Positive family history of prostate cancer (PCa) has been consistently associated with increased PCa risk [1]. It is widely used in clinical practice and is a strong indication, together with elevated prostate-specific antigen (PSA) levels, for prostate biopsy. An advantage of family history is that the information can be obtained without a laboratory test. However, rather than providing a direct measurement of the patient’s inherited risk, family history captures PCa information of the patient’s relatives. Consequently, family history is influenced by family size, age and survival status of male relatives, recall ability, family communication, and prevalence of the disease in populations. Several scenarios highlight these limitations: (1) Brothers are estimated to have exactly the same inherited risk for PCa based on family history, but in fact they share only 50% of their genetic makeup on average; (2) family history may change from negative to positive; and (3) in populations with low prevalence of PCa, such as among Asians, positive family history is rare and thus is not informative.
Genetic score is another recently proposed measurement of inherited risk for PCa. It is calculated based on genotypes of multiple PCa risk-associated genetic markers, weighted by their relative risk (RR) to PCa. A genetic score of 1.0 indicates an average risk in the general population. Genetic score is now feasible because approximately 40 PCa risk-associated single nucleotide polymorphisms (SNPs) have been identified from rigorously designed genomewide association studies [2]. Risk alleles of these SNPs are common in the white population, and although they confer modest risk to PCa individually, they have a stronger cumulative effect on PCa [3,4]. Importantly, all published studies to date demonstrate that genetic score based on combinations of these SNPs is informative in measuring individual risk and can serve as an independent predictor of PCa [5,6]. Genetic score requires a laboratory test; however, this is a trivial task with high-throughput and low-cost genotyping technology. For example, a panel of 33 PCa risk-associated SNPs can be assayed in 1 d from a blood or saliva sample at a cost similar to a PSA test [6].
The goal of this study was to compare, head-to-head, these two measurements of inherited risk for PCa in several study populations with different study designs (clinical trials, observational prospective cohort, and case-control) and diverse geographic regions (North America, Western Europe, Eastern Europe, and Sweden). In all study populations, family history of PCa in first- and second-degree relatives was obtained by questionnaire, and genetic score was calculated from 33 PCa risk-associated SNPs, as detailed elsewhere [6].
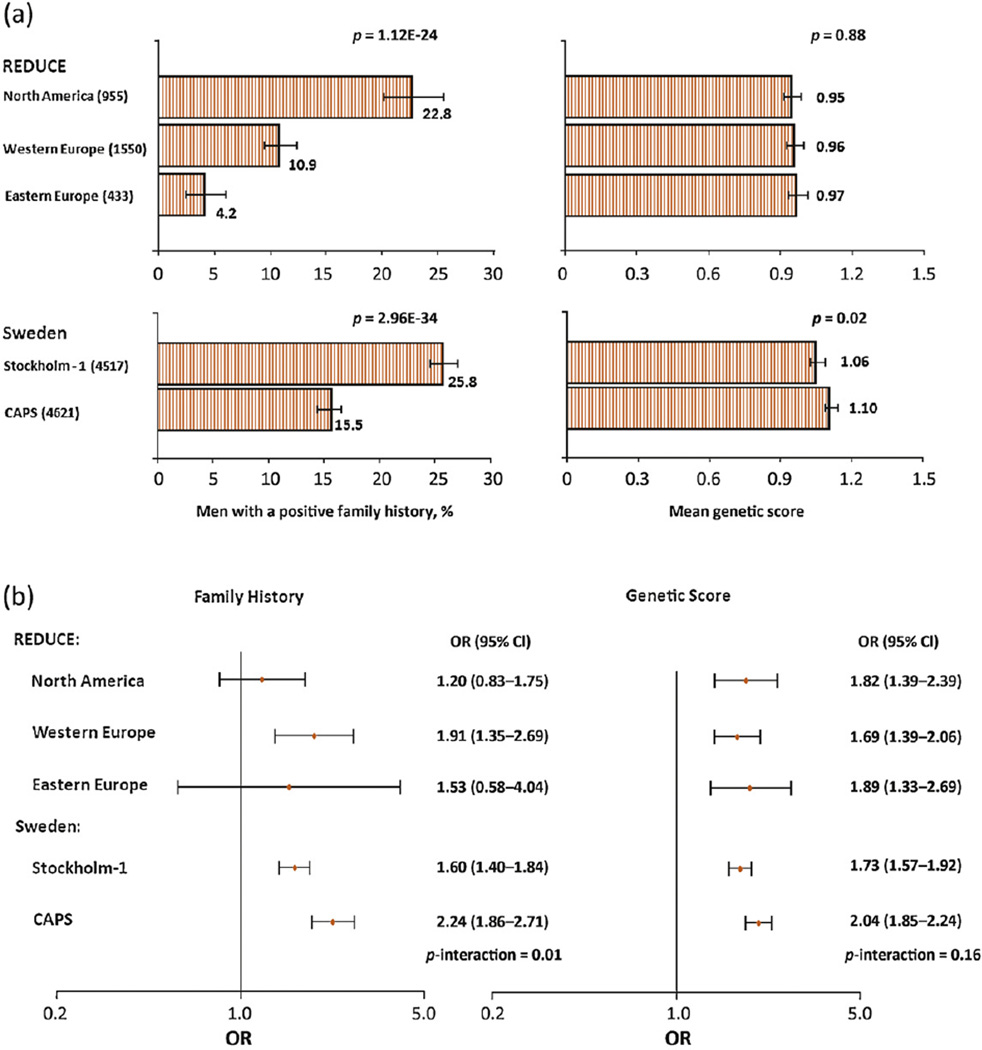
We found that the proportion of men with a positive family history of PCa differed considerably among study populations, whereas mean genetic score was similar (Fig. 1a). For example, among three geographic regions from the single clinical trial, Reduction by Dutasteride of Prostate Cancer Events (REDUCE), the proportion of positive family history was significantly different: 4.2%, 10.9%, and 22.8% in Eastern Europe, Western Europe, and North America, respectively (p < 0.001). These large differences were found even though the same protocol was used to obtain family history information. Furthermore, even for the two study populations from the same country (Sweden), the percentage of men with a positive family history differed substantially: 25.8% in a prospective cohort of patients that underwent prostate biopsy (Stockholm-1) and 15.5% in a case-control study (Cancer of the Prostate in Sweden [CAPS]) (p < 0.001). In contrast, the mean genetic score was similar among different geographic regions within the REDUCE study (0.95–0.97, p = 0.88) and between the two different studies in Sweden (1.06–1.10, p = 0.02). Considering that genetic susceptibility to PCa is likely to be similar among these white populations, the volatile estimates of inherited risk obtained from family history expose the weakness of this measurement. In contrast, genetic score is a more reliable measurement of inherited PCa risk.
Fig. 1.
(a) The distribution of family history and genetic score in five study populations: (left panel) the proportion of men with one family member or more affected with prostate cancer (PCa); (right panel) the mean genetic score. A chi-square test was used to test different proportions of positive family history between study populations and an analysis of variance was used to compare log-transformed mean genetic scores among different study populations. (b) The association of PCa risk with family history of PCa or genetic score in five study populations. The odds ratio (OR) was calculated from logistic regressions. Differences in OR among study populations were tested using an interaction term (family history by study populations or genetic score by study populations) in the logistic regression model. CI = confidence interval.
The effects of family history and genetic score on PCa risk are presented in Figure 1b. The RR of family history for PCa differed considerably among geographic regions within the REDUCE trial: 1.20, 1.53, and 1.91 in North America, Eastern Europe, and Western Europe, respectively. Similarly, the RR of family history for PCa differed significantly between the two studies in Sweden: RR was 1.60 in the prospective Stockholm-1 study and 2.24 in the case-control CAPS study. The differences in RR estimates among these five study populations were statistically significant (p = 0.01). In contrast, the RR estimates of genetic score for PCa were similar among these five study populations, from 1.69 to 2.04 (p = 0.16). These results suggest that genetic score has better precision for measuring the effect of inherited risk on PCa risk than family history.
The performance of family history and/or genetic score in discriminating PCa status in five study populations is shown in Table 1. The area under the receiver operating characteristic curve (AUC) of genetic score for predicting positive PCa biopsy was significantly higher (0.58–0.62) than family history (0.51–0.55) in each study population (p < 0.05). Furthermore, in each of the five study populations, the AUC of combined genetic score and family history was considerably higher than that of family history alone but was similar to that of genetic score alone. These results demonstrate that genetic score has a better predictive performance for biopsy outcome (PCa) than family history, and family history alone is not sufficient to capture inherited risk for PCa.
Table 1.
Performance of predicting prostate cancer outcome using family history and genetic score in five study populations
| Study population | No. of subjects | AUC | ||||
|---|---|---|---|---|---|---|
| Positive biopsy (or cases) | Negative biopsy (or controls) | All | FH | GS | FH + GS | |
| REDUCE | ||||||
| North America | 180 | 775 | 955 | 0.52 | 0.60 | 0.60 |
| Western Europe | 348 | 1202 | 1550 | 0.54 | 0.59 | 0.60 |
| Eastern Europe | 129 | 304 | 433 | 0.51 | 0.60 | 0.60 |
| Sweden | ||||||
| Stockholm-1 | 2226 | 2291 | 4517 | 0.55 | 0.58 | 0.60 |
| CAPS | 2899 | 1722 | 4621 | 0.55 | 0.62 | 0.64 |
AUC = area under the receiver operating characteristic curve; FH = family history; GS = genetic score.
It is possible that volatile estimates of positive family history and its effect on PCa risk observed in this study may be due to heterogeneous ascertainment of family history among different studies; this highlights the limitation of family history measurement. Collection of family history in real-world clinical settings is likely even more heterogeneous.
It is expected that additional PCa risk-associated SNPs may be identified in the future, and these SNPs can be incorporated into genetic score to further improve its predictive performance for PCa. However, due to the polygenic inheritance of PCa and the likelihood that common PCa risk-associated SNPs with the strongest effect have already been identified, the potential for improvement over the current set of SNPs is expected to be small [7]. In contrast, rare and high-penetrance PCa risk-associated mutations may significantly improve the predictive performance of PCa in small subsets of men, as demonstrated for the newly identified G84E mutation of the homeobox B13 (HOXB13) gene [8].
In conclusion, if family history is accepted and used by urologists and primary care physicians to assess an individual’s risk for PCa, we believe that genetic score should be added to family history to improve assessment of inherited risk for PCa.
Acknowledgments
Funding support: The study is partially supported by National Cancer Institute grants (CA148463 and CA140262) to Dr. Xu and a research contract by GSK to Dr. Xu. The study is also partially supported by the Swedish Research Council (grant no. K2010-70X-20430-04-3, and 70867901), the Swedish Cancer Foundation (grant no. 09-0677), the Hedlund Foundation, the Söderberg Foundation, the Enqvist Foundation, ALF funds from the Stockholm County Council, Stiftelsen Johanna Hagstrand och Sigfrid Linnér’s Minne, Karlsson’s Fund for urologic and surgical research, the Cancer Risk Prediction Center (CRisP; http://www.crispcenter.org), and a Linneus Centre award (Contract ID 70867902) financed by the Swedish Research Council.
Acknowledgment statement: We thank the patients enrolled in REDUCE who provided consent and genetic samples that enabled this study and the clinicians who contributed their expertise in recruiting study patients for the REDUCE clinical study. Dave Pulford, Jennifer Aponte, Jon Charnecki, and Mary Ellyn Volk participated in consent reconciliation and sample management to enable genetic sample selection for inclusion and genotype determination. Karen King provided data management support for this project. We appreciate the assistance of Lauren Marmor in coordinating the support of the Avodart Collaborative Research Team.
Footnotes
Conflicts of interest: Dr. Condreay is employed by and owns stock in GSK.
References
- 1.Johns LE, Houlston RS. A systematic review and meta-analysis of familial prostate cancer risk. BJU Int. 2003;91:789–794. doi: 10.1046/j.1464-410x.2003.04232.x. [DOI] [PubMed] [Google Scholar]
- 2.Goh CL, Schumacher FR, Easton D, et al. Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J Intern Med. 2012;271:353–365. doi: 10.1111/j.1365-2796.2012.02511.x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Sun J, Kader AK, et al. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69:1565–1572. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aly M, Wiklund F, Xu J, et al. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60:21–28. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kader AK, Sun J, Reck BH, et al. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: findings from the REDUCE trial. Eur Urol. 2012;62:953–961. doi: 10.1016/j.eururo.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Kader AK, Hsu FC, et al. Inherited genetic markers discovered to date are able to identify a significant number of men at considerably elevated risk for prostate cancer. Prostate. 2011;71:421–430. doi: 10.1002/pros.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson R, Aly M, Clements M, et al. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur Urol. doi: 10.1016/j.eururo.2012.07.027. In press. http://dx.doi.org/10.1016/j.eururo.2012.07.027. [DOI] [PubMed] [Google Scholar]