Abstract
Maintenance of the pluripotent state or differentiation of the pluripotent state into any germ layer depends on the factors that orchestrate expression of thousands of genes through epigenetic, transcriptional, and post-transcriptional regulation. Long noncoding RNAs (lncRNAs) are implicated in the complex molecular circuitry in the developmental processes. The ENCODE project has opened up new avenues for studying these lncRNA transcripts with the availability of new datasets for lncRNA annotation and regulation. Expression studies identified hundreds of long noncoding RNAs differentially expressed in the pluripotent state, and many of these lncRNAs are found to control the pluripotency and stemness in embryonic and induced pluripotent stem cells or, in the reverse way, promote differentiation of pluripotent cells. They are generally transcriptionally activated or repressed by pluripotency-associated transcription factors and function as molecular mediators of gene expression that determine the pluripotent state of the cell. They can act as molecular scaffolds or guides for the chromatin-modifying complexes to direct them to bind into specific genomic loci to impart a repressive or activating effect on gene expression, or they can transcriptionally or post-transcriptionally regulate gene expression by diverse molecular mechanisms. This review focuses on recent findings on the regulatory role of lncRNAs in two main aspects of pluripotency, namely, self renewal and differentiation into any lineage, and elucidates the underlying molecular mechanisms that are being uncovered lately.
Introduction
Long noncoding RNAs, previously thought as transcriptional noise, have recently come into the limelight as functional molecules taking part in epigenetic, transcriptional, and post-transcriptional regulation of gene expression [1–6]. After a decade of research, we are beginning to understand the functionality of this vast set of long noncoding transcripts. Recently, the release of ENCODE project has paved the way for identification and annotation of a large number of long noncoding RNAs [7,8]. The GENCODE consortium, under the ENCODE project [9], introduced a set of 14,880 manually curated, evidence-based lncRNA transcripts [10]. This set contains 9518 intergenic [which do not overlap with coding transcripts and hence referred to as long intergenic RNAs (lincRNAs)] and 5362 genic (that is either overlapped with a coding gene in sense or antisense or resides in an intron of a coding gene) transcripts. Many of the lncRNAs are epigenetic regulators of gene expression by acting as tethers or guides to the chromatin-modifying complexes [11]. The lncRNAs have a weaker evolutionary constraint and lower levels of expression compared to the protein-coding transcripts [12,13]. However, these noncoding transcripts exhibit more tissue-specific expression than the protein-coding genes, and many have been characterized with roles in epigenetic, transcriptional, and post-transcriptional gene regulation [14–19]. Different modes of gene regulation by lncRNAs have been reviewed extensively by Rinn and Chang recently [20].
In recent years, stem cells and their therapeutic applications have emerged as a rapidly developing field of research all over the world [21]. After decades of research, we have gained fragmented knowledge about stem cell biology and their actual therapeutic applications that needs to be assimilated to carry out useful implementations (as discussed by Parker and colleagues [22,23]). Pluripotent stem cells are a special type of stem cells that have the potential for differentiating into any of the three germ layers: endoderm, mesoderm, and ectoderm [24]. Owing to the capability of these pluripotent stem cells of self-renewal and of giving rise to cells from any lineage, these are thought to be the most potent cells that can be used for damage treatment for replacement of injured tissues and possibly whole organs [25,26]. In mammals, these pluripotent stem cells are derived from epiblasts in early embryos and are called embryonic stem cells (ESCs) [24]. Induced pluripotent stem cells (iPSCs) are engineered pluripotent cells that can be generated by reprogramming an adult somatic cell in such a way that they mimic the gene expression profile of an ESC. After the pioneering discovery of John Gurdon in 1958 for successful cloning of a frog using the intact nuclei from somatic cells of tadpoles [27], several advancements followed in the field of nuclear reprogramming [28–30]. However, it was in the year 2006 when reprogramming researches achieved a new height after the study of Yamanaka and colleagues, where the authors described the induction of the pluripotency-associated changes after the introduction of four key transcription factors overexpressed in ESCs, namely, Oct4, Sox2, Klf4, and c-Myc, into adult mouse fibroblasts [31]. This experiment was successfully repeated on humans [32]. However, a clear understanding of the cellular factors playing their parts in the maintenance of pluripotency is very important for successful therapeutic application of these pluripotent cells [33–37]. The transcriptional and epigenetic scenarios in the pluripotent state are very different from adult cells [38]. The distinctive gene expression signature of pluripotent cells is controlled by the key pluripotency-associated transcription factors acting in concert with other regulatory factors, such as noncoding RNAs [39]. This is likely because the widespread activation or repression of gene expressions defining the pluripotent state has to be controlled by some molecules that guide and bring different regulatory components close together.
MicroRNAs are known to be implicated in their regulatory roles in the developmental process [40–43]. Similarly, long noncoding RNAs are also identified as the key players in the developmental process, and many of them are differentially expressed during development [44–47]. Recent studies have identified hundreds of long noncoding RNAs showing significant high expression profiles in both embryonic as well as iPSCs [48]. Many of these lncRNAs have been identified to be transcriptionally activated or repressed by pluripotency-associated transcription factors. A recently published database [49] presents a comprehensive annotation of the transcription factor-binding sites in lncRNAs and transcriptional regulatory relationships of transcription factors and lncRNAs based on chromatin immunoprecipitation with the next-generation DNA sequencing (ChIP-Seq) data. This database includes lncRNAs whose promoters are found to have binding sites for pluripotency-associated transcription factors. There are also lines of evidence of lncRNAs binding with chromatin-modifying complexes to modulate the expression of pluripotency-associated factors to maintain pluripotency or to promote differentiation of pluripotent cells [40]. This review gives a brief account of pluripotency-associated lncRNAs, those acting in favor of maintaining pluripotency, and those facilitating differentiation and the possible molecular mechanisms underlying their regulatory effects.
Differential Expression of lncRNAs in Pluripotent Stem Cells
Mostly, long noncoding RNAs have cell type-specific expression signatures [10]. This selective expression signature is attributed to the epigenetic modifications such as DNA methylation, histone modification, and chromatin remodeling, similar to the protein-coding transcripts [50]. The study by Zhang and colleagues [51] of the lncRNA methylation status and expression profiles of lncRNAs in mouse ESCs, neuronal progenitor cells, and terminally differentiated fibroblasts revealed dynamic changes in the histone modification status for the lncRNA promoters in these three cell types. Moreover, they found EzH2-mediated histone modification H3K27 to play a key role in silencing lncRNAs in mouse ESCs [51]. Recent studies have identified many lncRNA transcripts to be differentially expressed in pluripotent cells, both in ESCs and in iPSCs [39,48,52], compared to fibroblasts or neuronal progenitors. We discuss in the following section the differential expression patterns of lncRNAs in both embryonic and iPSCs.
Differential expression in ESCs
A recent study sought to identify lncRNAs, which are differentially expressed in ESCs, fibroblast-derived iPSCs, and adult fibroblasts, from a pool of 900 lncRNAs. It uncovered over a 100 lncRNAs showing significant overexpression in ESCs and iPSCs compared to fibroblasts [48]. They also identified 104 lncRNAs significantly repressed in both ESCs and iPSCs. Previously, an RNA-seq based study identified 226 lncRNAs predominantly expressed in mouse ESCs [53]. The lncRNA microarray expression profiling study by Ng et al. [52] to find differentially expressed lncRNAs in human ESC and human neural progenitor cells (NPC) identified 36 lncRNAs showing significant overexpression in hESCs compared to NPCs, of which three lncRNAs were exclusively expressed in human ESCs and iPSCs. Two of these lncRNAs were identified to bind the epigenetic regulator SUZ12 and SOX2. A study of noncoding RNA expression profiles of mouse ESCs differentiating as embryoid bodies (EBs) over a 16-day timecourse identified many lncRNAs associated with each of the three stages of differentiation; pluripotency, primitive streak formation, and mesoderm differentiation [54]. Twelve lncRNA were found whose expression profiles correlated with the expressions of pluripotency markers Oct4, Nanog, and Sox2, while 7 lncRNAs showed correlated expression with EB differentiation marker genes Evx1 and brachyury (T).
The X-chromosome inactivator Xist, which represses one X-chromosome at the onset of differentiation of female ESCs, is found to be silenced in ESCs and only to be expressed at the onset of differentiation. Female ESCs are marked by the presence of two active X-chromosomes, one of which becomes silent at the verge of cell differentiation. This phenomenon is a landmark of initiation of differentiation and exits from the pluripotent state in female ESCs. Regulation of the phenomenon of X-chromosome inactivation (for differentiation) or reactivation (in case of iPSCs) is controlled by noncoding RNAs in the X-inactivation center (Xic), Xist being the central and most widely reviewed lncRNA in this area. Apart from Xist, other lncRNAs in Xic are also identified to be linked in the interplay of controlling X-inactivation (Xi), examples including lncRNAs Tsix, Jpx, and RepA [55–57]. These lncRNAs are in turn regulated by pluripotency-associated transcription factors such as Oct4, Nanog, and Sox2 in ESCs and iPSCs. A detailed account of this regulatory network in mouse ESCs and iPSCs can be found in the review by Kim et al. [58].
Besides this, many well-annotated lncRNAs are found to be differentially expressed in ESCs, such as NEAT1 or Rian [39]. The lncRNA NEAT1 was undetected in human ESCs (hESCs) and only induced upon differentiation of ESCs [59]. The lncRNA NEAT1 has been found to regulate the cytoplasmic export for the mRNAs containing inverted repeat (IRAlu) elements by facilitating nuclear paraspeckle formation in differentiated cells. Absence of the lncRNA NEAT1 in pluripotent hESCs seems to be responsible for the failure of nuclear paraspeckle formation in hESCs, which facilitates the cytoplasmic export of IRAlu-containing mRNAs [59]. One such example is the transcript of the pluripotency-associated gene Lin28. Lin28 is a very important factor for maintaining pluripotency and was one of the four factors used for reprogramming of mouse adult somatic cells into iPSCs [32]. On the contrary, NEAT1 lncRNA has been reported to be one of the mouse ESC-expressed lncRNAs that interact with a number of chromatin-binding protein/complexes in mouse ESCs [39]. Table 1 gives a list of annotated lncRNAs showing differential expression in ESCs and iPSCs.
Table 1.
LncRNA Transcripts Differentially Expressed in ESCs and iPSCs with Their Roles in Pluripotency
| LncRNA | Role in pluripotent cells | Molecular mechanisms |
|---|---|---|
| Gomafu | This nervous system-expressed lncRNA shows dynamic regulation during development of neural stem cells and also in mESCs. Harbors the Oct4 binding site at the promoter. Moreover, it appears to control Oct4 expression in a regulatory feedback loop. | Post-transcriptional (Gomafu can bind the splicing factor 1 [SF1] protein through its UACUAAC repeat sequences that results in the post-transcriptional regulation of splicing efficiency. The molecular mechanisms involving Oct4 expression control in mESCs remain elusive though.) |
| Mira | This lncRNA is expressed from the spacer DNA region (SDR) separating Hoxa6 and Hoxa7, which is transcriptionally silent in ESCs, but activated by retinoic acid treatment on mESCs, recruits the epigenetic activator MLL1 to the Mira gene locus triggers transcription of Hoxa6 and Hoxa7. These genes in turn trigger the expression of a number of genes resulting in germ layer specification and exit from pluripotent state. | Epigenetic (Mira plays a part in the epigenetic regulation of Hoxa6 and Hoxa7 by mediating a local rearrangement in chromatin structure to bring the Mira, Hox66, and Hoxa7 loci into a complex providing a mechanism for MLL1 to act on HoxA6/A7 after recruitment to the Mira locus.) |
| HOTAIRM1 | This mESC-expressed lncRNA from the HOXA locus was found to interact with a number of chromatin-binding proteins/complexes in mESCs, including PRC1, PRC2, and CBX1, to exert a cis-regulatory effect on the genes from the HOXA cluster. | Epigenetic (HOTAIRM1 interacts with a number of chromatin-binding proteins/complexes in mESCs, including PRC1, PRC2, and CBX1, to regulate the genes from the HOXA cluster) |
| Xist | In female undifferentiated mES and iPS cells, Xist is not expressed, and both X-chromosomes are active. Xist repress paternal X-chromosome in female ESCs to facilitate exit from the pluripotent state. Xist lncRNA is expressed from only the inactive X-chromosome and coats the entire chromosome in cis to epigenetically silence the gene expression from that chromosome. The epigenetic silencing of one X involves recruitment of polycomb group to confer silencing epigenetic mark H3K27Me3 on histone. | Epigenetic (Xist plays a vital role in the epigenetic silencing of the inactive X chromosome by binding to PRC2 via RepA and recruiting it on the Xi resulting in the deposition of the repressive chromatin mark H3K27me3 on the Xi.) |
| Tsix | Among many regulators of Xist within the X-inactivation center in pluripotent female ESCs, perhaps the most well known is lncRNA Tsix, which transcribes from both alleles. It mediates repression of Xist by physically associating with DNA methyltransferase Dnmt3a at the Xist promoter, leading to DNA methylation and silencing of Xist only on the active X-chromosome. lincRNA Tsix and its activator lncRNA Xite are identified to be direct targets of Oct4 in mESCs. | Epigenetic (Tsix epigenetically silences Xist expression by inhibiting RepA recruitment of polycomb complexes to maintain the active X-chromosome in females.) |
| Xite | lncRNA Xite is one of the functional lncRNAs in the X-inactivation center and is found to be the activator of lncRNA Tsix expression. It is identified to be direct targets of Oct4 in mESCs. A second pluripotency transcription factor Sox2 also binds Xite. | Not known yet. |
| Jpx | This lncRNA in the X inactivation center (Xic) is upregulated during both male and female ESC differentiation. It is Involved in the regulation of X-chromosome inactivation in mammals via activation of the Xist RNA, possibly by interfering with Tsix expression. | Not known yet. |
| NEAT1 | In mESCs, Neat1 has been reported to interact with a number of chromatin-binding protein/complexes in mESCs, including PRC1, PRC2,JARID1B, ESET, and SUV39H1, with the general pattern being interaction with repressors of gene expression [39]. In human, NEAT1 is not expressed in ESCs and it has been found to regulate mRNA cytoplasmic export for mRNAs containing inverted repeated Alu by facilitating nuclear paraspeckle formation in differentiated cells. |
Epigenetic/Post-transcriptional (Neat1 has been reported to interact with a number of chromatin-binding protein/complexes in mESCs, including PRC1, PRC2, JARID1B, ESET, and SUV39H1; while in human, NEAT1 is not expressed in ESCs, and it has been found to regulate the mRNA cytoplasmic export for mRNAs containing inverted repeated Alu by facilitating nuclear paraspeckle formation in differentiated cells. |
| Rian |
Rian transcripts bind to the PRC2-chromatin-modifying complex in mESCs. Rian was also found to interact with a number of other chromatin-binding proteins/complexes in mESCs, including PRC1, JARID1B, JARID1C, and CBX3, with the general pattern being interaction with repressors of gene expression [39]. This lncRNA is located in the imprinted Dlk1-Dio3 gene cluster on chromosome 12qF1, which is normally expressed in the ESCs, were found to be aberrantly silenced in most of the iPSC clones in a comparison of genetically identical mESCs and mouse iPSCs. These iPSCs failed to support development of entirely iPSC-derived animals. |
Epigenetic (Rian transcripts bind to the PRC2 chromatin-modifying complex in mESCs.) |
| Sox2ot | This mESC expressed transcript is downregulated upon induction of embryoid body (EB) differentiation like Sox2, but, unlike Sox2, is upregulated during EB late differentiation. | Not known yet |
| lncRNA-ES1-3 | These three hESC-specific lncRNAs are likely to be regulated by pluripotency-associated transcription factors OCT4 and/or NANOG, as these lncRNAs showed decreased expression after RNAi knockdown of NANOG and/or OCT4 in hESCs. | Not known yet |
| lincRNA-RoR (enriched in iPSCs) | This lincRNA is found enriched in both ESCs and iPSCs compared to the fibroblast and CD34 cells, but with increased level in iPSCs compared to ESCs, which suggest its potential role in reprogramming. This possibility was further supported by the fact that knockdown of this lincRNA resulted in reduced colony formation in iPSCs without affecting the cell number, whereas overexpression increased the number of iPSC colonies formed. It has a binding site for pluripotency-associated transcription factors Oct4, Sox2, and Nanog. Functions to promote reprogramming of differentiated cells to iPSCs. |
Not known yet |
| lincRNA-SFMBT2 (enriched in iPSCs) | This lincRNA is found enriched in both ESCs and iPSCs compared to the fibroblast and CD34 cells, but with an increased level in iPSCs compared to ESCs, which suggest its potential role in reprogramming. However, its knockdown did not affect generation of iPSC cells, suggesting that it is not essential to this process.It harbors the binding site for pluripotency transcription factors Oct4, Sox2, and Nanog. Knockdown of Oct4 or differentiation of stem cells caused downregulation of lincRNA-SFMBT2 expression, suggesting that expression is controlled by these transcription factors. | Not known yet |
| lincRNA-VLDLR (enriched in iPSCs) | This lincRNA is found enriched in both ESCs and iPSCs compared to the fibroblast and CD34 cells, but with increased level in iPSCs compared to ESCs, which suggest its potential role in reprogramming. However, its knockdown did not affect generation of iPSC cells, suggesting that it is not essential to this process. It harbors the binding site for pluripotency transcription factors Oct4, Sox2, and Nanog. Knockdown of Oct4 or differentiation of stem cells caused downregulation of lincRNA-SFMBT2 expression, suggesting that expression is controlled by these transcription factors. It is reported to have a synthetic correlate expressed in mESCs. |
Not known yet |
| MEG3 (expressed in ESCs, but aberrantly silenced in iPSCs) | Meg3 is important for proper growth and development and is a putative tumor suppressor. This lncRNA is located in the imprinted Dlk1-Dio3 gene cluster on chromosome 12qF1, which is normally expressed in the ESCs, were found to be aberrantly silenced in most of the iPSC clones in a comparison of genetically identical mESCs and mouse iPSCs. These iPSCs failed to support the development of entirely iPSC-derived animals. | Not known yet |
| MEG9 (expressed in ESCs, but aberrantly silenced in iPSCs) |
Meg9/Mirg is expressed in embryonic and extraembryonic tissue. Transcripts from the Meg9 locus bind to the PRC2 chromatin modification complex in mESCs. This lncRNA is located in the imprinted Dlk1-Dio3 gene cluster on chromosome 12qF1, which is normally expressed in the ESCs, were found to be aberrantly silenced in most of the iPSC clones in a comparison of genetically identical mESCs and mouse iPSCs. These iPSCs failed to support the development of entirely iPSC-derived animals. |
Epigenetic (Meg9 binds to the PRC2 chromatin modification complex in mESCs). |
ESCs, embryonic stem cells; iPSCs, induced pluripotent stem cells; mESC, mouse ESC.
Differential expression in iPSCs
The study by J. Rinn and colleagues identified 26 lncRNAs that were overexpressed in both human ESCs and iPSCs, but showed an elevated level in iPSCs compared to ESCs. They thought these lncRNAs as potential candidates important for reprogramming [48].
LncRNA transcripts Meg3, Meg9, and Rian in the imprinted Dlk1-Dio3 gene cluster on chromosome 12qF1, which is normally expressed in the ESCs [39] and found to interact with polycomb-repressing complex PRC2 in mouse ESCs [60], were found to be aberrantly silenced in most of the iPSC clones in a comparison of genetically identical mESCs and mouse iPSCs. These iPSCs failed to support the development of the entirely iPSC-derived animals [61]. Table 1 shows lncRNAs differentially expressed in iPSCs compared to ESCs.
LncRNAs Acting in Concert with Key Transcription Factors to Aid the Controlling of Pluripotency
LncRNAs are an integral part of the pluripotency-controlling network and a part of them are implicated in the maintenance of pluripotency and repressing differentiation and lineage programs [39], and on the other hand, some lncRNAs are found to act in the opposite way, by promoting differentiation of pluripotent cells. In the section below, we will discuss how lncRNAs act with pluripotency transcription factors in an orchestrated way to aid the controlling of pluripotency: being transcriptionally activated or repressed by pluripotency transcription factors and then regulating gene expressions to maintain the pluripotent state or promote differentiation of pluripotent cells. In addition, the evidence toward a potential regulatory feedback loop involving transcription factors and lncRNAs will be discussed.
Transcriptional activation or repression of lncRNAs by pluripotency-associated transcription factors
Transcription factors acting as key players for maintaining pluripotency control gene expression signature specific to pluripotent cells by transcriptionally activating thousands of genes in ESCs and also in iPSCs. As discussed in the previous section, hundreds of lncRNAs are differentially expressed in pluripotent cells, and many of them are likely to be regulated by the pluripotency-associated transcription factors. The recently published database ChIPBase [49] shows more than 8000 lncRNAs in mouse that harbor binding sites for at least one of nine key pluripotency transcription factors (including Oct4, Sox2, Nanog, c-Myc, n-Myc, Klf4, Zfx, E2F1, and Smad1) in their promoter region; identified by analyzing ChIP-Seq data. In the following section, we will discuss about lncRNAs activated or repressed by pluripotency-associated transcription factors in both ESCs and iPSCs.
In ESCs
A search for pluripotency transcription factors, the Oct4- and Nanog-binding sites in mouse ESC-expressed lncRNAs resulted in recognition of 10% of the Oct4-binding sites in the vicinity of the lncRNA genes [62]. Further search for the candidates with strong genomic conservation in the exonic sequences yielded four candidate lncRNAs, two of which, namely, Gomafu (AK028326) and AK141205 (cis-antisense to C18ORF22 homolog), were identified to be the direct targets of Oct4 and Nanog, respectively. shRNA-mediated knockdown of Oct4 and Nanog in mESCs resulted in a change in the expression levels of all four lncRNAs with putative binding sites for these two transcription factors. Of the two lncRNAs directly regulated by Oct4 and Nanog, lncRNA AK028326 is transcriptionally activated by Oct4, and AK141205 is transcriptionally repressed by Nanog. A recent study by Lander and colleagues identified that ∼75% of the all 226 lncRNAs expressed in mouse ESCs have binding sites for at least one of 9 pluripotency-associated transcription factors (Oct4, Sox2, Nanog, cMyc, nMyc, Klf4, Zfx, Smad, and Tcf3) they studied [39], and this observation was further validated by shRNA-mediated knockdown of 11 pluripotency-associated transcription factors, for which 60% mESC-expressed lincRNAs showed significant downregulation.
In the previously mentioned study by Ng et al. [52], the authors identified three human ESC-specific lncRNAs likely to be regulated by pluripotency-associated transcription factors Oct4 and/or Nanog, as these lncRNAs showed decreased expression after RNAi knockdown of Nanog and/or Oct4 in hESCs.
The phenomenon of X-chromosome inactivation at the onset of differentiation of female ESCs and the role of noncoding RNAs in X-inactivation center have been introduced in Section 1. The noncoding RNAs in this region are regulated by pluripotency-associated transcription factors such as Oct4, Sox2, and Nanog. The regulator of Xist transcription, lncRNA Tsix, and its activator lncRNA Xite are identified to be the direct targets of Oct4 in mESCs. A second pluripotency transcription factor Sox2 also binds Xite while indirectly interacting with Tsix by a looping interaction [63]. The lncRNA Xist itself contains Nanog binding sites in its intron 1, and co-occupancy of Nanog and Oct4 can reduce the Xist RNA level in undifferentiated mESCs, most likely in a Tsix-independent manner, as Nanog-deficient mESCs showed elevated levels of Xist, but no significant changes in the Tsix RNA level. Interestingly, Oct4 and Sox2 are found to be associated with Xist intron 1 in Nanog-depleted mESCs [64].
In iPSCs
Pluripotency-associated transcription factor-mediated activation of lncRNAs was also observed in iPSCs. The transcriptional signature of the ncRNAs in the X-inactivation center undergoes extensive changes after reprogramming mouse somatic cells to mouse iPSCs by introducing four key transcription factors: Oct4, Sox2, Klf4, and c-Myc. Xist becomes undetectable, and Tsix expression becomes biallelic while Xite is also expressed. These changes in the expression of lncRNAs in the X-inactivation center leads to reactivation of both X-chromosomes and eventually into a pluripotent state [65,66].
The study by Rinn and colleagues [48] of identifying lncRNAs playing key roles in reprogramming of somatic cells toward induced pluripotency detected three lncRNAs, overexpressed in iPSCs (compared to ESCs), to have the Oct4-binding sites in their promoters. The levels of all three lncRNAs, namely, lincRNA-SFMBT2, lincRNA-VLDLR, and lincRNA-ST8SIA3, fell after knockdown of Oct4 in the iPSC cell lines lincRNA-ST8SIA3 and lincRNA-SFMBT2, showing the strongest response.
Regulation of gene expression by lncRNAs to maintain pluripotency
LncRNAs are found to be an integral part of the regulatory network controlling the gene expression signature specific to pluripotency. In the lincRNA loss-of-function study of Lander and colleagues [39], 137 of 147 lincRNA, successfully targeted by shRNAs in mouse ESCs, induced changes in the gene expression profile in a significant level upon knockdown. Knockdown of each of these 137 lincRNAs affected expression of 175 genes in average, the range for individual lincRNAs being 20–936. This change in the gene expression signature was comparable to that obtained by knockdown of 40 known transcriptional or chromatin regulator genes playing a key part in pluripotency (38 of them induced expression changes for 207 genes in average, the range for individual regulator genes being 28–1187). Among the 147 lincRNAs expressed in mouse ESCs, 26 lincRNAs were identified that play key parts in the maintenance of pluripotency by inducing expression of pluripotency marker transcription factors such as Nanog and Oct4. Many of the lncRNAs studied here are seen to repress the lineage program in mESCs. Knockdown of 30 lincRNAs induced differentiation of mESCs into specific lineages, which in turn suggests that these lncRNAs are normally a hindrance to the differentiation programs.
A recent RNAi screening study by Buccholz and colleagues [67] identified three candidate lncRNAs in mouse ESCs (mESCs) with a potential role in maintaining pluripotency (based on loss-of-expression study of Oct4-driven GFP after transfection of the lnc-esiRNA library), and referred to them as pluripotency-associated noncoding transcripts 1–3 or Panct1–3. They further examined the role of Panct1 (as it exhibited the strongest phenotype) in mESC pluripotency by an RNAi-mediated loss-of-function study and found the pluripotency marker Oct4 and Nanog mRNA level to be reduced after Panct1 knockdown, while expression of lineage markers such as Gata6 (endoderm) and Fgf5 (ectoderm) increased. This gave an indication toward a role of the lncRNA Panct1 in maintaining the pluripotency in mESCs.
Rinn and colleagues [48] found iPSC-enriched lincRNA-ST8SIA3, named as linc-RoR (Regulator of Reprogramming), to have a functional role in iPSC colony formation by negatively regulating the proapoptotic pathways, such as the p53 pathway, DNA damage, and oxidative stress response pathway. That was indicated by the upregulation of genes involved in these proapoptotic pathways upon knockdown of lincRNA-ST8SIA3 in iPSC cells, which resulted in reduced iPSC colony formation [48].
Regulation of gene expression by lncRNAs to promote differentiation of pluripotent stem cells
Conversely, some lncRNAs are identified to promote differentiation. This class of lncRNAs includes the lncRNA Mistral (Mira), which directs germ layer differentiation in mESCs by transcriptional activation of the homeotic genes Hoxa6 and Hoxa7. Mira is located in the spacer region between Hoxa6 and Hoxa7 and is transcriptionally silent in ESCs. Upon retinoic acid-induced activation, Mira activates transcription of Hoxa6 and Hoxa7 by recruiting the epigenetic activator MLL1 to chromatin—which in turn induces expression of the genes involved in germ layer specification [68]. Another lncRNA HOTAIRM1, expressed from the HOXA cluster, undergoes a dramatic change in the expression level upon neuronal differentiation from iPSC and regulates genes from that cluster in cis [69].
The lncRNA–transcription factor feedback loop in pluripotency
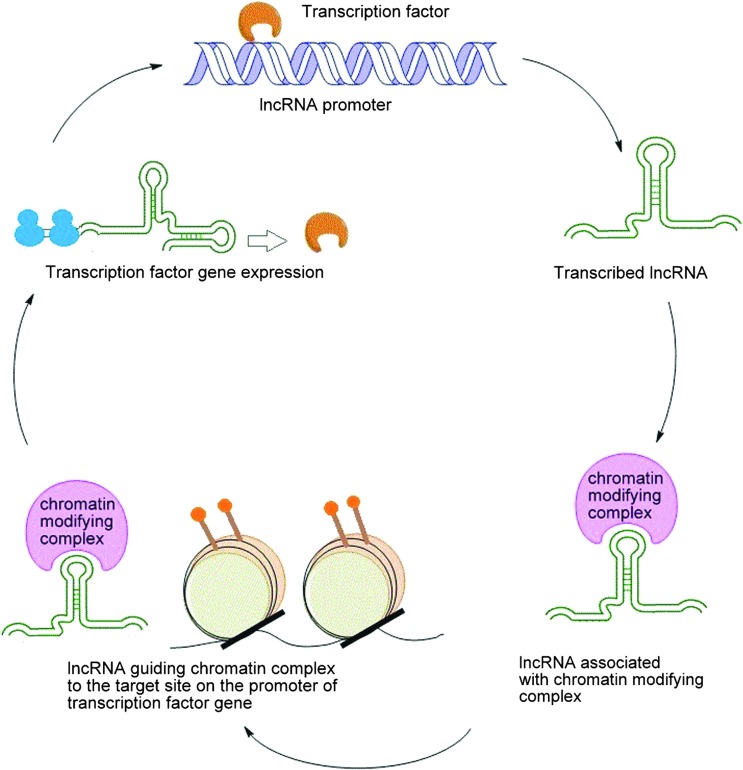
The circuitry controlling pluripotency in ESC and iPSCs have a complex interplay between pluripotency-associated factors such as transcription factors, chromatin-modifying complexes, and regulatory RNAs. These factors are interlinked by intriguing the potential feedback loops that maintain gene expression signatures specific to the pluripotent state, or promote differentiation under certain conditions. LncRNAs are also identified to be a part of such feedback loops with pluripotency-associated transcription factors that fine-tune their expression in pluripotent cells (see Fig. 1). The lncRNA Gomafu/Miat (AK028326) was identified to control Oct4 expression in a regulatory feedback loop in mouse ESCs [62]. This lncRNA is a direct target of Oct4 and is activated by Oct4 in mESCs. Knockdown of this lncRNA by siRNA resulted in decreased levels of Oct4 and Nanog in mESCs—which suggests that this lncRNA is involved in an autofeedback loop with its regulator Oct4 to maintain its level in mESCs and eventually promoting pluripotency [62].
FIG. 1.
LncRNA–transcription factor regulatory feedback loop in pluripotency. Color images available online at www.liebertpub.com/scd
Molecular Mechanisms for Regulation of Gene Expression by lncRNAs
LncRNAs are implicated in diverse regulatory roles in epigenetic, transcriptional, or post-transcriptional regulations of gene expression, and we are just beginning to understand the molecular mechanisms that enable the lncRNA molecules to perform such regulatory roles. In this context, a very interesting hypothesis by Guttman and Rinn [70] leads to an intriguing new direction for future investigations about lncRNA functionality. In this section, we will discuss about different modes of regulation by lncRNAs in pluripotent cells with specific examples.
Cis- or trans-acting lncRNAs in regulation of gene expression
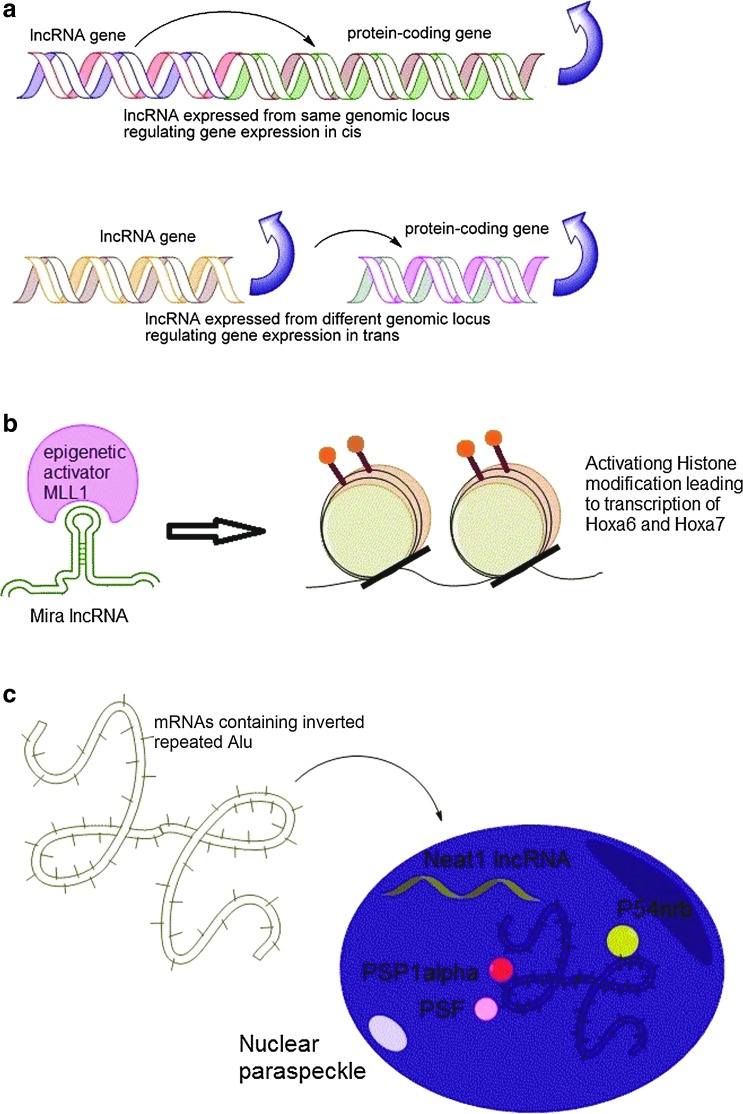
The mechanism of gene expression regulation by lncRNAs has been debated over the issue that whether they preferentially act in cis (regulating nearby genes) or in trans (regulating genes in distal loci) (see Fig. 2a). While some of the earlier studies claimed that lncRNAs mainly work in cis [71,72], this conclusion was contradicted by the study of Lander and colleagues [39], where they found only a small number of lincRNAs affect the expression of neighboring genes, as knockdown of only 8 lincRNAs (out of 137 mESC-expressed lincRNAs) had any effect on the expression of genes located within 300 kb of those lincRNAs. This finding was in contrast with an earlier report where 7 of the 12 lincRNAs studied were found to regulate the expression of the neighboring genes residing within 300 kb of the respective lincRNAs. Lander argued that this cis-regulatory behavior may be attributed to the shared upstream regulations or local transcriptional effects.
FIG. 2.
Molecular mechanisms of lncRNA regulation. (a) Cis-acting and trans-acting gene expression regulation. (b) lncRNA Mira acting as a guide to the chromatin-modifying epigenetic activator MLL1 for exerting activating histone marks on the Mira gene locus, leading to activation of Hoxa6 and Hoxa7 gene expression. (c) lncRNA Neat1 mediating post-transcriptional regulation of mRNAs by facilitating formation of nuclear paraspeckles and aiding in nuclear retention of mRNAs with inverted repeats in their 3′-UTR. Color images available online at www.liebertpub.com/scd
Recently, these conclusions were challenged by the findings of the GENCODE consortium under the ENCODE project [9]. From pairwise expression correlation study, they found that nearly 40% of the GENCODE-annotated lncRNA transcripts overlap the protein-coding genes, and these lncRNAs showed a significant positive correlation of expression with the intersecting protein-coding genes than the mRNA–mRNA control pairs [10]. Especially, the lncRNAs with antisense orientation to their intersecting mRNA exons showed more-significant positive correlation of expression, pointing toward the cis-antisense regulatory mechanism previously discovered for some lncRNAs. In the GENCODE release, they reported a total of 187 antisense lncRNAs whose expression strongly correlated with that of their respective intersecting genes.
Apart from the debate of whether lncRNAs preferentially act in cis or trans, there are several lines of evidence of both types of interactions in pluripotent cells. In addition to the previously mentioned 137 mESC-expressed lincRNA loss-of-function studies by Lander and colleagues [39], where most of the lincRNAs were found to affect the expression of distal genes, some of the other lincRNAs studied in mouse ESCs also showed that a trans-acting regulation of gene expression, such as lncRNA- AK028326 (Gomafu/Miat), was found to regulate Oct4 expression [62].
Antisense lncRNAs overlapping the Oct4 promoter and coding region and Oct4 pseudogene 5 (Oct4-ps5) coding region were found to regulate expression of these genes in a cis-regulatory manner, as depletion of the antisense lncRNAs in MCF7 cells resulted in increased expression of Oct4 and Oct4-ps5 [73].
LncRNAs emerging as effecter molecules in epigenetic regulation
The way in which differential gene expression profiles are maintained in different cell types is a matter of immense interest in biological research. Transcriptional activation or inactivation of genes is largely attributed to the epigenetic modulations such as chromatin remodeling, histone modification, or DNA methylation. Not only for tissue-specific gene expression, have epigenetic modifications played an important part in many diseases, including cancer. In a pluripotent state, chromatin exhibits a more permissive state that allows expression of genes that maintain pluripotency. The set of protein complexes that are responsible for chromatin modifications depends upon other cellular factors that help the complexes guide to their targets. In recent years, more and more numbers of lncRNAs are identified to mediate such epigenetic regulation by associating with chromatin-remodeling complexes. The role of lncRNAs functioning as guides to epigenetic modulator proteins is sufficiently well studied and well reviewed [15,20,74,75]. In this section, we are going to focus more on ESC- or iPSC-expressed lncRNAs that are identified to be associated with the chromatin-remodeling complexes and their roles in pluripotency and differentiation of pluripotent cells.
LncRNAs involved in epigenetic modulation of gene expression related to the maintenance of pluripotent cells
Lander and colleagues found many mESC-expressed lincRNAs to be physically associated with chromatin-remodeling proteins in mESCs [39]. By immunoprecipitation of the RNA–protein complexes in mESCs, they identified 24 lincRNAs among the set of 226 previously recognized mESC-expressed lincRNAs, to bind to the polycomb-silencing complex, which plays a critical role in the mESC pluripotency. Besides the polycomb complex, they identified 11 additional lincRNA-associated chromatin-modifying complexes, involved in reading (PRC1, Cbx1, and Cbx3), writing (Tip60/P400, PRC2, Setd8, ESET, and Suv39h1), and erasing (Jarid1b, Jarid1c, and HDAC1) histone modifications, and also a DNA-binding protein YY1 that is associated with chromatin. In total, 74 lincRNAs were identified to be associated with at least one of these 12 chromatin complexes. Moreover, many of these lincRNAs were found to interact with multiple chromatin-modifying complexes that are functionally interlinked with one another (a reader, writer, and eraser combination). They have found 17 lincRNAs associated with the PRC2 complex (writer of H3K27 repressive marks), PRC1 complex (reader of H3K27-repressive marks), and Jarid1b complex (eraser of H3K4-activating marks). These findings lead to a potential model of lncRNAs acting as scaffolds by interacting with two or more protein complexes via distinct binding domains, to bridge these functionally related protein complexes together for epigenetic regulation, which is consistent with the hypothesis by Guttman and Rinn [70].
LncRNAs involved in epigenetic modulation of gene expression related to differentiation of pluripotent cells
An example of lncRNA involved in promoting differentiation by epigenetic activation is the lncRNA Mistral (Mira) that recruits the epigenetic activator MLL1 to the Mira gene locus (see Fig. 2b) and triggers transcription of Hoxa6 and Hoxa7 [68] (discussed in Section 2).
The role of the lncRNA Xist in X-chromosome inactivation in female embryos has been studied broadly [76]. Xist lncRNA is expressed from only the inactive X-chromosome, and coats the entire chromosome in cis to epigenetically silence the gene expression from that chromosome [77]. The epigenetic silencing of one X involves the recruitment of the polycomb group to confer silencing epigenetic mark H3K27Me3 on histone. The polycomb-silencing complex PRC2 on the chromosome is recruited by a noncoding RNA originating from the 5′-end of Xist—called RepA [58]. RepA directly interacts with EzH2, a catalytic subunit of PRC2, via a secondary structure within repeat A. This ncRNA RepA is activated before X-chromosome inactivation and involved in aiding the activation of Xist. Among many regulators of Xist within the X-inactivation center in pluripotent female ESCs, perhaps the most well known is lncRNA Tsix, which transcribes from both alleles and has a 40-kb region enriched for H3K4 methylation and H4 acetylation in active state [78]. It mediates repression of Xist by physically associating with DNA methyltransferase Dnmt3a at the Xist promoter, leading to DNA methylation and silencing of Xist only on the active X chromosome [55,79,80]. Interplay of these lncRNAs with epigenetic modulators, along with other pluripotency factors, mediates the X-chromosome inactivation state in female ESCs, which is a marker of its differentiation status [14,58].
As discussed in the previous section, an lncRNA antisense to Oct4 pseudogene 5 (asOct4-pg5) was found to regulate expression of Oct4 in MCF7 cells, and possibly its regulatory function is mediated by chromatin modification at the Oct4 promoter. A loss-of-function study of this lncRNA as Oct4-pg5 to find its effect on the chromatin structure showed decreased levels of repressive histone marks H3K9me2, H3K27me3, and Ezh2 at the Oct4 promoter ([measured by chromatin immunoprecipitation (ChIP) after 72 h of transfection of siRNA targeting asOct4-pg5)]. This leads to the speculation that asOct4-pg5 may function by guiding the epigenetic silencer Ezh2 to the promoter of Oct4 [73] in MCF7 cells. Oct4 being a very important pluripotency factor, such findings also lead us to investigate about similar types of Oct4 regulation in pluripotent cells.
LncRNAs in transcriptional and post-transcriptional regulation of gene expression
Till now, most of the identified lncRNAs are implicated in the epigenetic regulatory process, acting as scaffolds for chromatin-modifying complexes. However, there exist other modes of actions for many lncRNAs, such as transcriptional or post-transcriptional regulation of gene expression. However, these types of functional roles of lncRNAs are much less studied, but our understanding is expanding.
Transcriptional regulation
There are several lines of evidence of lncRNAs acting in transcriptional regulation by directly associating with RNA pol II or transcription factors: by acting as decoys by binding to transcription factors and preventing them to bind to the promoter region of a gene [81] or blocking pol II or transcription factor binding and transcription initiation [82]. Detailed examples are discussed in the reviews by Chang and colleagues [15,20]. The above-discussed examples of lncRNA-mediated transcriptional regulations are not specific to pluripotent cells. However, another finding of transcriptional regulation of the pluripotency marker Oct4 gene by an antisense lncRNA as Oct4-pg5 (discussed in previous section) suggests possible roles for some of the ESC- or iPSC-expressed lncRNAs in the process of transcriptional regulation [73].
Post-transcriptional regulation
There are also examples of post-transcriptional regulation of gene expression by lncRNAs: by regulating the alternative splicing of the pre-mRNA transcripts, controlling mRNA export into the cytoplasm, or by interfering with the miRNA pathways. The lncRNA NEAT2 or MALAT1 is found to regulate pre-mRNA alternative splicing by modulating the cellular levels of active serine/arginine (SR) splicing factors [83].
Controlling nuclear retention of mRNAs
Another mode of regulation of gene expression by lncRNAs is controlling the nuclear retention of mRNAs with inverted repeats (Alu repeats in human). Often, in many types of cells, mRNAs containing the Alu elements in their 3′-UTRs are inefficiently transported to the cytoplasm and thus are retained to the nucleus, leading to their loss of expression. The phenomenon of nuclear retention of these mRNAs correlates with paraspeckle formation in the nucleus. The lncRNA NEAT1 expression was undetected in pluripotent human ESCs, but its expression was only found upon differentiation, and this correlates with the absence of nuclear paraspeckle formation in hESCs, while the differentiated cells show paraspeckle formation [59]. The absence of NEAT1 in hESCs resulted in an efficient export of some pluripotency-associated mRNAs such as Lin28 that contain inverted repeated Alu in their 3′-UTR (Fig. 2c).
LncRNAs interfering with miRNAs
LncRNAs can also influence post-transcriptional regulation by interfering with the miRNA pathways, by acting as competing endogenous RNAs (ceRNAs). These lncRNAs have miRNA-binding sites in them and control endogenous miRNAs available for binding their target mRNAs, thus reducing the repression of these mRNAs. This class of lncRNAs have been found to be an important regulator in cell cycle control and tumor suppression (e.g., PTEN-P1 blocking miR-19b and miR-20a from binding to PTEN tumor suppressor), as well as in developmental stages (e.g., linc-MD1-blocking miR-133 and miR-135 from binding to transcription factors involved in myogenic differentiation) [84,85]. Until now, there are no reports of this type of post-transcriptional regulation by lncRNAs in pluripotent cells; a detailed study in this direction may prove to be beneficial.
Discussion
LncRNAs, previously thought as junk products, are being identified as important regulatory molecules to control lineage-specific gene expression, and an important player in diseases such as cancer. The role of lncRNA in controlling the epigenetic landscape during development has been reviewed previously. The role of lncRNAs in maintenance of pluripotency has come under attention recently [86–88]. Our review goes a step beyond and elucidates the functions of lncRNAs in two quite distinct aspects of pluripotent stem cells, namely, their ability for maintenance of pluripotent state, and equally importantly their capability to differentiate into any germ layer. These two aspects taken together have profound potential in regenerative medicine. Some recent discoveries regarding lncRNA expression and their interplay with other cellular factors such as transcription factors and epigenetic modulators in pluripotent ESCs, as well as induced pluripotent cells, take us a step further in the process of understanding the intriguing cellular processes controlling pluripotency. The extent of lncRNA-mediated regulation and their molecular mechanisms are beginning to be understood. The number of functionally well-annotated lncRNAs was scarce until recently, but the recent release from the ENCODE project has cast light on the functioning of many previously unrecognized and unannotated large noncoding transcripts. The GENCODE consortium under the ENCODE project has already made available a list of 14,880 manually annotated lncRNAs in humans, with their expression profiles in 31 human tissues [10]. They have also analyzed these transcripts for their modes of action (cis or trans); their findings in this regard are summarized in this review.
As recent findings suggest, lncRNAs play an integral part in the circuitry controlling pluripotency along with specific transcription factors and chromatin complexes. They are activated by pluripotency-associated transcription factors, and some of them in turn regulate expression of these transcription factors through a feedback loop. Further experimentations are needed to conduct large-scale studies for understanding this kind of regulatory feedback loops in ESCs and iPSCs. Recently available vast datasets [49] of transcription factor binding sites in the lncRNA regulatory regions, identified by ChIP-Seq, can be explored and further investigated for actual regulatory relationships between pluripotency-associated transcription factors and lncRNAs in pluripotent cells. Association of the lncRNAs with chromatin-modifying complexes in pluripotent cells is comparatively well studied in comparison to their other regulatory mechanisms such as transcriptional or post-transcriptional regulation. More studies in the direction of finding potential roles of lncRNAs in transcriptional or post-transcriptional regulation of gene expression in pluripotent cells are needed.
Acknowledgments
This work was supported by intramural research funds of our host institutions.
Author Disclosure Statement
All authors have read and approved the manuscript, and hereby we declare that none of them has any competing interest.
References
- 1.Ponjavic J. Ponting CP. Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Bakel H. Nislow C. Blencowe BJ. Hughes TR. Most “dark matter”. transcripts are associated with known genes. PLoS Biol. 2009;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinger ME. Gascoigne DK. Mattick JS. The evolution of RNAs with multiple functions. Biochimie. 2011;93:2013–2018. doi: 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 4.Lipovich L. Johnson R. Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP. Oliver PL. Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M. Amit I. Garber M. French C. Lin MF. Feldser D. Huarte M. Zuk O. Carey BW, et al. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maher B. ENCODE: The human encyclopaedia. Nature. 2012;489:46–48. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- 8.Djebali S. Davis CA. Merkel A. Dobin A. Lassmann T. Mortazavi A. Tanzer A. Lagarde J. Lin W, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrow J. Frankish A. Gonzalez JM. Tapanari E. Diekhans M. Kokocinski F. Aken BL. Barrell D. Zadissa A, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien T. Johnson R. Bussotti G. Tanzer A. Djebali S. Tilgner H. Guernec G. Martin D. Merkel A, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caley DP. Pink RC. Trujillano D. Carter DR. Long noncoding RNAs, chromatin, and development. Sci World J. 2010;10:90–102. doi: 10.1100/tsw.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang KC. Frith MC. Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Marques AC. Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR. Dinger ME. Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 15.Wang KC. Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilusz JE. Sunwoo H. Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eddy SR. Noncoding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y. Liu N. Wang JP. Wang YQ. Yu XL. Wang ZB. Cheng XC. Zou Q. Regulatory long noncoding RNA and its functions. J Physiol Biochem. 2012;68:611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin L. Chang HY. Uncovering the role of genomic “dark matter” in human disease. J Clin Invest. 2012;122:1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinn JL. Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker GC. Stem cells and development in the Far East. Stem Cells Dev. 2008;17:609–611. doi: 10.1089/scd.2008.4167.ed. [DOI] [PubMed] [Google Scholar]
- 22.Di Nardo P. Parker GC. Stem cell standardization. Stem Cells Dev. 2011;20:375–377. doi: 10.1089/scd.2011.1500. [DOI] [PubMed] [Google Scholar]
- 23.Magnus T. Liu Y. Parker GC. Rao MS. Stem cell myths. Philos Trans R Soc Lond B Biol Sci. 2008;363:9–22. doi: 10.1098/rstb.2006.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 25.Matsui T. Akamatsu W. Nakamura M. Okano H. Regeneration of the damaged central nervous system through reprogramming technology: basic concepts and potential application for cell replacement therapy. Exp Neurol. 2012 doi: 10.1016/j.expneurol.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F. Citra F. Wang DA. Prospects of induced pluripotent stem cell technology in regenerative medicine. Tissue Eng Part B Rev. 2011;17:115–124. doi: 10.1089/ten.TEB.2010.0549. [DOI] [PubMed] [Google Scholar]
- 27.Gurdon JB. Elsdale TR. Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 28.Gurdon JB. From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Annu Rev Cell Dev Biol. 2006;22:1–22. doi: 10.1146/annurev.cellbio.22.090805.140144. [DOI] [PubMed] [Google Scholar]
- 29.Wilmut I. Schnieke AE. McWhir J. Kind AJ. Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 30.Tada M. Takahama Y. Abe K. Nakatsuji N. Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 34.Kim H. Kim D. Jang MJ. Han YM. Variations in the epigenetic regulation of lineage-specific genes among human pluripotent stem cell lines. Biochem Biophys Res Commun. 2012;424:331–337. doi: 10.1016/j.bbrc.2012.06.122. [DOI] [PubMed] [Google Scholar]
- 35.Bilic J. Izpisua Belmonte JC. Concise review: induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- 36.Muchkaeva IA. Dashinimaev EB. Terskikh VV. Sukhanov YV. Vasiliev AV. Molecular mechanisms of induced pluripotency. Acta naturae. 2012;4:12–22. [PMC free article] [PubMed] [Google Scholar]
- 37.Hussein SM. Nagy AA. Progress made in the reprogramming field: new factors, new strategies and a new outlook. Curr Opin Genet Dev. 2012;22:435–443. doi: 10.1016/j.gde.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz S. Diep D. Gore A. Panopoulos AD. Montserrat N. Plongthongkum N. Kumar S. Fung HL. Giorgetti A, et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci (USA) 2012;109:16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman M. Donaghey J. Carey BW. Garber M. Grenier JK. Munson G. Young G. Lucas AB. Ach R, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colas AR. McKeithan WL. Cunningham TJ. Bushway PJ. Garmire LX. Duester G. Subramaniam S. Mercola M. Whole-genome microRNA screening identifies let-7 and mir-18 as regulators of germ layer formation during early embryogenesis. Genes Dev. 2012;26:2567–2579. doi: 10.1101/gad.200758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallick B. Chakrabarti J. Ghosh Z. MicroRNA reins in embryonic and cancer stem cells. RNA Biol. 2011;8:415–426. doi: 10.4161/rna.8.3.14497. [DOI] [PubMed] [Google Scholar]
- 42.Hinton A. Hunter S. Reyes G. Fogel GB. King CC. From Pluripotency to Islets: miRNAs as critical regulators of human cellular differentiation. Adv Genet. 2012;79:1–34. doi: 10.1016/B978-0-12-394395-8.00001-3. [DOI] [PubMed] [Google Scholar]
- 43.Yang CS. Rana TM. Yang CS. Rana TM. Learning the molecular mechanisms of the reprogramming factors: let's start from microRNAs. Mol Biosyst. 2013;9:10–17. doi: 10.1039/c2mb25088h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amaral PP. Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 45.Lee TL. Xiao A. Rennert OM. Identification of novel long noncoding RNA transcripts in male germ cells. Methods Mol Biol. 2012;825:105–114. doi: 10.1007/978-1-61779-436-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meola N. Pizzo M. Alfano G. Surace EM. Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18:111–123. doi: 10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paralkar VR. Weiss MJ. A new “Linc” between noncoding RNAs and blood development. Genes Dev. 2011;25:2555–2558. doi: 10.1101/gad.183020.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loewer S. Cabili MN. Guttman M. Loh YH. Thomas K. Park IH. Garber M. Curran M. Onder T, et al. Large intergenic noncoding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang JH. Li JH. Jiang S. Zhou H. Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177–D187. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sati S. Ghosh S. Jain V. Scaria V. Sengupta S. Genome-wide analysis reveals distinct patterns of epigenetic features in long noncoding RNA loci. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu SC. Kallin EM. Zhang Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010;20:1109–1116. doi: 10.1038/cr.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng SY. Johnson R. Stanton LW. Human long noncoding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2011;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guttman M. Garber M. Levin JZ. Donaghey J. Robinson J. Adiconis X. Fan L. Koziol MJ. Gnirke A, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dinger ME. Amaral PP. Mercer TR. Pang KC. Bruce SJ. Gardiner BB. Askarian-Amiri ME. Ru K. Soldà G, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sado T. Wang Z. Sasaki H. Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 56.Tian D. Sun S. Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J. Sun BK. Erwin JA. Song JJ. Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DH. Jeon Y. Anguera MC. Lee JT. X-chromosome epigenetic reprogramming in pluripotent stem cells via noncoding genes. Semin Cell Dev Biol. 2011;22:336–342. doi: 10.1016/j.semcdb.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen LL. Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao J. Ohsumi TK. Kung JT. Ogawa Y. Grau DJ. Sarma K. Song JJ. Kingston RE. Borowsky M. Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stadtfeld M. Apostolou E. Akutsu H. Fukuda A. Follett P. Natesan S. Kono T. Shioda T. Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheik Mohamed J. Gaughwin PM. Lim B. Robson P. Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donohoe ME. Silva SS. Pinter SF. Xu N. Lee JT. The pluripotency factor Oct4 inter- acts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Navarro P. Chambers I. Karwacki-Neisius V. Chureau C. Morey C. Rougeulle C. Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 65.Maherali N. Sridharan R. Xie W. Utikal J. Eminli S. Arnold K. Stadtfeld M. Yachechko R. Tchieu J, et al. Directly reprogrammed fibroblasts show global epigenetic remodelling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Stadtfeld M. Maherali N. Breault DT. Hochedlinger K. Defining molecular cor- nerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chakraborty D. Kappei D. Theis M. Nitzsche A. Ding L. Paszkowski-Rogacz M. Surendranath V. Berger N. Schulz H, et al. Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat Methods. 2012;9:360–362. doi: 10.1038/nmeth.1894. [DOI] [PubMed] [Google Scholar]
- 68.Bertani S. Sauer S. Bolotin E. Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Lin M. Pedrosa E. Shah A. Hrabovsky A. Maqbool S. Zheng D. Lachman HM. RNA-Seq of human neurons derived from iPS cells reveals candidate long noncoding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guttman M. Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ponjavic J. Oliver PL. Lunter G. Ponting CP. Genomic and transcriptional co-localization of protein-coding and long noncoding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ørom UA. Derrien T. Beringer M. Gumireddy K. Gardini A. Bussotti G. Lai F. Zytnicki M. Notredame C, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hawkins PG. Morris KV. Transcriptional regulation of Oct4 by a long noncoding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flynn RA. Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev. 2012;22:172–178. doi: 10.1016/j.gde.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moran VA. Perera RJ. Khalil AM. Emerging functional and mechanistic paradigms of mammalian long noncoding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Penny GD. Kay GF. Sheardown SA. Rastan S. Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 77.Clemson CM. McNeil JA. Willard HF. Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JT. Davidow LS. Warshawsky D. Tsix, a gene antisense to Xist at the X- inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 79.Sado T. Hoki Y. Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 80.Nesterova TB. Popova BC. Cobb BS. Norton S. Senner CE. Tang YA. Spruce T. Rodriguez TA. Sado T. Merkenschlager M. Brockdorff N. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenet Chromatin. 2008;1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hung T. Wang Y. Lin MF. Koegel AK. Kotake Y. Grant GD. Horlings HM. Shah N. Umbricht C, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martianov I. Ramadass A. Serra Barros A. Chow N. Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 83.Tripathi V. Ellis JD. Shen Z. Song DY. Pan Q. Watt AT. Freier SM. Bennett CF. Sharma A, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cesana M. Cacchiarelli D. Legnini I. Santini T. Sthandier O. Chinappi M. Tramontano A. Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tay Y. Kats L. Salmena L. Weiss D. Tan SM. Ala U. Karreth F. Poliseno L. Provero P, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pauli A. Rinn JL. Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huo JS. Zambidis ET. Pivots of pluripotency: The roles of non-coding RNA in regulating embryonic and induced pluripotent stem cells. Biochim Biophys Acta. 2013;1830:2385–2394. doi: 10.1016/j.bbagen.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng SY. Stanton LW. Long non-coding RNAs in stem cell pluripotency. Wiley Interdiscip Rev RNA. 2013;4:121–128. doi: 10.1002/wrna.1146. [DOI] [PubMed] [Google Scholar]