFP-expressing cells after selection. Blank, histogram for the stained sample; Gray, control. (E) Expression of the stem cell markers CD133, nestin, Oct4, and Sox2 in GFP-expressing cells after selection.
Autofluorescence
It has been found that some types of stem cells exhibit intrinsic autofluorescence, which can be used to isolate and track stem cells [129–131]. Radovanovic and coworkers utilized intrinsic autofluorescence and distinctive morphology to isolate a subpopulation of CSCs from human gliomas. Excitation at 488 nm causes these cells to autofluoresce, with emission at approximately 520 nm (in the FL1 channel). These cells, termed FL1+ cells, possess many SC-like characteristics, such as a capacity for self-renewal in vitro and tumorigenesis in vivo, and these cells preferentially express the SC genes Nanog, Oct4, Sox2, and Notch1. Although the underlying mechanism remains unknown, these data suggest a novel approach for isolating and enriching CSCs from a tumor mass [132].
Adherent culture
Morphologic heterogeneity is a typical feature of malignant cells and has been attributed to genetic instability and clonal evolution [133]. Studies have revealed a close relationship between the clone morphology and stemness of cancer cells. After being plated at low densities, epithelial-derived malignant cells exhibit a spectrum of colony morphologies, ranging from round colonies of small, closely packed cells to irregularly shaped colonies consisting of larger, loosely packed cells; these different clone types are referred to as holoclones, meroclones, and paraclones [134]. The holoclones, which have a regular and compact shape, are enriched in CSCs and have the highest proliferative capacity. A similar phenomenon has been observed in head and neck squamous cell carcinoma and prostate carcinoma cells [135,136]. Recent studies have demonstrated that adherent culture is better than suspension culture because of the lower rate of spontaneous apoptosis and the high tumor-initiating ability in vivo [137,138]. Our observations also support the possibility of predicting “stemness” based on colony morphology in nonepithelial tumors. Tight clones formed by glioma cells also possess many CSC features, including the expression of SC markers (e.g., nestin, CD133, Sox2, Oct4, and CD44s), self-renewal, and multipotency [139].
By applying methodologies previously used for NSC culture in vitro [140], Pollard et al. demonstrated that CSCs can be grown as adherent monolayers using a simple modification of the suspension sphere culture. They obtained a monomorphic population that had many characteristics possessed by CSCs, such as the expression of SC markers (e.g., nestin, Sox2, and oligodendrocyte transcription factor 2), multipotency, genetic abnormalities consistent with the parental tumor, and tumorigenicity. Most importantly, these adherent glioma cells appear to be capable of long-term serial passaging; they contain a high percentage of true CSCs, with significantly fewer spontaneously differentiating or apoptotic cells; and they can be derived from nearly 100% of primary malignant gliomas [68]. However, what is interesting about this model is that these cells can recapitulate the original tumor phenotype after orthotopic injection. This model augments the previous validation of the serum-free de novo cell culture model [141,142].
Side population (SP) cells
A small population of cells, termed SP cells, was first described by Goodell et al. based on these cells' distinct ability to pump out Hoechst 33342. These cells were defined as a small subpopulation of cells with enriched SC activity, including clonogenic capacity, tumorigenicity, multipotency, long-term repopulating properties, and chemoresistance [143,144]. In recent years, some studies have indicated that the SP cells may be useful for the FACS-based identification and isolation of CSCs from human cancers [145–151]. However, it has been demonstrated that both sorted SP and non-SP cells from several cancer cell lines have a similar capacity for clone formation, tumorigenicity, and multipotency. Additionally, the phenotypes of these two cell fractions are interconvertible, and one cell type can give rise to the other [152]. Thus, one hypothesis is that the SP and non-SP populations, albeit phenotypically distinct, do not differ with respect to the number of SC-like cells or behavior. Similarly, although SP cells from the adrenocortical NCI-h295R cell line have a less differentiated phenotype, the proliferation rates of SP and non-SP cells are the same. Furthermore, both cell types have the capacity to give rise to the original SP-containing and non-SP-containing cell populations. SP cells exhibit no survival enhancement after exposure to cytotoxic agents commonly used to treat adrenocortical carcinomas. In addition, the growth kinetics of SP cells does not differ from those of the non-SP cells [153]. SP cells from the gastric cancer BGC-823 cell line also do not behave as CSCs do [154]. However, some studies have yielded conflicting results. Wu et al. isolated SP cells from the same cell line and demonstrated that these cells had stem-like properties [155]. In short, these results suggest that the concept of the SP phenotype as a universal CSC marker does not apply to some cancers. Additionally, the potential cytotoxicity of Hoechst 33342 is controversial, and some researchers have proposed that the differences in clonogenicity and tumorigenicity between SP and non-SP cancer cells might be the result of the dye itself [57]. However, other researchers have reported that this dye is not harmful to some cancer cells, such as small cell lung cancer cells [156].
Cancer cell division
SCs are defined by their ability to generate more SCs (“self-renewal”) and produce differentiating cells. These two tasks can be accomplished through a single mode of self-renewing mitotic division (“asymmetric self-renewing division”), in which one progeny cell retains the SC identity, and the other (progenitor) undergoes multiple rounds of division before entering a fully differentiated state. The two cells generated by the asymmetric division differ markedly in their proliferative potential; the SC remains quiescent or proliferates slowly, whereas the progenitor cell actively divides. This process ensures the production of a large number of differentiated progeny cells while maintaining a relatively small pool of long-lived SCs [157]. Although CSCs possess a greater ability to divide through symmetric self-renewing division than normal SCs, the cell division time is still shorter compared with their differentiated descendants.
PKH-26 is a fluorescent dye that binds to cell membranes and segregates in daughter cells after each cell division. Therefore, the intensity of staining correlates inversely with the rate of cell division [158]. The division rates of different subpopulations in a tumor mass vary during the same time span. Cicalese et al. established a novel strategy for isolating CSCs from breast cancer based on the PKH-26 staining intensity [13,159]. The PKH-26hi cells are highly enriched in CSCs (∼1:1), and represent the only cell subset capable of reconstituting a mammary tumor; thus, suggesting that breast CSCs undergo a limited number of divisions. Other researchers have also used this strategy to isolate CSCs from different types of human tumors, such as gastrointestinal cancers, glioblastomas, and nasopharyngeal carcinomas [160–163].
Carboxy fluorescein succinimidyl ester (CFSE) is a fluorescent dye that has been used to track the cell division frequency in several types of solid tumors. Recent studies have found that CFSE labeling can be used to identify and isolate a slow proliferating population of cells from glioblastomas. These dye-retaining brain tumor cell populations are enriched in CSCs, and the progeny that differentiate from these label-retaining cells exhibit all the pathological features of the primary disease [164,165]. Kamohara et al. used Hoechst 33342 and Pyronin Y to sort Huh7 cells into G0, G1, and G2/M fractions by FACS. They found that the G0 fraction was located within the neck of the SP fraction. These cells were capable of sphere formation and marked tumorigenesis. G0 cells, which do not express Ki67, were weakly positive for albumin expression and positive for keratin 19 expression. In contrast, the G1 cells were positive for Ki67 and albumin expression but negative for keratin 19. These findings suggest that the G0 Huh7 cell are promising CSC candidates [166]. Thus, most CSCs, if not all, possess slow proliferating characteristics. Furthermore, label-retaining techniques are unique methods for functionally identifying and isolating CSCs [167].
Cytotoxic and hypoxic resistance
It has been experimentally demonstrated that CSCs can resist apoptosis induced by cytotoxic drugs and radiation through multiple complicated mechanisms [4,5,168–178]. Enhanced chemoresistance is associated with a SC-like phenotype in tumor cells. These undifferentiated CSCs express high levels of specific drug resistance-associated proteins, such as ABC, that preferentially activate DNA damage checkpoint genes in response to radiation or cytotoxic drugs and can repair DNA damage more effectively than the progenitors and committed cells [5,177,178]. Additionally, CSCs have been shown to overexpress antiapoptotic genes [173,179] and downregulate some tumor suppressor genes [180]. This characteristic of functional CSCs has been confirmed by many studies in recent years and has been proposed to be the root cause of chemoresistance [181]. Moreover, the fraction of CSCs increases significantly following chemotherapy and radiation [5,149]. Furthermore, the proportion of CD133-positive cells in U251 glioma cells increases after the administration of rotenone and ethidium bromide [182]. Recent research has revealed that melanoma cells are enriched in ABCB5-expressing cells after temozolomide treatment and that these ABCB5-expressing cells exhibit enhanced tumorigenicity, with SC-like properties [183]. Ionizing radiation can activate stemness pathways in HCC cells and can result in the enrichment of a CSC subpopulation with higher resistance to radiotherapy [184]. Furthermore, methotrexate (MTX)-resistant U2OS/MTX300 osteosarcoma cells, termed osteosarcoma SCs, are enriched by chemotherapy. These cells have a greater ability to generate sarcospheres, can form the original tumor, and express SC surface markers, including CD117 and Stro-1 [185]. These data indicate that resistance to cytotoxic compounds could be used to enrich CSCs from a tumor mass [186–189]. However, Pajic et al. did not observe CSC enrichment in breast cancer 1 (BRCA1)- and p53-deficient breast cancers after cisplatin treatment [190].
The link between hypoxia and CSCs derived from solid tumors is fairly strong; hypoxia maintains the undifferentiated status and promotes the self-renewal capability of the SC population and a more stem-like phenotype in the nonstem population [180,191,192]. Several core stemness-related transcription factors, including Oct4, Nanog, c-myc, and Notch, are the direct or indirect targets of hypoxia-inducible factors [180,192]. More importantly, solid tumor CSCs are predominantly localized in hypoxic zones in vivo [180]. Regarding the observation that the proportion of the CD133-positive cells is increased under hypoxic conditions, another possibility may be that CSCs possess greater resistance to hypoxic conditions and can therefore, be enriched under these conditions. One study revealed that intratumoral hypoxia can enrich the CSC population in breast cancer and can limit the effect of antiangiogenic agents [193]. A similar result was found in another study, which identified a stem-like cell population in a metastatic breast cancer cell line subjected to repeated cycles of hypoxia and reoxygenation [194].
Invasiveness and adhesion
There is increasing evidence indicating that CSCs may possess a distinct potential for adherence, migration, and invasion. In glioma specimens, many nestin+/Sox2+ glioma cells, which can infiltrate into the surrounding normal tissue, are detected at the tumor-brain interface [141]. Huang et al. demonstrated that CSCs are more aggressive and invasive than core glioma cells [195]. Additionally, colorectal carcinoma cells with an enhanced migration and invasion capacity at the tumor's invading edge express SC markers [196]. Several studies have demonstrated that a CSC subset with the capacity for migration in the CD133+ CSC compartment express high CXCR4 or matrix metalloproteinase-2 (MMP-2) levels, and this high expression was correlated with metastatic potential [197,198]. A greater proportion of holoclone colonies with an indefinite proliferation capacity and enriched in CSCs is formed by early adherent cells [134]. Moreover, distinct CSCs were identified as having a CD133+ CXCR4+ phenotype at the invasive front of pancreatic tumors. The depletion of this subpopulation almost completely abrogated the metastatic phenotype of pancreatic tumors [28]. Many invasion-associated molecules are overexpressed in CSCs [4,168]. Extracellular signal-regulated kinase (ERK), N-cadherin, and integrinα6 have been found to play an important role during glioma stem cell invasion [199]. CD44+ prostate CSC-like cells possess a higher capacity for matrigel invasion, which is in contrast with the noninvasive nature of CD44− cells. More importantly, the genotype of the invasive cells closely resembles that of the CD44+CD24− prostate CSCs. These invasive cells are more tumorigenic than noninvasive cells [200]. In addition, transforming growth factor-β (TGF-β), snail and forkhead box protein C2 (FOXC2) -mediated EMT has been suggested to induce a CSCs phenotype accompanied with high metastatic potential [201–203]. Although many studies have demonstrated a relationship between CSCs in the primary tumor and metastasis in the secondary tumor, we still lack evidence for a causal relationship between the primary tumor and distant metastases. However, current studies and recent data strongly suggest that basement membrane invasion can be used as a tool to isolate and enrich CSCs from a tumor mass. In our previous study, we proposed a novel protocol to isolate and enrich CSCs from a tumor mass based on the heterogeneity of tumor cells with respect to invasiveness and adherence. Using this method, we successfully isolated SC-like subpopulations from glioma cell lines and resected samples [204].
Notably, based on the prediction that CSCs are readily detachable from tissue-culture plastic, Walia et al. established another interesting method to isolate CSCs. After dividing immortalized or transformed mammary epithelial cells into trypsin-sensitive and trypsin-resistant populations, these researchers demonstrated that the trypsin-sensitive cells were mesenchymal in morphology and expression profile, had enriched SC properties, and displayed mammosphere-forming properties, drug resistance, and CD44 expression. After several rounds of differential trypsinization, the trypsin-sensitive pool had an 80-fold higher mammosphere-forming ability than the trypsin-resistant population and a 20-fold higher ability than the starting population. Trypsin-sensitive cells are regarded as breast CSCs, and this method is faster, more affordable, and more efficient than other enrichment methods. Thus, differential adhesion may serve as the basis for an enrichment strategy to increase the size of the SC pool for subsequent manipulations for relatively differentiated epithelial cell types [205].
CSC immunoselection with natural killer cells
The responses of different tumor cell subpopulations to immune cells are heterogeneous. Reim et al. observed that sphere-forming MCF7 cells, which display a CD44hiCD24lo ‘‘CSC–like’’ phenotype, preferentially survived following treatment with natural killer (NK) cells. These surviving tumor cells displayed increased clonogenicity and tumorigenicity [206]. Similar results have been observed for glioma CSCs, suggesting that human leukocyte antigen-E (HLA-E) plays an important role in inhibiting NK cell-mediated lysis. HLA-E gene silencing in glioma CSCs can enhance the susceptibility to NK-cell mediated lysis [207]. Similarly, the CD44hiCD24lo population has also been found to be more immunoresistant in other breast cancer cell lines, such as SK-BR3, MDA-MB-231, and BT474 cells. Recently, CXCR4+ metastatic CSCs were shown to be induced by interferon-γ (IFN-γ) from NK resistant CSCs harbored in the highly tumorigenic CD44hiCD24lo subset [208]. Thus, a novel strategy based on immunoselection with NK cells could be used to isolate and enrich CSCs from cancer cell populations [209].
However, other studies have demonstrated that CSCs and non-CSCs derived from a melanoma cell line are equally susceptible to NK cell-mediated lysis [209]. Tseng et al. demonstrated that CSCs were more sensitive than their differentiated descendants. When NK cells were coincubated with primary oral squamous CSCs, increased cytotoxicity and augmented IFN-γ secretion were observed compared with differentiated cells [210]. Similar results for primary oral squamous carcinoma demonstrated that CSCs were significantly more susceptible to NK cell-mediated cytotoxicity than their differentiated counterparts or the parental cells [211]. Thus, it is questionable whether the immunoselection of tumor populations with NK cells can be used as a strategy for CSC isolation and enrichment.
Density gradient centrifugation
Recently, an interesting strategy has been developed to isolate CSCs from a primary rat HCC model using the physical properties of CSCs. Hepatic tumor cells were first isolated from the diethylnitrosamine-induced F344 rat HCC model by Percoll discontinuous gradient centrifugation (PDGC), followed by purification using differential trypsinization and differential attachment (DTDA). Among the four cell fractions (FI–FIV) obtained using this strategy, the third fraction (FIII) was enriched in cancer stem-like cells. This fraction had a higher nuclear-to-cytoplasmic ratio and expressed higher levels of SC markers, including alpha fetoprotein, EpCAM, and CD133. Furthermore, FIII exhibited a greater self-renewal capacity, multipotency, membrane invasiveness, and chemoresistance. The cells in this fraction were also found to form tumors in both the subcutaneous tissue and livers of nude mice in vivo. Taken together, the results of that study provide the basis for a novel strategy to isolate and enrich CSCs using the physical properties of these cells [212].
Conclusions and Perspectives
As mentioned above, there are many strategies for the isolation and/or enrichment of different types of CSCs (the suitable cancer types, CSC frequencies in the enriched populations, and references are summarized in Table 1). However, the scope, advantages, and limitations of these different strategies remain to be elucidated. In addition, the level of phenotypic similarity among cells obtained from the same starting material using different techniques remains to be determined. Thus, comparisons of different methods in the same lab using single tumors as the starting material are needed in the future. Kuch et al. performed an extensive cross-comparison of sphere formation capability, cell division, and putative CSC marker expression with the tumor-initiating properties of different melanoma and mammary tumor cells. Their observations suggest that it cannot be assumed that there is always a direct relationship between the sphere-forming ability, the expression of CSC markers, label retention, and the number of TICs in vivo. Indeed, there can be discrepancies in the identification of CSC-containing subpopulations when using different methods [213].
Table 1.
Strategies for the Isolation and Enrichment of Different Types of CSCs
| Strategy | Tumor type | Positive ratio (%) or fold enriching | CSC frequency (1:X) or tumorigenicity | Refs. |
|---|---|---|---|---|
| Surface markers | ||||
| CD34+CD38− | Leukemia | 0.02%–2.00% | 1:5,000 | [8,9] |
| CD44+CD24−/lo | Breast cancer | 1.32%–5.00% | 50 cells implanted, tumors detected in 4/7 mice | [11] |
| ESA+CD44+CD24−/loLin− | 0.60% | 1:100 | [10] | |
| CD133+ | 2.00%–5.90% | 50 cells implanted, tumors detected in 4/7 mice | [11] | |
| CD49f+DLL1hiDNERhi | Unknown | 1:48,249–1:97,134, well-differentiated tumor | [13] | |
| 1:15,708–1:30,840, poorly differentiated tumor | ||||
| Thy1+CD24+CD45− | 1.00%–4.00% | 1:213 | [14] | |
| CD44+CD49fhiCD133/2hi | 0.50%–2.00% | 250 cells implanted, tumors detected in 2/5 mice | [15] | |
| 250 cells implanted, tumors detected in 7/10 mice | ||||
| 250 cells implanted, tumors detected in 3/5 mice | ||||
| 50 cells implanted, tumors detected in 3/6 mice | ||||
| 50 cells implanted, tumors detected in 3/6 mice | ||||
| CD133+ | Colon cancer | 3.20%–24.50% | 1:262 | [29] |
| EpCAMhi/CD44+ | 0.03%–38.00% | 200 cells implanted, tumors detected in 5/10 mice | [31] | |
| 0.80%–38.00% | Unknown | |||
| EpCAMhi/CD166+ | 1.20%–16.00% | Unknown | ||
| ESAhi/CD166+ | Unknown | 4,000 cells implanted, tumors detected in 3/5 mice | ||
| ESAhi/CD44+ | Unknown | 2,000 cells implanted, tumors detected in 17/19 mice | ||
| ESAhi/CD44+/CD166+ | Unknown | 150 cells implanted, tumors detected in 1/2 mice | ||
| LGR5+ | 5.00%–10.00% | Unknown | [32] | |
| CD47+ | Leukemia | Unknown | Unknown | [33] |
| Beast cancer | ∼3.3-fold increase | 1×105 cells implanted, tumorigenicity unknown | [34] | |
| Colon cancer | 3.8×104 cells implanted, tumorigenicity unknown | |||
| Bladder cancer | 2,000 cells implanted, tumorigenicity unknown | |||
| Head & neck cancer | 1×105 cells implanted, tumorigenicity unknown | |||
| Ovarian cancer | 2.2×104 cells implanted, tumorigenicity unknown | |||
| Glioblastoma | 1×105 cells implanted, tumorigenicity unknown | |||
| Cripto-1+ | Embryonal cancer | 30.20%–60.60% | Unknown | [52] |
| CD44+CD133+ | Gallbladder cancer | 40.29% | 104 cells implanted, tumors detected in 4/4 mice, primary sample | [51] |
| 104 cells implanted, tumors detected in 5/5 mice, cell line | ||||
| CD44+ | Gastric cancer | 0%–94.00% | <1:33 | [25] |
| CD133+ | Brain tumor | 19.00%–29.00% | 1:100, glioma | [16] |
| 6.00%–21.00% | 1:100, medulloblastoma | |||
| SSEA-1+ | 26.60% | 1,000 cells implanted, tumors detected in 7/7 mice | [17] | |
| CD15+ | 34.00% | 104 cells implanted, tumors detected in 5/6 mice | [18] | |
| A2B5+ | Unknown | 5×105 cells implanted, tumors detected in 7/8 mice | [19] | |
| 61.70% | 1×104 cells implanted, tumors detected in 3/4 mice | [20] | ||
| EGFR | 4.00%–35.20% | Unknown | [21] | |
| CD44+ | Head and neck cancer | 5.47%–10.40% | 5,000 cells implanted, tumors detected in 6/7 mice | [24] |
| 53.00% | 1,000 cells implanted, tumors detected in 6/8 mice | [59] | ||
| CD133+ | Liver cancer | 0.05%–46.79% | 1×107 cells implanted, tumors detected in 4/4 mice | [45] |
| 0.10%–1.00% | 100 cells implanted, tumors detected in 3/6 mice | [46] | ||
| 1,000 cells implanted, tumors detected in 4/6 mice | ||||
| CD90+ | 0%–90.00% | 1,000 cells implanted, tumors detected in 2/2 mice | [47] | |
| CD13+ | 1.64% | 100 cells implanted, tumors detected in 2/4 mice | [48] | |
| CD166+ | Intestinal cancer | 8.10% | 100 cells implanted, tumors detected in 2/4 mice | [26] |
| Sca-1+CD45−PECAM-CD34+ | Lung cancer | 2.00%–16.00% | 100 cells implanted, tumors detected in 2/4 mice | [35] |
| CD133+ | 0.32%–22.00% | Unknown | [36] | |
| CD133+ABCG2+ | 6.05%–14.50% | Unknown | [37] | |
| 15.00–25.00% | 1:1×104 | |||
| CD133+CXCR4+ | 3.50%–7.50% | Unknown | ||
| CD133+ESA+ | 0.02%–35.00% | Unknown | ||
| CD133+ | 0.02%–36.00% | Unknown | ||
| CD133+CD44+ | 0.90% | Unknown | [38] | |
| CD166+ | Unknown | 1:5,175 | [39] | |
| CD20+ | Melanoma | 0.92% | Unknown, WM3539 cells | [40] |
| Unknown | Unknown, WM3523 cells | |||
| ABCB5+ | 1.60%–20.40% | 1:5,175 | [41] | |
| CD271+ | 2.50%–41.00% | Unknown | [42] | |
| ABCG2+CD133+ | 1.00% | 1:5,000 | [43] | |
| CD133+ | Osteosarcoma | 3.00%–5.00% | Unknown | [49] |
| CD133+CXCR4+ | 0.50%–80.00% | 100 cells implanted, tumors detected in 6/12 mice | [23] | |
| CD44+CD24+ESA+ | Pancreatic cancer | 0.20%–0.80% | Unknown | [27] |
| CD133+ | 1.09%–3.21% | Unknown | [28] | |
| Suspension culture | ||||
| CD15+ cells | Medulloblastoma | Unknown | 1×104 cells implanted, tumors detected in 5/6 mice | [18] |
| ALDH1+ cells | Esophageal cancer | 28.60% | Unknown | [67] |
| CD117+Stro-1+ cells | Osteosarcoma | 6.00%–14.00% | 200 cells implanted, tumors detected in 2/4 mice, K7M2 cells | [50] |
| 200 cells implanted, tumors detected in 2/5 mice, 318-1 cells | ||||
| Activity of intracellular enzymes | ||||
| ALDH1+ cells | Head and neck cancer | 1.00% | 250 cells implanted, tumors detected in 6/7 mice | [85] |
| Lung cancer | 0.60%–2.90% | 1:1,000 | [82] | |
| Breast cancer | 3.00%–10.00% | 5,000 cells implanted, tumors detected in 3/4 mice | [73] | |
| Melanoma | Unknown | 1:4 | [94] | |
| 26S proteasome | Glioma | <4.00% | 100 cells implanted, tumors detected in 1/4 mice | [87] |
| Breast cancer | Unknown | |||
| Concentration of ROS | Lung cancer | Unknown | Unknown | [105] |
| Unknown | Unknown | [115] | ||
| Breast cancer | Unknown | 100–1,000 cells implanted, tumors detected in 5/7 mice | [104] | |
| Mitochondrial membrane potential | Glioma | Unknown | Unknown | [116] |
| Promoter-driven system | ||||
| c-Myc-Ras-IκBα system | Breast cancer | 26.00% | 1:2,200 | [117] |
| LvPNanog-GFP system | Liver cancer | 2.40%–47.40% | 1 cell implanted, tumors detected in 2/6 mice | [128] |
| Autofluorescence | Glioma | 0.50%–3.20% | 1:1,000 | [132] |
| Adherent culture | ||||
| CD133+ cells | Head and neck cancer | 0.10%–0.18% | Unknown | [135] |
| CD44+ cells | 2.90%–12.30% | Unknown | ||
| CD29+ cells | 8.10%–17.80% | Unknown | ||
| CD44+CD133+ cells | 0.10% | Unknown | ||
| CD44+CD29+ cells | 0.80%–3.60% | Unknown | ||
| Sca-1+ cells | Breast cancer | 44.00%–6.97% | 1:865, 4T1 cells | [137] |
| 1:11,867, 66cl4 cells | ||||
| CD133+ cells | Melanoma | 0.03%–4.30% | 1:781, B16-F1 cells | |
| ALDH1+ cells | 12.30%–28.80% | 1:4,027, B16-F1 cells | ||
| 1:7,531, B16-F10 cells | ||||
| 1:13,567, Ret cells | ||||
| Adenocarcinoma | 0.03%–2.62% | Unknown | [153] | |
| Breast cancer | 0.20% | 1,000 cells implanted, tumors detected in 1/6 mice | [148] | |
| Colon cancer | 3.10%–17.00% | 100 cells implanted, tumors detected in 3/5 mice | [152] | |
| Hematopoietic tumor | 0.10% | Unknown | [143] | |
| Lung cancer | 1.50%–6.10% | 1,000 cells implanted, tumors detected in 2/4 mice | [145] | |
| 0.10%–4.00% | 50 cells implanted, tumors detected in 6/8 mice | [156] | ||
| Nasopharyngeal cancer | 0.10%–6.80% | 1×104 cells implanted, tumors detected in 3/3 mice | [146] | |
| Gastric cancer | 1.29%–1.69% | Unknown, Caco2 cells | [152] | |
| 0.08% | 1,000 cells implanted, tumors detected in 1/3 mice, MKN-45 cell | [154] | ||
| 0.18% | 5×104 cells implanted, tumors detected in 1/3 mice, BGC-823 cell | |||
| Glioma | 0.40% | Unknown | [147] | |
| 0.04%–0.10% | 1,000 cells implanted, tumors detected in 1/1 mouse | [148] | ||
| 12.00% | 1:1×104 | [186] | ||
| Neuroblastoma | 4.00%–37.00% | Unknown | [149] | |
| Ovarian cancer | 0.05% | Unknown | [148] | |
| 0.10%–1.70% | 1:500 | [151] | ||
| Osteosarcoma | 3.70%–10.20% | 500 cells implanted, tumors detected in 3/9 mice | [118] | |
| Glioma | 0.04%–0.10% | 100 cells implanted, tumors detected in 7/11 mice, U373 cells | [148] | |
| Breast cancer | 0–0.20 | 1,000 cells implanted, tumors detected in 1/6 mice, MCF7 cells | ||
| Ovarian cancer | 0.05% | Unknown | ||
| Prostate cancer | 0.07% | 100 cell implanted, tumors detected in 2/8 mice, LAPC-9 cells | ||
| Cell division | Breast cancer | 0.20%–0.40% | 1:26 | [13] |
| Unknown | 1:2 | [159] | ||
| Ovarian cancer | 7.00% | Unknown | [160] | |
| Gastrointestinal cancer | 20.00%–30.00% | Unknown | [161] | |
| 1.90%–2.70% | 10 cells implanted, tumors detected in 14/20 mice | [162] | ||
| Nasopharyngeal cancer | 0.60% | Unknown | [163] | |
| Glioma | 5.00% | 1:2×105 | [165] | |
| Liver cancer | 0.70% | 100 cells implanted, tumors detected in 2/4 mice | [166] | |
| Cytotoxic and hypoxic resistance | ||||
| CD133+ cells | Glioma | 2–10-fold increase | 1:1,000–5,000, IR treatment | [5] |
| 45.40% | Unknown, hypoxia selected | [182] | ||
| 37.00% | Unknown, rotenone treatement | |||
| ABCG2+ cells | 30.00% | Unknown, mitoxatrone treatement | [186] | |
| ALDH1+ cells | Esophageal cancer | 5.60% | Unknown, cisplatin treatment | [67] |
| ALDH1+ cells | Breast cancer | 5.60% | Unknown, ADR treatment | [188] |
| CD44+CD24− cells | 32.40% | 5,000 cells implanted, tumors detected in 1/2 mice, estrogen treatment | ||
| ALDH1+ cells | 2–4.8-fold increase | Unknown, sunitinib treatment, MDA-MB-231 cells | [193] | |
| 4.6-fold increase | Unknown, sunitinib treatment, SUM159 cells | |||
| 2-fold increase | Unknown, bevacizumab treatment, MDA-MB-231 cells | |||
| 2–3-fold increase | Unknown, hypoxia selected, SUM159 cells | |||
| CD44+CD24-ESA+ cells | 3-fold increase | 500 cells implanted, tumors detected in 4/5 mice, hypoxia selected, MDA-MB 231 cells | [194] | |
| 12-fold increase | 500 cells implanted, tumors detected in 4/5 mice, hypoxia selected, BCM2 F3 cells | |||
| CD44+CD24+ESA+ cells | Pancreatic cancer | 1.30% | 5×105 cells implanted, tumors detected in 8/8 mice, gemcitabine treatment | [181] |
| SP cells | Oral cancer | 20.80% | Unknown, 5-FU treatment | [179] |
| 28.00% | Unknown, CBDCA treatment | |||
| 12.60% | Unknown, 5-FU+CBDCA treatment | |||
| SP cells | Osteosarcoma | 0.11% | 5×106 cells implanted, tumors detected in all mice, U2OS cells, MTX treatment | [185] |
| CD177+Stro-1+ cells | 0.60% | |||
| ABCG2+ cells | 23.15% | Unknown, MG-63 cells, 3AB treatment | [187] | |
| Invasiveness and adhesion | ||||
| Direct selection | Glioma | 0.79%–41.02% | Unknown | [204] |
| Pancreatic cancer | 1.96%–47.20% | 1:500 | [28] | |
| TGF β treatment | Colon cancer | CD44 2.65% | Unknown | [201] |
| Sox2 5.25% | ||||
| Sca-1 1.90% | ||||
| CD24 1.90% | ||||
| FOXC2 treatment | Breast cancer | Unknown | 1:1,000 | [202] |
| Snail treatment | Colon cancer | CD44 4–17-fold increase | Unknown | [203] |
| CD133 2–3-fold increase | ||||
| Immunoselection | Breast cancer | 11.00%–23.50% | Unknown | [206] |
| Density gradient centrifugation | Liver cancer | 4.73% | 1×104 cells implanted, tumors detected in 1/6 mice | [212] |
In addition, some studies have indicated that CSCs in the same type of human cancer are heterogeneous [214]. This heterogeneity is not limited to the differential expression of surface markers but also involves various functional subsets of CSCs, which may be the result of genetic mutations and epigenetic modifications. For example, in pancreatic cancer, both CD133+CXCR4−and CD133+CXCR4+ populations were found to exhibit similar levels of tumorigenicity. However, the metastatic phenotype of the individual tumor was determined by the distinct subpopulation of CD133+CXCR4+ CSCs, and the metastatic phenotype, but not the tumorigenic potential, of pancreatic tumors was nearly abrogated after the depletion of this subset of cells [28]. In breast cancer, both ALDH1+ and CD44+CD24–/lo cells are thought to be CSCs. Interestingly, the overlapping subset of these two populations, ALDH1+CD44+CD24–/lo cells, exhibits greater tumorigenicity than the cells expressing either marker alone [73]. These results suggest that different phenotypes may exist in the overlapping tumorigenic population; thus, combination strategies rather than single strategies are needed to enrich or isolate CSCs.
In summary, research into CSCs is still in its infancy. CSC isolation is viewed as the foundation and starting point for better understanding the characteristics of this specific subpopulation. An ideal method for isolating CSCs should have the following characteristics. (i) Specificity: The isolated cells should not contain any nontumorigenic cells, progenitor cells, or differentiated cells and should be able to form the original tumor in mice using as few as one cell. (ii) Sensitivity: All CSC subgroups that can initiate tumor formation should be included in the isolated cells. (iii) Versatility: The method should be applicable for the isolation of CSCs from different types of tumors. (iv) Convenience: The isolation procedures should be simple, use limited resources (such as time, effort, and energy), and be user friendly. The obvious limitations include surface marker sorting, SP isolation, and sphere culture; however, many researchers have expended great effort to establish a simpler, universal method for isolating CSCs. It is reasonable to believe that additional reliable, efficient, and convenient methods for isolating CSCs from tumor masses will be developed in the near future.
Finally, it should be emphasized again that the CSC hypothesis is based on the functional identification of TICs in vivo using the serial xenotransplantation assay, which serves as the gold standard [6].
Acknowledgments
We would like to thank Prof. Peng Zhang, Dr. Zhi-hua Zhou, and Dr. Lang Yang for their constructive suggestions. This study was supported by grants from the National Natural Science Foundation of China (no. 81172071, 81272598), the National Basic Research Program of China (973 Program, no. 2010CB529400), and the National S&T Major Special Project on New Drug Innovation of China (no. 2011ZX09102-010-02).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Marusyk A. Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pine SR. Ryan BM. Varticovski L. Robles AI. Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc Natl Acad Sci U S A. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali AS. Ahmad A. Ali S. Bao B. Philip PA. Sarkar FH. The role of cancer stem cells and miRNAs in defining the complexities of brain metastasis. J Cell Physiol. 2013;228:36–42. doi: 10.1002/jcp.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu G. Yuan X. Zeng Z. Tunici P. Ng H. Abdulkadir IR. Lu L. Irvin D. Black KL. Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao S. Wu Q. McLendon RE. Hao Y. Shi Q. Hjelmeland AB. Dewhirst MW. Bigner DD. Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 6.Clarke MF. A self-renewal assay for cancer stem cells. Cancer Chemother Pharmacol. 2005;56(Suppl 1):64–68. doi: 10.1007/s00280-005-0097-1. [DOI] [PubMed] [Google Scholar]
- 7.Clarke MF. Dick JE. Dirks PB. Eaves CJ. Jamieson CH. Jones DL. Visvader J. Weissman IL. Wahl GM. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 8.Lapidot T. Sirard C. Vormoor J. Murdoch B. Hoang T. Caceres-Cortes J. Minden M. Paterson B. Caligiuri MA. Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet D. Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hajj M. Wicha MS. Benito-Hernandez A. Morrison SJ. Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright MH. Calcagno AM. Salcido CD. Carlson MD. Ambudkar SV. Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang-Verslues WW. Kuo WH. Chang PH. Pan CC. Wang HH. Tsai ST. Jeng YM. Shew JY. Kung JT, et al. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS One. 2009;4:e8377. doi: 10.1371/journal.pone.0008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pece S. Tosoni D. Confalonieri S. Mazzarol G. Vecchi M. Ronzoni S. Bernard L. Viale G. Pelicci PG. Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Cho RW. Wang X. Diehn M. Shedden K. Chen GY. Sherlock G. Gurney A. Lewicki J. Clarke MF. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 15.Meyer MJ. Fleming JM. Lin AF. Hussnain SA. Ginsburg E. Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70:4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SK. Hawkins C. Clarke ID. Squire JA. Bayani J. Hide T. Henkelman RM. Cusimano MD. Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Son MJ. Woolard K. Nam DH. Lee J. Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward RJ. Lee L. Graham K. Satkunendran T. Yoshikawa K. Ling E. Harper L. Austin R. Nieuwenhuis E, et al. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 2009;69:4682–4690. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
- 19.Tchoghandjian A. Baeza N. Colin C. Cayre M. Metellus P. Beclin C. Ouafik L. Figarella-Branger D. A2B5 cells from human glioblastoma have cancer stem cell properties. Brain Pathol. 2010;20:211–221. doi: 10.1111/j.1750-3639.2009.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogden AT. Waziri AE. Lochhead RA. Fusco D. Lopez K. Ellis JA. Kang J. Assanah M. McKhann GM, et al. Identification of A2B5+CD133- tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62:505–514. doi: 10.1227/01.neu.0000316019.28421.95. discussion 514–505. [DOI] [PubMed] [Google Scholar]
- 21.Mazzoleni S. Politi LS. Pala M. Cominelli M. Franzin A. Sergi Sergi L. Falini A. De Palma M. Bulfone A. Poliani PL. Galli R. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 22.Collins AT. Berry PA. Hyde C. Stower MJ. Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 23.Miki J. Furusato B. Li H. Gu Y. Takahashi H. Egawa S. Sesterhenn IA. McLeod DG. Srivastava S. Rhim JS. Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens. Cancer Res. 2007;67:3153–3161. doi: 10.1158/0008-5472.CAN-06-4429. [DOI] [PubMed] [Google Scholar]
- 24.Prince ME. Sivanandan R. Kaczorowski A. Wolf GT. Kaplan MJ. Dalerba P. Weissman IL. Clarke MF. Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takaishi S. Okumura T. Tu S. Wang SS. Shibata W. Vigneshwaran R. Gordon SA. Shimada Y. Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin TG. Powell AE. Davies PS. Silk AD. Dismuke AD. Anderson EC. Swain JR. Wong MH. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072–2082. doi: 10.1053/j.gastro.2010.08.053. e2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C. Heidt DG. Dalerba P. Burant CF. Zhang L. Adsay V. Wicha M. Clarke MF. Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 28.Hermann PC. Huber SL. Herrler T. Aicher A. Ellwart JW. Guba M. Bruns CJ. Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien CA. Pollett A. Gallinger S. Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 30.Ricci-Vitiani L. Lombardi DG. Pilozzi E. Biffoni M. Todaro M. Peschle C. De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 31.Dalerba P. Dylla SJ. Park IK. Liu R. Wang X. Cho RW. Hoey T. Gurney A. Huang EH, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schepers AG. Snippert HJ. Stange DE. van den Born M. van Es JH. van de Wetering M. Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 33.Majeti R. Chao MP. Alizadeh AA. Pang WW. Jaiswal S. Gibbs KD., Jr. van Rooijen N. Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willingham SB. Volkmer JP. Gentles AJ. Sahoo D. Dalerba P. Mitra SS. Wang J. Contreras-Trujillo H. Martin R, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim CF. Jackson EL. Woolfenden AE. Lawrence S. Babar I. Vogel S. Crowley D. Bronson RT. Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Eramo A. Lotti F. Sette G. Pilozzi E. Biffoni M. Di Virgilio A. Conticello C. Ruco L. Peschle C. De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 37.Bertolini G. Roz L. Perego P. Tortoreto M. Fontanella E. Gatti L. Pratesi G. Fabbri A. Andriani F, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang HZ. Lin XG. Hua P. Wang M. Ao X. Xiong LH. Wu C. Guo JJ. The study of the tumor stem cell properties of CD133+CD44+ cells in the human lung adenocarcinoma cell line A549. Cell Mol Biol (Noisy-le-grand) 2010;56(Suppl):OL1350–OL 1358. [PubMed] [Google Scholar]
- 39.Zhang WC. Shyh-Chang N. Yang H. Rai A. Umashankar S. Ma S. Soh BS. Sun LL. Tai BC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 40.Fang D. Nguyen TK. Leishear K. Finko R. Kulp AN. Hotz S. Van Belle PA. Xu X. Elder DE. Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 41.Schatton T. Murphy GF. Frank NY. Yamaura K. Waaga-Gasser AM. Gasser M. Zhan Q. Jordan S. Duncan LM, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boiko AD. Razorenova OV. van de Rijn M. Swetter SM. Johnson DL. Ly DP. Butler PD. Yang GP. Joshua B, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monzani E. Facchetti F. Galmozzi E. Corsini E. Benetti A. Cavazzin C. Gritti A. Piccinini A. Porro D, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 44.Curley MD. Therrien VA. Cummings CL. Sergent PA. Koulouris CR. Friel AM. Roberts DJ. Seiden MV. Scadden DT. Rueda BR. Foster R. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 45.Suetsugu A. Nagaki M. Aoki H. Motohashi T. Kunisada T. Moriwaki H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351:820–824. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- 46.Yin S. Li J. Hu C. Chen X. Yao M. Yan M. Jiang G. Ge C. Xie H, et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120:1444–1450. doi: 10.1002/ijc.22476. [DOI] [PubMed] [Google Scholar]
- 47.Ma S. Chan KW. Hu L. Lee TK. Wo JY. Ng IO. Zheng BJ. Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 48.Haraguchi N. Ishii H. Mimori K. Tanaka F. Ohkuma M. Kim HM. Akita H. Takiuchi D. Hatano H, et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326–3339. doi: 10.1172/JCI42550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tirino V. Desiderio V. d'Aquino R. De Francesco F. Pirozzi G. Graziano A. Galderisi U. Cavaliere C. De Rosa A. Papaccio G. Giordano A. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS One. 2008;3:e3469. doi: 10.1371/journal.pone.0003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adhikari AS. Agarwal N. Wood BM. Porretta C. Ruiz B. Pochampally RR. Iwakuma T. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010;70:4602–4612. doi: 10.1158/0008-5472.CAN-09-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi C. Tian R. Wang M. Wang X. Jiang J. Zhang Z. Li X. He Z. Gong W. Qin R. CD44+CD133+ population exhibits cancer stem cell-like characteristics in human gallbladder carcinoma. Cancer Biol Ther. 2010;10:1182–1190. doi: 10.4161/cbt.10.11.13664. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe K. Meyer MJ. Strizzi L. Lee JM. Gonzales M. Bianco C. Nagaoka T. Farid SS. Margaryan N, et al. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells. 2010;28:1303–1314. doi: 10.1002/stem.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehrazma M. Madjd Z. Kalantari E. Panahi M. Hendi A. Shariftabrizi A. Expression of stem cell markers, CD133 and CD44, in pediatric solid tumors: a study using tissue microarray. Fetal Pediatr Pathol. 2012 doi: 10.3109/15513815.2012.701266. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 54.Kemper K. Sprick MR. de Bree M. Scopelliti A. Vermeulen L. Hoek M. Zeilstra J. Pals ST. Mehmet H. Stassi G. Medema JP. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70:719–729. doi: 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- 55.Beier D. Hau P. Proescholdt M. Lohmeier A. Wischhusen J. Oefner PJ. Aigner L. Brawanski A. Bogdahn U. Beier CP. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 56.Meng X. Li M. Wang X. Wang Y. Ma D. Both CD133+ and CD133- subpopulations of A549 and H446 cells contain cancer-initiating cells. Cancer Sci. 2009;100:1040–1046. doi: 10.1111/j.1349-7006.2009.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng X. Shen G. Yang X. Liu W. Most C6 cells are cancer stem cells: evidence from clonal and population analyses. Cancer Res. 2007;67:3691–3697. doi: 10.1158/0008-5472.CAN-06-3912. [DOI] [PubMed] [Google Scholar]
- 58.Shmelkov SV. Butler JM. Hooper AT. Hormigo A. Kushner J. Milde T. St Clair R. Baljevic M. White I, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oh SY. Kang HJ. Kim YS. Kim H. Lim YC. CD44-negative cells in head and neck squamous carcinoma also have stem-cell like traits. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Karbanova J. Missol-Kolka E. Fonseca AV. Lorra C. Janich P. Hollerova H. Jaszai J. Ehrmann J. Kolar Z, et al. The stem cell marker CD133 (Prominin-1) is expressed in various human glandular epithelia. J Histochem Cytochem. 2008;56:977–993. doi: 10.1369/jhc.2008.951897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park SY. Lee HE. Li H. Shipitsin M. Gelman R. Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16:876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Z. Wang Z. Fan Y. Zheng Q. Expression of CD133 in SW620 colorectal cancer cells is modulated by the microenvironment. Oncol Lett. 2012;4:75–79. doi: 10.3892/ol.2012.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gopisetty G. Xu J. Sampath D. Colman H. Puduvalli VK. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene. 2012 doi: 10.1038/onc.2012.331. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pellacani D. Packer RJ. Frame FM. Oldridge EE. Berry PA. Labarthe MC. Stower MJ. Simms MS. Collins AT. Maitland NJ. Regulation of the stem cell marker CD133 is independent of promoter hypermethylation in human epithelial differentiation and cancer. Mol Cancer. 2011;10:94. doi: 10.1186/1476-4598-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reynolds BA. Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 66.Visvader JE. Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 67.Zhang G. Ma L. Xie YK. Miao XB. Jin C. Esophageal cancer tumorspheres involve cancer stem-like populations with elevated aldehyde dehydrogenase enzymatic activity. Mol Med Report. 2012;6:519–524. doi: 10.3892/mmr.2012.939. [DOI] [PubMed] [Google Scholar]
- 68.Pollard SM. Yoshikawa K. Clarke ID. Danovi D. Stricker S. Russell R. Bayani J. Head R. Lee M, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 69.Woolard K. Fine HA. Glioma stem cells: better flat than round. Cell Stem Cell. 2009;4:466–467. doi: 10.1016/j.stem.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 70.Pastrana E. Silva-Vargas V. Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krishnamurthy S. Nor JE. Orosphere assay: a method for propagation of head and neck cancer stem cells. Head Neck. 2012 doi: 10.1002/hed.23076. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida A. Hsu LC. Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1992;46:239–244. doi: 10.1159/000468794. [DOI] [PubMed] [Google Scholar]
- 73.Ginestier C. Hur MH. Charafe-Jauffret E. Monville F. Dutcher J. Brown M. Jacquemier J. Viens P. Kleer CG, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pearce DJ. Taussig D. Simpson C. Allen K. Rohatiner AZ. Lister TA. Bonnet D. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23:752–760. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- 75.Hess DA. Wirthlin L. Craft TP. Herrbrich PE. Hohm SA. Lahey R. Eades WC. Creer MH. Nolta JA. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chute JP. Muramoto GG. Whitesides J. Colvin M. Safi R. Chao NJ. McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Armstrong L. Stojkovic M. Dimmick I. Ahmad S. Stojkovic P. Hole N. Lako M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 78.Hess DA. Craft TP. Wirthlin L. Hohm S. Zhou P. Eades WC. Creer MH. Sands MS. Nolta JA. Widespread nonhematopoietic tissue distribution by transplanted human progenitor cells with high aldehyde dehydrogenase activity. Stem Cells. 2008;26:611–620. doi: 10.1634/stemcells.2007-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feldmann G. Dhara S. Fendrich V. Bedja D. Beaty R. Mullendore M. Karikari C. Alvarez H. Iacobuzio-Donahue C, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balicki D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell. 2007;1:485–487. doi: 10.1016/j.stem.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 81.Charafe-Jauffret E. Ginestier C. Iovino F. Tarpin C. Diebel M. Esterni B. Houvenaeghel G. Extra JM. Bertucci F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang F. Qiu Q. Khanna A. Todd NW. Deepak J. Xing L. Wang H. Liu Z. Su Y. Stass SA. Katz RL. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Neumeister V. Rimm D. Is ALDH1 a good method for definition of breast cancer stem cells? Breast Cancer Res Treat. 2010;123:109–111. doi: 10.1007/s10549-009-0656-y. [DOI] [PubMed] [Google Scholar]
- 84.Alison MR. Guppy NJ. Lim SM. Nicholson LJ. Finding cancer stem cells: are aldehyde dehydrogenases fit for purpose? J Pathol. 2010;222:335–344. doi: 10.1002/path.2772. [DOI] [PubMed] [Google Scholar]
- 85.Duarte S. Loubat A. Momier D. Topi M. Faneca H. Pedroso de Lima MC. Carle GF. Pierrefite-Carle V. Isolation of head and neck squamous carcinoma cancer stem-like cells in a syngeneic mouse model and analysis of hypoxia effect. Oncol Rep. 2012;28:1057–1062. doi: 10.3892/or.2012.1904. [DOI] [PubMed] [Google Scholar]
- 86.Korkaya H. Paulson A. Iovino F. Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vlashi E. Kim K. Lagadec C. Donna LD. McDonald JT. Eghbali M. Sayre JW. Stefani E. McBride W. Pajonk F. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009;101:350–359. doi: 10.1093/jnci/djn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan J. Zhang Q. Wang Y. You M. 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One. 2010;5:e13298. doi: 10.1371/journal.pone.0013298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiang Y. Zhu Z. Han G. Ye X. Xu B. Peng Z. Ma Y. Yu Y. Lin H. Chen AP. Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roesch A. Fukunaga-Kalabis M. Schmidt EC. Zabierowski SE. Brafford PA. Vultur A. Basu D. Gimotty P. Vogt T. Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Held M. Bosenberg M. A role for the JARID1B stem cell marker for continuous melanoma growth. Pigment Cell Melanoma Res. 2010;23:481–483. doi: 10.1111/j.1755-148X.2010.00726.x. [DOI] [PubMed] [Google Scholar]
- 92.Radberger P. Radberger A. Bykov VJ. Seregard S. Economou MA. JARID1B protein expression and prognostic implications in uveal melanoma. Invest Ophthalmol Vis Sci. 2012;53:4442–4449. doi: 10.1167/iovs.11-9296. [DOI] [PubMed] [Google Scholar]
- 93.Quintana E. Shackleton M. Sabel MS. Fullen DR. Johnson TM. Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shackleton MJ. Quintana E. Fullen DR. Sabel MS. Johnson TM. Melanoma: do we need a hatchet or a scalpel? Arch Dermatol. 2009;145:307–308. doi: 10.1001/archdermatol.2009.3. [DOI] [PubMed] [Google Scholar]
- 95.Quintana E. Shackleton M. Foster HR. Fullen DR. Sabel MS. Johnson TM. Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith J. Ladi E. Mayer-Proschel M. Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc Natl Acad Sci U S A. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ito K. Hirao A. Arai F. Matsuoka S. Takubo K. Hamaguchi I. Nomiyama K. Hosokawa K. Sakurada K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 98.Tsatmali M. Walcott EC. Crossin KL. Newborn neurons acquire high levels of reactive oxygen species and increased mitochondrial proteins upon differentiation from progenitors. Brain Res. 2005;1040:137–150. doi: 10.1016/j.brainres.2005.01.087. [DOI] [PubMed] [Google Scholar]
- 99.Ito K. Hirao A. Arai F. Takubo K. Matsuoka S. Miyamoto K. Ohmura M. Naka K. Hosokawa K. Ikeda Y. Suda T. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 100.Tothova Z. Kollipara R. Huntly BJ. Lee BH. Castrillon DH. Cullen DE. McDowell EP. Lazo-Kallanian S. Williams IR, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 101.Miyamoto K. Araki KY. Naka K. Arai F. Takubo K. Yamazaki S. Matsuoka S. Miyamoto T. Ito K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 102.Chen C. Liu Y. Liu R. Ikenoue T. Guan KL. Liu Y. Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Armstrong L. Tilgner K. Saretzki G. Atkinson SP. Stojkovic M. Moreno R. Przyborski S. Lako M. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 104.Diehn M. Cho RW. Lobo NA. Kalisky T. Dorie MJ. Kulp AN. Qian D. Lam JS. Ailles LE, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ye XQ. Li Q. Wang GH. Sun FF. Huang GJ. Bian XW. Yu SC. Qian GS. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer. 2011;129:820–831. doi: 10.1002/ijc.25944. [DOI] [PubMed] [Google Scholar]
- 106.Ishimoto T. Nagano O. Yae T. Tamada M. Motohara T. Oshima H. Oshima M. Ikeda T. Asaba R, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 107.Nesti C. Pasquali L. Vaglini F. Siciliano G. Murri L. The role of mitochondria in stem cell biology. Biosci Rep. 2007;27:165–171. doi: 10.1007/s10540-007-9044-1. [DOI] [PubMed] [Google Scholar]
- 108.Bavister BD. The mitochondrial contribution to stem cell biology. Reprod Fertil Dev. 2006;18:829–838. doi: 10.1071/rd06111. [DOI] [PubMed] [Google Scholar]
- 109.Lonergan T. Bavister B. Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–296. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parker GC. Acsadi G. Brenner CA. Mitochondria: determinants of stem cell fate? Stem Cells Dev. 2009;18:803–806. doi: 10.1089/scd.2009.1806.edi. [DOI] [PubMed] [Google Scholar]
- 111.Pietila M. Lehtonen S. Narhi M. Hassinen IE. Leskela HV. Aranko K. Nordstrom K. Vepsalainen A. Lehenkari P. Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells. Tissue Eng Part C Methods. 2010;16:435–445. doi: 10.1089/ten.tec.2009.0247. [DOI] [PubMed] [Google Scholar]
- 112.Heerdt BG. Houston MA. Wilson AJ. Augenlicht LH. The intrinsic mitochondrial membrane potential (Deltapsim) is associated with steady-state mitochondrial activity and the extent to which colonic epithelial cells undergo butyrate-mediated growth arrest and apoptosis. Cancer Res. 2003;63:6311–6319. [PubMed] [Google Scholar]
- 113.Heerdt BG. Houston MA. Augenlicht LH. The intrinsic mitochondrial membrane potential of colonic carcinoma cells is linked to the probability of tumor progression. Cancer Res. 2005;65:9861–9867. doi: 10.1158/0008-5472.CAN-05-2444. [DOI] [PubMed] [Google Scholar]
- 114.Heerdt BG. Houston MA. Augenlicht LH. Growth properties of colonic tumor cells are a function of the intrinsic mitochondrial membrane potential. Cancer Res. 2006;66:1591–1596. doi: 10.1158/0008-5472.CAN-05-2717. [DOI] [PubMed] [Google Scholar]
- 115.Ye XQ. Wang GH. Huang GJ. Bian XW. Qian GS. Yu SC. Heterogeneity of mitochondrial membrane potential: a novel tool to isolate and identify cancer stem cells from a tumor mass? Stem Cell Rev. 2011;7:153–160. doi: 10.1007/s12015-010-9122-9. [DOI] [PubMed] [Google Scholar]
- 116.Michelakis ED. Sutendra G. Dromparis P. Webster L. Haromy A. Niven E. Maguire C. Gammer TL. Mackey JR, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2:31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- 117.Wong DJ. Liu H. Ridky TW. Cassarino D. Segal E. Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang M. Yan M. Zhang R. Li J. Luo Z. Side population cells isolated from human osteosarcoma are enriched with tumor-initiating cells. Cancer Sci. 2011;102:1774–1781. doi: 10.1111/j.1349-7006.2011.02028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bourguignon LY. Wong G. Earle C. Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2012;287:32800–32824. doi: 10.1074/jbc.M111.308528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bai L. Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24:1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levings PP. McGarry SV. Currie TP. Nickerson DM. McClellan S. Ghivizzani SC. Steindler DA. Gibbs CP. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res. 2009;69:5648–5655. doi: 10.1158/0008-5472.CAN-08-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zbinden M. Duquet A. Lorente-Trigos A. Ngwabyt SN. Borges I. Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. Embo J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tamase A. Muraguchi T. Naka K. Tanaka S. Kinoshita M. Hoshii T. Ohmura M. Shugo H. Ooshio T, et al. Identification of tumor-initiating cells in a highly aggressive brain tumor using promoter activity of nucleostemin. Proc Natl Acad Sci U S A. 2009;106:17163–17168. doi: 10.1073/pnas.0905016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vermeulen L. De Sousa EMF. van der Heijden M. Cameron K. de Jong JH. Borovski T. Tuynman JB. Todaro M. Merz C, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 125.Wu F. Zhang J. Wang P. Ye X. Jung K. Bone KM. Pearson JD. Ingham RJ. McMullen TP. Ma Y. Lai R. Identification of two novel phenotypically distinct breast cancer cell subsets based on Sox2 transcription activity. Cell Signal. 2012;24:1989–1998. doi: 10.1016/j.cellsig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 126.Maksakova IA. Mager DL. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J Virol. 2005;79:13865–13874. doi: 10.1128/JVI.79.22.13865-13874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hotta A. Cheung AY. Farra N. Vijayaragavan K. Seguin CA. Draper JS. Pasceri P. Maksakova IA. Mager DL, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 128.Shan J. Shen J. Liu L. Xia F. Xu C. Duan G. Xu Y. Ma Q. Yang Z, et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004–1014. doi: 10.1002/hep.25745. [DOI] [PubMed] [Google Scholar]
- 129.Teisanu RM. Lagasse E. Whitesides JF. Stripp BR. Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells. 2009;27:612–622. doi: 10.1634/stemcells.2008-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ezzat T. Dhar DK. Malago M. Olde Damink SW. Dynamic tracking of stem cells in an acute liver failure model. World J Gastroenterol. 2012;18:507–516. doi: 10.3748/wjg.v18.i6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tacke F. Weiskirchen R. Update on hepatic stellate cells: pathogenic role in liver fibrosis and novel isolation techniques. Expert Rev Gastroenterol Hepatol. 2012;6:67–80. doi: 10.1586/egh.11.92. [DOI] [PubMed] [Google Scholar]
- 132.Clement V. Marino D. Cudalbu C. Hamou MF. Mlynarik V. de Tribolet N. Dietrich PY. Gruetter R. Hegi ME. Radovanovic I. Marker-independent identification of glioma-initiating cells. Nat Methods. 2010;7:224–228. doi: 10.1038/nmeth.1430. [DOI] [PubMed] [Google Scholar]
- 133.Weiss L. Metastasis of cancer: a conceptual history from antiquity to the 1990s. Cancer Metastasis Rev. 2000;19:I–XI. 193–383. [PubMed] [Google Scholar]
- 134.Locke M. Heywood M. Fawell S. Mackenzie IC. Retention of intrinsic stem cell hierarchies in carcinoma-derived cell lines. Cancer Res. 2005;65:8944–8950. doi: 10.1158/0008-5472.CAN-05-0931. [DOI] [PubMed] [Google Scholar]
- 135.Harper LJ. Piper K. Common J. Fortune F. Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36:594–603. doi: 10.1111/j.1600-0714.2007.00617.x. [DOI] [PubMed] [Google Scholar]
- 136.Li H. Chen X. Calhoun-Davis T. Claypool K. Tang DG. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res. 2008;68:1820–1825. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- 137.Kuch V. Schreiber C. Thiele W. Umansky V. Sleeman JP. Tumor-initiating properties of breast cancer and melanoma cells in vivo are not invariably reflected by spheroid formation in vitro, but can be increased by long-term culturing as adherent monolayers. Int J Cancer. 2013;132:E94–E105. doi: 10.1002/ijc.27785. [DOI] [PubMed] [Google Scholar]
- 138.Lim YC. Oh SY. Kim H. Cellular characteristics of head and neck cancer stem cells in type IV collagen-coated adherent cultures. Exp Cell Res. 2012;318:1104–1111. doi: 10.1016/j.yexcr.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 139.Zhou ZH. Ping YF. Yu SC. Yi L. Yao XH. Chen JH. Cui YH. Bian XW. A novel approach to the identification and enrichment of cancer stem cells from a cultured human glioma cell line. Cancer Lett. 2009;281:92–99. doi: 10.1016/j.canlet.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 140.Sun Y. Pollard S. Conti L. Toselli M. Biella G. Parkin G. Willatt L. Falk A. Cattaneo E. Smith A. Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Mol Cell Neurosci. 2008;38:245–258. doi: 10.1016/j.mcn.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 141.Lee J. Kotliarova S. Kotliarov Y. Li A. Su Q. Donin NM. Pastorino S. Purow BW. Christopher N, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 142.Li A. Walling J. Kotliarov Y. Center A. Steed ME. Ahn SJ. Rosenblum M. Mikkelsen T. Zenklusen JC. Fine HA. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 143.Goodell MA. Brose K. Paradis G. Conner AS. Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Van den Broeck A. Gremeaux L. Topal B. Vankelecom H. Human pancreatic adenocarcinoma contains a side population resistant to gemcitabine. BMC Cancer. 2012;12:354. doi: 10.1186/1471-2407-12-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ho MM. Ng AV. Lam S. Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 146.Wang J. Guo LP. Chen LZ. Zeng YX. Lu SH. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;67:3716–3724. doi: 10.1158/0008-5472.CAN-06-4343. [DOI] [PubMed] [Google Scholar]
- 147.Kondo T. Setoguchi T. Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Patrawala L. Calhoun T. Schneider-Broussard R. Zhou J. Claypool K. Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 149.Hirschmann-Jax C. Foster AE. Wulf GG. Nuchtern JG. Jax TW. Gobel U. Goodell MA. Brenner MK. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hadnagy A. Gaboury L. Beaulieu R. Balicki D. SP analysis may be used to identify cancer stem cell populations. Exp Cell Res. 2006;312:3701–3710. doi: 10.1016/j.yexcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 151.Hu L. McArthur C. Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer. 2010;102:1276–1283. doi: 10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Burkert J. Otto WR. Wright NA. Side populations of gastrointestinal cancers are not enriched in stem cells. J Pathol. 2008;214:564–573. doi: 10.1002/path.2307. [DOI] [PubMed] [Google Scholar]
- 153.Lichtenauer UD. Shapiro I. Geiger K. Quinkler M. Fassnacht M. Nitschke R. Ruckauer KD. Beuschlein F. Side population does not define stem cell-like cancer cells in the adrenocortical carcinoma cell line NCI h295R. Endocrinology. 2008;149:1314–1322. doi: 10.1210/en.2007-1001. [DOI] [PubMed] [Google Scholar]
- 154.Zhang H. Xi H. Cai A. Xia Q. Wang XX. Lu C. Zhang Y. Song Z. Wang H, et al. Not all side population cells contain cancer stem-like cells in human gastric cancer cell lines. Dig Dis Sci. 2013;58:132–139. doi: 10.1007/s10620-012-2330-1. [DOI] [PubMed] [Google Scholar]
- 155.Wu T. Li JY. Lu SX. [Isolation and characterization of side population cells in human gastric cancer cell line BGC-823] Zhonghua Zhong Liu Za Zhi. 2012;34:264–268. doi: 10.3760/cma.j.issn.0253-3766.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 156.Salcido CD. Larochelle A. Taylor BJ. Dunbar CE. Varticovski L. Molecular characterisation of side population cells with cancer stem cell-like characteristics in small-cell lung cancer. Br J Cancer. 2010;102:1636–1644. doi: 10.1038/sj.bjc.6605668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Morrison SJ. Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 158.Lanzkron SM. Collector MI. Sharkis SJ. Hematopoietic stem cell tracking in vivo: a comparison of short-term and long-term repopulating cells. Blood. 1999;93:1916–1921. [PubMed] [Google Scholar]
- 159.Cicalese A. Bonizzi G. Pasi CE. Faretta M. Ronzoni S. Giulini B. Brisken C. Minucci S. Di Fiore PP. Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 160.Kusumbe AP. Bapat SA. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009;69:9245–9253. doi: 10.1158/0008-5472.CAN-09-2802. [DOI] [PubMed] [Google Scholar]
- 161.Xue Z. Yan H. Li J. Liang S. Cai X. Chen X. Wu Q. Gao L. Wu K. Nie Y. Fan D. Identification of cancer stem cells in vincristine preconditioned SGC7901 gastric cancer cell line. J Cell Biochem. 2012;113:302–312. doi: 10.1002/jcb.23356. [DOI] [PubMed] [Google Scholar]
- 162.Xin HW. Hari DM. Mullinax JE. Ambe CM. Koizumi T. Ray S. Anderson AJ. Wiegand GW. Garfield SH. Thorgeirsson SS. Avital I. Tumor-initiating label-retaining cancer cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. 2012;30:591–598. doi: 10.1002/stem.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jiang QP. Yao KT. Isolation and detection of label-retaining cells in a nasopharyngeal carcinoma cell line. Chin J Cancer. 2010;29:572–574. doi: 10.5732/cjc.009.10420. [DOI] [PubMed] [Google Scholar]
- 164.Deleyrolle LP. Rohaus MR. Fortin JM. Reynolds BA. Azari H. Identification and isolation of slow-dividing cells in human glioblastoma using carboxy fluorescein succinimidyl ester (CFSE) J Vis Exp. 2012;62 doi: 10.3791/3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Deleyrolle LP. Harding A. Cato K. Siebzehnrubl FA. Rahman M. Azari H. Olson S. Gabrielli B. Osborne G. Vescovi A. Reynolds BA. Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain. 2011;134:1331–1343. doi: 10.1093/brain/awr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kamohara Y. Haraguchi N. Mimori K. Tanaka F. Inoue H. Mori M. Kanematsu T. The search for cancer stem cells in hepatocellular carcinoma. Surgery. 2008;144:119–124. doi: 10.1016/j.surg.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 167.Moore N. Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 20112011 doi: 10.1155/2011/396076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Salmaggi A. Boiardi A. Gelati M. Russo A. Calatozzolo C. Ciusani E. Sciacca FL. Ottolina A. Parati EA, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54:850–860. doi: 10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- 169.van Stijn A. van der Pol MA. Kok A. Bontje PM. Roemen GM. Beelen RH. Ossenkoppele GJ. Schuurhuis GJ. Differences between the CD34+ and CD34- blast compartments in apoptosis resistance in acute myeloid leukemia. Haematologica. 2003;88:497–508. [PubMed] [Google Scholar]
- 170.Dean M. Fojo T. Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 171.Eramo A. Ricci-Vitiani L. Zeuner A. Pallini R. Lotti F. Sette G. Pilozzi E. Larocca LM. Peschle C. De Maria R. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13:1238–1241. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 172.Ghods AJ. Irvin D. Liu G. Yuan X. Abdulkadir IR. Tunici P. Konda B. Wachsmann-Hogiu S. Black KL. Yu JS. Spheres isolated from 9L gliosarcoma rat cell line possess chemoresistant and aggressive cancer stem-like cells. Stem Cells. 2007;25:1645–1653. doi: 10.1634/stemcells.2006-0624. [DOI] [PubMed] [Google Scholar]
- 173.Wei C. Guo-min W. Yu-jun L. Apoptosis resistance can be used in screening the markers of cancer stem cells. Med Hypotheses. 2006;67:1381–1383. doi: 10.1016/j.mehy.2006.05.054. [DOI] [PubMed] [Google Scholar]
- 174.Yu F. Yao H. Zhu P. Zhang X. Pan Q. Gong C. Huang Y. Hu X. Su F. Lieberman J. Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 175.Li HZ. Yi TB. Wu ZY. Suspension culture combined with chemotherapeutic agents for sorting of breast cancer stem cells. BMC Cancer. 2008;8:135. doi: 10.1186/1471-2407-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Park CH. Bergsagel DE. McCulloch EA. Mouse myeloma tumor stem cells: a primary cell culture assay. J Natl Cancer Inst. 1971;46:411–422. [PubMed] [Google Scholar]
- 177.Singh A. Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nakai E. Park K. Yawata T. Chihara T. Kumazawa A. Nakabayashi H. Shimizu K. Enhanced MDR1 expression and chemoresistance of cancer stem cells derived from glioblastoma. Cancer Invest. 2009;27:901–908. doi: 10.3109/07357900801946679. [DOI] [PubMed] [Google Scholar]
- 179.Yajima T. Ochiai H. Uchiyama T. Takano N. Shibahara T. Azuma T. Resistance to cytotoxic chemotherapy-induced apoptosis in side population cells of human oral squamous cell carcinoma cell line Ho-1-N-1. Int J Oncol. 2009;35:273–280. [PubMed] [Google Scholar]
- 180.Li Z. Rich JN. Hypoxia and hypoxia inducible factors in cancer stem cell maintenance. Curr Top Microbiol Immunol. 2010;345:21–30. doi: 10.1007/82_2010_75. [DOI] [PubMed] [Google Scholar]
- 181.Hong SP. Wen J. Bang S. Park S. Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 182.Griguer CE. Oliva CR. Gobin E. Marcorelles P. Benos DJ. Lancaster JR., Jr. Gillespie GY. CD133 is a marker of bioenergetic stress in human glioma. PLoS One. 2008;3:e3655. doi: 10.1371/journal.pone.0003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chartrain M. Riond J. Stennevin A. Vandenberghe I. Gomes B. Lamant L. Meyer N. Gairin JE. Guilbaud N. Annereau JP. Melanoma chemotherapy leads to the selection of ABCB5-expressing cells. PLoS One. 2012;7:e36762. doi: 10.1371/journal.pone.0036762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ghisolfi L. Keates AC. Hu X. Lee DK. Li CJ. Ionizing radiation induces stemness in cancer cells. PLoS One. 2012;7:e43628. doi: 10.1371/journal.pone.0043628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tang QL. Liang Y. Xie XB. Yin JQ. Zou CY. Zhao ZQ. Shen JN. Wang J. Enrichment of osteosarcoma stem cells by chemotherapy. Chin J Cancer. 2011;30:426–432. doi: 10.5732/cjc.011.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bleau AM. Hambardzumyan D. Ozawa T. Fomchenko EI. Huse JT. Brennan CW. Holland EC. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Di Fiore R. Santulli A. Ferrante RD. Giuliano M. De Blasio A. Messina C. Pirozzi G. Tirino V. Tesoriere G. Vento R. Identification and expansion of human osteosarcoma-cancer-stem cells by long-term 3-aminobenzamide treatment. J Cell Physiol. 2009;219:301–313. doi: 10.1002/jcp.21667. [DOI] [PubMed] [Google Scholar]
- 188.Calcagno AM. Salcido CD. Gillet JP. Wu CP. Fostel JM. Mumau MD. Gottesman MM. Varticovski L. Ambudkar SV. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst. 102:1637–1652. doi: 10.1093/jnci/djq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yu SC. Ping YF. Yi L. Zhou ZH. Chen JH. Yao XH. Gao L. Wang JM. Bian XW. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265:124–134. doi: 10.1016/j.canlet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 190.Pajic M. Kersbergen A. van Diepen F. Pfauth A. Jonkers J. Borst P. Rottenberg S. Tumor-initiating cells are not enriched in cisplatin-surviving BRCA1;p53-deficient mammary tumor cells in vivo. Cell Cycle. 2010;9:3780–3791. doi: 10.4161/cc.9.18.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kim Y. Lin Q. Glazer PM. Yun Z. Hypoxic tumor microenvironment and cancer cell differentiation. Curr Mol Med. 2009;9:425–434. doi: 10.2174/156652409788167113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Heddleston JM. Li Z. McLendon RE. Hjelmeland AB. Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Conley SJ. Gheordunescu E. Kakarala P. Newman B. Korkaya H. Heath AN. Clouthier SG. Wicha MS. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci U S A. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Louie E. Nik S. Chen JS. Schmidt M. Song B. Pacson C. Chen XF. Park S. Ju J. Chen EI. Identification of a stem-like cell population by exposing metastatic breast cancer cell lines to repetitive cycles of hypoxia and reoxygenation. Breast Cancer Res. 2010;12:R94. doi: 10.1186/bcr2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Huang Z. Cheng L. Guryanova OA. Wu Q. Bao S. Cancer stem cells in glioblastoma—molecular signaling and therapeutic targeting. Protein Cell. 2010;1:638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brabletz T. Jung A. Spaderna S. Hlubek F. Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 197.Sampieri K. Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012;22:187–193. doi: 10.1016/j.semcancer.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 198.Asuthkar S. Velpula KK. Chetty C. Gorantla B. Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–1454. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Velpula KK. Rehman AA. Chelluboina B. Dasari VR. Gondi CS. Rao JS. Veeravalli KK. Glioma stem cell invasion through regulation of the interconnected ERK, integrin alpha6 and N-cadherin signaling pathway. Cell Signal. 2012;24:2076–2084. doi: 10.1016/j.cellsig.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 200.Klarmann GJ. Hurt EM. Mathews LA. Zhang X. Duhagon MA. Mistree T. Thomas SB. Farrar WL. Invasive prostate cancer cells are tumor initiating cells that have a stem cell-like genomic signature. Clin Exp Metastasis. 2009;26:433–446. doi: 10.1007/s10585-009-9242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zubeldia IG. Bleau AM. Redrado M. Serrano D. Agliano A. Gil-Puig C. Vidal-Vanaclocha F. Lecanda J. Calvo A. Epithelial to mesenchymal transition and cancer stem cell phenotypes leading to liver metastasis are abrogated by the novel TGFbeta1-targeting peptides P17 and P144. Exp Cell Res. 2013;319:12–22. doi: 10.1016/j.yexcr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 202.Hollier BG. Tinnirello AA. Werden SJ. Evans KW. Taube JH. Sarkar TR. Sphyris N. Shariati M. Kumar SV, et al. FOXC2 Expression Links Epithelial-Mesenchymal Transition and Stem Cell Properties in Breast Cancer. Cancer Res. 2013;6:1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fan F. Samuel S. Evans KW. Lu J. Xia L. Zhou Y. Sceusi E. Tozzi F. Ye XC. Mani SA. Ellis LM. Overexpression of Snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1:5–16. doi: 10.1002/cam4.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yu SC. Bian XW. Enrichment of cancer stem cells based on heterogeneity of invasiveness. Stem Cell Rev. 2009;5:66–71. doi: 10.1007/s12015-008-9047-8. [DOI] [PubMed] [Google Scholar]
- 205.Walia V. Elble RC. Enrichment for breast cancer cells with stem/progenitor properties by differential adhesion. Stem Cells Dev. 2010;19:1175–1182. doi: 10.1089/scd.2009.0430. [DOI] [PubMed] [Google Scholar]
- 206.Reim F. Dombrowski Y. Ritter C. Buttmann M. Hausler S. Ossadnik M. Krockenberger M. Beier D. Beier CP, et al. Immunoselection of breast and ovarian cancer cells with trastuzumab and natural killer cells: selective escape of CD44high/CD24low/HER2low breast cancer stem cells. Cancer Res. 2009;69:8058–8066. doi: 10.1158/0008-5472.CAN-09-0834. [DOI] [PubMed] [Google Scholar]
- 207.Wolpert F. Roth P. Lamszus K. Tabatabai G. Weller M. Eisele G. HLA-E contributes to an immune-inhibitory phenotype of glioblastoma stem-like cells. J Neuroimmunol. 2012;250:27–34. doi: 10.1016/j.jneuroim.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 208.Chen HC. Chou AS. Liu YC. Hsieh CH. Kang CC. Pang ST. Yeh CT. Liu HP. Liao SK. Induction of metastatic cancer stem cells from the NK/LAK-resistant floating, but not adherent, subset of the UP-LN1 carcinoma cell line by IFN-gamma. Lab Invest. 2011;91:1502–1513. doi: 10.1038/labinvest.2011.91. [DOI] [PubMed] [Google Scholar]
- 209.Pietra G. Manzini C. Vitale M. Balsamo M. Ognio E. Boitano M. Queirolo P. Moretta L. Mingari MC. Natural killer cells kill human melanoma cells with characteristics of cancer stem cells. Int Immunol. 2009;21:793–801. doi: 10.1093/intimm/dxp047. [DOI] [PubMed] [Google Scholar]
- 210.Tseng HC. Arasteh A. Paranjpe A. Teruel A. Yang W. Behel A. Alva JA. Walter G. Head C, et al. Increased lysis of stem cells but not their differentiated cells by natural killer cells; de-differentiation or reprogramming activates NK cells. PLoS One. 2010;5:e11590. doi: 10.1371/journal.pone.0011590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jewett A. Tseng HC. Arasteh A. Saadat S. Christensen RE. Cacalano NA. Natural killer cells preferentially target cancer stem cells; role of monocytes in protection against NK cell mediated lysis of cancer stem cells. Curr Drug Deliv. 2012;9:5–16. doi: 10.2174/156720112798375989. [DOI] [PubMed] [Google Scholar]
- 212.Liu WH. Wang X. You N. Tao KS. Wang T. Tang LJ. Dou KF. Efficient enrichment of hepatic cancer stem-like cells from a primary rat HCC model via a density gradient centrifugation-centered method. PLoS One. 2012;7:e35720. doi: 10.1371/journal.pone.0035720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kuch V. Schreiber C. Thiele W. Umansky V. Sleeman JP. Tumor-initiating properties of breast cancer and melanoma cells in vivo are not invariably reflected by spheroid formation in vitro, but can be increased by long-term culturing as adherent monolayers. Int J Cancer. 2013;132:E94–E105. doi: 10.1002/ijc.27785. [DOI] [PubMed] [Google Scholar]
- 214.Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]