Abstract
The dynamic interaction of chromatin-binding proteins with their nucleosome binding sites is an important element in regulating the structure and function of chromatin in living cells. Here we review the major factors regulating the intranuclear mobility and chromatin binding of the linker histone H1, the most abundant family of nucleosome-binding proteins. The information available reveals that multiple and diverse factors modulate the interaction of H1 with chromatin at both a local and global level. This multifaceted mode of modulating the interaction of H1 with nucleosomes is part of the mechanism that regulates the dynamics of the chromatin fiber in living cells.
The dynamic properties of the chromatin fiber result from the action of numerous nuclear proteins that modulate its structure and function. One of the most abundant and ubiquitous groups of chromatin-binding proteins is the histone H1 family of proteins. H1 proteins are viewed as major structural components of the chromatin fiber that are important in many chromatin-related activities. Numerous in vitro experiments have demonstrated that the interaction of H1 with nucleosome stabilizes the compact, higher-order chromatin structure and inhibits DNA-dependent activities such as transcription and replication. Histone H1 restricts nucleosome mobility, impedes the ability of regulatory factors to access their chromatin targets and inhibits the action of chromatin-remodeling complexes. The properties and function of these proteins have been extensively studied and reviewed in refs. 1–12.
More recent imaging studies of the interaction of H1 with chromatin in living cells and genetic analysis of various organisms in which H1 genes have been knocked out provide new information on the cellular function and mechanism of action of H1 (ref. 2). The imaging experiments reveal that H1 molecules are not stably associated with specific sites. In the living nucleus, H1 molecules are in constant motion and continuously exchange among chromatin binding sites13,14. These and additional observations suggest that H1 molecules function within the framework of a dynamic network of chromatin-binding proteins that continuously compete for chromatin binding sites2,15–19.
Genetic experiments in several organisms, including yeast and mice20–29, provide a new view on the biological function of H1 (ref. 2). Analysis of mice lacking various combinations of histone H1 variants has suggested that there is a high degree of functional redundancy among the variants and that the correct overall amount of H1, rather than the correct relative amounts of the various variants, is the most important factor in H1 function. The transcription profiles of wild-type and H1-knockout cells are very similar, indicating that loss of H1 affects only a limited number of genes and that H1 is not a general repressor of transcription (reviewed in ref. 2). Strikingly, in embryonic stem cells, a 50% reduction in H1 shortens the nucleosomal repeat but affects the expression of only 29 genes, reinforcing the notion that H1 proteins are not major global regulators of transcription2,29. Yet, a two-fold reduction in H1 content is not compatible with normal development, causing mutant embryos to die in midgestation with multiple defects27,29.
Phenotypic analyses of H1 knockouts provide information on the biological role of histone H1; the dynamic behavior of H1 intimates the underlying molecular mechanisms leading to the observed phenotypes2. Thus, studies on the factors that regulate the interaction of histone H1 with chromatin not only provide insights into mechanisms that regulate H1 function, but also are relevant to understanding the role of chromatin dynamics in regulating gene expression30. Here, we discuss the factors that determine the chromatin-binding characteristics and the dynamic behavior of histone H1 in living cells.
Histone H1 dynamics in living cells
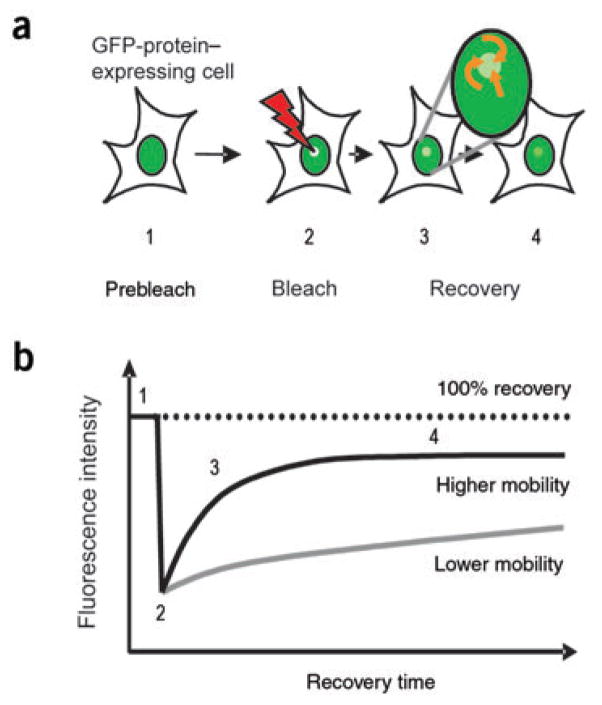
The dynamic properties of H1 are monitored by photobleaching experiments such as fluorescence recovery after photobleaching (FRAP; Fig. 1) or fluorescence loss in photobleaching (FLIP) that follow the mobility of H1 labeled with green fluorescent protein (GFP) in living cells, in real time. In the FRAP technique, a small area of a nucleus is irreversibly bleached with a laser beam and the rate at which the fluorescent signal in the photobleached area recovers is quantified. The rate of fluorescence recovery is indicative of the rate at which fluorescent molecules exchange with the photobleached molecules. The latter is directly proportional to the rate at which the molecules diffuse while migrating throughout the nucleus and inversely proportional to the time that the molecules reside at an immobile binding site such as chromatin. For H1 and other chromatin-binding proteins, the contribution of diffusion to the rate of recovery is negligible, and the observed fluorescence recovery rate (‘mobility’) is a direct reflection of their chromatin interactions, that is, their residence time at a particular chromatin binding site. Although in some experiments the position of the GFP tag in the H1 molecules affects its kinetic behavior31, several control experiments, including nuclear localization, in vitro binding assays, micrococcal nuclease digestion and salt extraction, have indicated that the GFP moiety does not substantially alter the interaction of H1 with chromatin13,32. Together with recent analyses of the in vivo and in vitro binding of H1 point mutants33, the data suggest that photobleaching studies of H1-GFP fusion proteins are useful for understanding the factors that regulate the interaction of H1 with chromatin.
Figure 1.
Analysis of protein dynamics in living cells by FRAP. (a) FRAP experiment. A small area in the nucleus of a cell expressing a fluorescent protein (1) is irreversibly photobleached with a laser beam (2), and the recovery of the fluorescence intensity due to entry of non-photobleached molecules (orange arrows) is continuously monitored (3 and 4).
(b) Hypothetical plots for fast and slow fluorescence recovery (high and low mobility, respectively). The rate of recovery is inversely related to the time that a protein is bound to chromatin13,66.
In living cells, H1 is mobile and the interaction of a specific H1 molecule with a particular nucleosome is transient. H1 molecules are continuously exchanged among chromatin binding sites in a ‘stop-and-go’ process in which H1 stays on a binding site for a limited time, then dissociates and moves rapidly and randomly to another binding site13,14. For H1 and other chromatin-binding proteins, the time spent in transit between nucleosomes (the ‘go’ stage) is substantially shorter than the time spent bound to a nucleosome. Thus, the observed mobility is a direct reflection of the strength of binding of H1 to chromatin33–35. Photobleaching results with mutants of histone H1 and other chromatin-binding proteins fully support this concept4,16,17,35.
In vivo cell-imaging studies have revealed that most of the time, most histone H1 molecules are associated with nucleosomes, and from a global view, histone H1 is continuously bound to chromatin. However, at the local level of the single nucleosome, the interaction of an H1 molecule with its binding sites is highly dynamic. H1 molecules constantly turn over on nucleosomes and continuously move throughout the entire nucleus. The movement of H1 in the nucleus is governed mostly by its interactions with chromatin. Compared to nucleosomal histones, H1 is highly mobile; the mean residence time of an H1 molecule at a binding site has been estimated to be approximately 3 minutes, whereas that of a core histone is several hours. However, compared to most other nuclear proteins, H1 is moving slowly, and its chromatin residence time is substantially longer than those of most other chromatin-binding proteins investigated so far, except Suv39h1 (refs. 17,36). For comparison, a non–chromatin-bound control such as GFP has an apparent mean residence time of less than 1 second, and fast-moving chromatin-binding proteins such as transcription factors or high-mobility group (HMG) proteins have mean residence times of 5–25 seconds17.
Studies on H1 mobility in various cell types exposed to various treatments have suggested that the dynamic interaction of H1 with chromatin is responsive to both internal and external regulatory signals. Modulation of H1-chromatin interactions seems to be one of the earliest events leading directly to changes in the structure and activity of the chromatin fiber. Photobleaching techniques are sensitive enough to detect small changes in protein mobility and therefore are powerful tools to explore the factors and mechanisms that regulate the interaction of H1 with chromatin in living cells.
Factors affecting the mobility of H1 in living cells
The key factors known to affect the interaction of H1 with chromatin in vitro also seem to be the major determinants of the chromatin residence time of H1 in living cells: (i) the structure of the H1 molecule, (ii) post-translational modifications of the H1 molecule or its chromatin binding site, and (iii) competition for chromatin binding sites.
Multiple chromatin-binding sites in H1 proteins
The typical structure of most somatic H1s consists of a globular, winged-helix domain flanked by positively charged regions (Fig. 2). The charge density and the number of hydrophobic amino acid residues in the unstructured flanks vary somewhat; these are most unusual in variants expressed in a developmental stage–specific or tissue-specific manner. Examples include H10, which is most abundant in terminally differentiated tissues, the oocyte-specific H1oo, the testis-specific H1t and the Xenopus laevis B4 protein. Notable exceptions to the typical structure are Tetrahymena thermophila HHO1, which contains only the C-terminal region, and Saccharomyces cerevisiae Hho1p, which contains two globular domains (Fig. 2). A discussion of the functional equivalency of the various H1s is beyond the scope of this review; however, these structural differences need to be taken into account when considering the major factors governing the dynamic properties of H1 molecules in living cells.
Figure 2.
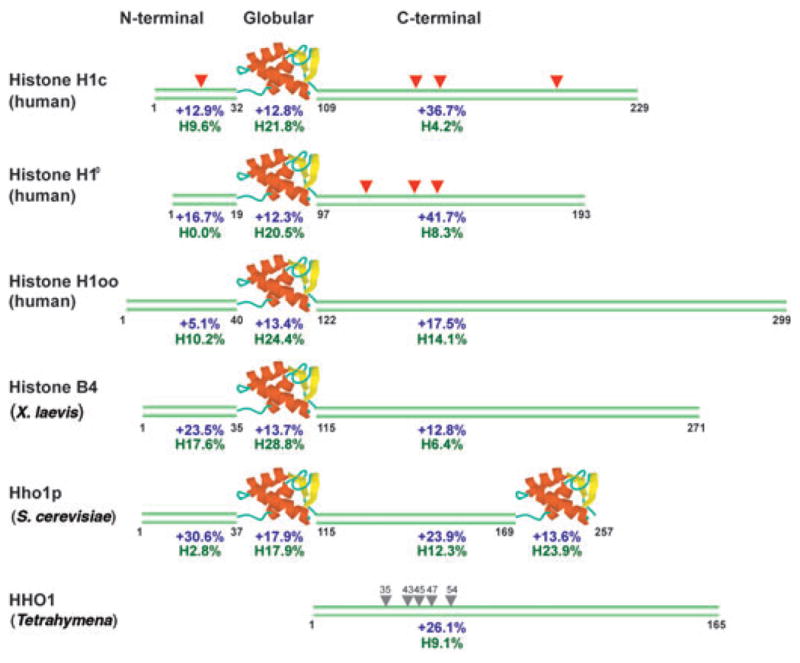
Structural features of H1 variants. Human H1c represents the typical somatic variant found in most cells. The amino acid positions at the boundaries of the domains are indicated under each sequence. For each domain, percentage of net positively charged residues is indicated in blue and percentage of hydrophobic residues (Val, Leu, Ile) in green. Red triangles, conserved SPTXK motifs; gray triangles, serine residues in Tetrahymena HHO1 protein, which have been shown to affect binding49.
Vertebrate cells contain up to eight distinct molecular species (variants) of H1, each encoded by a specific gene, which are expressed in a tissue-, differentiation- and cell replication–specific manner6. Some, but not all, FRAP analyses have detected differences in the affinities of the various H1 variants for chromatin binding sites13,33,37–40. The biggest differences in FRAP-recovery measurements were observed among the oocyte-specific variant H1oo-GFP and the somatic variants H10-GFP38 and H1c-GFP39. Differences in H1 variant mobility were seen in both oocytes and somatic cells, an indication that the binding characteristics of each H1 originate from an intrinsic property of the histone variant, rather than only from the cellular chromatin38.
As originally predicted from sequences41 and later demonstrated by in vitro experiments42, the positively charged C-terminal domain is a major determinant of H1’s chromatin residence time in living cells. Deletion of the C terminus of H10, H1c or H1a leads to a ten-fold reduction in the chromatin residence time of each of these variants14,17,31. Surprisingly, its high positive-charge density is not the crucial contribution of the C terminus to H1-chromatin interactions. Detailed FRAP analysis of H1a mutants points to the conserved S/TPXK motifs (where X is any residue) present in the C-terminal halves of somatic H1s as important elements for chromatin interactions. In vitro studies had already implicated these motifs as important for DNA binding and chromatin condensation43. FRAP analysis demonstrated that, in vivo, a single substitution of Thr152 with glutamate reduces the binding of H1 to chromatin to the same extent as does deletion of all of the 70 C-terminal residues, from position 152 to 222 (ref. 31). Further analysis of the effects of mutations at Thr152 and Ser183 showed the complexity and stringency of correct H1-chromatin interactions. Replacement of either Thr152 or Ser183 (but not both) with lysine increased chromatin binding, whereas replacement of both Thr152 and Ser183 with lysine, alanine or glutamate decreased binding, as measured by FRAP analysis in living cells. Mutations in the S/TPXK motif or phosphorylation at these sites (see below) may disrupt their ability to form β-turn motifs and bind the minor DNA groove44. Additional evidence that the structure of the C-terminal half is an important determinant for chromatin binding comes from comparative FRAP analysis of C-terminal domain swap mutants31,37. The contribution of the N-terminal flank to H1 binding has not been studied in detail; however, deletion of this region reduces the binding of the protein to chromatin31.
The globular domain seems to be the key region that regulates the binding of H1 to native chromatin. Analysis of the kinetic behavior of a battery of point mutants in this domain has identified several residues that are crucial for proper binding to chromatin. By mapping the positions of these residues onto the atomic structure of the H5 globular domain, Brown et al. have defined the nucleosome-interaction surface of the H1 globular domain in the chromatin of living cells33. Using this elegant approach, they have found that the globular region interacts with the nucleosomes through two distinct binding sites formed by several spatially clustered positively charged residues. One site interacts with one side of the DNA approximately one helical turn away from the dyad axis, whereas the second site interacts with one of the linker DNA strands approximately 15 base pairs from the end of the 147-base-pair nucleosomal core particle DNA (that is, it binds the end of chromatosomal DNA). These results are fully compatible with many previous in vitro studies and are among the first to provide detailed information on the interaction of a structural nuclear protein with unperturbed, native chromatin in living cells.
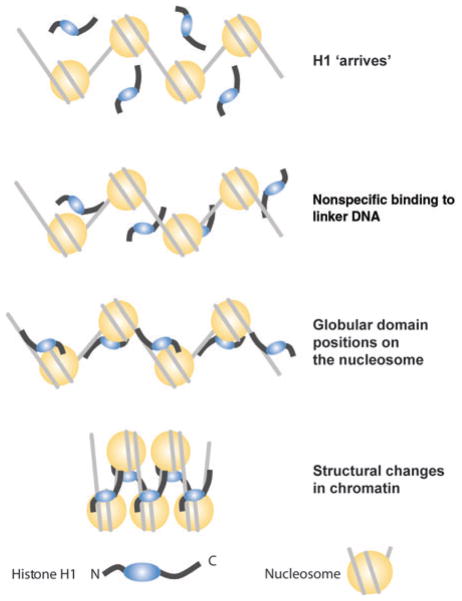
Thus, the unique structural features of H1 lead to a complex, multifaceted type of chromatin binding. In living cells, histone H1 interacts with chromatin through multiple sites, and deletions or modifications of a key site weakens, but does not abolish, its interaction. The high positive charge of the C-terminal region contributes to the strength of binding, but the interactions of this H1 domain with chromatin are governed by the amino acid sequence surrounding key residues. Much attention has been focused on S/TPXK motifs; however, the C-terminal domain has a relatively high content of prolines, and the positions of valine, isoleucine and leucine residues along the polypeptide chain are conserved. Given the known conformational effects of these hydrophobic residues, it is likely that they also have a role in the proper binding of H1 to chromatin. On the basis of all the information available from studies in living cells, Brown et al.33 suggest that the interaction of H1 with chromatin proceeds in several steps (Fig. 3). Initially, the C terminus of H1 binds weakly and nonspecifically to the linker DNA, mainly through charge interactions. This binding facilitates a specific and precise placement of the globular domain at the nucleosomal dyad axis, which stabilizes the nucleosome and leads to conformational changes in the C- and N-terminal flanks. These changes reposition the flanks, affect the nucleosomal repeat12 and stabilize higher-order chromatin structures.
Figure 3.
Dynamic binding of H1 to chromatin. According to Brown et al.33, the binding of H1 to chromatin is a multistep process initiated by a charge-based, low-affinity interaction of the C-terminal domain with the linker DNA, allowing the globular domain to ‘scan’ the nucleosome core for optimum placement. Proper placement of the globular domain induces conformational changes in the H1 flanks and leads to structural changes in chromatin.
Post-translational modifications in H1 and chromatin
Numerous biochemical and tissue-culture studies connect the reversible phosphorylation of H1 with various cellular events, including the progression of the cell cycle, chromatin decondensation and remodeling, replication and transcription4,6,11,45,46. Cell cycle–related chromatin decondensation and phosphorylation of H1 is associated with changes in the intranuclear mobility of the protein47,48. Thus, activation of H1 kinases increases, whereas inhibition of kinases decreases, H1 mobility in living cells14,48. Mutation of amino acid residues known to be phosphorylation targets decreases the mobility of H1 and abolishes the correlation between H1 mobility and the intracellular activity of kinases48. The Tetrahymena HHO1 lacks the H1 globular region and its N-terminal flank (Fig. 2) and therefore can serve as a model for events associated with the C-terminal domain of H1s. Although HHO1 is not necessary for viability, loss of this protein reduces the compaction of chromatin and alters gene expression. Analysis of multiple HHO1 mutant proteins suggests that phosphorylation of serines or threonines creates a patch of negative charges that reduces the binding of H1 to chromatin49,50.
FRAP experiments have also demonstrated that exposure of cells to histone deacetylase inhibitors reduces the binding of H1 to chromatin13. As deacetylase inhibitors such as trichostatin A lead to acetylation of the tails of core histones but not of H1, this increased dynamic exchange of H1 probably results from changes in chromatin (that is, in the H1-binding sites). The histone deacetylase–induced increase in H1 mobility could be due either to altered docking of H1 to nucleosomes containing acetylated histone tails or to acetylation-induced alterations in chromatin structure. As acetylation decreases the binding of H1 both in heterochromatin and in the less compact euchromatin13, it is possible that at least part of the increase in H1 mobility results from changes in the nucleosome.
In considering various mechanisms responsible for altering the mobility of H1, it is important to realize that the same cellular events that lead to modification of H1 also modify other nuclear components, including the core histones. In most cases, it is difficult to distinguish between the effects that are due to changes in H1 itself and those that are due to changes in the H1 binding target. As discussed above, the increase in H1 mobility upon treatment of cells with histone deacetylase inhibitors can be attributed to changes occurring mainly in chromatin, as so far only a single study has reported that H1 is acetylated51. In contrast, as specific point mutations abolish the effects of phosphorylation on H1 mobility, it is likely that these phosphorylation effects are due mostly to changes in the H1 molecule itself.
The mobility of H1 may also be modulated by its interaction with other nuclear components such as HP1. Interestingly, this interaction is modulated by post-translational modification of H1: a specific methylation increases, whereas a specific phosphorylation decreases, the interaction of HP1 with H1 (ref. 52). HP1 is known to interact with methylated H3, and therefore these findings suggest a mechanism to preferentially target H1 to methylated chromatin. Obviously, additional modifications to the template or to H1, such as DNA methylation or H1 ubiquitination, could also affect the interaction of H1 with chromatin; however, their effects in living cells have not been yet studied. Interestingly, studies with Tetrahymena HHO1 have identified an ATP-dependent process that increases H1 mobility in living cells without affecting H1 phosphorylation50. It is tempting to speculate that some of the known ATP-dependent chromatin-remodeling systems have a role in the process53.
In summary, the dynamics and chromatin interactions of H1 are modulated by post-translational modifications to both H1 and the chromatin template (Fig. 4). Modification-driven changes in H1 binding may reflect the synergistic effect of concurrent modifications in both H1 and chromatin. The multiplicity and continuous turnover of these modifications adds an additional layer of complexity to the dynamic interactions of H1 with chromatin.
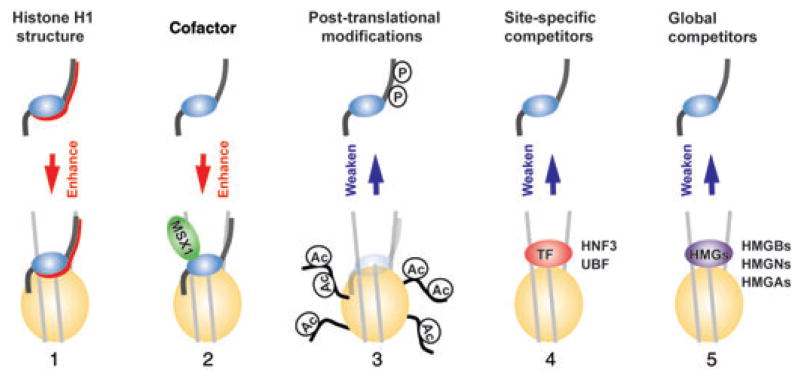
Figure 4.
Determinants of H1-nucleosome interactions. From left: the unique structural features of H1 are the major factor promoting its binding to chromatin (1); regulatory cofactors promote the binding of specific H1 variants to unique nucleosomes (2); post-translational modification of either H1 or chromatin generally reduces the binding of H1 to chromatin (3); site-specific transcription factors (TF) and similar regulatory factors compete with and reduce the binding of H1 to specific chromatin sites (4); non–sequence-specific nucleosome-binding proteins such as HMGs act as global competitors that weaken the interaction of H1 with nucleosomes throughout the entire chromatin (5).
Competition with chromatin-binding proteins
The temporary dissociation of H1 from its binding sites creates a window of opportunity for other factors to bind the vacated sites2,18. Therefore, competition for overlapping chromatin-binding sites could potentially be a major determinant of H1’s interaction with chromatin in living cells. Indeed, numerous in vitro studies have demonstrated that a wide range of nuclear proteins can affect the binding of H1 to chromatin. Broadly, these competitors can be subdivided into two general types: site-specific competitors that affect the binding of H1 to a specific chromatin site and non–site-specific competitors that act as general modulators of H1 binding throughout the entire chromatin fiber. Examples of site-specific modulators include HNF3, UBF, MeCP2 (studies cited in ref. 19) and parathymosin54. Individually, each of these affects H1 binding at only a few sites, but collectively, the site-specific H1 competitors could affect the binding of a sizeable fraction of the cellular H1s. One of the most studied non–site-specific competitors is the HMG superfamily of proteins. It has been known for some time that these proteins can compete with H1 for DNA binding sites55–58.
Photobleaching studies with HMG proteins have provided experimental evidence that, in living cells, chromatin-binding proteins affect the interaction of H1 with nucleosomes. The HMG superfamily consists of three families, HMGA, HMGB and HMGN, each with a unique molecular signature and a distinct chromatin-binding motif59,60. All HMGs are highly mobile and, like H1, bind only transiently to chromatin15–17,61. Members of the same HMG family compete with one another for chromatin binding sites, but members of different HMG families do not cross-compete, an indication that the each family targets distinct chromatin binding sites. All HMGs tested compete with H1 in a dose-dependent fashion, a typical feature of molecular competition. The dynamic interplay between H1 and HMGs is best explained by their transient binding to chromatin. During the transient dissociation of H1 from its binding site, the rapidly moving HMGs bind the vacated site and weaken the interaction of H1 with chromatin. When members of different HMG families are mixed, they synergize the displacement of H1 from chromatin, so that the effect is larger than the sum of individual dose-dependent effects15. The synergistic effects of HMG-H1 competition could be explained by assuming that each type of HMG competes for a distinct set of H1 binding sites.
These results demonstrate that the interaction of H1 with chromatin is modulated by the repertoire of chromatin-binding proteins in the nucleus. An increase in the cellular abundance of a site-specific competitor would affect the mobility of only a small fraction of H1s and might not be detected by FRAP, whereas changes in the concentration of a non–site-specific or ‘global’ H1 competitor would be detected. Such competitive interactions could be the underlying reason for differences in the mobility of H1 molecules in different cell types48. These interactions may be especially important in undifferentiated cells or at early developmental stages, when the abundances of HMG proteins are relatively high62,63. Indeed, the mobility of H1 is especially great in undifferentiated embryonic stem cells, suggesting that in these cells a relatively large fraction of H1 is loosely bound to chromatin or perhaps even found in an unbound state40. The increased mobility of H1 is required for embryonic stem cell differentiation, as an H1 mutant that binds strongly to chromatin inhibits differentiation. The decreased chromatin binding of H1 in embryonic stem cells is not necessarily due to changes in H1 itself, as the mobilities of other chromatin-binding proteins and even of core histones are greater in embryonic stem cells than in fully differentiated cells40. The decreased H1 chromatin residence in embryonic stem cells could result either from a yet-unidentified unique property of chromatin in these cells or from the presence of a large number of competing chromatin-binding proteins.
Thus, in living cells, H1 variants bind nucleosomes as members of a dynamic network of chromatin-binding proteins (Fig. 4). The network includes all the H1 variants, which cross-compete, members of all three HMG families and probably many of the other chromatin-binding proteins that have been shown to bind transiently to chromatin17. Furthermore, the network encompasses both site-specific and non–site-specific H1 competitors. Therefore, the in vivo interaction of H1 with chromatin is continuously modulated by a stochastic process determined by kinetic factors such as the concentrations and affinities of H1 and its competitors.
Perspective
The cellular function and mechanism of action of histone H1 is one of the most perplexing aspects of chromatin biology. Fully differentiated metazoan cells contain enough H1 to bind most of the nucleosomes, and numerous in vitro11 and gene-knockout studies indicate a role of H1 in chromatin structure12,24,29. In mice, a two-fold reduction in H1 content is not compatible with embryonic development, yet the expression of only 29 genes is markedly altered29; and the data available from gene-knockout experiments with living cells does not reveal an obvious role for H1 in the global regulation of transcription2. In fact, most of the H1 knockout results suggest that H1 variants may affect the expression of specific genes27,29,64, a finding that raises the possibility that the interaction of H1 with chromatin optimizes predetermined cell type–specific transcription patterns.
An understanding of the determinants that regulate the dynamic behavior of H1 variants in living cells may provide new insights into the cellular function and mechanism of action of H1. The information available reveals that multiple and diverse factors affect the interaction of H1 with chromatin at several levels. Reversible post-translational modifications, such as phosphorylation of a large fraction of H1s, lead to a global but temporary reduction in the association of H1 with chromatin. A network of structural proteins, such as the HMGs, continuously modulates the binding of H1 throughout the entire chromatin fiber. Changes in H1 binding at a specific site can be facilitated by changes in the concentration of a site-specific competitor, by modification of the H1 binding site or by the presence of a regulatory factor that recruits H1 to that site, such as Msx1 (ref. 65). This multifaceted mode of modulating the interaction of H1 with chromatin has a role in chromatin dynamics and is part of the mechanism that facilitates adequate cellular responses to the multitude of ever-changing internal and external signals to which cells are continuously exposed.
Acknowledgments
This work was supported by the Center for Cancer Research, NCI, through the Intramural Research Program of the US National Institutes of Health. We thank M. Wittenberger, G. Gerlitz and D. Landsman for helpful comments on the manuscript and T. Misteli for critical comments, for numerous discussions on the topic and for providing access to his manuscripts before their publication.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Harvey AC, Downs JA. What functions do linker histones provide? Mol Microbiol. 2004;53:771–775. doi: 10.1111/j.1365-2958.2004.04195.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown DT. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol. 2003;81:221–227. doi: 10.1139/o03-049. [DOI] [PubMed] [Google Scholar]
- 5.Parseghian MH, Hamkalo BA. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem Cell Biol. 2001;79:289–304. [PubMed] [Google Scholar]
- 6.Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene. 2001;271:1–12. doi: 10.1016/s0378-1119(01)00495-4. [DOI] [PubMed] [Google Scholar]
- 7.Kasinsky HE, Lewis JD, Dacks JB, Ausio J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 8.Georgel PT, Hansen JC. Linker histone function in chromatin: dual mechanisms of action. Biochem Cell Biol. 2001;79:313–316. [PubMed] [Google Scholar]
- 9.Thomas JO. Histone H1: location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 10.Wolffe AP, Khochbin S, Dimitrov S. What do linker histones do in chromatin? Bioessays. 1997;19:249–255. doi: 10.1002/bies.950190311. [DOI] [PubMed] [Google Scholar]
- 11.Van Holde KE. Chromatin. Springer-Verlag; New York: 1988. [Google Scholar]
- 12.Woodcock CL, Skoutchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 13.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 14.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 15.Catez F, et al. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phair RD, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roix J, Misteli T. Genomes, proteomes, and dynamic networks in the cell nucleus. Histochem Cell Biol. 2002;118:105–116. doi: 10.1007/s00418-002-0446-7. [DOI] [PubMed] [Google Scholar]
- 19.Zlatanova J, Caiafa P, Van Holde K. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 2000;14:1697–1704. doi: 10.1096/fj.99-0869rev. [DOI] [PubMed] [Google Scholar]
- 20.Ausio J. Are linker histones (histone H1) dispensable for survival? Bioessays. 2000;22:873–877. doi: 10.1002/1521-1878(200010)22:10<873::AID-BIES1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 21.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 22.Hellauer K, Sirard E, Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- 23.Lin Q, et al. Reductions in linker histone levels are tolerated in developing spermatocytes but cause changes in specific gene expression. J Biol Chem. 2004;279:23525–23535. doi: 10.1074/jbc.M400925200. [DOI] [PubMed] [Google Scholar]
- 24.Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 25.Takami Y, Nishi R, Nakayama T. Histone H1 variants play individual roles in transcription regulation in the DT40 chicken B cell line. Biochem Biophys Res Commun. 2000;268:501–508. doi: 10.1006/bbrc.2000.2172. [DOI] [PubMed] [Google Scholar]
- 26.Wierzbicki AT, Jerzmanowski A. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2004;169:997–1008. doi: 10.1534/genetics.104.031997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Y, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan Y, Skoultchi AI. Genetic analysis of H1 linker histone subtypes and their functions in mice. Methods Enzymol. 2004;377:85–107. doi: 10.1016/S0076-6879(03)77005-0. [DOI] [PubMed] [Google Scholar]
- 29.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 30.Mellor J. The dynamics of chromatin remodeling at promoters. Mol Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Hendzel MJ, Lever MA, Crawford E, Th’ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 32.Gunjan A, Alexander BT, Sittman DB, Brown DT. Effects of H1 histone variant overexpression on chromatin structure. J Biol Chem. 1999;274:37950–37956. doi: 10.1074/jbc.274.53.37950. [DOI] [PubMed] [Google Scholar]
- 33.Brown D, Izard T, Misteli T. Mapping the interaction surface of the linker H1 with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 35.Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- 36.Krouwels IM, et al. A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol. 2005;170:537–549. doi: 10.1083/jcb.200502154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Th’ng JP, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J Biol Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 38.Becker M, et al. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol Biol Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teranishi T, et al. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev Biol. 2004;266:76–86. doi: 10.1016/j.ydbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustin M, Rall SC, Stellwagen RH, Cole RD. Histone structure: asymmetric distribution of lysine residues in lysine-rich histone. Science. 1969;163:391–393. doi: 10.1126/science.163.3865.391-a. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, Hansen JC. Revisiting the structure and functions of the linker histone C-terminal tail domain. Biochem Cell Biol. 2003;81:173–176. doi: 10.1139/o03-041. [DOI] [PubMed] [Google Scholar]
- 43.Bharath MM, Ramesh S, Chandra NR, Rao MR. Identification of a 34 amino acid stretch within the C-terminus of histone H1 as the DNA-condensing domain by site-directed mutagenesis. Biochemistry. 2002;41:7617–7627. doi: 10.1021/bi025773+. [DOI] [PubMed] [Google Scholar]
- 44.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 45.Alexandrow MG, Hamlin JL. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J Cell Biol. 2005;168:875–886. doi: 10.1083/jcb.200409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horn PJ, et al. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol. 2002;9:263–267. doi: 10.1038/nsb776. [DOI] [PubMed] [Google Scholar]
- 47.Chen D, et al. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Contreras A, et al. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dou Y, Gorovsky MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell. 2000;6:225–231. doi: 10.1016/s1097-2765(00)00024-1. [DOI] [PubMed] [Google Scholar]
- 50.Dou Y, Bowen J, Liu Y, Gorovsky MA. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J Cell Biol. 2002;158:1161–1170. doi: 10.1083/jcb.200202131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 53.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 54.Martic G, et al. Parathymosin affects the binding of linker histone H1 to nucleosomes and remodels chromatin structure. J Biol Chem. 2005;280:16143–16150. doi: 10.1074/jbc.M410175200. [DOI] [PubMed] [Google Scholar]
- 55.Hill DA, Reeves R. Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varga-Weisz P, van Holde K, Zlatanova J. Competition between linker histones and HMG1 for binding to four-way junction DNA: implications for transcription. Biochem Biophys Res Commun. 1994;203:1904–1911. doi: 10.1006/bbrc.1994.2410. [DOI] [PubMed] [Google Scholar]
- 57.Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ner SS, et al. HMG-D and histone H1 interplay during chromatin assembly and early embryogenesis. J Biol Chem. 2001;276:37569–37576. doi: 10.1074/jbc.M105635200. [DOI] [PubMed] [Google Scholar]
- 59.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 61.Harrer M, Luhrs H, Bustin M, Scheer U, Hock R. Dynamic interaction of HMGA1a proteins with chromatin. J Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 62.Muller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med. 2004;255:332–343. doi: 10.1111/j.1365-2796.2003.01296.x. [DOI] [PubMed] [Google Scholar]
- 63.Furusawa T, et al. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol Cell Biol. 2006;26:592–604. doi: 10.1128/MCB.26.2.592-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alami R, et al. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci USA. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 66.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]