Abstract
Background
Variation in epigenetic modifications, arising from either environmental exposures or internal physiological changes, can influence gene expression, and may ultimately contribute to complex diseases such as asthma and allergies. We examined the association of asthma and allergic phenotypes with DNA methylation levels of retrotransposon-derived elements.
Methods
We used data from 704 men (mean age 73) in the longitudinal Normative Aging Study to assess the relationship between asthma, allergic phenotypes and DNA methylation levels of the retrotransposon derived elements Alu and LINE-1. Retrotransposons represent a large fraction of the genome (> 30%), and are heavily methylated to prevent expression. Percent methylation of Alu and LINE-1 elements in peripheral white blood cells was quantified using PCR pyrosequencing. Data on sensitization to common allergens by skin prick testing, asthma, and methacholine responsiveness was gathered approximately 8 years prior to DNA methylation analysis.
Results
Prior allergen sensitization was associated with increased methylation of Alu (β=0.32 [sensitized vs. non-sensitized], p value 0.003), in models adjusted for pack-years, BMI, smoking, air pollutants, percent eosinophils, white blood cell count and age. Of the men interviewed, 5 % of subjects reported diagnosis of asthma. Neither Alu, nor LINE-1 methylation was associated with asthma.
Conclusions
These data suggest that increased DNA methylation of repetitive elements may be associated with allergen sensitization, but does not appear to be associated with asthma. Future work is needed to identify potential underlying mechanisms for these relationships.
Keywords: allergen sensitization, DNA methylation, Alu, and LINE-1
Introduction
Allergic diseases are known to arise from both genetic and environmental causes. Epigenetics, or the study of changes to the genome peripheral to the DNA sequence itself (i.e. histone modifications, RNAi, and DNA methylation), is key to understanding how the environment alters the function of genes. DNA methylation, which occurs through the addition of methyl groups to cytosines located within CpG dinucleotides, is a reversible modification that can alter chromosome stability and suppress gene transcription. Epigenetic pathways, mediated by environmental exposures, may change gene expression profiles, ultimately resulting in a particular phenotype [1]. A number of environmental exposures, including air pollution and dietary factors, have been shown to influence the epigenome [2].
Epigenetic changes are important in immune system development, and may also have relevance for asthma and allergic disease etiology. The differentiation of T cells, a process necessary for normal immune function, is guided by changes in DNA/histone methylation and/or histone acetylation [3]. During differentiation of naïve T cells into Th1 cells, the promoter region in the IFN-γ gene is progressively de-methylated (thereby increasing the expression of this Th1 cytokine that enhances Th1 response) [4–8]. Epigenetic mechanisms are also known to influence Th2 differentiation, and are required for expression of forkhead box P3 (Foxp3) and T-regulatory cell function [9–11]. While the opportunity for epigenetic-driven changes in gene expression provides plasticity in immune function, it also makes the system susceptible to aberrant modifications that could ultimately give rise to an allergic disease phenotype. In fact, studies on epigenetic regulation of T-cells have led to the hypothesis that environmental exposures increasing DNA methylation may also increase allergic disease risk by suppressing pathways (Th1 and T regulatory cell differentiation) that inhibit differentiation of allergy-promoting Th2 cells [12, 13]. Consistent with this hypothesis, some [6, 14] but not all [15], human and animal studies show an association between dietary supplementation with folate (a methyl donor) during pregnancy and increased risk of asthma in offspring.
While DNA methylation has been investigated in relation to a variety of diseases, little is known about its relationship to asthma and allergic disease. A large portion of DNA methylation sites are located within retrotransposons, repetitive sequences which are generally not expressed. Demethylation of these sequences could potentially interfere with proper expression of other genes[16]. LINEs (Long interspersed nuclear elements) and SINEs (short interspersed nuclear elements) are retrotransposon-derived repetitive elements present in mammalian genomes. Retrotransposons utilize an RNA intermediary to create cDNA copies of themselves for reinsertion into the genome [17]. LINE-1 elements, which are 6000bp in their full form, comprise at least 17% of the human genome. Shorter Alu sequences, a type of SINE that occupies approximately 10% of repetitive elements, are also interspersed throughout the genome [17]. In addition to serving as markers of DNA methylation in a large portion (~30%) of the genome, LINE-1 and Alu repetitive elements may also directly influence asthma and allergic disease pathogenesis. Unmethylated CpG sequences in Alu repeats may mimic bacterial DNA CpG motifs [18], thereby stimulating the innate immune system and possibly mediating allergic disease response. Transcriptionally active LINE-1 elements are potential activators of interferon-γ (IFN-γ) and other pro-inflammatory mediators through toll like receptor (TLR) pathways [19], with possible implications for allergies and asthma.
In this study, we determined whether methylation of Alu and LINE-1 (two common categories of retrotransposon-derived elements) were associated with allergen sensitization, airway hyperresponsiveness or asthma. To our knowledge, this is the first epidemiological study to examine DNA methylation of Alu and LINE-1 and allergic disease.
Materials and Methods
Normative Aging Study
This study included 704 elderly men who, as of March 1999, were active participants in the Normative Aging Study (NAS). The NAS cohort was established by the Veterans’ Administration in 1961, which enrolled men 21 to 80 years of age from the greater Boston, MA area who were free of known chronic medical conditions [20]. Since the time of enrollment, participants have had comprehensive clinical examinations at 3- to 5-year intervals. In examinations that took place between March 1999 and November 2007, participants agreed to donate at least one blood sample for DNA. A physician elicited a complete medical history and smoking history using the American Thoracic Society questionnaire [21]. The questionnaire (ATS-DLD-78) [21] was administered to each participant to obtain information on smoking habits, respiratory symptoms, and illness, an average of 8 years prior to blood collection for DNA methylation (range 5 to 17.5 years). Of the 704 men with blood samples for DNA methylation data, 669 had questionnaire data for asthma. Men who reported a doctor’s diagnosis of asthma were classified as asthmatic. All participants gave written informed consent. This study was reviewed and approved by the institutional review boards of all participating institutions.
DNA methylation of Repetitive Elements
DNA was extracted from stored frozen buffy coat of 7 mL whole blood, using QiAmp DNA blood kits (QIAGEN). DNA methylation was quantitated using bisulfite- PCR and Pyrosequencing, as described previously [22, 23]. PCR primers were designed towards consensus LINE-1 and Alu sequences and allowed for the amplification of a representative pool of repetitive elements to serve as a surrogate for diffuse genomic DNA methylation changes. The degree of methylation was expressed as 5-methylated cytosines (%5mC) as a % of the sum of methylated and unmethylated cytosines. This method quantitatively assessed the proportion of methylated sites in LINE-1 and Alu repetitive elements dispersed throughout the genome. Non-CpG cytosine residues were used as built-in controls to verify bisulfite conversion. For both Alu and LINE-1, we measured the percentage of 5mC (%5mC) at each of three CpG dinucleotide positions that are repeated over the human genome with the sequence of interest. Each sample was tested in two replicates, and their average was used in the analysis.
Skin Testing and IgE
Skin prick testing was conducted an average of 8 years prior to blood sample collection. Skin testing was performed as previously described [24] by the prick method of Pepys.[25] Subjects were tested in double-blind fashion with four common aeroallergen preparations preserved in glycerin (ragweed, 1:20; mixed trees, 1:20; mixed grasses, 1:20; and house dust, 1:10) along with a glycerin control. Wheal reactions were measured at 20 minutes as the sum of the largest wheal diameter plus the perpendicular diameter divided by two. A positive skin test was defined as subjects having a wheal diameter of ≥ 5 mm to any of the above allergens, after subtraction of the glycerin control. Of the 704 men with DNA methylation data 591 (84%) had prior assessment for allergen sensitization. Serum total IgE was determined by paper radioimmunosorbent test (Pharmacia Diagnostics, Piscataway, New Jersey, USA) an average of 8 years prior to blood sample collection for DNA methylation (range 6 to 17.5 years). The mean of two determinations on each serum specimen was used for analysis.
Methacholine challenge protocol
Methacholine challenge testing was performed on the same day as skin prick testing. Sixty percent of men with DNA methylation assessment (423/704) also had methacholine challenge testing [24]. Protocols for methacholine challenge testing were performed as previously reported [26]. A positive methacholine challenge test (AHR) was defined as a 20% decline in forced expiratory volume in 1 second (FEV1) from the post-saline value at or before one inhalation of 25 mg/ml methacholine. Men with a 20% decline in FEV1 from the postsaline value at or before four inhalations of 5 mg/ml methacholine (equivalent to a cumulative dose of 8.58 μmol methacholine)were classified as responders.
Statistical Analysis
We used multiple linear regression models to determine the association between prior allergen sensitization and markers of DNA methylation of Alu and LINE-1. Distributions of Alu and LINE-1 were approximately normal, so these outcome variables were untransformed in the analyses. For models of allergen sensitization and DNA methylation, we used atopy (sensitization to any allergen), and specific allergen sensitization as predictors. Both univariate and multiple regression models for specific allergen sensitization were constructed. We also considered asthma and methacholine responsiveness as predictors of DNA methylation. Models were adjusted for BMI, age at blood sampling for DNA methylation analysis, pack-years of smoking, current smoking, air pollutants (7-day moving averages of PM2.5 and black carbon) percent eosinophils in peripheral blood cell count, and total white blood cell count. Air pollutant exposures at the time of blood sample collection were assessed using a stationary monitoring site 1 km from the examination site. We also examined dietary folate (estimated from a food frequency questionnaire) as a predictor of DNA methylation, and as a potential confounder in multiple regression models.
Results
The characteristics of the study population are shown in Table 1. The study subjects were older male individuals between the ages of 55.3 and 100.9 (mean age 72.7, SD =6.7). Mean blood DNA methylation levels, expressed as % 5mC (percentage of methylated cytosines) were 26.4 (SD =1.1, range= 22.8 to 32.2) for Alu, and 76.8 (SD=1.8, range = 70.1 to 84.6) for LINE-1 methylation.
Table 1.
Characteristics of NAS population at time of DNA methylation Analysis
| Age, years (n=704) | ||
| Mean, (SD) | 72.7 (6.7) | |
|
| ||
| BMI, kg/m2 (n=702) | ||
| Mean, (SD) | 28.5 (4.1) | |
|
| ||
| Smoking, n (%) | ||
| Never | 210 (30%) | |
| Quit | 463 (66%) | |
| Occasional | 2 (0.3%) | |
| Regular | 25 (4%) | 4 missing (0.6%) |
|
| ||
| Smoking, Pack-years (n=665) | ||
| (Median, range) | 14.0 (0 to 145.5) | |
|
| ||
| DNA Methylation, % 5mC | ||
|
(Mean, SD) Alu (n=689) |
26.4 (1.1) | |
| LINE-1 (n=681) | 76.8 (1.8) | |
Prior Allergen Sensitization and DNA methylation of Repetitive Elements
Twenty percent of the men tested for allergen sensitization were sensitized to at least one allergen (Table 2). In men sensitized to at least one allergen, mean DNA methylation levels were 26.6 (SD=1.2, range=23.6 to 30.7) for Alu and 76.9 (SD=1.8, range 71.1 to 81.0) for LINE-1; for unsensitized men mean levels were 26.2 (SD=0.96, range 22.8 to 30.1) for Alu and 76.8 (SD=1.8, range 70.1 to 84.6) for LINE-1 methylation. In multiple regression models, prior sensitization to at least one allergen was associated with a 0.31 increase in % 5mC methylation of Alu (p=0.004), but showed no association (p=0.67) with LINE-1 methylation (Table 3). Adjustment for potential confounders did not alter this effect. Dietary folate consumption was not associated with methylation of Alu or LINE-1, and did not confound the relationship between allergen sensitization and DNA methylation (data not shown). Adjustment for air pollutants (levels of PM2.5 and black carbon), did not change the association between allergen sensitization and DNA methylation. Alu methylation increased with each additional positive skin prick test (β=0.21, p<0.05 in adjusted models, Figure 1). Total IgE levels were not associated with methylation of either Alu or LINE-1 elements (p >0.20). Percent blood eosinophils were elevated in sensitized vs. non-sensitized individuals (4.0% vs. 3.4%, respectively; p value for t test= 0.03), but were not associated with DNA methylation. Of the men who tested positive for allergen sensitization, 6% were sensitized to tree allergens, 12% to grass, 9% to ragweed, and 2.5% to house dust extract. In univariate models specific allergies to tree and grass predicted increased methylation of Alu, but not LINE-1 (Table 4). However, in multiple regression models including a panel of specific allergies only sensitization to tree allergens was significantly associated with increased methylation of Alu. Specific allergen sensitization was not associated with LINE-1 methylation in multiple regression models (p=0.67). Percentage of methacholine responders was also similar in sensitized vs. non-sensitized subjects (22% vs. 25%; p value= 0.5).
Table 2.
Respiratory and Allergic Disease Outcomes in NAS cohort
| IgE, IU/ml (n=469) | 31.6 (4.2) |
| Geometric Mean, (SD) | |
|
| |
| Atopy (any SPT +, > 5mm) n (%) | 121 (20%) |
|
| |
| MeCH response, 20% FEV1 (n=423) | |
| n, (%) | |
| No | 323 (76) |
| Yes | 100 (24) |
|
| |
| Asthma, n (%) | |
| No | 634 (90%) |
| Yes | 35 (5%) |
Table 3.
Allergy as a predictor of Global DNA methylation
| Alu | LINE-1 | |||
|---|---|---|---|---|
| N=553 | N=552 | |||
| Model 1*: | β | P value | β | P value |
|
| ||||
| Prior positive Allergic Sensitization (SPT+) | 0.32 | 0.003 | 0.09 | 0.63 |
| Eosinophils (% of total WBC count) | 0.007 | 0.70 | 0.05 | 0.12 |
|
| ||||
| N=437 | N=435 | |||
| Model 2* | β | P value | β | P value |
|
| ||||
| Log 10 IgE (IU/ml) | 0.09 | 0.26 | 0.09 | 0.54 |
| Eosinophils (% of total WBC count) | 0.006 | 0.80 | 0.01 | 0.72 |
Model adjusted for age, BMI, pack-years smoking, current smoking, total white blood cell count (cells/mm3), and air pollutants (7-day moving average of PM2.5 and black carbon levels)
Figure 1. Alu % Methylation with Increasing Positive Skin Prick Tests.
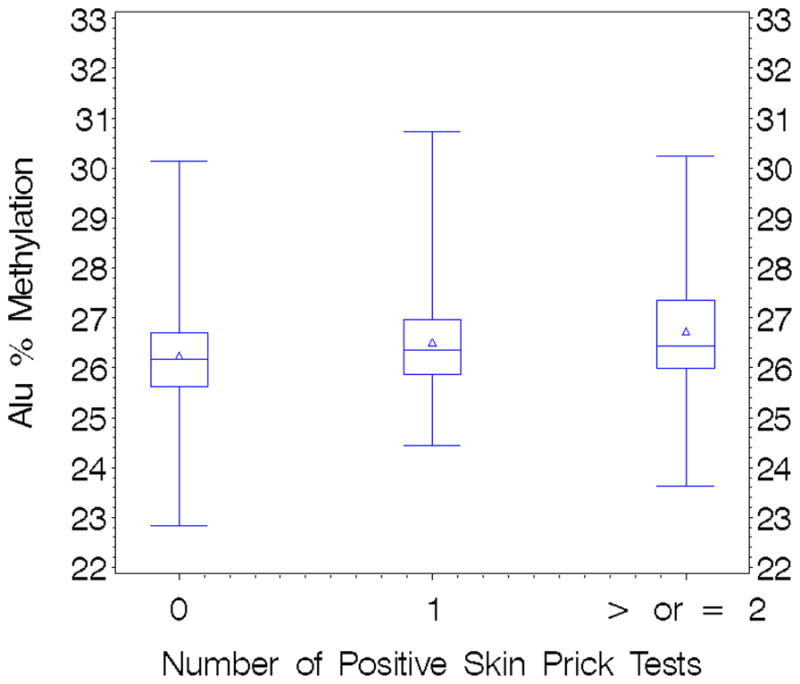
Whiskers = range; Boxes = quartiles; Triangle = mean
Table 4.
Specific Allergic Sensitization and global DNA methylation
| Univariate Models | Multiple Regression Models* | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Positive SPT for Allergic Sensitization | Alu | LINE-1 | Alu | LINE-1 | |||||
|
| |||||||||
| N (%) | β | P value | β | P value | β | P value | β | P value | |
|
|
|||||||||
| Tree | 37 (6%) | 0.48 | 0.007 | 0.35 | 0.26 | 0.38 | 0.05 | 0.43 | 0.21 |
|
|
|||||||||
| Grass | 68 (12%) | 0.24 | 0.07 | 0.08 | 0.72 | 0.19 | 0.18 | 0.03 | 0.89 |
|
|
|||||||||
| House Dust | 15 (2.5%) | 0.44 | 0.11 | 0.29 | 0.54 | 0.49 | 0.09 | −0.07 | 0.88 |
|
|
|||||||||
| Ragweed | 50 (8.5%) | 0.22 | 0.15 | −0.03 | 0.92 | −0.12 | 0.49 | −0.09 | 0.76 |
Model adjusted for age, BMI, pack-years smoking, current smoking, % eosinophils, total white blood cell count (cells/mm3), and air pollutants (7-day moving average of PM2.5 and black carbon levels)
Methacholine Responsiveness and DNA methylation of Repetitive Elements
One quarter of men tested for bronchial hyperresponsiveness showed ≥ 20% drop in FEV1 (Table 2). Methacholine responsiveness was not associated with methylation of Alu nor LINE-1 (Table 5). Similarly, methacholine responsiveness expressed as a continuous variable (log 10 slope of response) showed no significant relationship with DNA methylation [data not shown].
Table 5.
Respiratory outcomes and methylation of repetitive elements
| Alu | LINE-1 | |||
|---|---|---|---|---|
| N=609 | N=605 | |||
| Model 1*: | β | P value | β | P value |
|
| ||||
| Self Reported Asthma Diagnosis | −0.18 | 0.37 | 0.09 | 0.79 |
|
| ||||
| N=395 | N=391 | |||
| Model 2* | β | P value | β | P value |
|
| ||||
| Methacholine Responsiveness | 0.07 | 0.56 | 0.20 | 0.37 |
Models adjusted for bmi, age, pack-years, current smoking, percent eosinophils, white blood cell count (cells/mm3), and air pollutants (7-day moving average of PM2.5 and black carbon levels)
Asthma and DNA methylation of Repetitive Elements
Five percent (n=35) of men with DNA methylation data reported a doctor’s diagnosis of asthma. Thirty nine percent of self-reported asthmatics (12 out of 31 with skin prick testing) tested positive for allergen sensitization. Asthma was not associated with Alu and LINE-1 methylation levels in adjusted models (Table 5).
Discussion
Our study shows that prior allergen sensitization is associated with increased DNA methylation of the repetitive element Alu in a cohort of elderly men. No associations with allergic disease were observed for LINE-1. Hypermethylation of DNA from sensitized individuals suggests a potential underlying epigenetic mechanism for altered immune response in allergic disease. Although a considerable amount of epigenetic programming occurs during fetal development [27], the association between increased Alu methylation and allergen sensitization in this older population suggests that epigenetic changes in later life may also be relevant for allergy. Epigenetic modifications in adulthood may occur as a result of the aging process, or due to environmental exposures. In this cohort, aging [28], lead exposure [29], air pollution [30] and smoking [31] have all been associated with a decrease in methylation of repetitive elements in DNA. In contrast, dietary consumption of methyl donors may increase DNA methylation levels, potentially leading to an increased risk of allergic disease. Human and animal studies of prenatal exposures have shown a link between consumption of high levels of folate during pregnancy and increased risk of allergies and asthma in offspring [6, 14]. Folate consumption was not associated with DNA methylation in this cohort. However, data on folate supplementation and DNA methylation in humans is mixed [32–34], and the relationship between methyl donor consumption, allergies, and asthma in adults requires further study.
While prior allergen sensitization was associated with increased Alu methylation in this cohort, we did not find similar effects for total IgE levels or methacholine responsiveness. Total IgE as a marker of allergen sensitization is much less specific than skin prick testing, which could explain why we did not observe an effect for this predictor. Methacholine response showed a minimal overlap with sensitization, with only 20% of responders demonstrating sensitization to at least one allergen. This limited overlap may explain why a concomitant association was not detected for methacholine responsiveness and Alu methylation. An alternative explanation may relate to statistical power. Our sample size was larger for subjects with skin prick test data (n=605), as compared to the number subjects with IgE measurement (n=469) or methacholine responsiveness (n=423).
Since epigenetic patterns may be influenced by cell populations, we adjusted for percent eosinophils, which were elevated in subjects with allergen sensitization. The relationship between prior allergen sensitization and DNA methylation remained unchanged after adjustment for % eosinophils. While sensitization to any allergen was significantly associated with increased methylation of Alu, multiple regression models examining individual specific allergies showed tree sensitization as the strongest predictor.
Methylation of repetitive DNA sequences like Alu and LINE-1 may serve as a broad indicator of global DNA methylation (with potential implications for groups of specific genes involved in allergy, like those in T-cell differentiation and adaptive immunity), or the repetitive elements themselves may alter immune function in a way that influences allergy. While the majority of Alu copies are hypermethylated and therefore inactive, hypomethylation (as a result of vitamin B deficiency, treatment with medications such as 5-azacytidine, procainamide and hydralazine, or toxin exposure), results in transcription of Alu elements [18]. Transcripts of this transposon derived DNA are perceived as ‘foreign’ to the immune system, and may have immune stimulatory capabilities. Additionally, the unmethylated CpG rich Alu DNA fragments could potentially mimic the effects of bacterial CpG rich motifs, which are known to prime Th1 response, suppress IgE production, and initiate the release of the cytokines IL-12 and IFN-γ [35].
While this study showed a clear association between allergen sensitization and increased DNA methylation, there were some study limitations. The allergen sensitization data in the cohort was collected several years prior to DNA methylation data, therefore we were not able to assess the association between current allergen sensitization and DNA methylation. While sensitization to tree allergen seemed to drive the relationship between current allergen sensitization and methylation, the number of subjects within this subgroup was relatively small. Also, our analysis was limited to older, predominantly white men. Lastly, although methylation of repetitive elements may represent epigenetic modifications in a large portion of the genome, the significance of these epigenetic markers can be difficult to interpret.
In summary, this study suggests that increased methylation of repetitive elements may be associated with allergen sensitization, but not asthma, in this cohort. Future work is needed to identify potential underlying mechanisms for these relationships.
Acknowledgments
Funding/Support: Funded by NIH grants AG027214, ES015172, 014663, HL007427, HL089438, ES015172-01, ES00002, and Doris Duke Clinical Scientist Development Award.
We would like to thank the Normative Aging Study subjects for their participation. The Normative Aging Study is supported by the Cooperative Studies Program/ERIC of the U.S. Department of Veterans Affairs, and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). Dr. Sparrow is the recipient of a Research Career Scientist award from the VA Clinical Science Research & Development Service.
Footnotes
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 2.Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115:1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janson PC, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Biochim Biophys Acta. 2009;1790:906–919. doi: 10.1016/j.bbagen.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.White GP, Hollams EM, Yerkovich ST, Bosco A, Holt BJ, Bassami MR, Kusel M, Sly PD, Holt PG. CpG methylation patterns in the IFNgamma promoter in naive T cells: variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol. 2006;17:557–564. doi: 10.1111/j.1399-3038.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 5.White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO- T cells. J Immunol. 2002;168:2820–2827. doi: 10.4049/jimmunol.168.6.2820. [DOI] [PubMed] [Google Scholar]
- 6.Whitrow MJ, Moore VM, Rumbold AR, Davies MJ. Effect of supplemental folic acid in pregnancy on childhood asthma: a prospective birth cohort study. Am J Epidemiol. 2009;170:1486–1493. doi: 10.1093/aje/kwp315. [DOI] [PubMed] [Google Scholar]
- 7.Winders BR, Schwartz RH, Bruniquel D. A distinct region of the murine IFN-gamma promoter is hypomethylated from early T cell development through mature naive and Th1 cell differentiation, but is hypermethylated in Th2 cells. J Immunol. 2004;173:7377–7384. doi: 10.4049/jimmunol.173.12.7377. [DOI] [PubMed] [Google Scholar]
- 8.Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL. Effect of promoter methylation on the regulation of IFN-gamma gene during in vitro differentiation of human peripheral blood T cells into a Th2 population. J Immunol. 2003;171:2510–2516. doi: 10.4049/jimmunol.171.5.2510. [DOI] [PubMed] [Google Scholar]
- 9.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 10.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 12.Bousquet J, Jacot W, Yssel H, Vignola AM, Humbert M. Epigenetic inheritance of fetal genes in allergic asthma. Allergy. 2004;59:138–147. doi: 10.1046/j.1398-9995.2003.00359.x. [DOI] [PubMed] [Google Scholar]
- 13.Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. 2008;177:567–573. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, Bailey N, Potts EN, Whitehead G, Brass DM, Schwartz DA. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–3469. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Magdelijns FJ, Mommers M, Penders J, Smits L, Thijs C. Folic Acid use in pregnancy and the development of atopy, asthma, and lung function in childhood. Pediatrics. 2011;128:e135–44. doi: 10.1542/peds.2010-1690. [DOI] [PubMed] [Google Scholar]
- 16.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest AR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 17.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 18.Goldberg B, Urnovitz HB, Stricker RB. Beyond danger: unmethylated CpG dinucleotides and the immunopathogenesis of disease. Immunol Lett. 2000;73:13–18. doi: 10.1016/s0165-2478(00)00191-7. [DOI] [PubMed] [Google Scholar]
- 19.Crow MK. Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity. 2010;43:7–16. doi: 10.3109/08916930903374865. [DOI] [PubMed] [Google Scholar]
- 20.Park SK, O’Neill MS, Vokonas PS, Sparrow D, Schwartz J. Effects of air pollution on heart rate variability: the VA normative aging study. Environ Health Perspect. 2005;113:304–309. doi: 10.1289/ehp.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris BG. Epidemiology Standardization Project (American Thoracic Society) Am Rev Respir Dis. 1978;118:1–120. [PubMed] [Google Scholar]
- 22.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 23.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sparrow D, O’Connor G, Colton T, Barry CL, Weiss ST. The relationship of nonspecific bronchial responsiveness to the occurrence of respiratory symptoms and decreased levels of pulmonary function. The Normative Aging Study. Am Rev Respir Dis. 1987;135:1255–1260. doi: 10.1164/arrd.1987.135.6.1255. [DOI] [PubMed] [Google Scholar]
- 25.Pepys J. Skin tests. Br J Hosp Med. 1984;32:120, 122, 124. [PubMed] [Google Scholar]
- 26.Chatham M, Bleecker ER, Norman P, Smith PL, Mason P. A screening test for airways reactivity. An abbreviated methacholine inhalation challenge. Chest. 1982;82:15–18. doi: 10.1378/chest.82.1.15. [DOI] [PubMed] [Google Scholar]
- 27.Szyf M. The early life environment and the epigenome. Biochim Biophys Acta. 2009;1790:878–885. doi: 10.1016/j.bbagen.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, Pesatori AC, Bonzini M, Apostoli P, Costa G, Bertazzi PA, Chow WH, Schwartz J, Baccarelli A. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2010 doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pufulete M, Al-Ghnaniem R, Khushal A, Appleby P, Harris N, Gout S, Emery PW, Sanders TA. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54:648–653. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiepers OJ, van Boxtel MP, de Groot RH, Jolles J, Kok FJ, Verhoef P, Durga J. DNA methylation and cognitive functioning in healthy older adults. Br J Nutr. 2011:1–5. doi: 10.1017/S0007114511003576. [DOI] [PubMed] [Google Scholar]
- 34.Basten GP, Duthie SJ, Pirie L, Vaughan N, Hill MH, Powers HJ. Sensitivity of markers of DNA stability and DNA repair activity to folate supplementation in healthy volunteers. Br J Cancer. 2006;94:1942–1947. doi: 10.1038/sj.bjc.6603197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaung HC. CpG oligodeoxynucleotides as DNA adjuvants in vertebrates and their applications in immunotherapy. Int Immunopharmacol. 2006;6:1586–1596. doi: 10.1016/j.intimp.2006.06.001. [DOI] [PubMed] [Google Scholar]


