Abstract
Deep-sea subsurface sediments are the most important archives of marine biodiversity. Until now, these archives were studied mainly using the microfossil record, disregarding large amounts of DNA accumulated on the deep-sea floor. Accessing ancient DNA (aDNA) molecules preserved down-core would offer unique insights into the history of marine biodiversity, including both fossilized and non-fossilized taxa. Here, we recover aDNA of eukaryotic origin across four cores collected at abyssal depths in the South Atlantic, in up to 32.5 thousand-year-old sediment layers. Our study focuses on Foraminifera and Radiolaria, two major groups of marine microfossils also comprising diverse non-fossilized taxa. We describe their assemblages in down-core sediment layers applying both micropalaeontological and environmental DNA sequencing approaches. Short fragments of the foraminiferal and radiolarian small subunit rRNA gene recovered from sedimentary DNA extracts provide evidence that eukaryotic aDNA is preserved in deep-sea sediments encompassing the last glacial maximum. Most aDNA were assigned to non-fossilized taxa that also dominate in molecular studies of modern environments. Our study reveals the potential of aDNA to better document the evolution of past marine ecosystems and opens new horizons for the development of deep-sea palaeogenomics.
Keywords: ancient DNA, deep-sea sediments, palaeogenomics, microfossils, next-generation sequencing
1. Introduction
Deep-sea cores are commonly used to study palaeoceanographical changes based on rich microfossil record composed of agglutinated, calcareous or siliceous skeletons of Foraminifera and Radiolaria. Both groups also comprise non-fossilized taxa, such as allogromiid Foraminifera or Acantharia that have been shown to be particularly diverse and abundant according to recent metagenetic surveys [1,2]. These and other environmental DNA studies confirm that the deep-sea floor is a molecular repository for virtually all groups of organisms living in the open ocean [3,4]. Environmental DNA is mainly extracellular and its concentration is high, reaching 0.3 g m–2 [5]. Theoretically, the stable abiotic conditions at the deep-sea bottom, including low temperature, great pressure and small disturbance, enhance DNA preservation [6]. Indeed, 125 000-year-old eukaryotic ancient DNA (aDNA) sequences were successfully amplified from Mediterranean sapropels [7]. However, the preservation of aDNA in the deep sea has been demonstrated only in exceptional, anoxic settings. Here, we show that eukaryotic aDNA can also be recovered from oxic, subsurface sediments at abyssal depths. We focused on Foraminifera and Radiolaria, comparing their fossil record with aDNA sequences obtained from down-core sediment samples. Our results provide evidence for the preservation of aDNA in deep-sea sediments, and demonstrate that subsurface sampling of abyssal sediments could be envisioned for palaeogenomic studies.
2. Material and methods
Deep-sea sediment was sampled at abyssal depth using a multicorer in two stations during the RV Meteor cruise M79–1 to the South Atlantic (see the electronic supplementary material, table S1). For each station, two cores (550/552 and 600/601) showing no evident sign of bioturbation were subsampled every 10 cm. Volumetric grain size distribution and microfossil assemblages were analysed from each subsample. Sediment age was determined using accelerator mass spectrometry 14C on planktonic foraminifer tests from the 30-cm subsamples of cores 550 and 601.
Rigorous precautions were taken to avoid DNA cross-contaminations from surface sediment and to ensure DNA preservation: gloves were worn at all time; the central part of the core was subsampled with sterile spoons and flame-decontaminated spatulas; subsamples were directly frozen at −20°C after manipulation in a cold room (less than 4°C). For each layer, total DNA was extracted (blank extraction ratio 1 : 2) from approximately 0.5 g of sediment using the MoBio PowerSoil Kit in laboratories with no history of Radiolaria or Foraminifera work (DTAMB and PALGENE platforms; Lyon, France). Aliquots from controlled extracts were discarded in the event of contamination. Controlled PCR amplifications realized in dedicated hoods (blank ratio 1 : 2 to 1 : 3) and using specific primers targeting the small subunit of the ribosomal RNA gene (SSU rDNA) of radiolarians (approx. 270 bp) or foraminiferans (68–196 bp) were prepared, cloned and Sanger sequenced as in Lejzerowicz et al. [8] or using the next-generation sequencing (NGS) technology Illumina (MiSeq instrument). Additional PCR amplifications were conducted using primer sets targeting foraminiferal SSU rDNA fragments of approximately 400 and approximately 1000 bp (see the electronic supplementary material, table S2). Sanger sequences were assigned as previously (foraminiferans, [8]; radiolarians, [9]) and clustered into operational taxonomic units (OTUs) as in Lejzerowicz et al. [8] at 3 per cent divergence. Illumina sequences were treated separately and clustered to Sanger OTUs at 5 per cent divergence. For methodological details, see the electronic supplementary material. DNA sequences were deposited to Dryad (doi:10.5061/dryad.b5m0j).
3. Results
(a). Sediment type and age
The sediments are deep-sea muds composed mainly of silt and classified as clay-bearing and clayey nanofossil oozes. They contain 30–55% of very fine silt and clay fractions, and up to 4 per cent of sand, mainly of biogenic origin. In all cores, the grain size decreases with increasing depth (see the electronic supplementary material, table S3 and figures S1 and S2).
The age of sediments at 30 cm core depth was 11 870 ± 70 14C years BP (12 790–13 245 calibrated years BP) and 28 950 ± 260 14C years BP (32 050–33 230 calibrated years BP) for cores 550 and 601, respectively (see the electronic supplementary material, table S4). The calculated averaged linear sediment accumulation rates are on the order of 2.3 cm kyr−1 in core 550 and 0.9 cm kyr−1 in core 601.
(b). Microfossils
In all cores, diverse and well-preserved radiolarian and foraminiferal microfossil assemblages were encountered (see the electronic supplementary material, figures S2 and S3). A clear down-core trend of decreasing radiolarians and increasing foraminiferans is observed both quantitatively and from qualitative inspection of the glass slides with the less than 20 µm fraction (see the electronic supplementary material, tables S5 and S6 and figure S4). Regarding the planktonic Foraminifera (Globigerinida), none was found at the sediment surface except in core 600 but they appear in considerable numbers below 20 cm. Moreover, in the lower parts of cores 600 and 601, a coarser mineral material (greater than 20 µm) is dominated by disintegrated tests of planktonic Foraminifera (see the electronic supplementary material, figure S4d), whereas in the higher latitude cores 550 and 552, mineral particles are not biogenic (see the electronic supplementary material, figure S4b).
(c). Molecular data
DNA sequences were obtained from almost all down-core samples, but the DNA showed a clear pattern of degradation with depth. The size of successfully amplified fragments with diverse Foraminifera-specific primers decreased from approximately 1000 bp at the surface to approximately 350 bp in the 10 cm layer and to approximately 100 bp in lower layers (see the electronic supplementary material, figure S5).
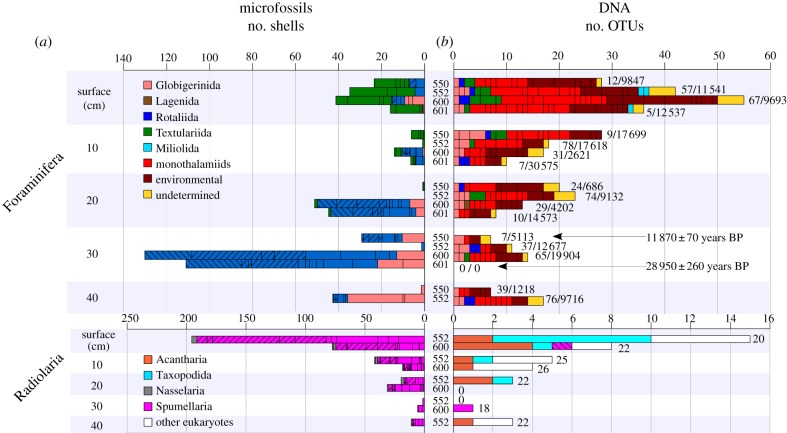
We obtained 627 and 256 925 foraminiferal sequences using cloning and NGS, of which 361 and 77 221 were from samples below 10 cm, respectively (figure 1). All sequences clustered into 169 OTUs, belonging mainly to non-fossilized monothalamous taxa (77.5%). The diversity of foraminiferal assemblages decreased with depth, as measured by diverse local diversity indices (see the electronic supplementary material, figure S6). In agreement with micropalaeontological observations, calcareous benthic species (Rotaliida) were nearly absent at the surface, but also rare in deeper layers in spite of the abundance of tests. Nevertheless, an OTU assigned to the common deep-sea species Epistominella exigua was detected in the two deepest layers of core 552, exclusively among Illumina sequences. NGS data also allowed the detection of large numbers of sequences assigned to the planktonic Globorotalia inflata/crassaformis (Globigerinida) in subsamples below 10 cm (see the electronic supplementary material, figure S7). This and other planktonic species such as Globigerinita uvula were also detected when targeting very short fragments (82 and 89 bp, respectively). In addition, only a short sequence (84 bp) of the deep-sea rotaliid Pullenia subcarinata abundant in the fossil record of core 600 was retrieved from the corresponding layer using species-specific primers.
Figure 1.
Taxonomic composition of foraminiferans and radiolarians based on (a) micropalaeontological counts and (b) DNA sequences. For each sediment layer, the cores are sorted from top to down: 550, 552, 600 and 601. Only cores 552 and 600 were investigated for Radiolaria and only cores 550 and 552 have a 40 cm layer. Black arrows indicate the radiocarbon-dated subsamples. The bars are labelled with the number of sequences generated (Sanger/NGS) and are split by assigned taxa. The proportions of dominant microfossils were reduced (hatched bars; Oridorsalis, Pullenia and Osangularinella: 50% displayed, Epistominella: 10% displayed). Among Spumellaria, the Spongodiscidae are displayed in hatched, pink bars.
In the case of planktonic radiolarians, we retrieved 21 OTUs from 109 sequences assigned to Taxopodida (10 OTUs), Acantharia (nine OTUs) and Spumellaria (two OTUs). Other major eukaryotic groups were also detected among 46 sequences (e.g. Cercozoa, Stramenopiles). The radiolarian diversity decreased from 15 OTUs at the surface to less than five OTUs below 10 cm. Both the non-fossilized Taxopodida and Acantharia were absent in micropalaeontological observations but dominated at the surface with eight and six OTUs, respectively. None of these OTUs appeared down-core where two distinct OTUs for both these groups appeared in station 552, at 10 and 20 cm for Taxopodida and at 20 and 40 cm for Acantharia. One OTU detected in the oldest radiocarbon-dated layer under study (core 600, 30 cm) was assigned to Spumellaria, the major group of Radiolaria observed in microfossils (figure 1).
4. Discussion
Authenticity of aDNA sequences is guaranteed by the use of dedicated palaeogenomic platforms and remote laboratories for DNA extractions and amplifications, stringent blank controls and precautions during sampling. We avoid common contaminants by using specific primers targeting selected groups of marine protists. Finally, the absence of high molecular weight DNA amplification in down-core samples argues against cross-contamination or leaking of DNA from surface sediments. This PCR amplification pattern corroborates the distribution of infaunal foraminiferans in deep-sea sediments (ca 10–15 cm; [10]).
The finding of sequences assigned to diverse, well-known planktonic and benthic species also found in the fossil record provides compelling evidence for aDNA preservation in deep-sea down-core sediments. However, the taxonomic composition inferred from aDNA sequences do not mirror the microfossil assemblage (figure 1). Within the Foraminifera, only Rotaliida, Textulariida and Globigerinida were observed as microfossils while the molecular repository was dominated by sequences of non-fossilized monothalamids (74.4%), mainly undetermined, environmental phylotypes (66.1%). In radiolarians, Spumellaria dominate the fossil assemblage, but Acantharea and Taxopodida were mainly sequenced. These discrepancies could be explained by the overall dominance of small-sized, non-fossilized taxa also reported in environmental DNA surveys of modern samples [1,2]. Biases related to the specificity of the PCR primers could not be excluded, as 29.6 per cent of the sequences of the radiolarian dataset were assigned to other eukaryotes. These limitations could be minimized by designing highly specific primers targeting fossil species (Pullenia) or by using NGS to increase the sequence sampling (G. inflata; [11]).
Our study shows that aDNA is a promising tool to detect both fossilized and non-fossilized taxa in oxic, subsurface abyssal sediments. However, the preservation of DNA in ancient sediments may be limited, depending on biotic as well as abiotic factors. Some radiolarians such as Acantharia produce heavy mineralized cysts that rapidly sink to the sea floor, accelerating the burial of fresh DNA in the sediment. Deep-sea monothalamids may squat other foraminiferal empty tests, avoiding predation and protecting their DNA. Among abiotic factors, the composition of the sediment substrate is of prime importance. Clayey sediments (e.g. deep-sea ‘red clays’) present the highest capacity to adsorb and hence protect DNA from nucleases [12]. However, this fraction represents less than half of the poorly sorted sediments we studied. We therefore suppose that aDNA could be amplified from much older deep-sea sediment, provided that it has a higher content of phyllosilicates.
Further investigations of aDNA in deep-seas will require larger sediment volumes to improve the detection of rare aDNA sequences. Prior to sequencing, highly specific primers could help to amplify marine cryptic species and to refine past distribution patterns of palaeoceanographical indicators (e.g. planktonic Foraminifera [13]). Alternatively, universal eukaryotic primers could better document faunal changes by providing extensive data on non-fossilized taxa. In the context of recent palaeoclimatic studies, analyses of micro-eukaryotic aDNA from deep-sea subsurface sediment could complement micropalaeontological research to understand climate change over the past hundred thousand years.
Acknowledgements
The authors thank captain and crew of R/V Meteor for help during sampling, Michał Rzeszewski for grain size analysis, Roland Marmeisse, Marc Lemaire and Jérôme Briolay for infrastructure support in Lyon and Catherine Hänni for providing access to the PALGENE platform and Fasteris SA for sequencing services. This study was supported by the Swiss National Science Foundation grant no. 31003A-140766.
References
- 1.Lecroq B, Lejzerowicz F, Bachar D, Christen R, Esling P, Baerlocher L, Østerås M, Farinelli L, Pawlowski J. 2011. Ultra-deep sequencing of foraminiferal microbarcodes unveils hidden richness of early monothalamous lineages in deep-sea sediments. Proc. Natl Acad. Sci. USA 108, 13 177–13 182 10.1073/pnas.1018426108 (doi:10.1073/pnas.1018426108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Not F, Del Campo J, Balagué V, De Vargas C, Massana R. 2009. New insights into the diversity of marine picoeukaryotes. PLoS ONE 4, e7143. 10.1371/journal.pone.0007143 (doi:10.1371/journal.pone.0007143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bik HM, Sung W, De Ley P, Baldwin JG, Sharma J, Rocha-Olivares A, Thomas WK. 2011. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Mol. Ecol. 21, 1048–1059 10.1111/j.1365-294X.2011.05297.x (doi:10.1111/j.1365-294X.2011.05297.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawlowski J, Christen R, Lecroq L, Bachar L, Shahbazkia HR, Amaral-Zettler L, Guillou L. 2011. Eukaryotic richness in the abyss: insights from pyrotag sequencing. PLoS ONE 6, e18169. 10.1371/journal.pone.0018169 (doi:10.1371/journal.pone.0018169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dell'Anno A, Danovaro R. 2005. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309, 2179. 10.1126/science.1117475 (doi:10.1126/science.1117475) [DOI] [PubMed] [Google Scholar]
- 6.Corinaldesi C, Barucca M, Luna GM, Dell'Anno A. 2011. Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol. Ecol. 20, 642–654 10.1111/j.1365-294X.2010.04958.x (doi:10.1111/j.1365-294X.2010.04958.x) [DOI] [PubMed] [Google Scholar]
- 7.Boere AC, Rijpstra WIC, de Lange GJ, Sinninghe Damsté JS, Coolen MJL. 2011. Preservation potential of ancient plankton DNA in Pleistocene marine sediments. Geobiology 9, 377–393 10.1111/j.1472-4669.2011.00290.x (doi:10.1111/j.1472-4669.2011.00290.x) [DOI] [PubMed] [Google Scholar]
- 8.Lejzerowicz F, Voltsky I, Pawlowski J. 2013. Identifying active Foraminifera in the Sea of Japan using metatranscriptomic approach. Deep-Sea Res. 86, 214–220 10.1016/j.dsr2.2012.08.008 (doi:10.1016/j.dsr2.2012.08.008) [DOI] [Google Scholar]
- 9.Decelle J, Suzuki N, Mahé F, de Vargas C, Not F. 2012. Molecular phylogeny and morphological evolution of the Acantharia (Radiolaria). Protist 163, 435–450 10.1016/j.protis.2011.10.002 (doi:10.1016/j.protis.2011.10.002) [DOI] [PubMed] [Google Scholar]
- 10.Corliss BH. 1985. Microhabitats of benthic Foraminifera within deep-sea sediments . Nature 314, 435–438 10.1038/314435a0 (doi:10.1038/314435a0) [DOI] [Google Scholar]
- 11.Andersen K, Bird KL, Rasmussen M, Haile J, Breuning-Madsen H, Kjaer KH, Orlando L, Gilbert MTP, Willerslev E. 2011. Meta-barcoding of ‘dirt’ DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 21, 1966–1979 10.1111/j.1365-294X.2011.05261.x (doi:10.1111/j.1365-294X.2011.05261.x) [DOI] [PubMed] [Google Scholar]
- 12.Cai P, Huang Q, Zhang X, Chen H. 2006. Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol. Soil Biol. Biochem. 38, 471–476 10.1016/j.soilbio.2005.05.019 (doi:10.1016/j.soilbio.2005.05.019) [DOI] [Google Scholar]
- 13.Darling KF, Kucera M, Wade CM. 2007. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc. Natl Acad. Sci. USA 104, 5002–5007 10.1073/pnas.0700520104 (doi:10.1073/pnas.0700520104) [DOI] [PMC free article] [PubMed] [Google Scholar]