Abstract
Eukaryotic translation initiation begins with assembly of a 43S preinitiation complex. First, Met-tRNAiMet, eukaryotic initiation factor 2 (eIF2) and GTP form a ternary complex (TC). The TC, eIF3, eIF1 and eIF1A cooperatively bind to the 40S subunit yielding the 43S preinitiation complex, which is ready to attach to mRNA and start scanning to the initiation codon. Scanning on structured mRNAs additionally requires DHX29, a DExH-box protein that also binds directly to the 40S subunit. Here, we present a cryo-electron microscopy structure of the mammalian DHX29-bound 43S complex at 11.6Å resolution. It reveals that eIF2 interacts with the 40S subunit via its α-subunit and supports Met-tRNAiMet in a novel P/I orientation (eP/I). The structural core of eIF3 resides on the back of the 40S subunit establishing two principal points of contact, whereas DHX29 binds around helix 16. The structure provides insights into eukaryote-specific aspects of translation, including the mechanism of action of DHX29.
Keywords: eukaryotic translation initiation, ribosome, eIF2, eIF3, DHX29, cryo-EM
INTRODUCTION
Translation initiation on most eukaryotic mRNAs occurs by the scanning mechanism, which involves the complex functional interplay between multiple initiation factors (Jackson et al., 2010). The process begins with ribosomal recruitment of Met-tRNAiMet by eIF2, which forms an eIF2/GTP/Met-tRNAiMet ternary complex (TC). In cooperation with eIF3, eIF1 and eIF1A, the TC binds to the 40S ribosomal subunit, yielding the 43S preinitiation complex. This complex initially attaches to the 5’-proximal region of mRNA after the latter has been unwound by eIF4A, eIF4B and eIF4F. eIF4F comprises eIF4E (a capbinding protein), eIF4A (a DEAD-box RNA helicase), and eIF4G (a scaffold for eIF4E and eIF4A, which also binds to eIF3). eIF4F’s position on the 43S complex has not been determined and it is therefore not known whether mRNA slots directly into the entire length of the mRNA-binding cleft of the 40S subunit or is instead threaded into it through the mRNA entrance. After attachment to the mRNA, the 43S complex scans to the initiation codon, whereupon it forms a 48S initiation complex with established codon-anticodon base-pairing. Scanning on structured mRNAs in higher eukaryotes additionally requires the DExH-box protein DHX29, which also binds directly to the 40S subunit (Pisareva et al., 2008; Parsyan et al., 2009; Abaeva et al., 2011; Yu et al., 2011; Dhote et al., 2012). Following start codon recognition, eIF5 and eIF5B promote hydrolysis of eIF2-bound GTP, release of eIFs and joining of a 60S subunit to the 48S complex. Together with eIF1A, eIF1 plays a key role in maintaining the fidelity of initiation, discriminating against codon-anticodon mismatches and preventing premature eIF5-induced hydrolysis of eIF2-bound GTP and Pi release (for review see Hinnebusch, 2011).
eIF2 consists of α, and γsubunits. Its complete structure is not known but it can be inferred from the structure of the highly homologous archaeal aIF2 (Marintchev and Wagner, 2004), which can even substitute for it in 43S complex formation and scanning (Dmitriev et al., 2011). The e/aIF2γ subunit comprises the GTP-binding domain (DI) and β-barrel domains DII and DIII, and is structurally homologous to elongation factor EF-Tu (Schmitt et al., 2002). The rigid core of aIF2 is formed by aIF2γ, the C-terminal domain D3 of aIF2α and the N-terminal α-helix of aIF2β. The core is flexibly connected to the rest of aIF2β and to aIF2α’s D1 and D2 domains (Yatime et al., 2006, 2007; Stolboushkina et al., 2008). In the crystal structure of the aIF2/Met-tRNAiMet/GDPNP ternary complex (Schmitt et al., 2012), tRNA is bound by the γ and α subunits: the acceptor stem interacts with aIF2γ-DI, aIF2γ-DII and aIF2α-D3, whereas the elbow contacts aIF2α-D1 and aIF2α-D2. However, eIF2α’s role in initiation is controversial. Although mutations in its D1 domain reduce the fidelity of initiation (Cigan et al., 1989), eIF2α is not essential in yeast, and, in contrast to its activity in archaea, makes only a minor contribution to the Met-tRNAiMet-binding activity of yeast eIF2 (e.g. Naveau et al., 2013). The structure of the 40S-bound eIF2 ternary complex has not been determined, but recent directed hydroxyl radical cleavage (HRC) experiments indicated that in yeast 48S complexes, eIF2γ’s DIII faces the 40S subunit in the vicinity of helix (h) 44 (Shin et al., 2011). However, the ribosomal positions of eIF2α and eIF2β have remained unknown.
The ~800-kDa mammalian eIF3 consist of 13 subunits (eIF3a-eIF3m), six of which (eIFs 3a, 3c, 3e, 3k, 3l, 3m) contain PCI (proteasome, COP9/signalosome, eIF3) domains that consist of helical repeats followed by a winged helix domain, and two of which (eIF3f, eIF3h) contain MPN (Mpr1 -Pad1 - N-terminal) domains that typically consist of a β-barrel surrounded by α helices and additional β strands, and function to promote assembly of multiprotein complexes (e.g. Pena et al., 2007; Enchev et al., 2010). This PCI/MPN core, which forms a stable octamer, also occurs in the 26S proteasome lid and the COP9/signalosome and in each instance adopts a similar five-lobed structure (Enchev et al., 2010; Sun et al., 2011; Beck et al., 2012). The remaining subunits of eIF3 contain RRM domains (eIF3b, eIF3d, eIF3g), WD β-propellor domains (eIF3b, eIF3i) and a putative Zn-binding motif (eIF3g) (e.g. Marintchev and Wagner, 2004), and are likely flexibly linked to the PCI/MPN scaffold (Sun et al., 2011). Negative-stain EM reconstruction of the ‘native’ 40S subunit (Srivastava et al., 1992) and modeling of the eIF3/40S interaction on the basis of the eIF3/hepatitis C virus (HCV) IRES and 40S/HCV IRES complexes (Siridechadilok et al., 2005) indicate that eIF3 resides on the 40S subunit’s solvent side. eIF3 is involved in nearly all stages of initiation, including ribosomal subunit antiassociation, promoting binding of the TC to the 40S subunit, ribosomal attachment to mRNA by interacting with the eIF4G subunit of eIF4F and scanning, and in addition was also shown to interact with eIF1, eIF1A, eIF2, eIF4B and eIF5 (for review, see Hinnebusch, 2006; Jackson et al., 2010; Valasek, 2012).
eIF1 and eIF1A are monomers, which bind cooperatively to the 40S subunit (Maag and Lorsch, 2003) and reside at the subunit interface: eIF1 to the P site between the platform and Met-tRNAiMet (Lomakin et al., 2003; Rabl et al., 2011), and eIF1A primarily to the A site (Yu et al., 2009). Their binding to yeast 40S subunits induces opening of the mRNA entry channel latch formed by h18 in the body and h34 and rpS3e in the beak (Passmore et al., 2007), which was implicated in the mechanism by which these factors ensure the fidelity of initiation and stimulate scanning. The establishment of codon–anticodon basepairing leads to tightening of the eIF1A-40S interaction and dissociation of eIF1 (Maag et al., 2005; 2006). In cooperation with eIF1A, eIF1 also promotes ribosomal recruitment of the TC.
DHX29 belongs to the SF2 DEAH/RHA helicase family of proteins and comprises a unique N-terminal region, central catalytic RecA1/RecA2 domains, and the C-terminal part, which includes winged-helix (WH), ratchet and OB domains that are characteristic of DEAH/RHA helicases (Dhote et al., 2012). DHX29 promotes unwinding of stable stems at the mRNA entrance, ensuring that mRNA is subjected to linear inspection. Although in DHX29’s absence, intact stems are not prevented from entering the mRNA-binding channel, they cannot be threaded through its exit portion (Abaeva et al., 2011). This results in incorrect positioning of mRNA upstream of the P site, which makes 48S complexes formed on AUGs downstream of intact stems susceptible to dissociation by eIF1. When 48S complexes form on AUGs preceding stable stems, the stem and the adjacent mRNA region are accommodated in the A site, and such complexes are dissociated by DHX29. DHX29 stimulates 48S complex formation most strongly when it is present in substoichiometric amounts relative to 43S complexes and can participate in multiple rounds of initiation (Pisareva et al., 2008), which suggests that DHX29 may not be present on the 43S complex before attachment to mRNA, and might join it only during scanning. Although foot-printing analysis suggested that DHX29 binds near the mRNA entrance in the vicinity of h16 (Pisareva et al., 2008), the exact ribosomal position of DHX29 and its orientation relative to the mRNA entrance have not been determined, and it is therefore not clear whether DHX29 itself unwinds mRNA before it enters the mRNA-binding cleft or acts indirectly by remodeling the ribosomal complex.
The absence of a structure for the 43S complex and the very limited nature of current structural data concerning the interactions of even individual eIFs with the 40S subunit (see above) constitute a significant impediment to further detailed characterization of the mechanism of key steps in eukaryotic initiation, in particular ribosomal attachment to mRNA and scanning. Here, we present the first structure of the mammalian DHX29-bound 43S complex at 11.6Å resolution, which provides insights into several specific aspects of eukaryotic translation.
RESULTS AND DISCUSSION
The overall structure of the DHX29-bound 43S complex
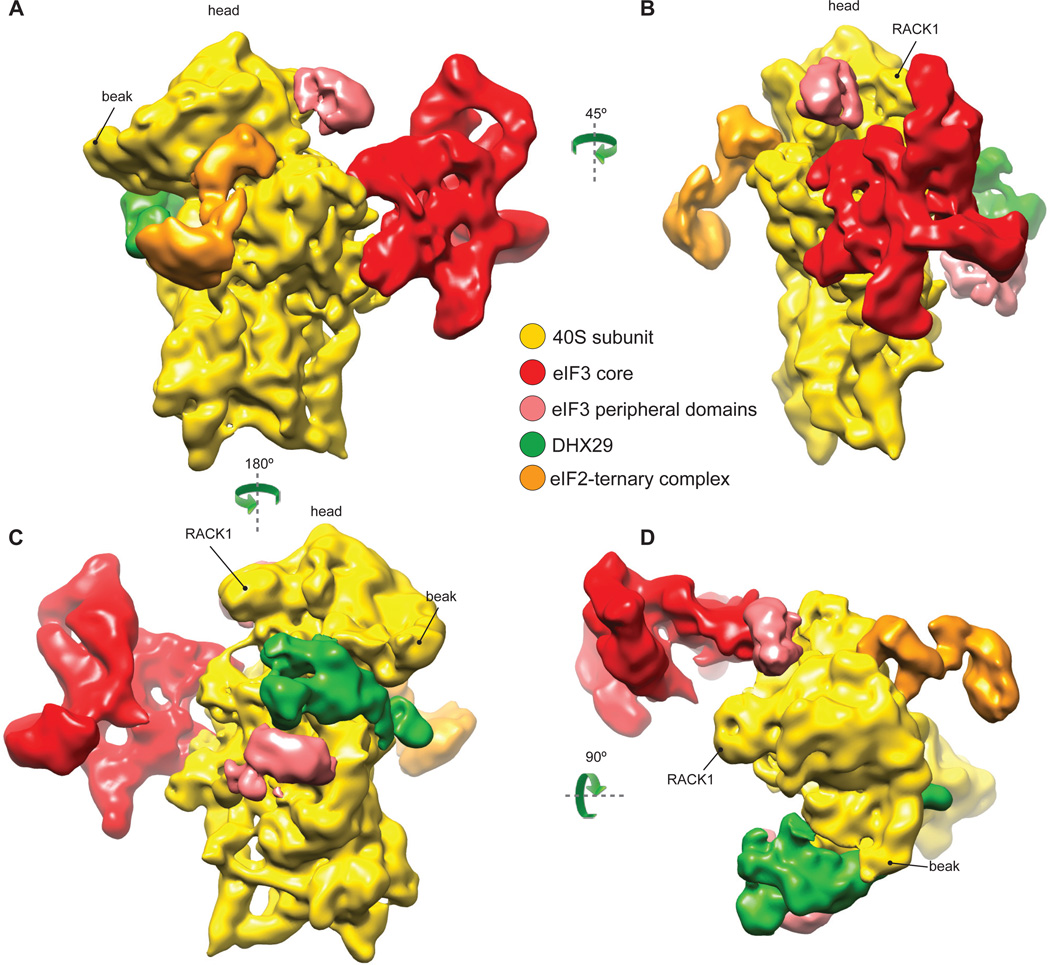
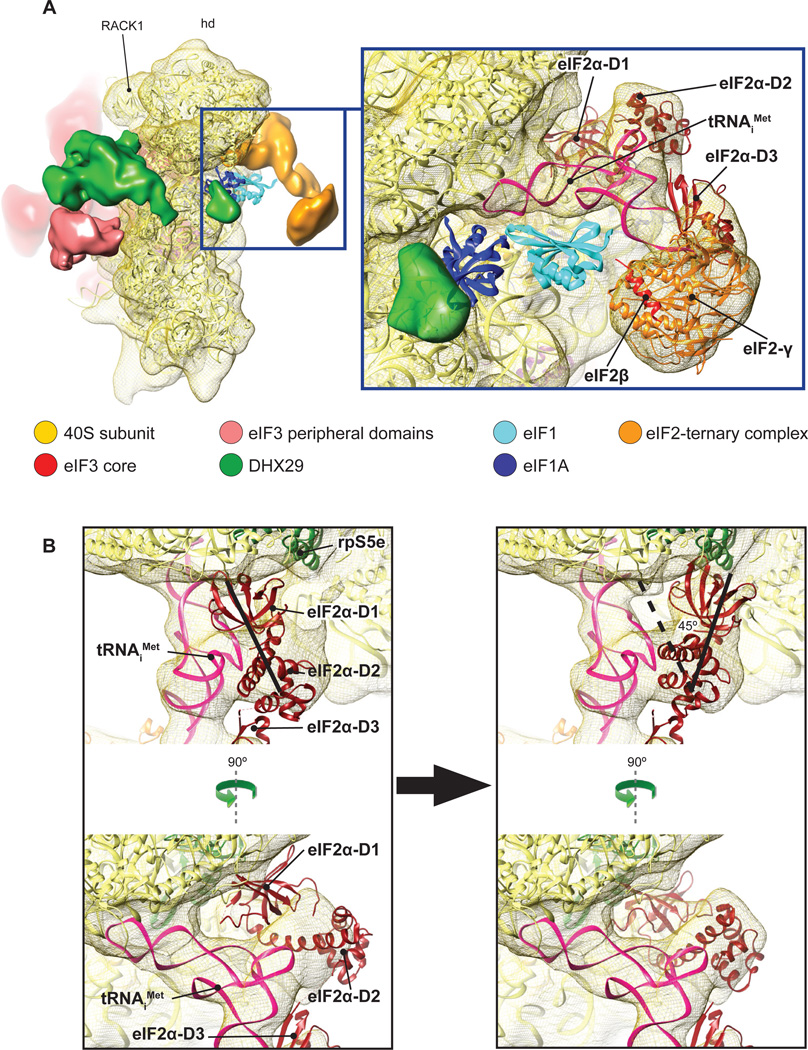
DHX29-bound 43S complexes were assembled in vitro by incubating individual native 40S subunits, eIF2 and eIF3 (lacking the j subunit) that had been purified from rabbit reticulocyte lysate (RRL), recombinant eIF1, eIF1A and DHX29, and Met-tRNAiMet in the presence of GDPNP, and were then purified by gel-filtration on a Superose 6 column. The composition of the resulting complexes was verified by SDS-PAGE followed by fluorescent SYPRO staining (eIF2, eIF3, DHX29) or western blotting (eIF1 and eIF1A) (Fig. S1 A–C). Cryo-EM images of DHX29-bound 43S complexes were collected at 120 kV, yielding highly contrasted images with clearly visible 43S particles (Fig. S2A). Images of particles were processed with SPIDER (Frank et al., 1996) and pySPIDER (Langlois and Frank, unpublished; see Materials and Methods), then classified and reconstructed with RELION (Scheres, 2012). Several classes containing different sets of components were identified (Fig. S3A). The present analysis focused on the class with the fullest complement of initiation factors, yielding an 11.6Å reconstruction (see Supplementary Information, Fig. S2B and S5 for details regarding the resolution). This reconstruction reveals three large distinct masses of density on the 40S subunit: (i) at the back of the subunit, (ii) at the subunit interface in the area of the P site, and (iii) near h16 of 18S rRNA (Fig. 1). The mass at the back (red in Fig. 1) was attributed to the structural core of eIF3, based on its reported shape and location (Srivastava et al., 1992; Siridechadilok et al., 2005). The density at the subunit interface (orange) was interpreted as the TC (Shin et al., 2011). The density around h16 and a small weakly connected mass at the subunit interface near the A site (green) were assigned to DHX29, based on foot-printing analysis (Pisareva et al., 2008). The position of DHX29 was also verified by HRC experiments, in which [Fe(II)-BABE]-derivatized wt full-length DHX29 induced cleavage in the apex loop of h16 (Fig. S1 D and E). Two additional densities, on the solvent side next to DHX29 and on the head behind RACK1 (light red in Fig. 1), were attributed to peripheral domains of eIF3. As in the cryo-EM reconstruction of the yeast 40S-eIF1-eIF1A complex (Passmore et al., 2007), no density corresponding to eIF1 or eIF1A was found at their known locations (Lomakin et al., 2003; Rabl et al., 2011; Yu et al., 2009) even though they were firmly associated with assembled DHX29-43S complexes (Fig. S1 B and C). This suggests that, consistent with their cooperativity in binding (Maag and Lorsch, 2003), eIF1 and eIF1A both dissociated during grid preparation, likely due to shear forces and/or contact with the air-water interface (Taylor and Glaser, 2008).
Fig.1. Cryo-EM structure of the DHX29-bound 43S preinitiation complex.
The map was segmented and colored variably. In all panels: 40S subunit is displayed in yellow, eIF2-tRNAiMet-GDPNP TC in orange, DHX29 in green, eIF3 structural core in red and its peripheral domains in light red. (A) The preinitiation complex viewed from the intersubunit face, (B) back, (C) solvent face and (D) top. Green arrows indicate the spatial relationship between views. See also figures S1, S2 and S3.
Location and structure of eIF3 on the 40S subunit
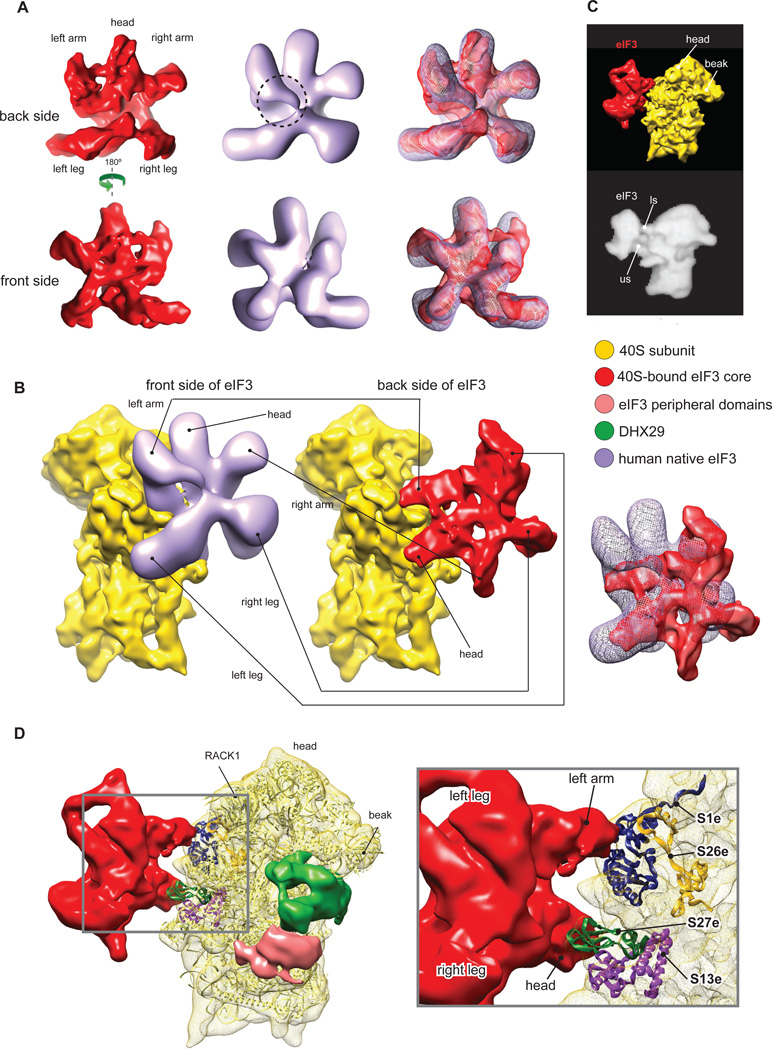
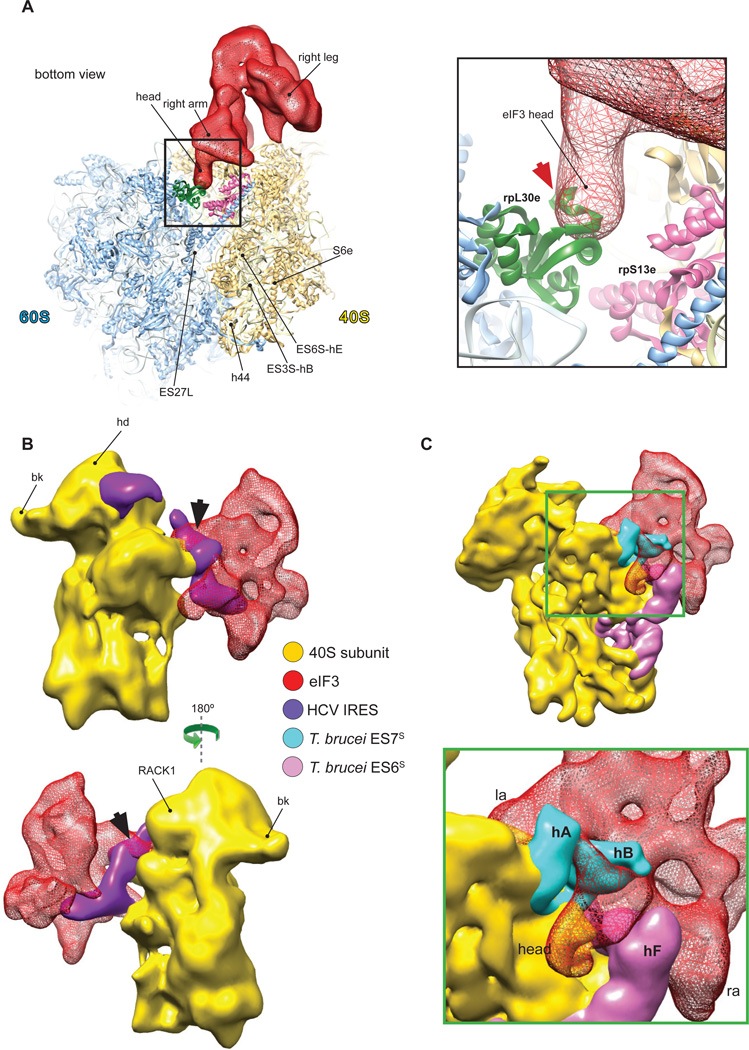
In the context of the 43S complex, the structural core of eIF3 adopts a five-lobed conformation similar to that in the 30Å cryo-EM structure of individual native eIF3 from HeLa cells (Siridechadilok et al., 2005) (Fig. 2A). One notable difference is that in our reconstruction, the left arm has a shorter density at the center of the protein on the back face (Fig. 2A, dashed circle). In contrast to eIF3 from HeLa, eIF3 from RRL contains a truncated eIF3a subunit (Fig. S1A) due to endoproteolytic cleavage, and although this does not affect the activity of eIF3 (Pisarev et al., 2007), it could account for the observed structural difference if one assumes that eIF3a forms part of the left arm. However, although the shape and the overall ribosomal location of eIF3 in our reconstruction are similar to those modeled in Siridechadilok et al. (2005), its orientation on the 40S subunit is quite different. In our reconstruction, eIF3 is rotated and flipped, compared to the previous model, so that its back rather than front side faces the 40S subunit (Fig. 2B). In the previous model (Siridechadilok et al., 2005; Fig 2B, left), eIF3 was predicted to cover a rather large area on the back of the 40S subunit. In addition, its left leg was proposed to bind below the platform near the 60S subunit interface, which would cover ribosomal protein (rp) S13e (rp15p) that participates in the formation of intersubunit bridge B4 (Ben-Shem et al., 2010). The basis for eIF3’s ribosomal anti-association activity was therefore proposed to involve the disruption of this bridge (Siridechadilok et al., 2005). Our reconstruction revealed that eIF3 contacts a relatively small area of the 40S subunit, interacting in two principal regions via its head and left arm (Fig. 2B, right). Thus, eIF3’s head protrudes into the subunit interface underneath the platform and interacts with the eukaryote-specific N-terminal domain of rpS13e and the eukaryote-specific rpS27e, whereas its left arm contacts the eukaryote-specific rpS1e and rpS26e (Fig. 2D). In fact, a similar position of eIF3 on the 40S subunit was observed in the negative-stain EM reconstruction of 40S complexes purified from rabbit reticulocytes (Srivastava et al., 1992; Fig. 2C). This position on the ribosome is also consistent with the cross-links of eIF3 with the region of mRNA upstream of the P site (positions −8 to −17) and eIF3’s potential to form an extension of the mRNA-binding channel (Pisarev et al., 2008). In its location near rpS13e, the tip of eIF3’s head would clash with rpL30e of the 60S subunit, a component of the intersubunit bridge B4 (Fig. 3A). Thus, our reconstruction also indicates that eIF3’s ribosomal anti-association activity could involve disruption of bridge B4, as suggested previously (Siridechadilok et al., 2005), but this disruption would be mediated by the head rather than the left leg of eIF3.
Fig.2. Structure and ribosomal interactions of eIF3.
The map was segmented and colored variably. In all panels: 40S subunit, DHX29, eIF3 structural core and its peripheral domains are colored as in Fig. 1. (A) Comparison of the 30Å structure of individual eIF3 from HeLa cells (light purple) (Siridechadilok et al., 2005) with the 11.6Å structure of ribosome-bound eIF3 from RRL (red; present study). A small area of potential difference between structures is highlighted by a dashed circle. (B) Side-by side comparison of eIF3-40S subunit interactions as modeled previously (left) (Siridechadilok et al., 2005) and reconstructed in the present study (middle). Right panel shows an overlay of eIF3. (C) Comparison of eIF3-40S subunit interactions determined in the present study (top) and in a 48Å negative stain reconstruction of a ‘native’ 40S subunit (bottom) (Srivastava et al., 1992), in which eIF3 connections with the 40S that are called ls and us correspond to eIF3’s left arm and head, respectively. (D) Ribosomal proteins involved in interaction of eIF3 with the 40S subunit. Left, global view showing the T. thermophila 40S subunit crystal structure (light yellow ribbon) (Rabl et al., 2011) rigid-body fitted into our segmented 40S subunit Cryo-EM map (yellow mesh). Right, close-up view on the left arm and head regions of eIF3 showing the ribosomal proteins contacting eIF3 (colored ribbons). See also figure S4.
Fig.3. Compatibility of the ribosomal position of eIF3 with the 60S subunit, HCV IRES and expansion segments ES6S and ES7S of the T. brucei40S subunit.
(A) Structural basis for the ribosomal anti-association activity of eIF3. eIF3 from our reconstruction (red mesh) was fitted on the 40S subunit in the crystal structure of yeast 80S ribosome (left) (Ben-Shem et al., 2011). Close-up view (right) showing the potential clash (red arrow) between eIF3’s head, covering rpS13e, and rpL30e of the 60S subunit. (B) Compatibility of eIF3 and the HCV IRES on the 40S subunit. eIF3 from our reconstruction (red mesh) was fitted on the 19.8Å cryo-EM structure of the 40S/HCV IRES complex (purple) (Spahn et al., 2001). A small clash between eIF3’s left arm and the pseudoknot/domain IIIf area of the IRES is indicated by the black arrow. (C) Compatibility of eIF3 with the T. brucei 40S subunit. Fitting of eIF3 from our reconstruction onto the 5Å cryo-EM structure of the T. brucei 40S subunit (Hashem et al., 2013), showing shape complementarity between eIF3 (red mesh) and the T. brucei expansion segments ES6S (hot pink) and ES7S (cyan).
To further corroborate the ribosomal location of eIF3 determined here, we assessed its compatibility with a well-characterized ribosomal ligand, the HCV IRES, and with divergent exposed elements of 40S subunits from Trypanosoma brucei. During translation initiation on HCV mRNA, its IRES must interact specifically with the 40S subunit and eIF3 (Pestova et al., 1998). Comparison of eIF3’s ribosomal position in reconstructed 43S complexes with the ribosomal position of the IRES (Spahn et al., 2001) revealed a clash involving eIF3’s left arm and the pseudoknot/IIIf domain region of the IRES (Fig. 3B). The simultaneous presence of eIF3 and the IRES on the 40S subunit would thus require a rearrangement either of the IRES or of eIF3 at that location.
The structure and position of mammalian eIF3 are complementary to peripheral unique features of the 40S subunits of kinetoplastids such as T. cruzi (Gao et al., 2005) and T. brucei (Hashem et al., 2013), which are characterized by extraordinarily large expansion segments 6S and 7S (Fig. 3C). Thus, the trypanosome-specific ES6S-hF penetrates the space between the right arm and the head of eIF3, ES7S-hB protrudes between the left arm and the head, while ES7S-hA wraps around the left arm from the other side without clashing. Importantly, T. brucei eIF3 is well conserved in comparison with the mammalian factor (Ivens et al., 2005). The structural complementarity between eIF3 and the T. brucei 40S subunit suggests a role for the large ES6S and ES7S in kinetoplastids in stabilizing the eIF3-40S subunit interaction.
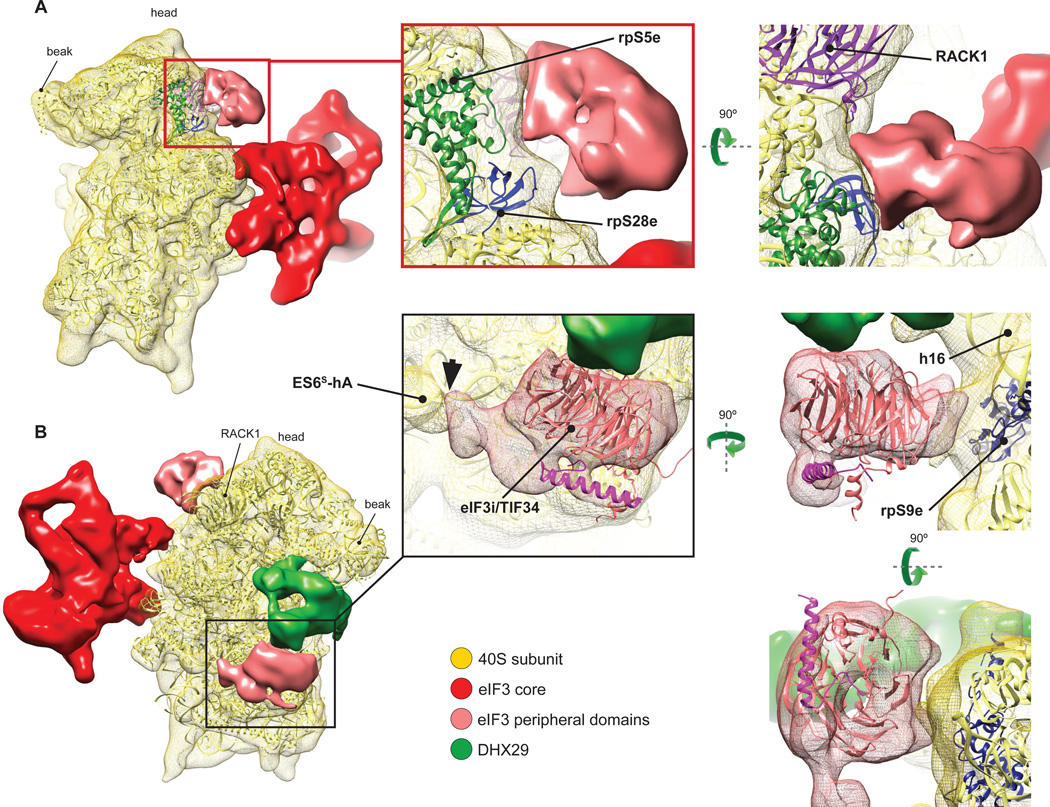
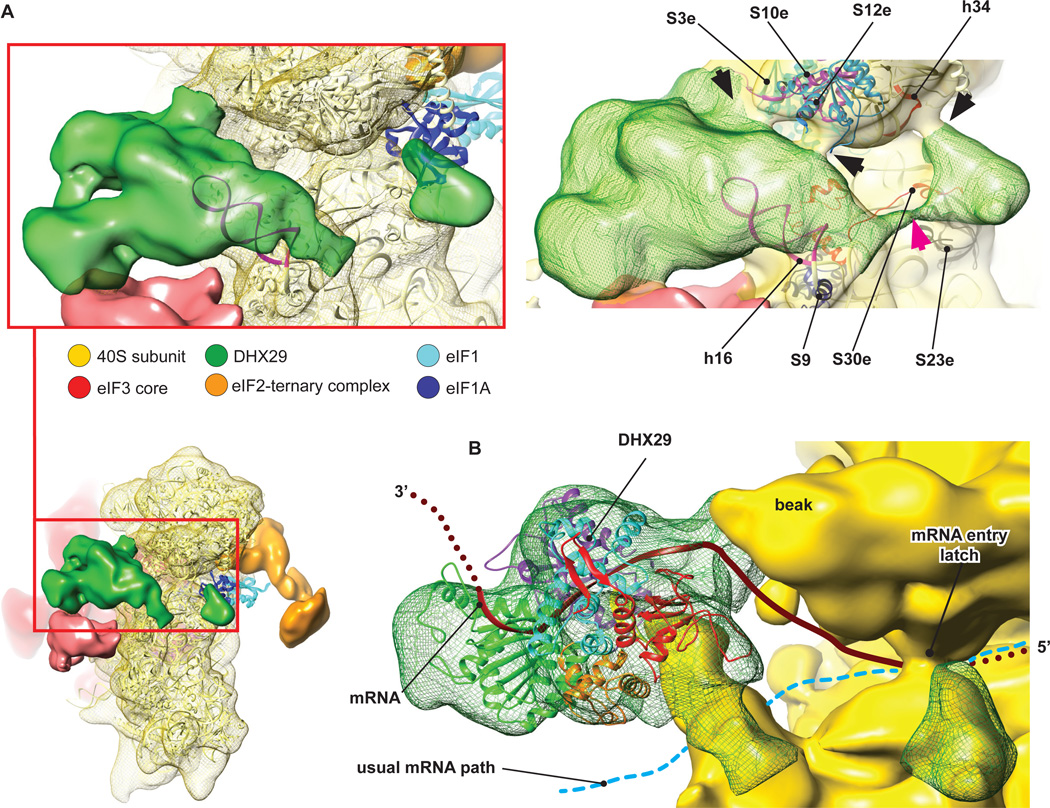
Interestingly, the ~800kD native eIF3 (Siridechadilok et al., 2005), a ~700kD in vitro reconstituted 12-subunit eIF3 (lacking j and containing truncated a* and c* subunits), and a ~400kD in vitro reconstituted PCI/MPN eIF3 octamer composed of a*, c*, and full-length e, k, l, m, f, and h subunits showed very similar structures (Sun et al., 2011). This led to the suggestion that significant parts of b, d, g and i subunits constitute flexible regions of eIF3 that were averaged out during reconstruction, but which together with the flexible regions of eIF3a and eIF3c may protrude from the structural core and envelope the 40S subunit during 43S complex formation (Sun et al., 2011). In our reconstruction, the structure of eIF3 is also very similar to that of the reconstituted PCI/MPN eIF3 octamer. Moreover, comparison of our higher-resolution structure of eIF3 with the near-atomic resolution cryo-EM structure of the 26S proteasome lid (Beck et al., 2010) (Fig. S4) confirms previously noted similarities between the lowresolution structures of eIF3, the COP9/signalosome and the 26S proteasome lid (Enchev et al., 2010). The continuous mass on the back of the 40S subunit cannot account for the entire eIF3, and two additional masses, next to DHX29 and on the head behind RACK1 (Figs. 1 and 4), were therefore attributed to peripheral domains of eIF3 that are flexibly linked to its structural core. The attribution of these additional masses to eIF3 is supported by the comparison with reconstructions from other classes lacking eIF3 core (Fig. S3A), namely classes 1 and 2 that also lack these additional masses. In contrast, the presence of DHX29 is independent of eIF3. The mass on the 40S head is asymmetrical (Fig. 4A) and displays a lower intensity than the eIF3 core, probably because of residual conformational heterogeneity as the lower local resolution and higher variance at this location suggest (Fig. S5). This mass contacts the eukaryote-specific N-terminal domain of rpS5e, rpS28e and possibly RACK1, and is also weakly connected with the left arm of eIF3 (Fig. 4A). Notably, N-terminal deletions of rpS5e reduced association of eIF3 with 40S subunits in yeast (Lumsden et al., 2010). The other mass below DHX29 is positioned on the C-terminal domain of rpS9e (rp4p) and contacts ES6S-hA and a large area on DHX29 (Fig. 4B). The large interaction surface of this doughnut-like shaped mass with DHX29 suggests that the latter may stabilize this domain of eIF3 at the observed position. Domains of eIF3 subunits that could potentially account for this mass of density include the WD β-propeller domains of eIF3b and eIF3i (TIF34 in yeast) (Marintchev and Wagner, 2004). eIF3i/TIF34 interacts with the C-terminal α-helix of eIF3b, and the crystal structure of this complex (Herrmannová et al., 2012) could be fitted to the density next to DHX29 with a cross-correlation coefficient (CCC) of ~0.90 (Fig. 4B) and cross-FSC value between the model and the map of 21Å (Fig. S2C), consistent with its lower local resolution and higher variance (Fig. S5). However, the structures of eIF3b’s WD domains are not known, and we can therefore not exclude that this mass might instead belong to eIF3b.
Fig.4. Peripheral domains of eIF3.
(A) eIF3 peripheral domain (light red) located on the head of the 40S subunit, behind RACK1 (left). Close-up views (middle, right) showing interactions of this domain with ribosomal proteins (colored ribbons). (B) eIF3 peripheral doughnut-shaped domain (light red) located next to DHX29 (green) (left). Close-up views (middle, right) showing shape and size complementarity of the cryo-EM density of this domain with the crystal structure of eIF3i/TIF34 (Herrmannová et al. 2012). In all panels, the crystal structure of the T. thermophila 40S subunit (light yellow ribbon) (Rabl et al., 2011) was rigid-body fitted into our segmented 40S cryo-EM map (yellow mesh). See also figure S5.
Although genetic and biochemical experiments have implicated several ribosomal proteins (including rps S0A, S2e, S3e and S20e) in interactions with yeast eIF3 (for review, see Valasek, 2012), we did not observe a direct interaction of the eIF3 core with these ribosomal proteins. There was also no eIF3 density located in the vicinity of eIF2 or near the ribosomal positions of eIF1 or eIF5 (Lomakin et al., 2003; Rabl et al., 2011; Luna et al., 2012), even though biochemical and genetic data indicate that eIF3 interacts with these factors (for reviews see Hinnebusch 2006; Valasek 2012). The interactions of eIF3 with ribosomal proteins and eIFs mentioned above are thus likely established by its flexible domains.
Location and conformation of the eIF2-ternary complex on the 40S subunit
The high homology of mammalian eIF2 with archaeal aIF2 justifies the use of the 5Å resolution crystal structure of the ternary complex formed by aIF2, E. coli Met-tRNAiMet and GDPNP (Schmitt et al., 2012) for rigid-body fitting into the density mass that was attributed to the TC. Met-tRNAiMet, aIF2γ, and the aIF2γ-interacting α-helix of eIF2β and aIF2α-D3 fit very well (Fig. 5A, right), with a CCC of 0.93 and a cross-FSC of 12.9Å (Fig. S2C). No clash between Met-tRNAiMet and eIF1 would occur if the latter occupied its reported position (Lomakin et al., 2003; Rabl et al., 2011). Whereas the anticodon of Met-tRNAiMet in DHX29-43S complexes is positioned essentially in the P site, its elbow is tilted towards the E site compared to the classical P/P orientation (Fig. S6A), as was previously described for the P/I and P/I1 orientations of initiator tRNA in cryo-EM structures of prokaryotic initiation complexes (Allen et al., 2005; Simonetti et al., 2008; Julián et al., 2011). Although the orientation of Met-tRNAiMet in the present complex is closer to the P/I state (in which the acceptor end is shifted in the direction of the E site; Allen et al., 2005; Simonetti et al., 2008) than to the P/I1 state (where the CCA end is oriented towards the A site; Julián et al., 2011), it also differs from the former in that the elbow is lifted up, in the direction of the head of the 40S subunit (Fig. S6B). To differentiate the state of initiator tRNA in our structure from the P/I orientation observed in prokaryotic initiation complexes, we termed it eukaryotic P/I or eP/I. Although eIF2γ faces h44, which is generally consistent with the results of HRC experiments done using yeast 48S complexes (Shin et al., 2011), it nevertheless does not contact it (or any other region of the 40S subunit) directly (Fig. 5A): the distance between the closest domain of eIF2γ (domain III) and h44 is ~34Å. Although the distance between eIF2γ and h44 would permit medium-intensity HRC in this helix from the surface of eIF2γ (Lancaster et al., 2002), which was observed in 48S complexes (Shin et al., 2011), eIF2 in our reconstruction shows conformational variability, indicating flexibility of eIF2, which could potentially allow eIF2γ to approach the 40S subunit more closely in 48S complexes after establishment of codon-anticodon interaction.
Fig.5. The ribosomal position of the eIF2-ternary complex.
(A) The location of the TC (orange) at the 40S subunit interface in the area of the P-site (left). The structures of eIF1 (cyan ribbon) and eIF1A (dark blue ribbon) were docked onto our map based on (Rabl et al., 2011) and (Battiste et al., 2000; Carter et al., 2001), respectively. Close-up views (middle, right) showing rigid body fitting of the crystal structure of the archaeal aIF2/Met-tRNAiMet/GDPNP complex (Schmitt et al., 2012) on the density assigned to the TC. (B) (Left) Poor accommodation of eIF2α D1–D2 domains after rigid-body fitting of the crystal structure of the archaeal aIF2-ternary complex (Schmitt et al., 2012) on the cryo-EM density of the DHX29-bound 43S complex. (Right) A different orientation of the aIF2αD1–D2 domains relative to Met-tRNAiMet when better fitted into density after rotation by 45 degrees and displacement further towards the 40S subunit neck by ~15 Å. The thick black lines in both panels highlight the main axis of the D1–D2 domain. See also figures S5 and S6.
Whereas the position of aIF2α-D3 domain within the aIF2 TC crystal structure (where it interacts with eIF2γ and the acceptor stem of Met-tRNAiMet) fits well into the mass attributed to TC, the position of aIF2α’s N-terminal D1–D2 domains appears to be inconsistent with it (Fig. 5B, left). In order to accommodate D1–D2, which are flexibly connected to D3 (Ito et al., 2004), they had to be rotated toward the subunit platform by ~45° and shifted by ~15Å in the direction of the neck, to be inserted very close to the E site (Fig. 5B, right). In this orientation, D2 would interact strongly with the elbow of Met-tRNAiMet, whereas D1 would contact rpS5e (rp7p) (Fig. 5B, right). This position of D1 is consistent with the reported interaction of eIF2α with the -3 position of mRNA in 48S complexes (Pisarev et al., 2006). There was no density that could be attributed to eIF2β, except for a mass relating to its eIF2γ-interacting C-terminal α-helix. However, a high variance region, which is consistent with the presence of a flexible domain at that location, can be found in the area where eIF2β binds on eIF2γ according to the crystal structure of aIF2 (Stolboushkina et al., 2008) (Fig. S5B, cyan arrows). Stabilization of the ribosomal position of eIF2β might therefore require the presence of eIF5, with which it interacts directly (Asano et al., 1999). Despite its extensive interaction with Met-tRNAiMet, eIF2α is not essential for stable association of yeast eIF2 with Met-tRNAiMet (Nika et al., 2001; Naveau et al., 2013). However, our observation that the only stable contact of eIF2 with the 40S subunit is mediated by eIF2α suggests that it should have an important role in ribosomal recruitment of the mammalian TC and stabilization of initiation complexes. In this respect it should be noted that eIF2 lacking its α subunit had substantially reduced activity in 48S complex formation, particularly in the presence of eIF1 (Pisarev et al., 2006).
Location of DHX29 and implications for the mechanism of DHX29’s action
The results of HRC experiments, in which medium-intensity cleavage from DHX29 occurred between nucleotides 530 and 533 in the apical loop of h16 (Fig. S1 D and E), allowed us to unambiguously assign the large mass of density near h16 to DHX29 (Fig. 1). The main DHX29 mass is located around the tip of h16, where it bridges h16 with the beak by interacting with rpS3e, rps10e and rpS12e (Fig. 6A, right, black arrows), and establishes extensive contact with the peripheral doughnut-shaped domain of eIF3 that we suggest might be eIF3i/TIF34 (Fig. 6A, left). This mass also extends down along the frontal face of h16, reaching the C-terminal helix of rpS9e (rp4p), and then enters the subunit interface through the flexible linker (Fig. 6A, right, magenta arrow) where it forms a small domain next to the eIF1A binding site. This small domain interacts with rpS30e, rpS23e and weakly with h34, thereby bridging the body with the beak just outside the A site, near the mRNA entry channel latch (Fig. 6A, right, black arrow). This intersubunit domain of DHX29 would not seem to be in a position to clash with eIF1A, if the latter were present in its reported location (Yu et al., 2009). However, the presence of this small domain could potentially account for the inability of DHX29 to bind efficiently to 80S ribosomes (Pisareva et al., 2008), and for DHX29’s activity in dissociating aberrant ribosomal complexes containing intact mRNA hairpins in the A site (Abaeva et al., 2011).
Fig.6. DHX29 binding on the 40S subunit.
(A) Density assigned to DHX29 (green) showing that it mainly binds around h16, with a small domain residing at the subunit interface near the A-site (left). eIF1 (cyan ribbon) and eIF1A (dark blue ribbon) were docked as in Fig. 5. (Right) DHX29 density at a lower threshold, displaying several contacts with the 40S subunit beak (highlighted by black arrows), formed by interaction with ribosomal proteins and rRNA (colored ribbons). The magenta arrow indicates the weak linker between the main DHX29 density and the small domain. (B) Atomic model of DHX29 aa 551–1302 (colored to show its different domains, as in (Dhote et al., 2012)) rigid-body fitted into the DHX29 density. The brown ribbon shows the path of mRNA through DHX29, modeled on the basis of the crystal structure of single-stranded DNA bound to the structurally related DExH-box DNA helicase Hel308 (Büttner et al., 2005), and the thick cyan dashed line shows the conventional mRNA path. See also figure S5 and S6.
Although DHX29 by itself is not a processive helicase (Pisareva et al., 2008), the possibility that association with 43S complexes might enhance DHX29’s helicase activity cannot be strictly excluded. Therefore the principal question concerning the mechanism of action of DHX29 continues to be whether it directly unwinds mRNA before it enters the mRNA-binding cleft, or whether it acts indirectly by inducing conformational changes in the 40S subunit. DHX29 is not located in a way that mRNA would have to pass through it in order to enter the mRNA-binding channel. Although the crystal structure of DHX29 has not been determined, the structure of the C-terminal two thirds of the protein, which comprise its RecA1/RecA2, WH, ratchet and OB domains, has recently been modeled (Dhote et al., 2012) on the basis of the crystal structure of the DExH-box helicase Prp43p (He et al., 2010). This model could be fitted reasonably well into a part of the density in such a way that unwound single-stranded mRNA (modeled on the basis of the crystal structure of DNA-bound helicase Hel308, which is structurally related to DHX29; Büttner et al., 2007) would be able to enter the mRNA-binding cleft with the correct 5’-3’ polarity (Fig. 6B). However, in this position of DHX29, which is the most favorable for the ‘unwinding’ mechanism, mRNA would not be threaded through the entire mRNA-binding channel and would avoid a large part of its entry portion (Fig. 6B), and it is difficult to suggest a mechanism that would direct mRNA to enter through DHX29 rather than the classical mRNA entrance.
On the other hand, the position of DHX29, bridging h16 with the beak, is favorable for induction of conformational changes in the 40S subunit, which, depending on the NTP/NDP-bound state of DHX29, could involve opening and closing of the mRNA entrance, which in turn could indirectly help 43S complexes to unwind entering mRNA stems. In addition, closure by DHX29 of the mRNA-binding cleft at two positions, in the entry portion by bridging h16 and rps S3/S10/S12, and near the A site by bridging rpS30e/rpS23e with h34, would also contribute to the processivity of scanning 43S complexes by proper fixation of mRNA in the mRNA-binding channel and by creating a physical barrier to prevent unwound mRNA stems from entering. An additional function of DHX29 might be to stabilize ribosomal association of the doughnut-like peripheral domain of eIF3, which, as suggested above, might be formed by eIF3i/TIF34. Although eIF3i is dispensable for assembly of active mammalian eIF3 in vitro (Masutani et al., 2007), TIF34 has recently been implicated in ribosomal scanning in yeast (Cuchalová et al., 2010).
In conclusion, the ribosomal position of DHX29 would be more consistent with an indirect mechanism of action. Further clarification of this mechanism can be anticipated from determination of the structure of the 43S complex associated with DHX29 in its NDP-bound form.
Conformation of the 40S ribosomal subunit in the DHX29-bound 43S complex
Binding of eIF1 and eIF1A to yeast 40S subunits induces opening of the mRNA entry channel latch formed by h18 in the body and h34 and rpS3e in the beak, and establishment of a new head-body connection on the solvent side between h16 and rpS3e (Passmore et al., 2007). However, in this 40S-eIF1-eIF1A reconstruction, eIF1 and eIF1A themselves are not seen. It is conceivable, as an explanation, that the conformation of the 40S subunit could be maintained after dissociation of eIFs 1 and 1A, because grids are prepared at +4°C, and at this temperature, the 40S subunit likely remains trapped in its conformation (at least for a short time) even after dissociation of some factors (see e.g. Fischer et al., 2010). In our reconstruction of the DHX29-bound 43S complex, the latch is fully closed (Fig. S6C). We believe that it is not the dissociation of eIF1 and eIF1A that is responsible for the closure of the latch, but rather the binding of DHX29. This view is further supported by the fact that DHX29 establishes several contacts in the region of the mRNA channel latch bridging the beak to the body just outside the A site by the intersubunit domain of DHX29 (Fig. 6A, right).
The position of eIF4F in the 43S complex is not known, and the mechanism of attachment to the 43S complex to mRNA remains obscure. If eIF4F were located at the E-site side of the 40S subunit, then mRNA could bind directly by slotting into the mRNA-binding cleft, in which case opening of the latch would be conducive for its attachment. However, subsequent scanning might require stronger fixation of mRNA in the mRNA-binding channel. By closing the latch, DHX29 would trap mRNA inside the channel, which in turn could contribute to DHX29’s activity in increasing the scanning processivity of 43S complexes.
We believe that the DHX29/GMPPNP-bound 43S complex investigated in the present study can be considered to mimic one of the states of the scanning 43S complex if the reasonable assumption is made that the presence of mRNA would not drastically alter its conformation. However, the current structure would represent only one of the states of the scanning complex, because DHX29’s activity requires NTP hydrolysis, which would cause at least localized switching of DHX29 between different conformations.
Interestingly, the multiple ribosomal contacts established by eIFs and DHX29 in DHX29-43S complexes did not cause any major conformational changes in the 40S subunit. Only small differences were noted in the conformation of ES6S compared to that in the crystal structure of the Tetrahymena thermophila 40S subunit (Rabl et al., 2011). Thus, both ES6S-hA and ES6S-hB are oriented toward the bottom, being displaced by 5–10Å relative to their position in the T. thermophila 40S subunit (Fig. S6D). Conformational changes in ES6S are consistent with its reported flexibility (Ben-Shem et al., 2011). However, the conformational change in ES6S-hA could be due to its interaction with the DHX29-bound peripheral domain of eIF3.
In conclusion, the model of the DHX29/GMPPNP-bound 43S complex (which represents one of its conformations during scanning since DHX29 has to hydrolyze NTP) provides the first structural context for the canonical eukaryotic translation initiation mechanism and will serve as a framework for further elucidation of individual steps in this complex process.
MATERIALS AND METHODS
Purification of 40S ribosomal subunits, initiation factors, DHX29 and aminoacylation of tRNAiMet
Native eIF2, eIF3, DHX29 and 40S ribosomal subunits were purified from rabbit reticulocyte lysate, and recombinant human eIF1, eIF1A and DHX29 were expressed and purified from E. coli as described (Pisarev et al., 2007; Pisareva et al., 2008; Skabkin et al., 2010). In vitro transcribed tRNAiMet (Pestova and Hellen, 2001) was aminoacylated using recombinant E. coli methionyl tRNA synthetase as described (Lomakin et al., 2006).
Assembly and purification of 43S-DHX29 preinitiation complexes
DHX29-bound 43S complexes were assembled by incubating 30 pmol 40S subunits, 90 pmol eIF2, 60 pmol eIF3, 100 pmol eIF1, 100 pmol eIF1A, 100 pmol Met-tRNAiMet and 50 pmol recombinant DHX29 in 100 µl of buffer A (20 mM Tris pH 7.5, 100 mM KAc, 2 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine) supplemented with 1 mM GDPNP for 10 min at 37°C and then purified by FPLC on a Superose 6 gel filtration column equilibrated with buffer A containing 5 mM MgCl2 and supplemented with 1 mM GDPNP prior to applying them onto grids. Composition of the complex was verified by SDS-PAGE (Fig. S1 A–C).
Electron Microscopy
3 µL of each sample was applied to holey carbon grids (carbon-coated Quantifoil 2/4 grid, Quantifoil Micro Tools GmbH) containing an additional continuous thin layer of carbon (Grassucci et al., 2007). Grids were blotted and vitrified by rapidly plunging into liquid ethane at −180°C with a Vitrobot (FEI) (Dubochet et al., 1988; Wagenknecht et al., 1988). Data acquisition was done under low-dose conditions (12 e−/Å2) on a FEI Tecnai F20 (FEI, Eindhoven) operating at 120 kV with a Gatan 914 side-entry cryo-holder. The dataset was collected with the automated data collection system Leginon (Suloway et al., 2005) at a calibrated magnification of 51,570x on a 4k × 4k Gatan Utrascan 4000 CCD camera with a physical pixel size of 15 µm, thus making the pixel size 2.245 Å on the object scale.
Image Processing
The data were preprocessed using pySPIDER (Langlois and Frank, unpublished). Arachnid is a Python-encapsulated version of SPIDER, replacing SPIDER batch files with Python. It also contains novel procedures such as Autopicker, which was used for the automated particle selection, yielding a total number of particles of ~650,000. Those particles were classified with RELION (Scheres, 2012) and a class of 29,000 particles with all factors present was isolated. This class was further refined to a resolution of 11.6Å, estimated with the gold standard Fourier Shell Correlation (FSC) = 0.143 (Fig. S2B) (Henderson et al., 2012; Scheres, 2012). To assess the quality of our reconstruction, we performed a reference-free 2D classification using RELION (Scheres, 2012) and compared the obtained class averages with projections generated from our final reconstruction (Fig. S3B).
Density maps segmentation and display
Cryo-EM reconstructions were segmented using the SEGGER module (Pintilie et al., 2010) implemented in UCSF Chimera (Pettersen et al., 2004). Segments counting less than 10,000 voxels were discarded. Segments were refined manually using the VOLUME ERASER module implemented in UCSF Chimera. Finally, the obtained segments were smoothed using a Gaussian filter in the VOLUME FILTER module also implemented in Chimera. The final maps were displayed and rendered with Chimera.
Fitting of atomic structures into EM maps
To determine the relative position of the TC to eIF1, the 40S+eIF1 crystal structure of Tetrahymena thermophila (Rabl et al., 2010) was rigid-body fitted into the 43S+DHX29 reconstruction using UCSF Chimera (Pettersen et al., 2004). The relative position of the TC to eIF1A was assessed by fitting the NMR structure of eIF1A (PDB: 1D7Q) (Battiste et al., 2000) based on the position of its prokaryotic homolog IF1 on the 30S subunit (PDB: 1HR0) (Carter et al., 2001). In all models, optimal fit was guided by the highest CCC.
An atomic model of the TC was obtained by rigid-body fitting the crystal structure of the archaeal aIF2/Met-tRNAiMet/GDPNP ternary complex (PDB: 3V11) (Schmitt et al., 2012) to the 43S-DHX29 reconstruction. Rigid-body fitting accommodated the structure well except for the D1–D2 domains of aIF2α, which displayed a different orientation relative to Met-tRNAiMet. In order to accommodate the aIF2αD1–D2 domains into the density, they had to be rotated by 45 degrees and displaced further towards the 40S by ~15 Å. It was thus possible to fit the obtained model of the aIF2/Met-tRNAiMet/GDPNP ternary complex with a CCC of ~0.93.
A homology model of DHX29 aa 551–1302 (Dhote et al., 2012) based on the crystal structure of the related DExH box protein Prp43p (He et al., 2010) was fitted into the DHX29 density by rigid body docking with a CCC of ~0.94. The crystal structure of the eIF3i/Tif34 WD domain (PDB: 3ZWL) (Herrmannová et al., 2012) was fitted into its putative density near DHX29 with a CCC = 0.90.
The proteasome lid atomic model (Beck et al. 2012) was rigid-body fitted into eIF3 core segmented density using the protocol described above.
Supplementary Material
Research Highlights.
The eIF3e core binds the 40S subunit via ribosomal proteins S1e, S13e, S26e and S27e
eIF2 binds the 40S via interaction of the eIF2a-D1 domain with ribosomal protein S5e
Initiator tRNA adopts a novel P/I orientation close to that in prokaryotic initiation
In DHX29-43S complexes, DHX29 binds h16 of 18S rRNA and the mRNA latch is closed
AKNOWLEDGEMENTS
We thank Melissa Thomas for assistance in preparation of figures, and Assen Marintchev and Trevor Sweeney for critical reading of the manuscript. This work was supported by both HHMI and NIH R01 GM29169 (to J.F.) and NIH R01 GM59660 (to T.V.P.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Cigan AM, Pabich EK, Feng L, Donahue TF. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc. Natl Acad. Sci. USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchalová L, Kouba T, Herrmannová A, Dányi I, Chiu WL, Valásek L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 2010;30:4671–4686. doi: 10.1128/MCB.00430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Stolboushkina EA, Terenin IM, Andreev DE, Garber MB, Shatsky IN. Archaeal translation initiation factor aIF2 can substitute for eukaryotic eIF2 in ribosomal scanning during mammalian 48S complex formation. J. Mol. Biol. 2011;413:106–114. doi: 10.1016/j.jmb.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Dhote V, Sweeney TR, Kim N, Hellen CU, Pestova TV. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3150–E3159. doi: 10.1073/pnas.1208014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Enchev RI, Schreiber A, Beuron F, Morris EP. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure. 2010;18:518–527. doi: 10.1016/j.str.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Gao H, Ayub MJ, Levins MJ, Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10206–10211. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryoelectron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Fu J, Buss SN, Jossinet F, Jobe A, Zhang Q, Liao HY, Grassucci RA, Bajaj C, et al. High-resolution cryo-EM structure of the Trypanosoma brucei eukaryotic ribosome. Nature. 2013;494:385–389. doi: 10.1038/nature11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmannová A, Daujotyte D, Yang JC, Cuchalová L, Gorrec F, Wagner S, Dányi I, Lukavsky PJ, Valásek LS. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic Acids Res. 2012;40:2294–2311. doi: 10.1093/nar/gkr765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Marintchev A, Wagner G. Solution structure of human initiation factor eIF2alpha reveals homology to the elongation factor eEF1B. Structure. 2004;12:1693–1704. doi: 10.1016/j.str.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Ivens IC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Milon P, Agirrezabala X, Lasso G, Gil D, Rodnina MV, Valle M. The cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biology. 2011;9:e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Kiel MC, Kaji A, Noller HF. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden T, Bentley AA, Beutler W, Ghosh A, Galkin O, Komar AA. Yeast strains with N-terminally truncated ribosomal protein S5: implications for the evolution, structure and function of the Rps5/Rps7 proteins. Nucleic Acids Res. 2010;38:1261–1272. doi: 10.1093/nar/gkp1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo HC, et al. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep. 2012;1:689–702. doi: 10.1016/j.celrep.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveau M, Lazennec-Schurdevin C, Panvert M, Dubiez E, Mechulam Y, Schmitt E. Roles of yeast eIF2α and eIF2β subunits in the binding of the initiator methionyl-tRNA. Nucleic Acids Res. 2013;41:1047–1057. doi: 10.1093/nar/gks1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nika J, Rippel S, Hannig EM. Biochemical analysis of the eIF2beta gamma complex reveals a structural function for eIF2alpha in catalyzed nucleotide exchange. J. Biol. Chem. 2001;276:1051–1056. doi: 10.1074/jbc.M007398200. [DOI] [PubMed] [Google Scholar]
- Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, Mathonnet G, Tettweiler G, Hellen CU, Pestova TV, et al. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:22217–22222. doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key-3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl R, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Blanquet S, Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Panvert M, Lazennec-Schurdevin C, Coureux PD, Perez J, Thompson A, Mechulam Y. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat. Struct. Mol. Biol. 2012;19:450–454. doi: 10.1038/nsmb.2259. [DOI] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE. Initiation factor eIF2γ promotes eIF2-GTP-Met-tRNAi(Met) ternary complex binding to the 40S ribosome. Nat. Struct. Mol. Biol. 2011;18:1227–1234. doi: 10.1038/nsmb.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT -1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Stolboushkina E, Nikonov S, Nikulin A, Bläsi U, Manstein DJ, Fedorov R, Garber M, Nikonov O. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the alpha- and beta-subunits. J. Mol. Biol. 2008;382:680–691. doi: 10.1016/j.jmb.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol. 1992;226:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–50. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audí J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3) Proc. Natl. Acad. Sci. U.S.A. 2011;108:20473–20478. doi: 10.1073/pnas.1116821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 2008;163:214–223. doi: 10.1016/j.jsb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valásek LS. 'Ribozoomin'--translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs) Curr. Protein Pept. Sci. 2012;13:305–330. doi: 10.2174/138920312801619385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. Direct localization of the tRNA--anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J. Mol. Biol. 1988;203:753–760. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structural switch of the gamma subunit in an archaeal aIF2 alpha gamma heterodimer. Structure. 2006;14:119–128. doi: 10.1016/j.str.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18445–18450. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.