Abstract
Staphylococcus aureus is one of the major causes of community and hospital-acquired infections. Bacteriophage considered as a major risk factor acquires S. aureus new virulence genetic elements. A total number of 119 S. aureus isolated from different specimens obtained from (RKH) were distinguished by susceptibility to 19 antimicrobial agents, phage typing, and PCR amplification for mecA gene. All of MRSA isolates harbored mecA gene, except three unique isolates. The predominant phage group is belonging to the (mixed group). Phage group (II) considered as an epidemiological marker correlated to β-lactamase hyper producer isolates. MRSA isolates indicated high prevalence of phage group (II) with highly increase for phage types (Ø3A), which were correlated to the skin. Phage types (Ø80/Ø81) played an important roll in Community Acquired Methicillin Resistant S. aureus (CAMRSA). Three outpatients MRSA isolates had low multiresistance against Bacitracin (Ba) and Fusidic acid (FD), considered as CAMRSA isolates. It was detected that group I typed all FD-resistant MSSA isolates. Phage groups (M) and (II) were found almost to be integrated for Gentamycin (GN) resistance especially phage type (Ø95) which relatively increased up to 20% in MRSA. Tetracycline (TE) resistant isolates typed by groups (II) and (III) in MSSA. Only one isolate resistant to Sulphamethoxazole/Trimethoprim (SXT) was typed by (III/V) alone in MSSA. MRSA isolates resistant to Chloramphenicol (C) and Ba were typed by all groups except (V). It could be concluded that (PERSA) S. aureus isolates from the wound that originated and colonized, and started to build up multi-resistance against the topical treatment antibiotics. In this study, some unique sporadic isolates for both MRSA and MSSA could be used as biological, molecular and epidemiological markers such as prospective tools.
Keywords: Community, MRSA, MSSA, Phage typing, mecA, Prospective tools, Multiresistance, Hospital, Acquired infections
1. Introduction
Staphylococcus aureus is a versatile important human pathogen causing a number of variety medical infections. In the other wise, the fact that 90% of hospital staff are carriers of S. aureus portends serious for the epidemiology and pathogenesis of Staphylococcal infections (Fey et al., 2003; Lacey et al., 1984). The wide spread of antibiotic resistance among S. aureus strains is a major concern in the treatment of Staphylococcus infections. These strains often resistance to multiple antibiotics with attendant increased morbidity; the surveillance and control of these strains was highly desirable (Mathur and Mehudiratt, 2000; Lyon and SKurray, 1987). The increasing importance of MRSA as a cause of nosocomial infections can be inferred from several recent studies (Schmitz et al., 1997). There is relatively little information on the diversity of strains causing infection. It is important to have knowledge of the most common strains associated with human infections and their sources in each episodes and environment in order to improve our understanding of the epidemiology of this pathogen and solve the problems. Although bacterial interaction is a well recognized phenomenon, there has been surprisingly little research with respect to MRSA and MSSA. The mechanism/s responsible for this phenomenon is not readily apparent. Gopal Rao and Wong (2003) concluded that there is a complex relationship between various strains of EMRSA and MSSA especially in the skin. This interaction may have an important bearing on colonization of patients with MRSA. It may explain some of the epidemiological and clinical observations as well as understanding the methods for the movement of resistant genes, like: Transduction (phages) – Plasmids – Integrons – Transposons.
In Saudi Arabia, significant increase of MRSA was noticed from 6.9% to 33% starting in the last decade from 1995 up to 2004 alarming it remarkably, as an outbreak of EMRSA and EMSSA. Several investigations were done concerning the screening and emergence of MRSA in the Kingdom. Belkum et al. (1997) identified that MRSA Saudi isolates all belonging to phage group III. It was clear that consensus SmaI pattern observed for Saudi strains was different from other non-related isolates.
Kishan et al. (1998) reported for the most β-lactamase-producing isolates of S. aureus belonging to other phage groups. Skov et al. (1995) have reported that all phage group II isolates recovered prior to harbored blaz on the chromosome. The final event in multiplication of phage was lysis of the host cell by murein hydrolyses of various substrate specificities (Young, 1992; Loessner et al., 1995, 1998).
The genetic studies that showed that drug-resistance genes were easily transduced among S. aureus cells by prophage (Mitsuhashi et al., 1965). This suggested a relationship between the intracellular state of the drug-resistance genes and temperate phages. Thus, they could show that some of the temperate phages transduced the drug-resistance genes.
The aim of this study is to approach questions related to the spread of MRSA in Saudi cohort by examining phenotypic (resistance to antibiotics, phage typing) and the existed genetic backgrounds mecA gene. As well as defined the epidemic drift of phage-types within originated susceptible wild type S. aureus population as a microbial biomarkers for monitoring the usage of antibiotics. And analyze the contribution of the phage typing, as a phenotypic marker and answer question such as: are the phage types between isolates similarly or differently?
-
•
What is the homogeneity and heterogeneity between MSSA and MRSA?
-
•
Which of S. aureus isolates carrying the mecA gene?
-
•
What are the antimicrobial resistance determinants that phage could contribute?
2. Materials and methods
2.1. Bacterial isolates
A total numbers of 119 isolates of S. aureus from different patients were collected over a period of one year from 2003 to 2004 from microbiology laboratory in Riyadh Armed Forces Hospital (RAFH). The following reference strains of bacterial species were used as controls:
Methicillin resistant S. aureus (MRSA) (NCTC 10442), methicillin sensitive S. aureus (MSSA) (ATCC 25923)
Coagulase Negative Staphylococcus epidermidis (CNS) (ATCC 12228).
Identification of S. aureus isolates
The suspected colonies were identified according to the following criteria:
2.2. Colonial morphology
All isolates were streaked for purity growth on Blood Agar plates (Blood Agar Base.Oxoid.Code: CM55) for over night incubated at 37 °C (Collee et al., 1989).
2.3. Microscopic examination
Gram stain smears from a Staph Culture showed gram positive cocci in clusters.
2.4. Catalase test
This test detects the presence of cytochrome oxidase enzymes in Micrococcaceae according to Konerman et al. (1992).
2.5. Coagulase test
Slide agglutination test (Staphurex) based on the detection of clumping factor and protein A. The test was performed by Slidex Staph-kit (BioMereux, Chabonnieres-Bain France) according to MacFaddin (1980) and Baron et al. (1994).
Tube coagulase test measures the production of free coagulase (Staphylocoagulase). The test (Remel Coagulase plasma. Rabbit plasma w/EDTA) was performed according to Fairbrother and Chapman (1940), Piper et al. (1988) and Luijendijk et al. (1996).
2.6. Manitol fermentation
All isolates were streaked onto Manitol Salt Agar (MSA) (Oxoid.Code: CM85). If the manitol was fermented to produce acid, the phenol red in then medium changes color from red to yellow. If this color change exists, it can be presumed that the isolate is a strain of S. aureus.
2.7. Deoxyribonuclease (DNase) test
This test was performed by heavily spot-inoculating several colonies on nutrient agar media (Oxoid.Code: CM3) containing DNA. Colonies of S. aureus that produced DNase were surrounding by a clear zone where the DNA had been depolymerized and hydrolyzed as done by Franki and Murray (1986) and Duguid (1989).
2.8. Detection of methicillin resistance in S. aureus
Kirby–Bauer technique of disk diffusion method was used to detect the methicillin resistance. The isolates were tested by employing Mueller Hinton Agar (MHA) (Oxoid.Code: CM337). The 1 μg oxacillin (OX) and cefoxitin (FOX) 30 μg disk diffusion were used with MHA media supplemented with 4% NaCl was used to detect MRSA according to the NCCLS National Committee for Clinical Laboratory Standards (2004). Disk diffusion testing by FOX disk was performed to simultaneously evaluate its value in predicting OX resistance. Commercial antimicrobial agents containing disks were placed on the plates by multidispenser (Oxoid).
2.9. Genotypic method for identification of S. aureus isolates and detection of methicillin resistance
Triplex Polymerase Chain Reaction (PCR) method to amplify three different genes was used according to Al-Shammary (2005).
16SrRNA: a specific gene for Staphylococci species.
nuc: for detection of S. aureus.
mecA: to detect methicillin resistance strains.
2.10. DNA templet extraction
Two to four colonies overnight blood agar subculture of S. aureus strains were taken and resuspended into 200 ml of PCR water (Molecular Biology Grade Water, eppendorf) in an eppendorf tube, vortex and then centrifuged at 13,000 rpm for one min (just to form a pellet) then the DNA can be obtained directly from the supernatant.
2.11. Primers
Oligonucleutide primers are listed in Table 1. The primers were obtained from MWG-Biotech AG (Berlin, Germany).
Table 1.
Sequences of oligonucleotide primers for identification of S. aureus and detection of MRSA strains.
| Gene | Primer | Oligonucleotide sequence | bp | Quality control |
|---|---|---|---|---|
| mecA | mecA1 | 5′CTT TGC TAG AGT AGC ACT CG 3′ | 531 | NCTC 10442 |
| mecA2 | 5′GCT AGC CAT TCC TTT ATC TTG 3′ | |||
| 16s RNA | 16sf | 5′GTA GGT GGC AAG CGT TAT CC 3′ | 218 | ATCC 12228 |
| 16sr | 5′CGC ACA TCA GCG TCA G 3′ | |||
| nuc | nuc1 | 5′GCG ATT GAT GGT GAT ACG GTT 3′ | 280 | ATCC 25923 |
| nuc2 | 5′AGC CAA GCC TTG ACG AACTAA AGC 3′ |
2.12. PCR reaction mix
A total volume 25 μl of reaction mix was prepared by adding 1.5 μl of primer mix, the primer mix was done by adding 1 μl from each mecA primers, 0.25 μl from each nuc primers and from each 16srRNA primers. 4 mM MgCl2 (stock 25 mM), 12.5 μl Ready Mix™ (Ready Mix PCR, Master Mix Con.2X. AB gene), {when Ready Mix™ used as 1× it will contain a concentration of 1.25U Thermo prime plus DNA polymerase, 75 mM tris HCl [ph:8.8], 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% (v/v) Tween® 20, 0.2 mM dNTPs, precipitant and red dye for electrophoresis} 2 μl PCR water and 5 μl Template DNA. Each run contained the MRSA, MSSA and CNS standard strains with and two negative controls (blank reagent and PCR water).
2.13. The cycling protocol
The reaction was carried by using the Thermal Cycler (TC) from Gene Amp PCR system 9700 (Perkin–Elmer). The cycling protocol consisted of one cycle for 5 min at 94 °C, followed by 30 cycles consisting of denaturation for 1 min at 94 °C, annealing for 1 min at 55 °C, and elongation for 2 min at 72 °C then one cycle of final elongation for 4 min at 72 °C. After the final cycle, the products were kept at 4 °C till the analysis done.
2.14. Products analysis
Ten microlitres of the product was loaded onto 2% Pulsed Field Certified Agarose and electrophoresis was done at 120 V for 2 h (GT UVTP Gel tray 15 × 25 cm. GT-Gel casting system. Sub-Cell® GT system Comb size 20 × 0.75 mm. Power supply, Bio-Rad. Triss Borate EDTA (TBE), buffer.10X. USB). After electrophoresis, the DNA visualized by staining with ethidium bromide solution (Plus one. Pharmacia Biotech. 1 μg/ml). The gel staining for 15–30 min. Viewed under UV transillumination (Bio-Rad) and photograph using gel documentation system, print image (Bio-Rad P68V paper. 110 mm × 21 mm high density type) and store it. Destain for 15–30 min in distilled water. The DNA molecular size marker was used (100 Base-Pair ladder 1 μg/μl. Amersham Pharmacia Biotech Inc.). The analysis of the DNA PCR products was evaluated according to the following controls:
MRSA control NCTC 10442 (mecA, 16Sr RNA and nuc positive)
MSSA control ATCC 25923 (mecA negative, 16SrRNA and nuc positive)
CNS control ATCC 12228 (mecA and nuc negative, 16Sr RNA positive)
PCR water (Negative control) (mecA, 16SrRNA and nuc negative)
2.15. Isolates stored
S. aureus isolates were inoculated on blood agar plates and a colonies from pure cultures were stored on nutrient agar slopes at 4 °C with subculture them every six months. In addition, the isolates were also stored at −70 °C in nutrient broth contained 15% glycerol. Several tubes of each culture were prepared and only one were used for sub culturing each year. Before being tested the isolates were removed from storage and streaked onto blood agar plates and incubated under aerobic conditions at 37 °C for 18–24 h. All inoculate were prepared from these subcultures (Miller, 1987).
2.16. Antibiotic susceptibility tests
The sensitivity methods were performed as described by Clinical Microbiology procedures Handbook (Hindler, 1992). A guide to sensitivity testing (Phillips et al., 1993) and Manual of Clinical Microbiology (Woods and Washington, 1995). Mueller Hinton plates (without salt) were inoculated in the same way which was used for OX and FOX, by
Kirby–Bauer technique. The sensitivity was recorded according to the NCCLS guidelines.
Modified stocks technique were performed by both the test and control organisms were inoculated on the same plate. Susceptibility categories were interpreted according the stock’s technique criteria. Reading the reaction of the test organism to each antibiotic as ‘sensitive’ or ‘resistant’ as follows:
Sensitive (S): Zone radius wider than, equal to or not more than 3mm smaller than the control. Resistant (R): No zone of inhibition or zone radius measure 2 mm or less.
2.17. Phage typing
According to Laboratory of Hospital Infections, Central Public Health Laboratory, Colindale. London which resembled An International Human Staphylococcal Phage Typing Set (IPS), containing 23 phages was used for discriminatory isolates.
2.18. Propagation the phages
Suspend the dried phage in 1.0 ml of broth or we can use 1.0 ml of stock phages, and dilute in 10-fold steps to 1 in 1,000,000. Apply one 0.02 ml drop of each dilution to a dried lawn of propagating strain and incubate overnight at 30 °C, then we determine the titer and RTD. The RTD (routine test dilution) is that dilution of phage which just fails to give confluent lysis (i.e. gives semi-confluent lysis) on its propagating strain. After that either soft agar or broth propagation methods might be used depend on the phage, as recommended in the phage set but two methods were performed for all the phages as follows:
2.19. Broth propagation
An overnight broth culture of the propagating strain at a dilution of 1/100 was added to nutrient broth containing calcium. Phage suspension was added to give a final dilution equivalent of 1 × RTD/ml, if this does not give a satisfactory propagation then input phage of 1/RTD and/or 10 × RTD/ml may also be tried. The mixture was incubated at 37 °C for 6 h, with shaking. After incubation, the lysate was centrifuged, the supernatant pipetted off and titrated (in some cases it may be desirable to filter at this point). The titration plate was incubated overnight at the temperature appropriate for the species; the supernatant stored overnight at 4 °C. If the titer is high enough – RTD should be at least 1 in 10,000 the supernatant is filtered through a 0.45 μl filter, tested for sterility and re-titrated, Sterility testing involves incubating aliquots of phage filtrate in nutrient broth for a week. The filtrate is then distributed into screw-capped bottles and stored at 4 °C without preservative. It is permissible to use chloroform to sterilize S. aureus phage 79.
2.20. Soft agar propagation
A slope culture of the propagating strain was prepared on nutrient agar slope in a test tube. Fourteen centimetre Petri dishes containing nutrient agar to a depth of approx 5 mm were used. Prepared nutrient agar (soft agar) containing 0.5% agar which melted and cooled to 45 °C. Calcium chloride (sterile) was added to give a final calcium concentration of 400 μg/ml. A suspension of the propagating strain, prepared by suspending the growth from the nutrient agar slope in 2 ml nutrient broth, is added to give a final dilution of 1/100. Add sufficient phage to give near-confluent lysis after overnight incubation. This phage concentration varies between phages, but was usually between 105 and 106 pfu/ml of soft agar. After mixing, 7.5 ml of the cell-phage suspension was layered on the surface of each nutrient agar plate. The plates are incubated, usually for 18 h at the optimum temperature varies with phage. Most yield higher titer propagation at 37 °C. Broth was added per plate and the soft agar layer scraped off with a sterile, bent glass or plastic rod. Both broth and soft agar are taken off. The agar lumps were broken up by rapid pipetting or shaking, the mixture centrifuged and the supernatant was removed and titrated.
2.21. Phage typing method
A two-hour, lightly inoculated, nutrient broth culture grown with shaking at 37 °C was used for typing. The grown culture is used to flood a plate containing appropriate agar, and the excess culture removed. The flooded plate is dried open and this can take several hours, a minimum of 2 h is necessary. Phages, appropriate for the species, are applied at 100× RTD by using multi-head inoculators. Lids are replaced as soon as the phages suspension has been applied. After drying, the plates are incubated overnight at 30 °C. Phage reaction were read, by eye, and recorded on the conventional phage scales according to Public Health Laboratory, Colindale, London.
| 1 | 1–5 plaques |
| 2 | 6–19 plaques |
| 3 | 20–49 plaques |
| 8 | 50 ors more plaques |
| 9 | Confluent lysis |
| 0 | Inhibition reaction (inhibition, or thinning of bacterial growth) |
| – | No reaction |
3. Results
3.1. S. aureus isolates identification
The colonized 119 isolates of S. aureus on BA plates showed smooth, slightly domed colonies with 1–2 mm diameters. All isolates were positive to catalase, Staphurex, manitol fermentation, tube coagulase and DNase tests. All isolates were contain the 16sRNA and nuc genes which identified as S. aureus by genotypic method in Fig. 1.
Figure 1.
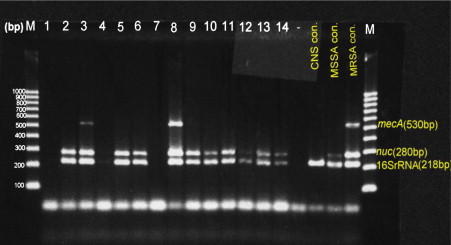
Amplification of three genes: mecA for MRSA, nuc for S. aureus and 16SrRNA for Staphylococci genus using the specific primers for each. Lanes: 1–14 showed the PCR products of S. aureus studied DNA samples compared to the mentioned positive controls. Lane (−) is a negative control. Lanes: 3 and 8 harbored mecA gene, while the others proved to be MSSA lacking mecA. The last three lanes exhibit positive controls. M: is a 1000 bp DNA marker.
3.2. Detection of methicillin resistance in S. aureus
OX and FOX disks diffusion methods were applied for detection of MRSA according to NCCLS, that recommended the use of the cefoxitin disk test as a surrogate marker for the detection of oxacillin resistance in Staphylococcal isolates. Seventy-seven isolates were identified as MSSA strains and 42 isolates were characterized as MRSA strains. The results were confirmed by PCR detection for mecA gene. Thirty-nine out of 42 MRSA isolates were positive for mecA gene in Fig. 1.
3.3. Antimicrobial susceptibility patterns of S. aureus
Antibiotics susceptibility among S. aureus isolates is summarized in Tables 2 and 3, Figs. 2–4.
Table 2.
Antibiotic susceptibility tests for 77 MSSA isolates against 19 antibiotics.
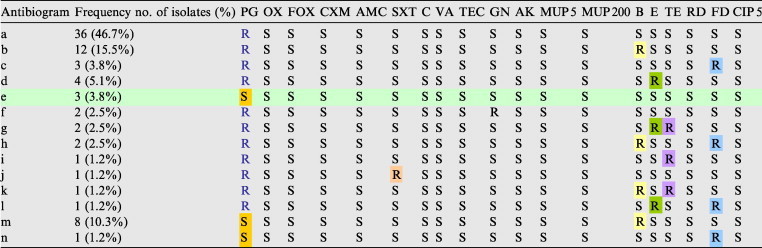
Table 3.
Antibiotic susceptibility tests for 42 MRSA isolates against 19 antibiotics.
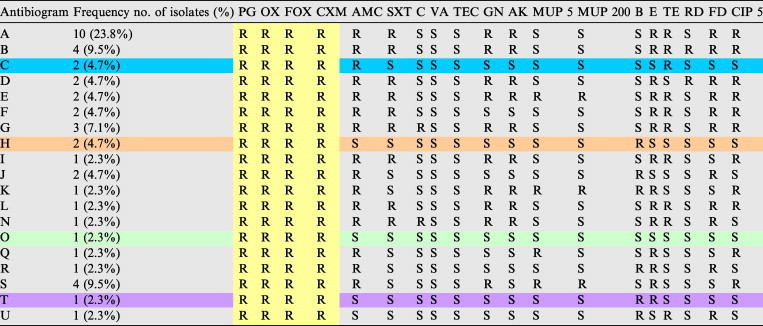
R: Resistant to, S: Sensitive to, PG 10: Penicillin, OX: Oxacillin, FOX: Cefoxitin, CXM: Cefuroxime, AMC: Augmantin, SXT: Sulphamethoxazole/Trimethoprim, C: Chloramphenicol, VA: Vancomyc, TEC: Ttechoplanin, GN: Gentamycin, AK: Amikacin, MUP 5: Mupirocin 5, MUP 200: Mupirocin 200, Ba: Bacitracin, E: Erythromycin, TE: Ttetracycline, RD: Rifambin, FD: Fusidic acid, CIP 5: Ciprofloxacin.
Figure 2.
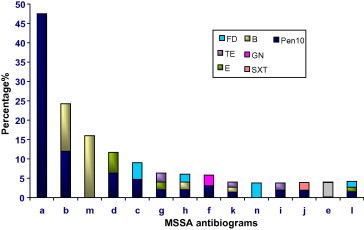
Emergence of resistance between MSSA isolates.
Figure 3.
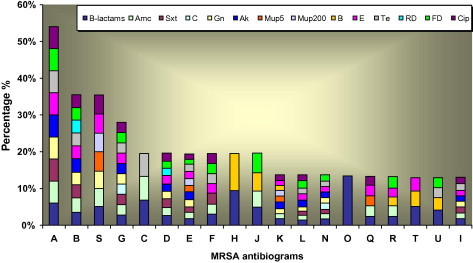
Prevalence of multi resistance in MRSA isolates.
Figure 4.
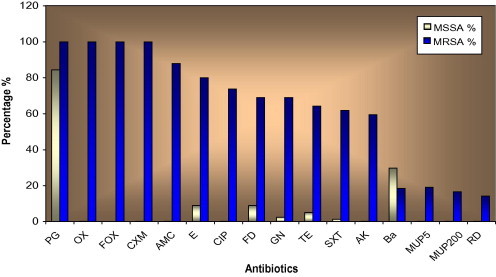
Antibiotics susceptibility among MSSA and MRSA isolates.
The 77 MSSA isolates were classified into 14 various antibiograms from (a) to (n). 42 MRSA isolates were classified into 19 antimicrobial agents were (from A to U) in Table 3 and Fig. 3.
For β-lactam antibiotic+β-lactamase inhibitor (AMC), all MSSA isolates demonstrated susceptible to it. While 88% of MRSA isolates were resistant. For other antibiotics, the resistance rates were significantly increased in MRSA isolates compared with the MSSA. In contrast, the resistance to Ba antibiotic in MSSA (Table 2) was higher than that was found in MRSA which was 32.4% in the former and 19% in the latter in Table 3. So, all isolates (both MSSA and MRSA) demonstrated susceptibility to glycopeptides (VA and TEC) (Fig. 4).
It was observed a group of 29 isolates emerged out of 77 of MSSA had resistant for two or three of the antibiotics, which occurred in Table 2.
3.4. Phage typing
3.4.1. Distribution of phage types
The distribution of studied S. aureus isolates in the different phage groups are shown in Tables 4 and 5 and Figs. 5 and 6. A 76.4% of the total isolates were typable by IPS (83.1% in MSSA and 64.2% in MRSA strains) and 30.7% (16.8% in MSSA – 35.7% in MRSA) were nontypable. There were 55 different phage typing patterns among 64 MSSA isolates, while 21 various patterns of 27 typable MRSA isolates. The predominant phage group in the study belonged to the Mixed group (64.8%) followed by group III (see Fig. 7).
Figure 5.
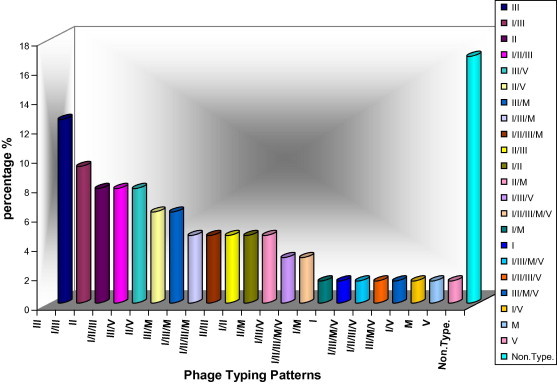
Phage typing patterns of MRSA isolates.
Figure 6.
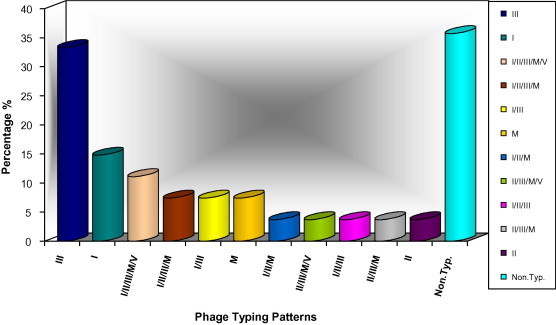
Phage typing patterns of MRSA isolates.
Figure 7.
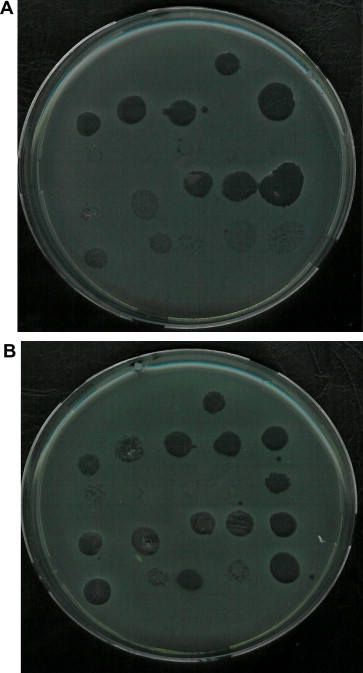
Phage typing of S. aureus isolates in different reactions. (A) MRSA isolate and (B) MSSA isolate that highly sensitive to phage types in different reactions (strong reaction–weak reaction).
Some isolates showed identical phage pattern. In MSSA, the most common identical phage typing patterns were seven in 15 isolates as follows:
(79/80/6/42E/47/53/54/75/77/83A/52A/29. n = 2 – No. 60 and 56),
(3C/55/71. n = 2 – No. 87 and 102),
(55/94/96. n = 2 – No. 111 and 119),
(47/94/96. n = 2 – No. 109 and 110),
(81/71. n = 2 – No. 84 and 108),
(79/3A/3C/55/71. n = 2 – No. 34 and 80 from wound), and
(54. N = 3 – No. 79, 93 and 33).
These data are exhibited in Table 4 and Fig. 5. The prevalence of the same phage typing patterns in MRSA (Table 5 and Fig. 6). These repetitions were shown for MRSA isolates which typed only with one phage as follows:
(29. n = 3 – No. 22, 70 and 4), (85. n = 2 – No. 101 and 112), (81. n = 2 – No. 15 and 48) and (75. N = 2 – No. 17 and 40).
It was observed identical phage typing patterns between MSSA and MRSA isolates: (81. No. 97 in MSSA_No. 15 and 48 in MRSA) and (54/85. No. 78 in MSSA_No. 85 in MRSA).
3.5. Phage lytic groups and antibiotics
In MSSA isolates, it was observed the PG resistance isolates were distributed in all phage lytic groups, but group III was the most common (52.3%). All nontypable isolates exhibited resistant toPG, in addition another four isolates were multiresistant to other antibiotics (No. 39) was resistant to TE, Ba, (No. 82) was resistant to TE, E, and (No. 6 and 76) were resistant toBa. In MRSA isolates, β-lactams resistant isolates were distributed in all phage lytic groups, but the most common one was III (40.4%). Also for the AMC, SXT, TE,CIP and MUP resistance isolates, it was observed that the group III was the most common.
In MSSA isolates, both GN resistance isolates exhibited typability by groups II and M.
TE resistant isolate typed by II and III. One isolate which resistant to SXT was typed by group III/V alone. Both E and Ba resistance isolates distributed in all lytic groups, but group III was the most in E resistant isolates (57.1%) whereas groups I, II and III had the similar high percentage (60.8%) in Ba resistance isolates. FD resistance isolates typed by I, II, III and V. Group I typed all FD resistant isolates. Whereas the group I (34.4%) and III (37.9%) were the most in FD resistance MRSA isolates. AK resistance MRSA isolates were typed by group III (36%) and II (32%) as a common lytic group. Ba resistant isolates typed by all groups except V lytic group and C resistant isolates typed by II and M groups. All nontypable isolates distributed among various antibiotics.
4. Discussion
S. aureus could be one of the most dexterous bacteria in exchanging useful genetic information with other bacterial species, also with phages that have genomic DNA materials. As a clear example, the GC content of Staphylococcal Chromosomal Cassette mec (SCC mec) (which have the mecA gene that responsible for resistance to methicillin) is non Staphylococcal origin suggested that it may have been acquired long ago in evolutionary terms. S. aureus could be infected by both polyvalent or/and host restricted phages so-called staphages belonging to families: Myoviridae and Siphoviridae (Francki et al., 1991). Useful transducer genetic elements or jumping genes could be transferred horizontally in S. aureus population. The originated and colonized bacteria from Hospital (RAFH) reflected the epidemiological phenotypic and genotypic determinant markers for S. aureus group in Saudi cohort. It could help for setting up an evaluated antimicrobial usage strategy.
Clinical microbiology laboratories testing for antimicrobial agents susceptibility among S. aureus is important to monitor for emerging resistance patterns (Fabiana et al., 2004). The emergence of antibiotic resistance was one of the most serious phenomena of the last 20 years and several strategies have been proposed to try to tackle it. The selective pressure resulting from the extensive use of antibiotics has lead to the acquisition and spread of antimicrobial resistance determinants (Goni et al., 2004).
In general, MRSA isolates were more resistant to the antibiotics than MSSA that may due to: (1) All MRSA strains have SCC mec which act as harbor for the different Staphylococcal genes, especially resistant encoded gene. (2) Most MRSA isolates were from inpatients which may expose to different antibiotics causing emerging the resistance or by gain the resistance from the hospital strains.
MRSA often displays a resistance to a wide variety of antimicrobial agents including non-β-lactams, which makes it difficult to treat of MRSA infections and leads to a high mortality rate in immuno compromised hosts (Lee et al., 2001).
Phenotypic and genotypic disseminating analysis were performed in this study using antimicrobial agents, international phage typing (IPT), mecA gene revealed diversity between the 119 clinical S. aureus epidemiologically unrelated isolates from one Hospital (RAFH).
4.1. Could phage alter bacterial susceptibility to antibiotics?
In the present study, it was clinically crucial to determine rapidly, whether S. aureus methicillin resistant or not. Data revealed that mechanism expressing mecA gene as an important one against methicillin in S. aureus, differed among the studied 119 clinical isolates. A 77 of them were disseminated as MSSA lacking mecA and 42 were MRSA showed phenotypic resistance to methicillin. A 39 of them harbored mecA gene except three isolates. The characteristic cards of these isolates are:
-
•
No. 58 – out p., wound, I/II/III/M/V, antibiogram (A)
-
•
No. 30 – in p., nose I/III, antibiogram (A)
-
•
No. 31 – in p., swab, nontypable, antibiogram (H)
The structural gene for methicillin resistance, mecA, encodes a novel penicillin binding protein (PBP)-2′, which has reduced affinity for β-lactam antibiotics (Hartman and Tomasz, 1984). SCCmec is found in another staphylococcus species from which is presumed to have been transferred; however the original donor of mecA to Staphylococci is unknown, as the element has not yet been identified outside this genus. Staphylococcus sciuri has an intrinsic PBP that shares 87.8% amino acid homology with PBP-2′ (Wu et al., 1996). It means that mecA gene mechanism is diverse, and the presented studied S. aureus group is heterogeneous and homogeneous population.
These strains can arise because of hyper production of β-lactamase, production of normal PBP with altered binding capacity, and/or other as yet unidentified factors (Chambers, 1997). The increase of phage group II had been linked to a frequent use of beta-lactam antibiotics (Renneberg and Rosdahl, 1992).
Phage-typing data on MRSA strains indicated high special prevalence of phage type II with a increase for phages 3A and 3C. Phage type II on MSSA considered as an epidemiologic marker with frequent strong reaction typability (79.1%) individually or mixed with other phage groups compared to group III (67.2%) and phage group I (56%). Contrarily, Blouse et al. (1979) found weak lytic reaction with group II phages. The increase of phage group II had been linked to a frequent use of β-lactams.
Many MRSA strains showed heterogeneously resistance to β-lactams (Al-Shammary, 1997; Salyers and Whitt, 1994). Blair and Carr (1961) successfully induced and isolated temperate phages from drug-resistance and proved the roll of the integrated prophage. Recently, from Saudi, Al-Digs (2004) studied another episode of S. aureus population from the same hospital (RAFH), induced and classified 8 new very virulent and two temperate staphages from highly incidence prophages (3–4) inside the individual S. aureus. Phage group III resembled the highest typability for S. aureus followed by group I that started to appear. Remarkably, phage group II did not exist mostly in that previous study.
The investigators suggested that the conflict findings could be due to a variable and complex expression of resistance among the different MRSA sub-populations or strains (de Lencastre et al., 1991). Besides, some MRSA isolates may have incomplete regulator genes (mecI and/or mecR1) and/or high genomic diversity as was recently reported.
Numbers of antibiograms for MSSA isolates were less than MRSA as revealed in Figs. 2 and 3. It was observed in MSSA that resisted only one antibiotic as different antibiograms (a, m and n) and was the most resistant antibiogram that resembled PEN (46.7%). MSSA antibiograms started to show sporadic of antimicrobial resistance for some isolates. The emergence of individual antibiotic/antibiogram resistance in MSSA isolates such as B, FD were observed in Fig. 2 as a serious phenomena. It seems to have few isolates in the beginning within MSSA population exhibited horizontally distributed resistance only to one antibiotic and continued building up this resistance to be more than one vertically in MRSA population for the same of them such as mentioned for FD, B and E in Fig. 3. Crisostomo et al. (2001) found that two of the early MSSA strains exhibiting resistance only to PG, were genetically similar to the properties of two internationally spread MRSA. Also, showed resistance only to β-lactams and another contemporary multi resistant MRSA widely spread. The pattern of multi drug resistance that characterizes the nosocomial strains may be result of the exposure to multiple antibiotics. Theses increased susceptibility patterns among CMRSA strains was consistent with other investigations (Herold et al., 1998).
Alternative drugs should be used when resistance is high (French, 2001), this was clear for the increasing resistance of MSSA to a topical Ba (29.8%) rather than MRSA (19%). In this study, the resistance rate for MSSA had increase to 9% for FD antibiotic comparing with the other studies (Turnidge and Collignon, 1999). In the Australian study found rates of 1–3%. Similar low rates similar that in Canada (0.6%) in 1995 (Tomazs and Barriault, 1995). Also in MRSA isolates, this rate was highly in present study 69%, while it was reported a decrease to 31% in previous study (Al-Digs, 2004). The reasons of this selection were due to the widely use FD in unreasonable quantities that led to the emergence of resistance rapidly. Manson and Howard (2004) found significant relationship between the use of topical FD and the isolation of MSSA resistant organism at the individual patient level and support the hypothesis that the observed increase in resistance is causally associated with the increased use of topical FD.
The emergence and global spread of MRSA may be viewed as a process of accelerated evolution (Tomasz, 1998), which took place in the contemporary clinical environment (Crisostomo et al., 2001).
Type 95, a phage type has been shown to be strong and stabile colonizer (Vintov et al., 2003). This type 95 had emerged and another phage group II had noticed to be important in the present study. In addition, the importance of this type 95 was obviously noticed with a percentage of (18%) for MSSA had lowest existence comparing to the other phages. The change in phage type pattern showed that the increase was caused by the introduction and spread of many resistant strains of type 95 and group V, and an increased occurrence e of resistant strains of phage group II. These could suggest the undergoing similar changes in the distribution of types, probably also influenced by selective pressures through the use of PG and other drugs (Vintov et al., 2003). The usage of TE for MSSA promoted the resistance strains 82 and 78 (PG, TE and E) for antibiogram g in Fig. 2. As well as another isolate in antibiogram i and k showed resistant for (PG, TE) and (PG, TE, B). These four isolates had different in phage types. Three of them showed that phage type 80 (group I) was eradicated as a marker phage for frequency resistance of PG, except isolate 78 that typed by it. The promotion also extended to have another type of phage 83A (group III) that was mentioned by Vintov et al. (2003), who proved a link of the new mentioned antibiogram and the emerged phage due to the multiple resistant. Among MSSA isolates, increased variability to antimicrobial agents. However, the persistence of one predominant clone of MSSA was shown several different strains of both MSSA and MRSA were capable of maintaining persistent colonization (Trzcinski et al., 1997). A Japanese study, conducted in 1999 and 2000, of 229 S. aureus isolates from a variety of skin infections in outpatients reported a 21% prevalence of MRSA (Nishijima and Kurokawa, 2002). When GN was used, physicians should be alert to the possibility of the emergence and potential spread of resistance (Lewis and Altemeier, 1978). In the present study two isolates of MSSA (No. 84 and 108) started to emerge a resistant to GN and PG. Also correlated to phage type 81/71 (II/M).
Sunderrajan and Kale (1984) concluded the correlation with the penicillinase production and the isolates phage groups. He noticed that multidrug-resistance and penicillinase production was higher in isolates from operation theatres. Crossley and Archer (1997), stated that the particularly virulent, penicillinase-producing strain of phage type 80/81, in which the isolates were sensitive only to phages 80 and 81, became persistently prevalent in multiple countries during the1950s. After several years, outbreaks of penicillinase-producing isolates with phage type 52/52A/80/81 also appeared. It was determined lately; the prototype 80/81 strain carried a cryptic defective prophage (designated 80′) that conferred resistance to phages 52 and 52A. All of the isolates that were sensitive to group I, II, or group V phages were sensitive to methicillin. Of the isolates that were sensitive to group III phages, 96% were methicillin resistant, and 70.5% of them were sensitive to phage 75 and 85 (Samra et al., 2001). Changing in phage type pattern showed that the increase was caused by the introduction and spread of many resistant strains of type 95 and the 94, 96-complex (Vintov et al., 2003). During the last decade, resistance to GN showed high performance through MRSA isolates 90% for MRSA and 5.7% for MSSA (Al-Shammary, 1997) compared to ours which was 69% for MRSA while it was 2.5% for MSSA.
The distribution of various phage types could be considered as an indicator for the outbreak of infection associated with a predominant phage type, this could demonstrate the spread of MRSA in Saudi hospitals. In our situation, if phage type 80/81/75 was prevalence in the population and introduced by phage group M or/V such as phage type 95 as mixed such as group or individually, that could be phage type indicator for an emergence resistance threatening.
Phage type 95 and 29 were remarkably increased in the MRSA population compared to MSSA.
Depending on a phage marker (85 or 54) which was predominant in isolates. It was clear to discriminative isolates into a major group was typed by this phage indicator. That had 42.8% prevalence of this marker.
In 64 typed MSSA isolates, was observed to lack the similarity of phage typing patterns within about 55. It means different distribution of number and type of phage for each isolate which was unrelated.
Typically, the response to particular phage was consistent for isolates representing the same strain, and thus a panel of diverse phages could be used to identify and differentiate of S. aureus. MRSA isolates reflected somehow of homogeneity concerning phage typing. The nontypable isolates were 15.1% in S. aureus may reflect a variety of circumstances, including the absence of an appropriate cell surface receptor for the phage restriction–modification systems that prevent replication of phage DNA, or the presence of an incompatible lysogenic phage. The phage type assigned to an isolate indicates those phages to which the isolate was determined to be sensitive, typable) (Crossley and Archer, 1997).
It could be concluded from the study, that we do have an accumulating multiresistant isolates especially for FD, B, E, and TE. This mainly originated from wound and nose and the phage could integrate especially groups I and II in increasing the virulent factors in MRSA. As well as studying the genetic back ground of MSSA isolates by combining the phenotypic as well as genotypic may reflect the behavior of S. aureus for acquiring the source of resistance from its origin. In addition of using each individual special isolates for the evaluation of antibiotic susceptibility.
In recent years, there has been an alarming increase in nosocomial Staphylococcal infections by strains with multiple drug resistance. At present, this situation is leading to evaluation of Staphylococcal pathogens potentially resistant to any available antibiotic.
5. Conclusion
It is important for Clinical microbiology laboratories that testing for antibiotics among S. aureus to monitor for emerging resistance patterns (EMSSA).
Phenotypic and genotypic analysis performed in this study revealed diversity in the epidemiologically between MSSA/MRSA studied isolates.
Heterogeneity inside MSSA for having virulent factors was so wide, while it is homogenized for MRSA, i.e. the variations between MSSA isolates seemed to be horizontally, while in MRSA were vertically.
The emergence of virulence strains was contributed by existence strains that had resistance to FD.
Alternative drugs should be used when resistance is high, this was clear for the increasing resistance of MSSA to Ba.
The resistance rate for FD due to the widely use of FD in unreasonable quantities that led to the emergence of resistance rapidly.
Three MSSA isolates showed fully sensitive seemed to be native wild type.
All MRSA showed fully resistant to β-lactams.
Phage type Ø95 is a new phage type, strong and stabile colonizer.
The changes in phage typing pattern showed that the increase was caused by the introduction and spread of many resistant strains.
The phage marker Ø85 or/and Ø54 which common in most isolates.
MRSA isolates reflected somehow of homogeneity concerning phage typing rather than MSSA.

References
- Al-Digs, E.K., 2004. The Role of Staphage in the Treatment of Methicillin Resistant S. aureus Infection. Ph.D. Microbiology Department, College of Science, King Saud University.
- Al-Shammary, M., 2005. Comparison between Traditional and Molecular Methods of Typing Isolates of Methicillin Resistant S. aureus. Ph.D. Microbiology Department, College of Science, King Saud University.
- Al-Shammary, M., 1997. Methicillin Resistant S. aureus (MRSA) in Riyadh. Mater thesis. Microbiology Department, College of Science, King Saud University.
- Baron E.J., Peterson L.R., Finegold S.M. ninth ed. St. Louis Missouri; Mosby: 1994. Baily and Scott’s Diagnostic Microbiology. [Google Scholar]
- Blair J.E., Carr M. Lysogenic Staphylococci. J. Bacteriol. 1961;82:984–993. doi: 10.1128/jb.82.6.984-993.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouse L.E., Brockett R.M., Steele N.P., Ward E.R. Colonization and infection of newborn infants caused by bacteriophage-group II S. aureus strains. J. Clin. Microbiol. 1979;5:604–606. doi: 10.1128/jcm.10.4.604-606.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H.F. Methicillin resistance in Staphylococci: molecular and biochemical basis and clinical implications. J. Clin. Microbiol. Rev. 1997;10(4):781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collee J.C., Duguid J.P., Fraser A.G., Marmion B.P. Staphylococcus cluster-forming gram-positive cocci. In: Macki, McCartney, editors. Practical Medical Microbiology. third ed. Churchill Living Stone; New York: 1989. pp. 303–324. [Google Scholar]
- Crisostomo, M.L., Westh, H., Tomasz, A., Chung, M., Oliveira, D.C., Delencastre, H., 2001. Evolution of Methicillin Resistance in S. aureus: Similarity of Genetic Backgrounds in Historically Early Methicillin-Susceptible and Resistant Isolates and Contemporary Epidemic Clones. Department of Clinical Microbiology Laboratory of Molecular Genetics, 98 (17), 9865–9870. [DOI] [PMC free article] [PubMed]
- Crossley K.B., Archer G.L. Churchill Livingstone; New York: 1997. The Staphylococci in Human Disease. p. 261. [Google Scholar]
- de Lencastre H., Sa-Figueiredo A.M., Urban C., Rahal J., Tomasz A. Methicillin resistance and improved methods for detection in clinical isolates of S. aureus. Antimicrob. Agents Chemother. 1991;35:632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid J.P. Staphylococcus: cluster-forming gram-positive cocci. In: Collee J.G., Duguid J.P., Fraser A.G., Marmion B.P., editors. Mackie and Mccartney, Practical Medical Microbiology. 13th ed. Churchill Livingstone; UK: 1989. pp. 303–316. [Google Scholar]
- Fabiana B.M., Darlene M., Marcus S.M., Ander C.R., Eduardo C.A. Ciprofloxacin and levofloxacin resistance among methicillin-sensitive S. aureus isolates from keratitis and conjuntivis. J. Am. Ophthalmol. 2004;137(3):453–458. doi: 10.1016/j.ajo.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Fairbrother L., Chapman M. Study the Staphylococcal affinity to fibrinogen by passive hemagglutination: a tool for the S. aureus identification. Zbl. Bacteriol. Hyg. Orig. 1940;251:171–176. [PubMed] [Google Scholar]
- Franki H., Murray P.P. Medium dependence for rapid determination of thermonuclear activity. J. Clin. Microbiol. 1986;24:482–483. doi: 10.1128/jcm.24.3.482-483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki, R.I.B., Fauquet, C.M., Knudson, D.L., Brown, F., 1991. Classification and nomenclature of viruses. Fifth Report of the International Committee on Taxonomy of Viruses. Springer-Verlag Wien, New York.
- Fey P.D., Salim B.S., Rupp M.E., Hinrichs S.H., Boxrud D.J., Davis C.C., Kreiswirth B.N., Schlievert P.M. Comparative molecular analysis of community or hospital acquired methicillin-resistant S. aureus. J. Antimicrob. Agents Chemothe. 2003;47(1):196–203. doi: 10.1128/AAC.47.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French G. Linezolid. J. Int. Clin. Prect. 2001;55:59–63. [PubMed] [Google Scholar]
- Goni P., Vergara Y., Ruiz J., Albizu I., Vila J., Gomez-Lus R. Antibiotic resistance and epidemiological typing of S. aureus strains from ovine and rabbit mastitis. J. Int. Antimicrob. Agent. 2004;23:268–272. doi: 10.1016/j.ijantimicag.2003.07.016. [DOI] [PubMed] [Google Scholar]
- Gopal Rao G., Wong J. Interaction between methicillin resistant S. aureus (MRSA) and methicillin sensitive S. aureus (MSSA) J. Hosp. Infect. 2003;55(2):116–118. doi: 10.1016/s0195-6701(03)00287-1. [DOI] [PubMed] [Google Scholar]
- Hartman B.J., Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in S. aureus. J. Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold B.C., Immergluck L.C., Maranan M.C., Lauderdale D.S., Gaskin R.E., Boyle-Varva S., Leitch C.D., Daum R.S. Community acquired methicillin-resistant S. aureus in children with no identified predisposing risk. JAMA. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- Hindler J. Antimicrobial susceptibility testing. In: Isenberg H.D., editor. Clinical Microbiology Procedures Handbook. American Society of Microbiology; Washington, DC: 1992. pp. 5.0–5.251. [Google Scholar]
- Kishan R., Voladri R., Kernodle D.S. Characterization of a chromosomal gene encoding type B β-lactamase in phage group II isolates of S. aureus. J. Antimicrob. Agents Chemother. 1998;42(12):3163–3168. doi: 10.1128/aac.42.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konerman E.W., Allen S.D., Janda W.M., Schreckenberger P.C., Winn W.C. fourth ed. J.B. Lippincott Company; Philadelphia: 1992. Color Atlas and Textbook of Diagnostic Microbiology. [Google Scholar]
- Lacey R.W., Keyworth N., Lincoln C. Staphylococci in the UK: a review. J. Antimicrob. Chemother. 1984;14:19–25. doi: 10.1093/jac/14.suppl_d.19. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Suh J.T., Lenz W., Sickim Y., Schaal K.P., Bierbaum G. Typing and antimicrobial susceptibilities of methicillin-resistant S. aureus (MRSA) strains isolated in a Hospital in Korea. J. Korean Med. Sci. 2001;16:381–385. doi: 10.3346/jkms.2001.16.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.A., Altemeier W.A. Emergence of clinical isolates S. aureus to gentamicin and correlation of resistance with bacteriophage type. J. Infect. Dis. 1978;137:3. doi: 10.1093/infdis/137.3.314. [DOI] [PubMed] [Google Scholar]
- Loessner M.J., Gaeng S., Wendliger G., Mair S.K., Scherer S. The two-component lysis system of S. aureus bacteriophage twort: a large TTG-start holing and an associated amidase endolysin. FEMS Mirobiol. Lett. 1998;162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- Loessner M.J., Wendliger G., Scherer S. Heterogeneous endolysin in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holing genes within the siphoviral lysis cassettes. Mol. Microbiol. 1995;16:1231–1241. doi: 10.1111/j.1365-2958.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- Luijendijk A.D., Van Belkum A., Verburgh H., Kluytmans J. Comparison of five tests for identification of S. aureus from clinical samples. J. Clin. Microbiol. 1996;34(9):2267–2269. doi: 10.1128/jcm.34.9.2267-2269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon B.R., SKurray R. Antimicrobial resistance of S. aureus genetic basis. J. Microbiol. Rev. 1987;51(1):88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFaddin, J.F., 1980. Biochemical Tests for Identification of Medical Bacteria, 2Ed, PP64-77 Baltimore. Williams and Wilkins, Keiichi, H.
- Manson B.W., Howard A.J. Fusidic acid resistance in community isolates of methicillin susceptible S. aureus and the use of topical fusidic acid: a retrospective case-control study. J. Int. Antimicrob. Agents. 2004;23(3):299–302. doi: 10.1016/j.ijantimicag.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Mathur M.D., Mehudiratt P.L. Characterization of methicillin resistant S. aureus strains by a set of MRSA phages. Indian Journal of Medical Research, New Delhi. 2000;111(March):77–81. [PubMed] [Google Scholar]
- Miller H. Practical aspects of preparing phage and plasmid DNA: growth, maintenance and storage of bacteria and bacteriophage. Method Enzymol. 1987;152:145. doi: 10.1016/0076-6879(87)52016-x. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S., Hashimoto H., Kono M., Morimura M. During resistance of Staphylococci. II. Joint elimination and joint transduction of the determinants of penicillinase production and resistance to macrolide antibiotics. J. Bacteriol. 1965;89:988–992. doi: 10.1128/jb.89.4.988-992.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards, 2004. Performance Standards for Antimicrobial Susceptibility Testing; Fourteenth Informational Supplement. vol. 24(1), Standard M100-S14. Natl. Comm. Clin. Lab. Stand, Wayne, Pa.
- Nishijima S., Kurokawa I. Antimicrobial resistance of S. aureus isolated from skin infections. Int. J. Antimicrob. Agents. 2002;19:241–243. doi: 10.1016/s0924-8579(01)00496-4. [DOI] [PubMed] [Google Scholar]
- Phillips I., Andews J.M., Bridson E., Cooke E.M., Holt H.A., Spencer R.C., Wise R., Wise A.J., Brown D.F.J., Greenwood D., King A., William R.J. The British Society for Antimicrobial Chemotherapy, Academic Press Ltd.; UK: 1993. A Guide to Sensitivity Testing. [Google Scholar]
- Piper J., Hadfield T., McClesky F., Evans M., Friedstrom S., Lauderdale P. Efficacies of rapid agglutination tests for identification of methicillin-resistant staphylococcal strains as S. aureus. J. Clin. Mirobiol. 1988;26(9):1907–1909. doi: 10.1128/jcm.26.9.1907-1909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renneberg J., Rosdahl V.T. Epidemiological studies of penicillin resistance in Danish S. aureus strains in three period 1977–1990. Scand. J. Infect. Dis. 1992;24:401–409. doi: 10.3109/00365549209052624. [DOI] [PubMed] [Google Scholar]
- Salyers A.A., Whitt D.D. ASM Press; Washington, DC: 1994. Bacterial Pathogenesis. [Google Scholar]
- Samra Z., Gadba R., Ofir O. Antibiotic susceptibility and phage typing of methicillin-resistant and sensitive S. aureus clinical isolates at three periods during 1991–1997. Eur. J. Clin. Microbiol. Infect. Dis. 2001;20(6):425–427. doi: 10.1007/pl00011284. [DOI] [PubMed] [Google Scholar]
- Schmitz F.J., Mackenzie C.R., Geisel R., Wangers S., Idel H., Verhoef J., Hadding V., Heinz H.P. Enterotoxin and toxic shock syndrome toxin-1 production of methicillin resistant and methicillin sensitive S. aureus strain. Eur. J. Epidemiol. 1997;13:699–708. doi: 10.1023/a:1007357206672. [DOI] [PubMed] [Google Scholar]
- Skov R.L., Williams T.J., Pallesen L., Rosdahl V.T., Espersen F. Beta-lactamase production and genetic location in S. aureus: introduction of a β-lactamase plasmid in strains of phage group II. J. Hosp. Infect. 1995;30:111–124. doi: 10.1016/0195-6701(95)90151-5. [DOI] [PubMed] [Google Scholar]
- Sunderrajan P.P., Kale W. Correlation of antibiogram, phage typing and penicillinase production of S. aureus isolated from clinical material. J. Postgraduate Med. 1984;30(1):33–37. [PubMed] [Google Scholar]
- Tomasz A. Neth. J. Med. 1998;52:219–227. doi: 10.1016/s0300-2977(98)00035-7. [DOI] [PubMed] [Google Scholar]
- Tomazs E., Barriault D. Antimicrobial activity of fusidic acid and disk diffusion susceptibility testing criteria for gram-positive cocci. J. Clin. Microbiol. 1995;33:1712–1715. doi: 10.1128/jcm.33.7.1712-1715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzcinski K., Hryniewicz W., Kluytmans J., Van Leeuwen W., Sijmons M., Dulny G., Verbrug H., Van Belkum A. Simultaneous persistence of methicillin resistant and methicillin susceptible clones of S. aureus in neonatal ward of a Warsaw Hospital. J. Hosp. Infect. 1997;36:291–303. doi: 10.1016/s0195-6701(97)90056-6. [DOI] [PubMed] [Google Scholar]
- Turnidge J., Collignon P. Resistance to fusidic acid. J. Int. Antimicrob. Agents. 1999;12(2):35–44. doi: 10.1016/s0924-8579(98)00072-7. [DOI] [PubMed] [Google Scholar]
- Vintov J., Aarestrup F.M., Zinn Ch.E., Olsen J.E. Association between phage types and antimicrobial resistance among Bovine S. aureus from 10 countries. J. Veterinary Microbiol. 2003;95:133–147. doi: 10.1016/s0378-1135(03)00156-1. [DOI] [PubMed] [Google Scholar]
- Woods G.L., Washington J.A. Antimicrobial susceptibility tests: dilution and disk diffusion methods. In: Murray P.R., Baron E.J., Pfaller M.A., Tenover F.C., Yolken R.H., editors. Manual of Clinical Microbiology. sixth ed. American Society of Microbiology Press; Washington, DC: 1995. pp. 1327–1341. [Google Scholar]
- Wu S., Piscitelli H., Lencastre De, Tomasz A. Tracking the evolutionary origin of the methicillin susceptible strain of S. sciuri. Microbiol. Drug. Resist. 1996;2:435–441. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- Young R. Bacteriophage lysis: mechanisms and regulation. Microbiol. Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Francki, R.I.B., Fauquet, C.M., Knudson, D.L., Brown, F., 1991. Classification and nomenclature of viruses. Fifth Report of the International Committee on Taxonomy of Viruses. New York.
- Tyler W.I. Catalyse test as an aid to the identification of Enterobacteriaceae. Appl. Microbiol. 1972;24:58–61. doi: 10.1128/am.24.1.58-61.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintov J., Aarestrup F.M., Zinn Ch.E., Olsen J.E. Phage types and antimicrobial resistance among Danish Bovine S. aureus isolates since the 1950s. J. Veterinary Microbiol. 2003;97:63–72. doi: 10.1016/s0378-1135(03)00186-x. [DOI] [PubMed] [Google Scholar]


