Abstract
A new class of glycan-reactive HIV-neutralizing antibodies, including PG9 and PG16, has been recently discovered that appear to recognize novel glycopeptide epitopes on HIV-1 gp120. However, further characterization and reconstitution of the precise neutralizing epitopes are complicated by the heterogeneity of glycosylation. We report here the design, synthesis, and antigenic evaluation of novel cyclic V1V2 glycopeptides carrying defined N-linked glycans at the conserved glycosylation sites (N160 and N156/N173) derived from gp120 of two HIV-1 isolates. Antibody binding studies confirmed the necessity of a Man5GlcNAc2 glycan at N160 for recognition by PG9 and PG16, and further revealed a critical role of a sialylated N-glycan at the secondary site (N156/N173) in the context of glycopeptides for antibody binding. In addition to defining the glycan specificities of PG9 and PG16, the identified synthetic glycopeptides provide a valuable template for HIV-1 vaccine design.
Characterization of the epitopes for broadly neutralizing antibodies (bNAbs) is a critical step in HIV vaccine design1–4. Extensive N-linked glycosylation of the HIV-1 envelope glycoprotein gp120 constitutes a strong defense mechanism for the virus to evade host immune surveillance because of the generally weak immunogenicity of the viral N-glycans5,6. For a long time, 2G12 was the only known carbohydrate-reactive broadly neutralizing antibody (bNAb), which had evolved a special domain-swapped structure to recognize a novel cluster of high-mannose type N-glycans on gp120 7,8. Recently, a new class of glycan-reactive bNAbs, including PG9/PG16, CH01-CH04, and the PGT series antibodies represented by PGT121 and PGT128, has been discovered from HIV-infected individuals9–12. These antibodies neutralize primary HIV-1 strains with remarkable breadth and potency, and share a common feature of antigen recognition: they all target glycan-reactive quaternary epitopes located primarily in the first, second, and third variable regions (V1V2 and V3) of gp120. Epitope mapping via mutational and biochemical analysis indicates that PG9 and PG16 recognize two conserved N-glycans in the V1V2 domain, one at the N160 (HXB2 numbering) glycosylation site and the other at N156 (for the majority of HIV-1 strains) or N173 (for ZM109 strain)10. Recent structural study of PG9 antigen-binding fragment (Fab) and its complex with a scaffolded V1V2 domain reveals a novel antigen recognition mode for PG9, showing that a Man5GlcNAc2 N-glycan at N160 provides the major contacts for the antibody, with additional contributions from another N-glycan at N156 (CAP45 strain) or N173 (ZM109 strain) and a strand of V1V2 peptide13. In the ZM109 co-crystal structure, only the protein proximal GlcNAc of the N173 glycan is resolved, whereas in the CAP45-bound structure, the complete Man5-linked structure at N156 is visualized. Interestingly, the N-glycans at the N156 and N173 sites are located at a spatially identical position binding to the same pocket. These structural studies indicate that a conserved glycopeptide antigen in the V1V2 domain might constitute the neutralizing epitope of PG9. X-ray structural studies on antibodies PGT127 and PGT128 Fabs and their complexes with a recombinant gp120 outer domain also show a similar antigen recognition mode, with two glycans and a peptide motif in the V3 domain as the essential components of the epitopes of PGT127/128 14.
Despite these impressive structural studies, the precise nature of the neutralizing epitopes, particularly the fine structures of the N-glycans at the respective sites, remain to be characterized. Further analysis of the epitopes is complicated by the complexity and heterogeneity of HIV-1 gp120 glycosylation15–17. Indeed, to facilitate crystallization by minimizing glycosylation heterogeneity, the scaffolded V1V2 and the glycosylated gp120 outer domain used for these structural studies were expressed in GnTI−/− mammalian cells that lack β-N-acetylglucosaminyl transferase I, a key enzyme essential for processing high-mannose N-glycans to hybrid and complex type N-glycan structures. As a result, the glycoforms generated might not represent the actual glycosylation patterns found in native gp120. To further characterize the neutralizing epitopes of antibody PG9 and PG16, we launched a project aiming to reconstitute the minimal antigenic glycopeptide structures through a synthetic chemistry approach. In this paper, we report the design, synthesis and antigenicity of a series of homogeneous cyclic glycopeptides corresponding to the V1/V2 domain, in which defined N-glycans were installed selectively at the pre-determined glycosylation positions (N160 and N156/N173). More than 25 V1V2 glycopeptides containing high-mannose or complex type N-glycans and their combinations were synthesized by a novel chemoenzymatic method. Antibody binding studies confirmed that a Man5GlcNAc2 glycan at the N160 position was essential for PG9 and PG16 recognition. Surprisingly, our data also revealed a critical role of a terminal sialylated complex type N-glycan at the secondary glycosylation site (N156 or N173) for recognition by PG9 and PG16, which was not revealed by previous structural and biochemical studies.
RESULTS
Design of cyclic V1V2 glycopeptides as putative epitopes
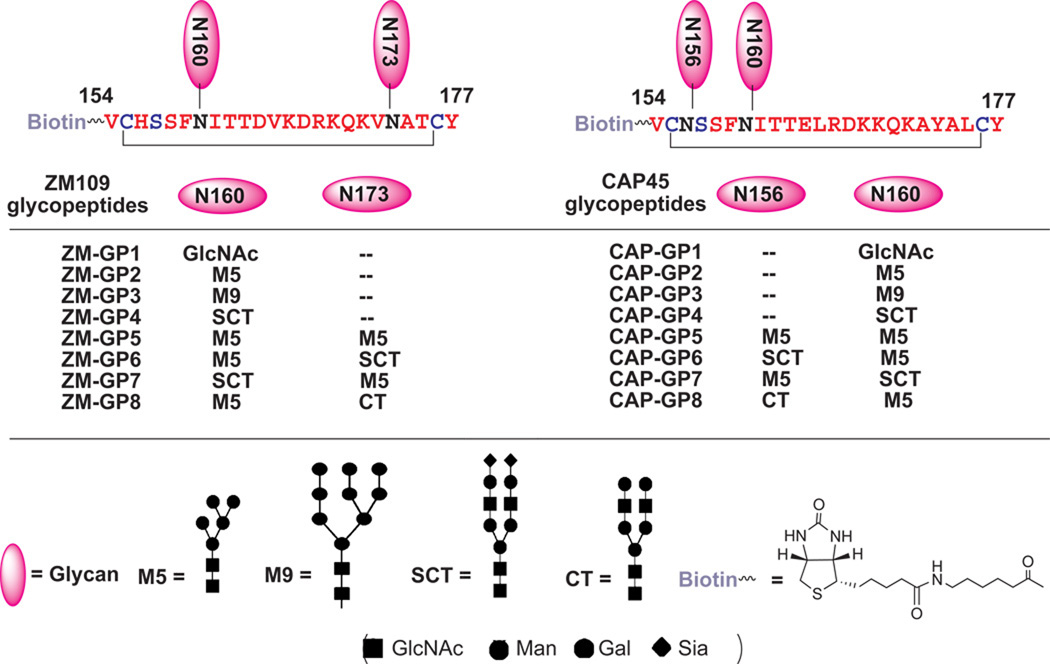
The recently solved crystal structure of PG9 in complex with a scaffolded V1V2 domain suggests that the putative PG9 epitope consists of the glycans attached to N160 and N156/N173 and a strand of V1V2 peptide (aa154–177) 13. On the basis of this recognition mode, we designed V1V2 cyclic glycopeptides (aa154–177) derived from two HIV-1 strains (ZM109 and CAP45) as the first synthetic targets to mimic this epitope (Fig. 1). A disulfide bond was engineered into the V1V2 sequence at positions K155 and F176 to stabilize the β-hairpin found in the crystal structure. In glycopeptide series of GP2–4, different types of N-glycans were installed at the N160 position. The second set of glycopeptides (GP5–8) carried two N-glycans, one at N160 and the other at N156 (CAP45 strain) or N173 (ZM109 strain) sites. A biotin tag was selectively introduced at the N-terminus of the glycopeptides to facilitate oriented immobilization of the glycopeptides on streptavidin chips, which may minimize the disruption of conformational epitopes in antibody binding and detection.
Figure 1. Structures of the designed V1V2 glycopeptides derived from HIV-1 ZM109 and CAP45 strains.
Chemoenzymatic synthesis of the V1V2 glycopeptides
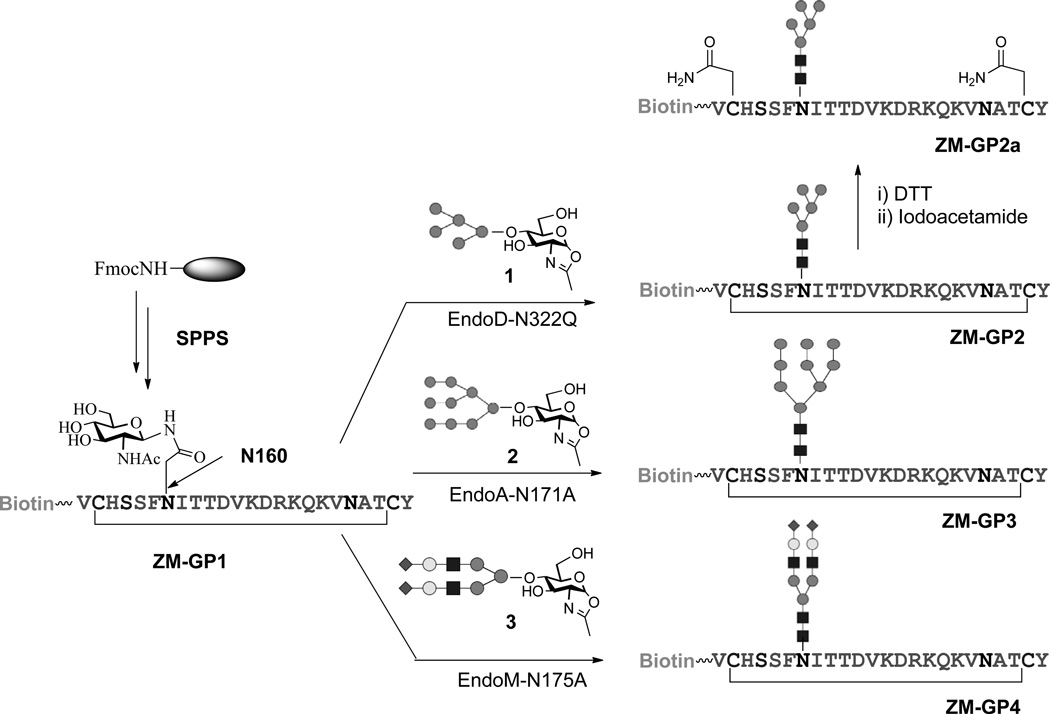
Synthesis of large, biologically relevant glycopeptides carrying complex natural N-glycans is still a challenging task18,19. Here we employed a chemoenzymatic method that we have developed for synthesis of complex glycopeptide/glycoprotein 20,21 to construct the designed V1V2 glycopeptides. In this approach, a monosaccharide moiety (e.g., GlcNAc) is installed at the pre-determined glycosylation site in peptide/protein during solid-phase peptide synthesis (SPPS) and then a synthetic glycan, in the form of activated glycan oxazoline, is transferred to the GlcNAc moiety by an endoglycosynthase mutant to provide a homogeneous glycopeptide/glycoprotein 22–33. This enzymatic ligation method is highly convergent, allows independent synthetic manipulation of the glycan and protein portions, and results in the formation of the native β-1,4-glycosidic linkage between the two core GlcNAc moieties found in all natural N-glycans. The use of novel glycosynthase mutants ensures high-yield synthesis without product hydrolysis. The synthesis of the ZM109 V1V2 glycopeptides carrying an N-glycan at N160 (ZM-GP2, ZM-GP3, and ZM-GP4) was shown in Fig. 2. Briefly, the precursor polypeptide (ZM-GP1) carrying an Asn-linked GlcNAc residue at N160 and a biotin tag at the N-terminus was synthesized by automated SPPS, using Fmoc-(Ac3GlcNAc)Asn-OH as the building block to introduce the Asn-linked GlcNAc moiety to give ZM-GP1 after deprotection, cyclization, and RP-HPLC purification. For the synthesis of Man5GlcNAc2-containing glycopeptides, we developed a semi-synthesis of the donor substrate, Man5GlcNAc oxazoline (1) using bovine ribonuclease B (RNase B), a glycoprotein carrying Man5–9GlcNAc2 glycan, as the starting material. Thus, trimming RNase B with a recombinant α-1,2-mannosidase34 followed by treatment with Endo-D, an endo-β-N-acetylglucosaminidase from S. pneumoniae32 that can hydrolyze specifically the Man5GlcNAc2 glycan released the Man5GlcNAc oligosaccharide. This glycan was efficiently converted to 1 via a single-step transformation in aqueous solution35,36 (see Online Methods and Supplementary Results, Supplementary Fig. 1a). The purity and identity of 1 were confirmed by MS, HPAEC-PAD, and 1H-NMR analysis (Supplementary Fig. 1b–1f).
Figure 2. Chemoenzymatic synthesis of ZM109 V1V2 glycopeptides carrying defined N-glycans at the N160 site.
The synthesis of glycopeptide ZM-GP1 was achieved by the reaction between ZM-GP1 and 1 under the catalysis of an endoglycosidase mutant, EndoD-N322Q that we have recently reported 32. We found that the EndoD-N332Q mutant was highly efficient for transferring Man5GlcNAc oxazoline and gave an essentially quantitative transformation when a 3-fold excess of oxazoline was used, although the Endo-D mutant was inactive for the larger high-mannose and complex type glycan oxazolines because of its strict substrate specificity. The excess glycan oxazoline was efficiently recovered as free glycan after reaction and was reused. The synthesis of glycopeptide ZM-GP3 carrying a Man9GlcNAc2 glycan at N160 was accomplished by an EndoA-N171A catalyzed glycosylation of ZM-GP1 with Man9GlcNAc-oxazoline (2) 26. Similarly, glycopeptide ZM-GP4 carrying a sialylated complex type N-glycan was synthesized by transglycosylation of ZM-GP1 with complex type glycan oxazoline (3) 36 under the catalysis of EndoM-N175A 25 (Fig. 2). A linear (acyclic) glycopeptide carrying Man5GlcNAc2 at N160 site, ZM-GP2a, was also prepared in order to evaluate the importance of the cyclization for antibody binding. The purity and identity of the synthetic glycopeptides (ZM-GP2, ZM-GP3, and ZM-GP4) were confirmed by their HPLC and ESI-MS analysis (Supplementary Fig. 2). It should be pointed out that the regio- and stereo-specificity of the endoglycosidase-catalyzed transglycosylation ensures the formation of the native β-1,4-glycosidic linkage between the two core GlcNAc moieties found in all natural N-glycans, as confirmed by detailed NMR and enzymatic conversion analysis 23,26,37. The corresponding glycopeptides of the HIV-1 CAP45 strain carrying a defined N-glycan at the N160 site were synthesized in the same manner as for the preparation of the ZM109 glycopeptides (Supplementary Fig. 3).
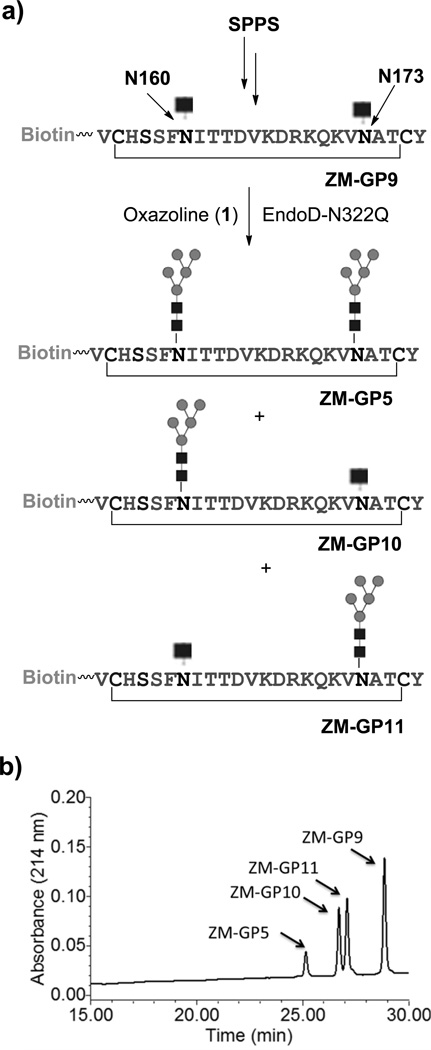
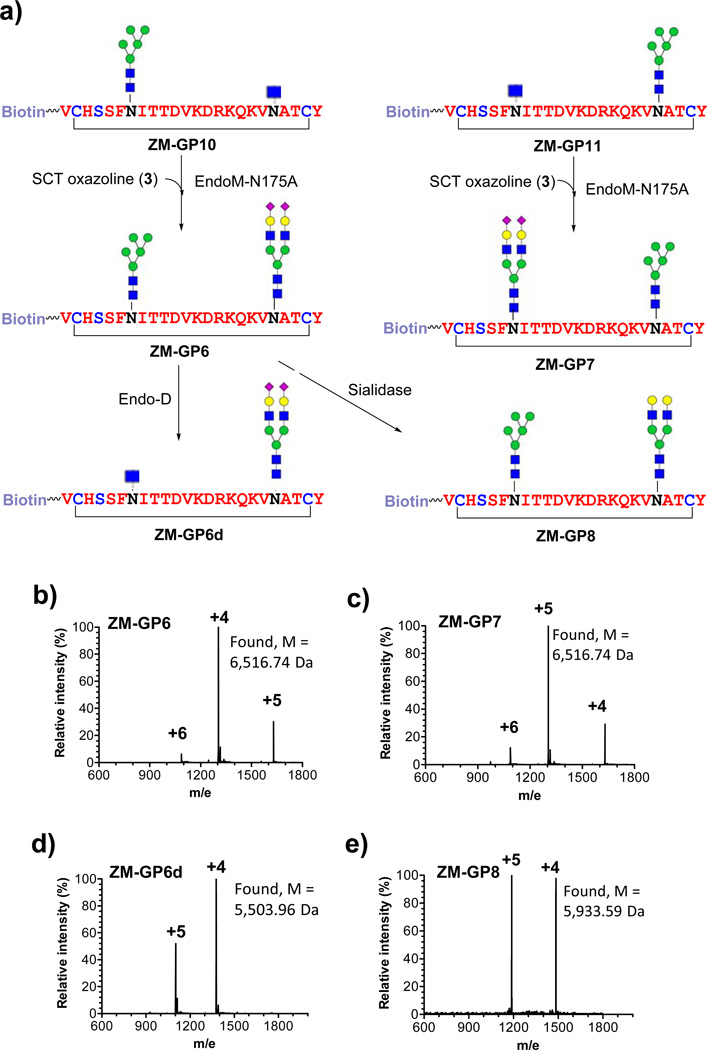
The synthesis of the ZM109 glycopeptides carrying two N-glycans commenced with the preparation of the precursor glycopeptide, ZM-GP9, in which two GlcNAc moieties were introduced at the N160 and N173 glycosylation sites by SPPS (Fig. 3a). To synthesize the glycopeptide carrying two Man5GlcNAc2 moieties (ZM-GP5), an excess (5 equivalent) of 1 was used for enzymatic glycosylation of ZM-GP9 with EndoD-N322Q, giving ZM-GP5 in excellent yield. However, the synthesis of glycopeptides carrying two distinct N-glycans (at N160 and N173) would be more challenging. To address this issue, we performed a controlled transglycosylation of ZM-GP9 using 2.5 molecular equivalent of 1, leading to the formation of two mono-glycosylated isomeric compounds, ZM-GP10 and ZM-GP11, together with the double-glycosylated compound ZM-GP5. Fortunately, the three glycopeptides could be readily separated by RP-HPLC (Fig. 3b). ESI-MS analysis of ZM-GP5, ZM-GP9, ZM-GP10, and ZM-GP11 indicated that ZM-GP10 and ZM-GP11 were isomers with a Man5GlcNAc2 at either the N160 or the N173 position (Supplementary Fig. 4a–d). Further characterization of the identity of the two isomers was achieved by site-specific digestion with chymotrypsin (specific for phenylalanine amide bond) and trypsin (specific for lysine and arginine amide linkage), followed by LC-MS analysis of the fragments (Supplementary Fig. 5). The results revealed that ZM-GP10 carried the Man5GlcNAc2 glycan at N160 while ZM-GP11 carried the Man5GlcNAc2 at N173 site. Upon separation of the two mono-glycosylated peptides, a sialylated complex type N-glycan was installed on the remaining GlcNAc moiety by glycosylation with 3 under the catalysis of EndoM-N175A to give ZM-GP6 and ZM-GP7, respectively, in which the two conserved N-glycosylation sites carry distinct N-glycans (Fig. 4a). Treatment of ZM-GP6 with Endo-D selectively removed the Man5GlcNAc glycan to give a truncated glycoform, ZM-GP6d, useful for probing how a sialylated N-glycan at N173 alone contributes to antibody binding. In addition, treatment of ZM-GP6 with a bacterial sialidase gave another new glycoform, the asialylated derivative (ZM-GP8). All the compounds were purified by RP-HPLC and characterized by ESI-MS (Fig. 4b–4e). The CAP45 series glycopeptides carrying two N-glycans at N160 and N156 positions were synthesized following the same strategy. Briefly, the monoglycosylated derivatives (CAP-GP10 and CAP-GP11) were synthesized by controlled glycosylation of CAP-GP9 with 1 (Supplementary Fig. 6). The glycopeptide products were readily separated by RP-HPLC and characterized by ESI-MS (Supplementary Fig. 7). Further characterization of the site of glycosylation in the two isomers was achieved by digestion with chymotrypsin followed by LC-MS analysis of the fragments, which unambiguously confirmed that the CAP-GP10 carried a Man5GlcNAc2 glycan at N160 while the CAP-GP11 carried a Man5GlcNAc2 glycan at N156 position (Supplementary Fig. 8). Finally, further enzymatic glycosylation of the remaining GlcNAc moiety with 3 gave CAP-GP6 and CAP-GP7, respectively, which carry two distinct N-glycans at the N160 and N156 positions (Supplementary Fig. 9). Treatment of CAP-GP6 with Endo-D selectively removed the Man5GlcNAc glycan to give CAP-GP6d. Enzymatic desialylation of CAP-GP6 gave the asialylated glycopeptide (CAP-GP8). Again, the purity and identity of the final glycopeptide products were confirmed by HPLC and ESI-MS analysis (Supplementary Fig. 10).
Figure 3. Controlled glycosylation and HPLC separation of mono- and doubly glycosylated ZM-glycopeptides.
a) synthetic scheme; b) the HPLC separation of the mono- and doubly glycosylated ZM-glycopeptides. The reverse phase HPLC was performed on a C18 column with a linear gradient of 15–30% MeCN in 30 min.
Figure 4. Chemoenzymatic synthesis and ESI-MS characterization of the doubly glycosylated ZM-glycopeptides.
a) the synthetic scheme; b) ESI-MS of ZM-GP6; c) ESI-MS of ZM-GP7; d) ESI-MS of ZM-GP6d; and e) ESI-MS of ZM-GP8.
SPR binding studies with PG9/PG16 Fab
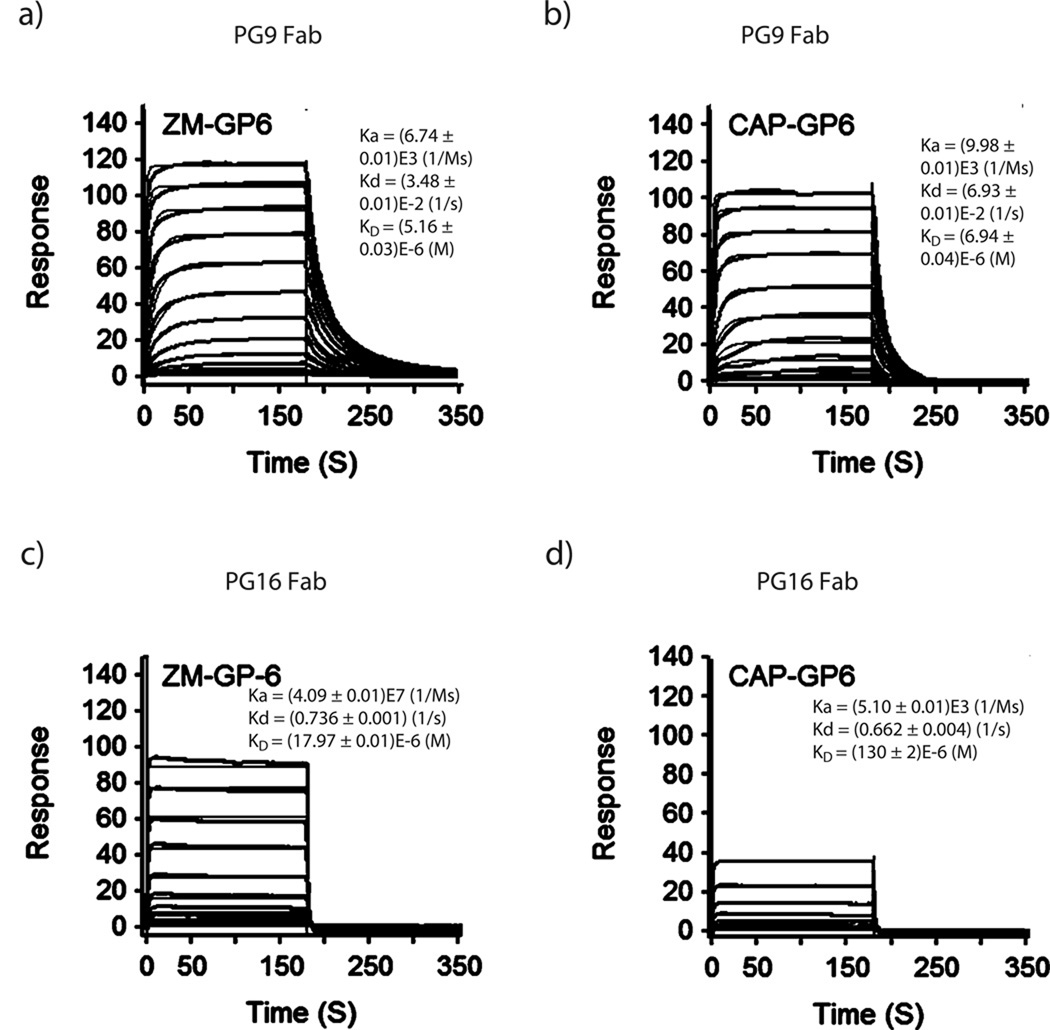
To assess the affinity of antibody-antigen interactions, we used PG9/PG16 Fab instead of the bivalent whole IgG in order to simplify the kinetic assessment of affinity between Fab and antigen interaction by SPR analysis. The biotin-tagged glycopeptides were immobilized on streptavidin chips and antibody Fabs were used as the analytes. The binding responses provided a quick assessment of the affinity of these synthetic glycopeptides for the antibody Fabs. The SPR sensorgrams for the binding of ZM-GP6 and CAP-GP6 with PG9/PG16 Fabs were shown in Fig. 5 (for binding of additional compounds, see Supplementary Fig. 11; the KD values were summarized in Supplementary Table 1). Our data revealed that PG9 Fab showed apparent affinity only for those glycopeptides carrying a Man5GlcNAc2 glycan at the N160 position. The cyclic peptides without an N-glycan, or with a GlcNAc, a Man9GlcNAc2, or a complex type N-glycan at the N160 position, all did not show binding at up to 100 µM. This result was consistent with the observation from previous structural study showing that a Man5GlcNAc2 at N160 made the major contact with PG9 Fab13. Comparison of the cyclic glycopeptide ZM-GP2 (KD = 106 µM) and the linear glycopeptide ZM-GP2a (KD = 330 µM) indicated that the cyclic glycopeptide ZM-GP2 had at least 3-fold higher affinity for PG9 Fab than that of the corresponding linear (acyclic) glycopeptide, ZM-GP2a, suggesting that cyclization played a positive role in enhancing the affinity by constraining the conformations. Surprisingly, installation of an additional Man5GlcNAc2 at the second glycosylation site, N156 (for the CAP45 strain) or N173 (for the ZM109 strain), as implicated by the crystal structure,13 neither enhanced nor decreased the affinity for PG9 Fab. However, attachment of a sialylated complex type N-glycan at N156 or N173 resulted in a substantial enhancement of the affinity. As estimated by the dissociation constant (KD), the affinity of PG9 Fab for ZM-GP6 (KD = 5.1 µM) carrying a sialylated N-glycan at N173 and a Man5GlcNAc2 at N160 was about 20-fold higher than that of PG9 Fab for the ZM-GP2 (KD = 101 µM) carrying two Man5GlcNAc2 glycans. The importance of sialylation at the N173 glycan for the PG9 binding was confirmed by the fact that desialylation of ZM-GP6 reduced the affinity of PG9 Fab by 34-fold (ZM-GP8, KD = 176 µM). This tendency was also found for the CAP45 series glycopeptides, where the CAP-GP6 ((KD = 6.9 µM), which carries a Man5GlcNAc2 and a sialylated N-glycan at the N160 and N156 positions, respectively, showed about 20-fold higher affinity than that of the CAP-GP5 (KD = 130 µM) carrying two Man5GlcNAc2 glycans. On the other hand, the ZM-GP6d and CAP-GP6d, in which the Man5GlcNAc was removed by Endo-D did not show binding at up to 100 µM, suggesting that a sialylated N-glycan at N173 alone was not enough for efficient recognition by PG9. For the PG16 Fab, it was found that a Man5GlcNAc2 glycan at the N160 position alone did not seem to provide sufficient affinity for the antibody, as no binding was detected at up to 100 µM for those glycopeptides carrying only a Man5GlcNAc2 at N160. However, installation of an additional sialylated N-glycan at the N156 (for the CAP45 strain) or the N173 position (for the ZM109 strain) in combination with a Man5GlcNAc2 glycan at N160 resulted in significant enhancement in affinity for PG16 Fab. These results suggest that a Man5GlcNAc2 at the N160 position is essential for PG9 and PG16 recognition, while an additional sialylated N-glycan at the secondary glycosylation site (N156/N173) is critical for a much tighter interaction. The synthetic glycopeptides, ZM-GP6 and CAP-GP6 had a comparable affinity as that of the recombinant, 1FD6-scaffolded ZM109 V1V2 domain for PG9 Fab (KD = 5.6 µM) 13. Nevertheless, the affinity of ZM-GP6 was still about 16- and 54-fold lower than that of another scaffolded V1V2 domain (1JO8-ZM109V1V2, KD = 0.32 µM) and the ZM109 gp120 (KD = 0.097 µM), respectively (Supplementary Table 1). These results may suggest that the 1JO8-protein scaffold and the gp120 protein domain could present the V1V2 glycopeptide antigenic structures better than the minimal synthetic glycopeptide epitopes by confining the favorable β-hairpin conformations that PG9 recognizes. It is likely that having all four strands of V1/V2 (as present in the scaffolded V1V2 domain and gp120) would increase the stability of the β-sheet and keep strands B and C in the proper orientation13.
Figure 5. SPR analysis of the binding of synthetic V1V2 glycopeptides to PG9/PG16 Fabs.
Biotinylated glycopeptides were immobilized on streptavidin chips and antibody PG9 Fab or PG16 Fab flowed through as analytes. The surface-plasmon resonance sensorgrams were recorded with 2-fold serial dilutions starting at the highest concentration of 100 µM. a) PG9 Fab and ZM-GP6; b) PG9 Fab and CAP-GP6; c) PG16 Fab and ZM-GP6; d) PG16 Fab and CAP-GP6ZM-GP6. The fitted curves were shown in orange color. The binding of ZM-GP2, ZM-GP5, ZM-GP8, CAP-GP2, CAP-GP5 and CAP-GP8 was shown in Supplementary Fig. 11. The following ZM- and CAP-glycopeptides did not show apparent binding responses at up to 100 µM: ZM-GP1, ZM-GP3, ZM-GP4, ZM-GP7, ZM-GP9; CAP-GP1; CAP-GP7, CAP-GP9; and those non-glycosylated peptides.
Glycopeptides as coating antigens for detecting PG9/PG16
We examined the feasibility of using the synthetic biotinylated V1V2 glycopeptides as coating antigens to detect PG9 and PG16 IgG antibodies. The biotinylated glycopeptides were immobilized on streptavidin microplates and titrated against PG9 and PG16 monoclonal antibodies at serial dilutions. We found that the V1V2 glycopeptides having a Man5GlcNAc2 glycan at the N160 position (the GP2, GP5, GP6, and GP8 of the ZM109 and CAP45 strains) could detect PG9 with a clear dose response (Supplementary Fig. 12 a–b). The presence of a sialylated N-glycan at the N173 (for the ZM109 strain) or the N156 position (for CAP45 strain) significantly enhanced the sensitivity of detection. When the apparent EC50 values were used as an estimate of the detection sensitivity (Table 1), the sialylated glycopeptide (ZM-GP6, EC50 = 0.008 µg/mL, 0.05 nM) was about 50-, 40- and 30-fold more sensitive for PG9 detection than the glycopeptides without a glycan at N173 (ZM-GP2, EC50 = 0.41 µg/mL), with a second Man5GlcNAc2 at N173 (ZM-GP5, EC50 = 0.34 µg/mL), or with a desialylated N-glycan at the N173 position (ZM-GP8, EC50 = 0.24 µg/mL), respectively. The same trend was also found for the CAP45 series of glycopeptides. In the case of antibody PG16, the dependence on a sialylated glycan at the secondary site was apparent, as revealed by the fact that only the glycopeptides (ZM-GP6 and CAP-GP6) that carry both a Man5GlcNAc2 at N160 and a sialylated N-glycan at N173/N156 were sensitive for detection of PG16 (Supplementary Fig. 12 c and Table 1). These ELISA results were consistent with the SPR binding data. In comparison with the 1FD6-scaffolded V1V2 recombinant glycoproteins (ZM109 V1V2 and CAP45 V1V2)13, glycopeptide ZM-GP6 (EC50 = 0.008 µg/mL) in the current ELISA format was more sensitive (12-fold) than the 1FD6-scaffoled ZM109 V1V2 (EC50 = 0.1 µg/mL) for PG9 detection, and the CAP-GP6 glycopeptide (EC50 = 0.027 µg/mL) was also more sensitive than the corresponding 1FD6-scaffolded CAP45 V1V2 (EC50 = 0.2 µg/mL) 13. For detection of PG16, the ZM-GP6 (EC50 = 0.22 µg/mL) was more sensitive (15-fold) than the scaffolded ZM109 V1V2 (EC50 = 3.4 µg/mL), while the CAP-GP6 (EC50 = 20 µg/mL) was about 2-fold less sensitive than the scaffolded CAP45 V1V2 domain (EC50 = 9.1 µg/mL) 13. However, it should be pointed out that the two ELISA studies used different methods for antigen immobilization, which may result in difference in orientation and density of the coating antigens.The synthetic glycopeptides were immobilized via their biotin handles onto a streptavidin–coated plate, whereas the scaffolded antigens were coated directly onto the plate. The clustering and site-specific orientation of glycopeptides on the immobilized streptavidin in the present ELISA format could enhance the avidity of PG9/PG16 binding, which may explain the enhanced sensitivity of antibody detection observed in the present ELISA. Taken together, these results suggest that the well-defined synthetic V1V2 glycopeptides could serve as efficient coating antigens for sensitive detection of glycopeptide-specific, PG9/PG16-like broadly neutralizing antibodies in biological samples.
Table 1.
EC50 (µg/mL) of PG9 and PG16 IgG binding to synthetic ZM109/CAP45 glycopeptides and scaffolded V1V2 domains
| Abs | ZM-GP1 | ZM-GP2 | ZM-GP3 | ZM-GP4 | ZM-GP5 | ZM-GP6 | ZM-GP7 | ZM-GP8 | 1FD6 ZM109 V1V2 |
|---|---|---|---|---|---|---|---|---|---|
| PG9 | -- | 0.41 (2.7 nM) |
-- | -- | 0.34 (2.3 nM) |
0.008 (0.05 nM) |
-- | 0.24 (1.6 nM) |
0.1* (0.7 nM) |
| PG16 | -- | -- | -- | -- | -- | 0.22 (1.5 nM) |
-- | -- | 3.4* (23 nM) |
| Abs | CAP-GP1 | CAP-GP2 | CAP-GP3 | CAP-GP4 | CAP-GP5 | CAP-GP6 | CAP-GP7 | CAP-GP8 | 1FD6 CAP45 V1V2 |
|---|---|---|---|---|---|---|---|---|---|
| PG9 | -- | 0.27 (1.8 nM) |
-- | -- | 0.75 (5.0 nM) |
0.027 (0.18 nM) |
-- | 3.3 (22 nM) |
0.2* (1.4 nM) |
| PG16 | -- | -- | -- | -- | -- | 19.0 (126 nM) |
-- | -- | 9.1* (61 nM) |
“--“, no binding at 20 µg/mL;
adapted from ref. 13
DISCUSSION
Characterization of the fine epitopes of the newly discovered, glycan-reactive broadly neutralizing antibodies such as PG9 and PG16 is significant for HIV vaccine design. However, a major difficulty in this pursuit is the structural heterogeneity of glycosylation of envelope glycoprotein gp120 15–17. Protein N-glycosylation is a complex co- and post-translational modification that involves a series of glycan processing steps and often results in production of mixtures of glycoforms38. It is currently impossible to install a structurally well-defined N-glycan at selected glycosylation sites in gp120 by cell-based expression systems. In the present study, we address this challenge by design and synthesis of a series of homogeneous HIV-1 V1V2 glycopeptides, in which well-defined N-glycans are installed at the pre-determined glycosylation sites (N160 and N156/N173). More than 25 V1V2 glycopeptides belonging to two different HIV-1 strains were synthesized by an efficient chemoenzymatic method. SPR binding and ELISA analysis indicated that the presence of a Man5GlcNAc2 glycan at the N160 position was essential for the recognition by PG9 and PG16. Moreover, our data also revealed a critical role of a terminal sialylated N-glycan at the secondary glycosylation site (N156 or N173). The importance of the terminal sialic acid in the secondary N-glycan was apparent by the fact that desialylation resulted in more than 30-fold decrease in affinity for PG9 and PG16 binding (ZM-GP6 vs. ZM-GP8 and CAP-GP6 vs. CAP-GP8). Glycopeptide ZM-GP6 showed a comparable affinity for PG9 Fab as the 1FD6-scaffolded ZM109 V1V2 domain. However, the fact that the affinity of the minimal synthetic V1V2 glycopeptide (ZM-GP6) for PG9 was still 50-fold lower than that of the corresponding HIV-1 ZM109 gp120 suggests that further constraining of the glycopeptide in the favorable β-hairpin structure may enhance the affinity of the synthetic glycopeptide epitopes. Our data showed that both the nature and the location of the N-glycans in the context of the V1V2 polypeptide were critical for recognition by the antibodies, as N-glycans or V1V2 peptides alone were not sufficient for the antibody binding, neither did the antibody bind to those glycopeptides where the right N-glycans were misplaced. Our results point to an exciting opportunity to identify and reconstitute well-defined glycopeptide neutralizing epitopes for HIV-1 vaccine design. Further improvement on the glycopeptide antigens is possible by a combination of synthesis and structure-activity relationship studies.
The dependence of a sialylated complex type N-glycan at the secondary glycosylation site for antigen recognition by PG9 and PG16 was not revealed by previous structural and biochemical studies, partially due to the difficulty to control the glycosylation at individual sites for the recombinant glycoprotein antigens. A recent crystal structural study on PG16 Fab in complex with a scaffolded V1V2 domain expressed in 293F cells in the presence of swainsonine, which results in glycoforms carrying both Man5GlcNAc2 and hybrid N-glycans, showed that a sialylated hybrid N-glycan at the secondary site (N156/N173) did provide critical contacts with residues in PG16 (Peter Kwong et al, unpublished data). These new structural data are consistent with our findings on the glycan specificity of PG9 and PG16. A relevant broadly neutralizing antibody, PGT121, the neutralizing activity of which appears to depend on glycans at N332 and N301 sites in the V3 domain, can bind to complex type N-glycans as revealed by crystal structure and glycan microarray analysis39. On the other hand, antibodies PGT127 and PGT128 have been shown to be specific for high-mannose type (Man8GlcNAc2 or Man9GlcNAc2) glycans at N332 and/or N301 in the V3 domain14. These have raised important questions on what types of N-glycans are actually present at the N332 and N301 glycosylation sites of gp120 of most HIV-1 strains and what glycan structures, in the context of V3 domain, are required for the recognition by PGT121, PGT127 and PGT128. The chemoezymatic synthesis approach described here should be also applicable for further characterization and reconstitution of glycopeptide eptitopes of these bNAbs. The cyclic V1V2 glycopeptide epitopes of PG9 and PG16 identified in the present study should be highly valuable for HIV-1 vaccine design. For example, the synthetic glycopeptide epitopes can be conjugated and displayed on a virus-like particle platform such as bacteriophage Qβ 40 to provide a vaccine candidate. In this case, a dense, highly ordered repetitive epitope display pattern on Qβ is likely important for enhancing the immunogenicity and the possibility to raise PG9-like antibodies, as a recent structural study indicates that PG9 binds preferentially native gp120 trimer by recognizing an additional N160 glycan on the adjacent gp120 molecule in the trimeric spike41. Finally, the synthetic biotinylated V1V2 glycopeptides would be valuable as coating antigens for detecting PG9/PG16-like bNAbs from sera of HIV-infected individuals or those involved in HIV vaccine trials.
ONLINE METHODS
Peptide synthesis
Peptides were prepared via solid-phase peptide synthesis (SPPS) on a Pioneer automatic peptide synthesizer (Applied Biosystems) using the Fmoc approach. Fluorenylmethyloxycarbonyl (Fmoc)-protected amino acids were used as building blocks, 2-(1-H-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) was used as the coupling reagent, and a CLEAR amide resin was used as the solid support. To install a GlcNAc moiety at a pre-determined site, Fmoc-Asn(Ac3GlcNAc)-OH was used as the building block to replace the corresponding Fmoc-Asn(Trt)-OH at the site during SPPS. The Fmoc deprotection was achieved with 20% piperidine. A biotin tag was added at the N-terminus of the peptide at the end of the peptide synthesis by treatment of the resin-bound N-terminal deprotected peptide with succinymidyl-6-(biotinamido)hexanoate in the presence of DIPEA. The Peptides were cleaved from the resin by treatment with Cocktail R (TFA/thioanisole/EDT/Anisole, 90/5/3/2), followed by precipitation with cold ether. In the case of GlcNAc-containing peptides, the crude peptides were treated with 2.5% aqueous hydrazine to remove the O-acetyl groups from the GlcNAc moiety, which also led to simultaneous cyclization via a disulfide bond formation between the two cysteine residues. The peptides or GlcNAc-peptides were purified by reverse phase HPLC (RP-HPLC). The purity and identity of the peptides were analyzed by RP-HPLC and ESI-MS.
HPLC
Analytical RP-HPLC for peptide/glycopeptide analysis was performed on a Waters 626 HPLC instrument with a C18 column (3.5 µm, 4.6 × 250 mm) at 40°C with a flow rate of 0.5 mL/min. The column was eluted with a suitable gradient of aq. MeCN containing 0.1% TFA. Preparative RP-HPLC was performed with a Waters 600 HPLC instrument on a Waters C18 column (5.0 µm, 10 × 250 mm). The column was eluted with a suitable gradient of aq MeCN containing 0.1%TFA at a flow rate of 4 mL/min.
Mass spectrometry
ESI-MS spectra were measured on a LXQ linear ion trap mass spectrometer (Thermo Scientific). MALDI-TOF MS spectra were measured on a MALDI-TOF/TOF 4800 spectrometer (Applied Biosystems) with 2,5-dihydroxybenzoic acid as the matrix and Cal 4700 standard peptide mixture was used (Applied Biosystems) as internal standard.
Preparation of Man5GlcNAc and Man5GlcNAc oxazoline (1)
A solution of bovine ribonuclease B (RNase B) (200 mg) in a phosphate buffer (5 mL, 10 mM, pH 6.5, containing 5 mM of CaCl2) was incubated with a recombinant murine α-1,2-mannosidase (40 µg) for 23 h. LC-MS monitoring indicated that more than 90% of the high-mannose N-glycans (Man5–9GlcNAc2) on RNase B was converted into the Man5GlcNAc2 glycoform together with a minor fraction of the Man6GlcNAc2 glycoform. The mixture was subjected to gel filtration on a Sephadex G-25 column and the glycoprotein fractions were pooled and lyophilized. The glycoproteins were then dissolved in a phosphate buffer (5 mL, 50 mM, pH 7.2) and treated with Endo-D (an endo-β-N-acetylglucosaminidase from Streptococcus pneumoniae (100 µg), which can specifically release the Man5GlcNAc without hydrolysis of the Man6GlcNAc2 glycoform.. The solution was incubated for 3 h at 23 °C and the released Man5GlcNAc was then separated from the protein portions by preparative RP-HPLC using a gradient of 5–25% aq. acetonitrile containing 0.1% TFA. The proper fractions containing Man5GlcNAc were lyophilized to obtain the free glycan Man5GlcNAc (10.6 mg), the purity and identity were characterized by HPAEC, MS and NMR analyses (supplementary Fig. 1). HRMS MALDI-TOF MS: calcd for C38H65NO31, 1031.354, found 1054.349 [M + Na]+.
To a solution of Man5GlcNAc (10.3 mg, 100 µmol) in H2O (0.5 mL) was added Et3N (42 µL, 0.3 mmole) and 2-chloro-1,3-dimethylimidazolinium chloride (DMC) (17 mg, 0.10 mmole) at 0 °C. The reaction mixture was stirred for 0.5 h and the reaction was monitored by DIONEX HPAEC-PAD which indicated that the free oligosaccharide was converted into a new product that was eluted earlier than the reducing sugar under the HPAEC conditions (supplementary Fig. 1). The product was purified by gel filtration on a Sephadex G-10 column using 0.1% TEA (aq.) as the eluent to afford the Man5GlcNAc oxazoline (1) (9.6 mg, 94%) as a white solid after lyophilization. The purity and identity of the Man5GlcNAc oxazoline were confirmed by HPAEC and NMR analysis (Supplementary Fig. 1a and Fig. 1f).1H NMR (400 MHz, D2O): δ 1.98 (3H, CH3), 3.29–4.30 (36H, m), 4.67 (1H, s, overlapped with H-OD signal), 4.79 (2H, br s, overlapped with H-OD signal), 4.98 (1H, s), 5.02 (1H, s), 6.00 (1H, d, J = 6.8 Hz, H-1, oxazoline).
Chemoenzymatic synthesis of glycopeptides
Full-size glycopeptides were synthesized by enzymatic transglycosylation of the chemically synthesized GlcNAc-containing peptide precursors with respective glycan oxazolines as the donor substrates. Representative procedures for the enzymatic transfer of Man5GlcNAc, Man9GlcNAc, and a complex type N-glycan were exemplified by the synthesis of ZM-GP2, ZM-GP3, and ZM-GP4, respectively. Other glycopeptides carrying one or two of the respective natural N-glycans were prepared by the chemoenzymatic method in a similar manner.
Synthesis of glycopeptide ZM-GP2 carrying a Man5GlcNAc2 moiety: A solution of the GlcNAc- peptide (ZM-GP1) (2.42 mg, 0.73 mmol) and 1 (3.00 mg, 2.19 mmol) in a phosphate buffer (0.97 mL, 50 mM, pH 7.2) was incubated with EndoD-N223Q (final concentration, 40 ng/µL). The reaction was monitored by RP-HPLC. After 0.5 h, the reaction was quenched with 0.1% aq. TFA. The transglycosylation product was purified by RP-HPLC to give ZM-GP2 (2.79 mg, 89%). ESI-MS of ZM-GP2: calcd M = 4310.48; found, 863.13 [M + 5H]5+, 1078.23 [M + 4H]4+, 1437.56 [M + 3H]3+. Deconvolution data, M = 4310.65 ± 0.33.
Synthesis of glycopeptide ZM-GP3 carrying a Man9GlcNAc2 moiety: A solution of the GlcNAc- peptide (ZM-GP1) (2.01 mg, 0.61 mmol) and the Man9GlcNAc -oxazoline (2) (3.04 mg, 1.83 mmol) in a phosphate buffer (0.81 mL, 50 mM, pH 7.2) was incubated with EndoA-N171A (final concentration, 0.1 µg/µL). The reaction was monitored by RP-HPLC. After 3 h, the reaction was quenched with 0.1% aq. TFA. The transglycosylation product was purified by RP-HPLC to give ZM-GP3 (2.14 mg, 71%). ESI-MS of ZM-GP3: calcd M = 4958.90; found 828.89 [M + 4H]4+, 992.78 [M + 5H]5+, 1240.67 [M + 4H]4+, 1654.25 [M + 3H]3+. Deconvolution mass 4958.74 ± 0.11.
Synthesis of glycopeptide ZM-GP4 carrying a sialylated complex type N-glycan: A solution of the GlcNAc-peptide (ZM-GP1) (2.17 mg, 0.66 mmol) and the (NeuGalGlcNAc)2Man3GlcNAc-oxazoline (3) (3.96 mg, 1.98 mmol) in a phosphate buffer (0.74 mL, 50 mM, pH 7.2) was incubated with EndoA-N171A (final concentration, 0.5 µg/µL). The reaction was monitored by RP-HPLC. After 3 h, the reaction was quenched with 0.1% aq. TFA. The transglycosylation product was purified by RP-HPLC to give ZM-GP4 (2.89 mg, 83%). ESI-MS of ZM-GP4: calcd M = 5299.04; found 1060.77 [M + 5H]5+, 1325.68 [M + 4H]4+, 1767.13 [M + 3H]3+. Deconvolution mass 5299.19 ± 1.07.
Synthesis of acyclic glycopeptide ZM-GP2a: the cyclic glycopeptide carrying a N160 Man5GlcNAc2 glycan (ZM-GP2) (410 µg, 95 nmole) was dissolved in 7M guanidine HCl (0.2 mL) followed by addition of DTT (0.5 mg, 3.25 µmole) to reduce the disulfide bond. The mixture was stirred for 1h and then idoacetamide (2.4 mg, 13 µmole) was added to block the free cysteine. The mixture was stirred for 0.5 h in dark and the crude product was purified by preparative HPLC to afford the acyclic title compound (361 µg, 86% yield); analytical RP-HPLC, tR = 19.8 min (gradient, 0–90% aq. MeCN containing 0.1% TFA for 30 min; flow rate, 0.5 mL/min); ESI-MS: calcd M = 4,426.60; found 886.37 [M + 5H]5+, 1107.56 [M + 4H]4+, 1476.09 [M + 3H]3+. Deconvolution mass, 4,426.38 ± 0.12.
Surface plasmon resonance (SPR)
SPR measurements were performed on a Biacore T100 instrument (GE Healthcare) at 25 °C. Biotinylated glycopeptides were immobilized on streptavidin-coated sensor chips (SA) in a solution of 1X HBS-P buffer (0.1 M HEPES, 0.15 M NaCl, 0.5% v/v surfactant P20, pH 7.4) by injecting the samples manually until 20–30 RU (low loading) or 300–330 RU (high loading) was achieved. PG9 Fab or PG16 Fab was injected over four cells at 2-fold increasing concentrations with a flow rate of 50 µl/ min for 3 min and allowed to dissociate for another 5 min. Regeneration was performed by injection of 3 M MgCl2 with a flow rate of 50 µL/min for 3 min followed by injection of 1X HBS-P buffer with a flow rate of 50 µl/min for 5 min. Data were collected at the rate of 10 Hz. T-100 Biacore Evaluation software was utilized to subtract appropriate blank references and to fit the sensorgrams globally applying a 1:1 Langmuir binding model. Mass transfer effects were checked by the tc values displayed by the T-100 Biacore evaluation software. No significant mass transportation effects were observed.
Enzyme-linked immunosorbent assay (ELISA)
The 96-well ELISA microtiter plates were first coated with 10 µg/mL streptavidin in PBS and incubated at 4°C overnight. After washings with PBS/0.5% Tween-20, nonspecific binding was blocked with 5% sodium caseinate (w/v) in PBS at room temperature for 1 h. Plates were washed three times, and then 2 µg/mL of the respective biotinylated glycopeptide antigen in 1% casein was added. Plates were incubated at 37 °C for 1 h. Then plates were again washed, and titrated against 1:3 serial dilutions of human monoclonal antibodies (PG9 IgG and PG16 IgG) in 1% sodium caseinate starting at 20 µg/mL. The plates were incubated at 37°C for 2 h. After washing, a solution (100 µL) of 1:3000 diluted horseradish peroxidase (HRP)-conjugated goat anti-human IgG in 1% PBS was added to the plates. The plates were kept 1 h at 37 °C and then the plates were washed and a solution of 3,3′,5,5′-tetramethylbenzidine (TMB) was added. Color was allowed to develop for 5 min, and then the reaction was quenched by adding a solution of 0.5 M H2SO4 (200 µL) to each well. The readout was measured at a wavelength of 450 nm.
Supplementary Material
Acknowledgments
We thank Mr. Shuquan Fan for providing the recombinant endoglycosidase D (Endo-D), and Dr. Kelley Moremen and Dr. Yong Xiang for providing the recombinant murine α-1,2-mannosidase. This work is supported in parts by grants from the National Institute of Allergy and Infectious Diseases (NIAID) (NIH grant 1R21AI101035 to LXW), the International AIDS Vaccine Initiative’s Neutralizing Antibody Consortium, and by the Intramural Research Program of the Vaccine Research Center, NIAID/NIH.
Footnotes
Author Contributions
M.N.A., J.S.M., W.H., P.D.K. and L.X.W. designed the research and analyzed the data; M.N.A., J.S.M., W.H., and J.O. performed the research; L.X.W. conceived the idea and supervised the research; D.R.B. and W.A.K. contributed PG9 and PG16 antibodies; L.X.W. and M.N.A. wrote the manuscript; all authors contributed to revisions of the manuscript.
Competing Financial Interests Statement
None declared.
References
- 1.Burton DR, et al. A blueprint for HIV vaccine discovery. Cell host & microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwong PD, Mascola JR, Nabel GJ. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harb Perspect Biol. 2011;3:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr. Opin. Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 6.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 7.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 9.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard CK, et al. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 16.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV- gp120 expressed in CHO cells. Biochemistry. 2000;39:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 17.Go EP, et al. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamblin DP, Scanlan EM, Davis BG. Glycoprotein synthesis: an update. Chem. Rev. 2009;109:131–163. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 19.Schmaltz RM, Hanson SR, Wong CH. Enzymes in the synthesis of glycoconjugates. Chem. Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- 20.Wang LX. Chemoenzymatic synthesis of glycopeptides and glycoproteins through endoglycosidase-catalyzed transglycosylation. Carbohydr. Res. 2008;343:1509–1522. doi: 10.1016/j.carres.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LX, Lomino JV. Emerging technologies for making glycan-defined glycoproteins. ACS Chem. Biol. 2012;7:110–122. doi: 10.1021/cb200429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, et al. Chemoenzymatic synthesis of HIV-1 V3 glycopeptides carrying two N-glycans and effects of glycosylation on the peptide domain. J. Org. Chem. 2005;70:9990–9996. doi: 10.1021/jo051729z. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Zeng Y, Hauser S, Song H, Wang LX. Highly efficient endoglycosidase-catalyzed synthesis of glycopeptides using oligosaccharide oxazolines as donor substrates. J. Am. Chem. Soc. 2005;127:9692–9693. doi: 10.1021/ja051715a. [DOI] [PubMed] [Google Scholar]
- 24.Ochiai H, Huang W, Wang LX. Expeditious chemoenzymatic synthesis of homogeneous N-glycoproteins carrying defined oligosaccharide ligands. J. Am. Chem. Soc. 2008;130:13790–13803. doi: 10.1021/ja805044x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umekawa M, et al. Mutants of Mucor hiemalis endo-beta-N-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J. Biol. Chem. 2008;283:4469–4479. doi: 10.1074/jbc.M707137200. [DOI] [PubMed] [Google Scholar]
- 26.Huang W, et al. Glycosynthases enable a highly efficient chemoenzymatic synthesis of N-glycoproteins carrying intact natural N-glycans. J. Am. Chem. Soc. 2009;131:2214–2223. doi: 10.1021/ja8074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Zhang X, Ju T, Cummings RD, Wang LX. Expeditious chemoenzymatic synthesis of CD52 glycopeptide antigens. Org. Biomol. Chem. 2010;8:5224–5233. doi: 10.1039/c0ob00341g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umekawa M, et al. Efficient glycosynthase mutant derived from Mucor hiemalis endo-beta-N-acetylglucosaminidase capable of transferring oligosaccharide from both sugar oxazoline and natural N-glycan. J. Biol. Chem. 2010;285:511–521. doi: 10.1074/jbc.M109.059832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz F, et al. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat. Chem. Biol. 2010;6:264–266. doi: 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou G, et al. Chemoenzymatic synthesis and Fcgamma receptor binding of homogeneous glycoforms of antibody Fc domain. Presence of a bisecting sugar moiety enhances the affinity of Fc to FcgammaIIIa receptor. J. Am. Chem. Soc. 2011;133:18975–18991. doi: 10.1021/ja208390n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amin MN, Huang W, Mizanur RM, Wang LX. Convergent Synthesis of Homogeneous Glc(1)Man(9)GlcNAc(2)-Protein and Derivatives as Ligands of Molecular Chaperones in Protein Quality Control. J. Am. Chem. Soc. 2011;133:14404–14417. doi: 10.1021/ja204831z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan SQ, Huang W, Wang LX. Remarkable transglycosylation activity of glycosynthase mutants of Endo-D, an endo-beta-N-acetylglucosaminidase from Streptococcus pneumoniae. J. Biol. Chem. 2012;287:11272–11281. doi: 10.1074/jbc.M112.340497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J. Am. Chem. Soc. 2012;134:12308–12318. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lal A, et al. Substrate specificities of recombinant murine Golgi alpha1, 2-mannosidases IA and IB and comparison with endoplasmic reticulum and Golgi processing alpha1,2-mannosidases. Glycobiology. 1998;8:981–995. doi: 10.1093/glycob/8.10.981. [DOI] [PubMed] [Google Scholar]
- 35.Noguchi M, Tanaka T, Gyakushi H, Kobayashi A, Shoda SI. Efficient synthesis of sugar oxazolines from unprotected N-acetyl-2-amino sugars by using chloroformamidinium reagent in water. J. Org. Chem. 2009;74:2210–2212. doi: 10.1021/jo8024708. [DOI] [PubMed] [Google Scholar]
- 36.Huang W, Yang Q, Umekawa M, Yamamoto K, Wang LX. Arthrobacter endo-beta-N-acetylglucosaminidase shows transglycosylation activity on complex-type N-glycan oxazolines: one-pot conversion of ribonuclease B to sialylated ribonuclease C. ChemBioChem. 2010;11:1350–1355. doi: 10.1002/cbic.201000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Ochiai H, Zhang X, Wang LX. Introducing N-glycans into natural products through a chemoenzymatic approach. Carbohydr. Res. 2008;343:2903–2913. doi: 10.1016/j.carres.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 39.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 41.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. USA. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.